Abstract
Expression quantitative trait locus (eQTL) studies in human liver are crucial for elucidating how genetic variation influences variability in disease risk and therapeutic outcomes and may help guide strategies to obtain maximal efficacy and safety of clinical interventions. Associations between expression microarray and genome-wide genotype data from four human liver eQTL studies (n = 1,183) were analyzed. More than 2.3 million cis-eQTLs for 15,668 genes were identified. When eQTLs were filtered against a list of 1,496 drug response genes, 187,829 cis-eQTLs for 1,191 genes were identified. Additionally, 1,683 sex-biased cis-eQTLs were identified, as well as 49 and 73 cis-eQTLs that colocalized with genome-wide association study signals for blood metabolite or lipid levels, respectively. Translational relevance of these results is evidenced by linking DPYD eQTLs to differences in safety of chemotherapy, linking the sex-biased regulation of PCSK9 expression to anti-lipid therapy, and identifying the G-protein coupled receptor GPR180 as a novel drug target for hypertriglyceridemia.
An expression quantitative trait locus (eQTL)1 is a genetic variant that can affect gene expression through mechanisms including alterations in gene transcription and transcript stability. Gene expression represents a mechanism underlying variation in drug response and susceptibility to disease.2,3 Approximately 90% of variants associated with complex traits are located in noncoding regions of the genome,1 suggesting the effects of these variants may be mediated through gene expression.
The liver is critical to the maintenance of homeostasis and health. Estimates indicate that 75% of the 200 most widely prescribed drugs are eliminated through liver metabolism or biliary excretion.4 While it is widely accepted that genetic and environmental variation influences drug efficacy and adverse events, the majority of variation in drug response remains unexplained. Increased knowledge of the contribution of genetic variation to the variability in liver gene expression, especially the identification of novel regulatory variants in genes of drug response, can provide the basis for translating genetic variations into clinically relevant tools.
Sexual dimorphism has also been shown to contribute to differences in disease susceptibility and drug efficacy and toxicity.5,6 For example, a lower incidence of coronary artery disease (CAD) in women has been linked to sex-biased differences in lipoprotein pathophysiology and is consistent with reports of sex-biased gene enrichment in studies of dyslipidemia and CAD.5 Lipid-lowering therapy has been reported to exhibit reduced efficacy in women,7,8, suggesting that sex-biased differences in the expression of drug metabolizing enzymes and/or therapeutic targets may contribute to sex-biased clinical outcomes. Thus, a systematic understanding of the role of liver eQTLs in sex-biased traits is of great clinical relevance.
Circulating metabolite levels serve as direct readouts of cellular processes and represent intermediate phenotypes. As such, metabolic profiles are used for clinical risk assessment, diagnosis, prognosis, and evaluation of treatment efficacy. Disruption in metabolic processes is associated with many chronic diseases, such as type 2 diabetes, and genome-wide association studies (GWAS) have identified numerous loci associated with serum concentrations of metabolites such as glucose and lipids. Identification of genetic variants that are associated with alterations in the homeostasis of key metabolites will be the basis for explaining the genetics of chronic diseases. For example, the identification of novel liver eQTLs associated with serum lipoprotein levels could lead to insights into the mechanisms by which genetic variants drive the risk of dyslipidemia and cardiovascular disease, leading to novel drug targets.
In eQTL mapping, sample size has been reported to greatly affect the probability of discovering novel eQTLs, in particular ones with smaller effect sizes.9,10 The most comprehensive eQTL work published to date, the Genotype-Tissue Expression (GTEx) project,10 included only 153 liver specimens. We aim to overcome these limitations by performing an analysis of four human liver eQTL data sets, totaling 1,183 livers. We present discoveries on genes of drug response, metabolism, and sex-biased regulation of gene expression. As has been demonstrated by many examples of the genetics of complex traits, genetic variation is one of many factors that can influence translation into medicine.11 We show evidence of the translational impact of novel eQTLs, including DPYD (related to the risk of severe toxicity from cancer chemotherapy), PCSK9 (related to anti-lipid therapy), and GPR180 (related to triglyceride levels).
RESULTS
Identification of liver cis-eQTLs
Four data sets which included genome-wide DNA genotyping and RNA transcriptome analysis from nondiseased human liver tissues were combined (Table 1). After quality control (Supplementary Methods and Results, Figures S1 and S2), the data sets included 145–555 unrelated samples, totaling 1,183 livers from individuals of genetic European ancestry. False discovery rate Q values < 0.05 were considered statistically significant. Unless otherwise stated, the number of eQTLs reported has not been pruned for linkage disequilibrium (LD). In the four individual data sets, 156,182–1,872,669 cis-eQTLs were identified (Table 2), and our combined analysis increased the number of cis-eQTLs by more than 20% to 2,391,948 cis-eQTLs for 15,668 genes (Figure 1, Table 2). Approximately 75% of genes included in our analysis were associated with at least one cis-eQTL, consistent with previous reports suggesting that expression of nearly all genes is influenced by genetic variation.10
Table 1.
Description of the four data sets and patient demographics. Number of samples from each study utilized in the combined analysis following removal of individuals with non-European genetic ancestry, sex mismatches, and related samples within and between data sets
| Data sets | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| N | 145 | 161 | 322 | 555 |
| Sex | ||||
| Male | 68 (46.9%) | 105 (65.2%) | 150 (46.6%) | 152 (27.4%) |
| Female | 77 (53.1%) | 56 (34.8%) | 172 (53.4%) | 403 (72.6%) |
| Age (years) | ||||
| ≤ 1 | 0 (0%) | 2 (1.2%) | 1 (0.3%) | 0 (0%) |
| 2–19 | 2 (1.4%) | 35 (21.7%) | 12 (3.7%) | 5 (0.9%) |
| 20–59 | 76 (52.4%) | 81 (50.3%) | 198 (61.5%) | 496 (89.4%) |
| > 59 | 67 (46.2%) | 43 (26.7%) | 111 (34.5%) | 54 (9.7%) |
| Mean (range) | 58.3 (7–85) | 41.4 (1–81) | 50.9 (0–94.3) | 45.5 (18–75) |
| Genotyping | Illumina HumanHap300-Duo v2.0 (GEO: GSE39036) | Illumina Human610-Quad v1.0 (GEO: GSE26105) | Affymetrix GeneChip Human Mapping 500K | HumanHap 650Y |
| Expression microarray | Illumina Human Whole Genome-6 v2.0 (GEO: GSE32504) | Agilent-014850 Whole Human Genome 4x44K (GEO: GSE25935) | Agilent Technologies (Custom ~40K transcripts) (GEO: GSE9588) | Agilent Technologies (Custom ~40K transcripts) (GEO: GSE24293) |
Table 2.
Summary of cis-eQTLs (expression quantitative trait loci) in each data set and in the combined analysis. cis-eQTLs were considered statistically significant at a false discovery rate Q value ≤ 0.05. The number of genes with at least one significant cis-eQTL at false discovery rate Q values ≤ 0.1 and ≤ 0.2 are reported for reference
| Data set | cis-eQTLs | Q value ≤ 0.05 | Q value ≤ 0.1 | Q value ≤ 0.2 | ||
|---|---|---|---|---|---|---|
| cis-eQTLs associated with a single gene | cis-eQTLs associated with > 1 gene | Genes with at least one cis-eQTL | Genes with at least one cis-eQTL | Genes with at least one cis-eQTL | ||
| 1 | 322,007 | 282,066 | 39,941 | 5,998 | 8,544 | 12,402 |
| 2 | 156,182 | 139,640 | 16,542 | 5,902 | 8,323 | 11,961 |
| 3 | 681,087 | 541,382 | 139,705 | 7,748 | 10,627 | 14,609 |
| 4 | 1,872,669 | 1,265,593 | 607,076 | 14,743 | 17,318 | 18,669 |
| Combined analysis | 2,391,948 | 1,531,501 | 860,447 | 15,668 | 17,966 | 19,378 |
Figure 1.
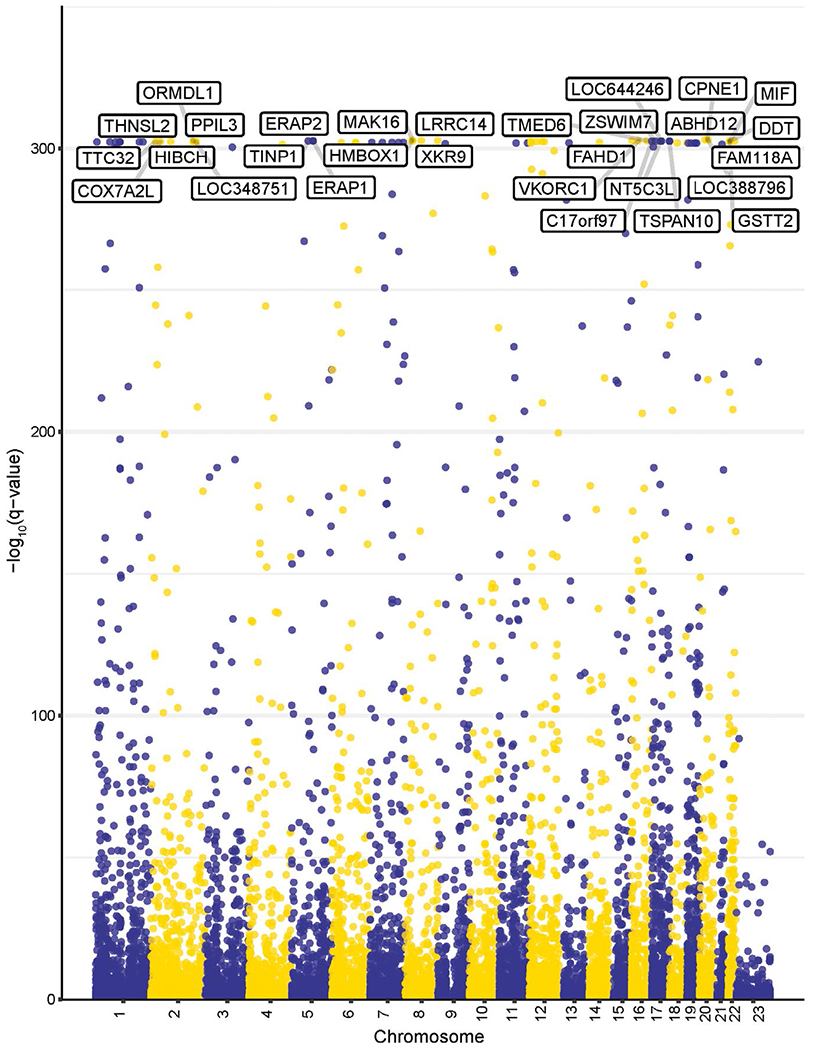
Manhattan plot of cis-eQTL (expression quantitative trait locus) associations in the liver. Each point on the graph represents a variant–gene pair. Gene names are shown for representative cis-eQTLs with a Q value < 1 E-300.
Identification of cis-eQTLs in genes of drug response
The cis-eQTLs identified above were filtered against a list of 1,496 genes of drug response (Table S1). In the four individual data sets, there were 9,072-149,712 cis-eQTLs (Table S2), and our analysis increased the number of cis-eQTLs by more than 20% to 187,829 cis-eQTLs for 1,191 genes (Table S2, Figure 2). Table S3 lists the cis-eQTLs with the lowest P value for each of 300 drug response genes with the most significant eQTL associations.
Figure 2.
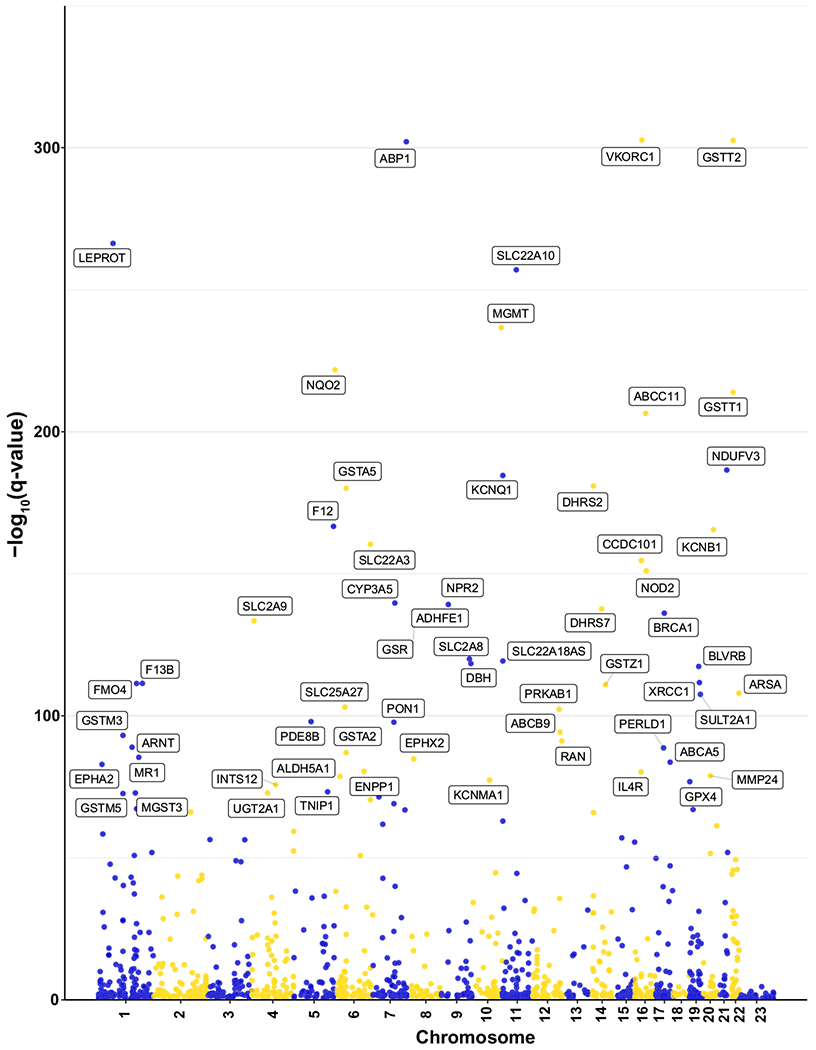
Manhattan plot of cis-eQTLs (expression quantitative trait loci) in 1,496 genes of drug response. The most significant cis-eQTL association in each drug response gene is plotted. Gene names are shown for cis-eQTLs with a Q value ≤ 1 E-75.
Translational evidence of cis-eQTLs for drug response: the example of DPYD
The dihydropyrimidine dehydrogenase (DPD) gene (DPYD) codes for the enzyme that inactivates fluoropyrimidines, including 5-fluorouracil (5-FU) and capecitabine. In our study, rs59353118 was the most significant cis-eQTL in a haplotype block associated with DPYD expression (Q value = 1.00 E-10, T statistic = −7.26), with the minor allele associated with reduced expression (Figure 3a, Figure S3A, Figure S4). When rs75017182, a splice variant in the HapB3 haplotype reducing DPYD expression,12 was included as a covariate in a conditional analysis, the effect of rs59353118 was independent and even stronger (Q value = 4.08 E-17, T statistic = −8.88). Two variants in high LD with rs59353118 (r2 > 0.94), rs72728438 and rs12022243, have been associated with decreased DPD activity in mononuclear cells13 and increased risk of capecitabine toxicity,14 respectively. No study has shown how these variants affect the expression of DPD in the liver where the inactivation of fluoropyrimidines occurs.
Figure 3.
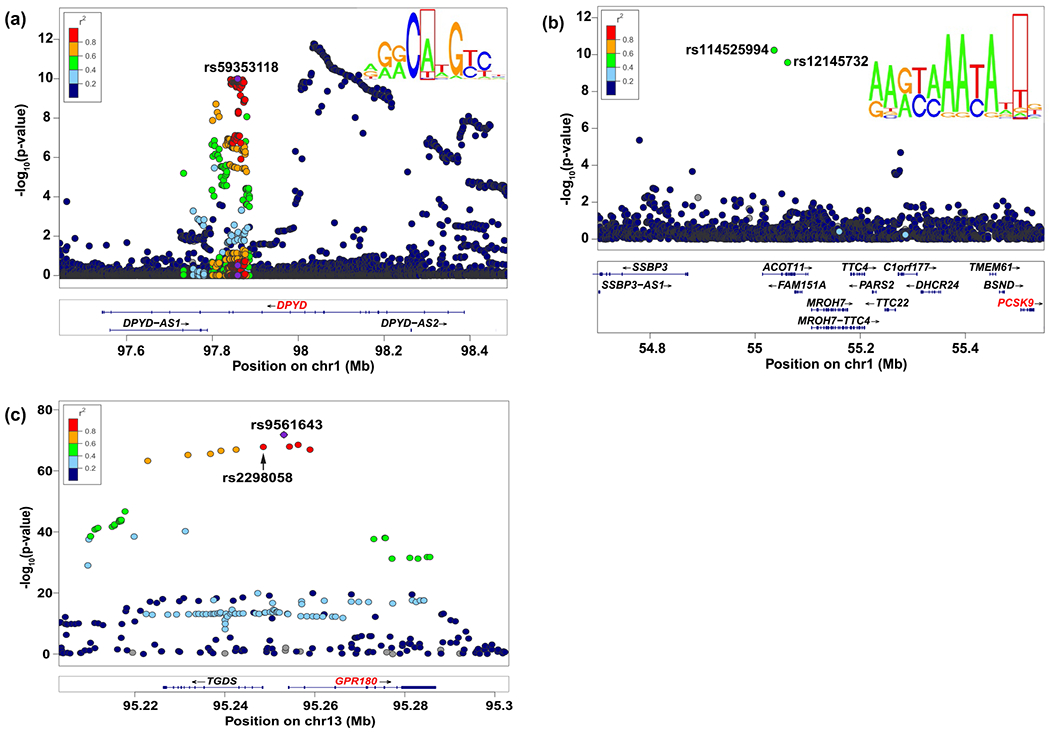
LocusZoom plots of cis-eQTLs (expression quantitative trait loci) in DPYD, PCSK9, and GPR180. (a) DPYD: rs59353118 (located in DPYD intron 14), rs75017182 (a splice variant in the HapB3 haplotype), and p53 transcription factor binding motif. (b) Two sex-biased cis-eQTLs in PCSK9: rs12145732 (r2 = 0.8 with rs114525994) is located ~ 450 kb 5′ of PCSK9 and predicted to alter FOXA transcription factor binding; FOXA transcription factor binding motif. (c) GPR180: rs9561643 is located ~ 1 kb 5′ of GPR180 and colocalized with rs2298058, a MVP (Million Veteran Program) GWAS (genome-wide association study) variant associated with blood triglyceride levels. chr, chromosome; FOXA, forkhead box protein A; Mb, megabase.
Identification of sex-biased cis-eQTLs
In the four individual data sets, there were 58–38,508 sex-biased cis-eQTLs (Table S4), and our combined analysis resulted in 1,683 sex-biased cis-eQTLs for 460 genes (Table S4, Figure 4a). Substantially more sex-biased cis-eQTLs were identified in data set 2 than in any of the other individual data sets or in our combined analysis. Data set 2 consisted of nearly twice as many males as females and this unbalanced sex ratio combined with the small sample size possibly contributed to more false positives in this data set. Filtering for the list of 1,496 genes of drug response, 116 sex-biased cis-eQTLs were identified for 42 genes (Figure 4b). Table S5 lists the sex-biased cis-eQTLs with the lowest P value for each of 300 genes with the most significant eQTL associations.
Figure 4.
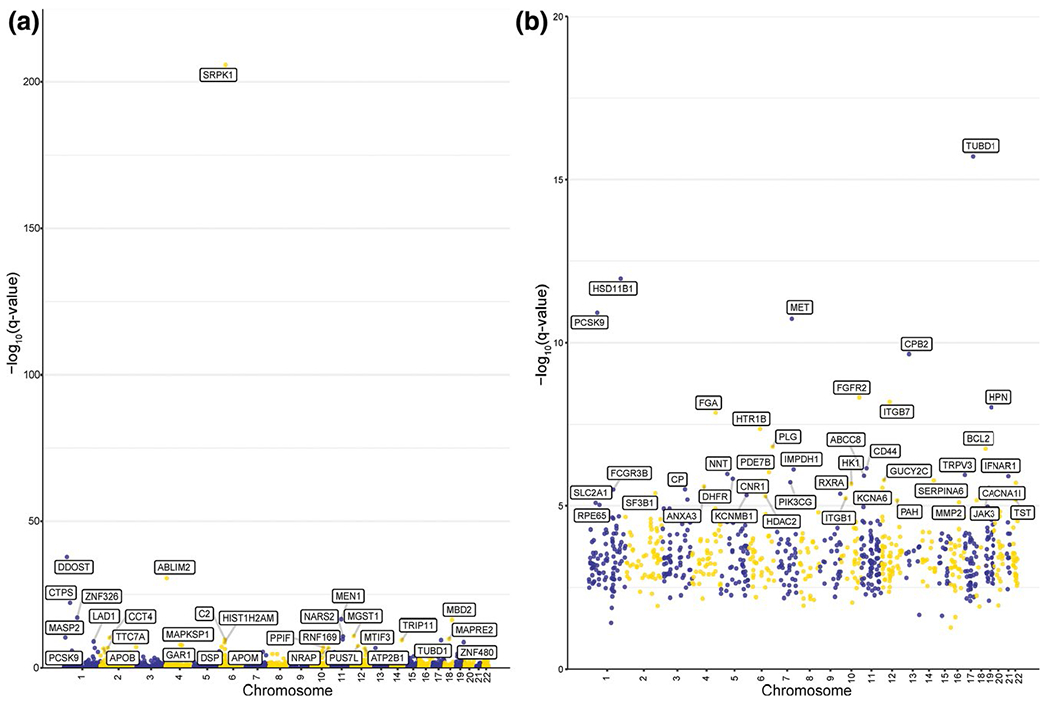
Manhattan plots of (a) sex-biased cis-eQTLs and (b) sex-biased cis-eQTLs (expression quantitative trait loci) in 1,496 genes of drug response. Genotype-by-sex interaction Q values for the most significant cis-eQTL (expression quantitative trait locus) associations in each gene are plotted. Gene names are shown for cis-eQTLs with a Q value ≤ 1 E-5.
Translational evidence of sex-biased cis-eQTLs: the example of PCSK9
PCSK9 codes for proprotein convertase subtilisin/kexin type 9, a key regulator of circulating low density lipoprotein cholesterol (LDL-C). PCSK9 is produced in the liver, and sex-biased differences in its regulation and function have been demonstrated.15,16 Estrogens have been shown to attenuate the effects of PCSK9 on LDL-C, while androgens augment the effects.17–19 Our analysis has shown two sex-biased cis-eQTLs in PCSK9 (Figure 3b): rs114525994 (Q value = 6.74 E-6, T statistic = 6.55) and rs12145732 (Q value = 2.82 E-5, T statistic = 6.32, Figure S3B) are in moderately high LD (r2 = 0.74) and associated with higher PCSK9 expression in males but not in females. Sex-biased differences in PCSK9 expression may help explain a decreased response to PCSK9 inhibitors and inability to reach optimal plasma LDL levels observed in some women (Figure S5).
Colocalization analysis of cis-eQTLs and GWAS variants associated with lipid traits
Cis-eQTLs were compared with variants associated with individual variability in lipid traits (triglycerides, LDL-C, high-density lipoprotein cholesterol (HDL-C), and total cholesterol) reported in a GWAS by the Million Veteran Program (MVP).20 Based on MVP GWAS lead variant LD (r2 > 0.8) with the most significant cis-eQTL, 73 MVP GWAS variants colocalized with eQTLs for 84 genes (some liver eQTLs associated with more than one gene, Table S6). Figure 5 shows the cis-eQTL association of the GWAS effect allele with liver gene expression of the 84 genes. Of the 73 MVP GWAS variants that colocalized with a cis-eQTL, 23 were novel associations with lipid traits identified by the MVP GWAS and represent novel candidate genes at these loci.
Figure 5.
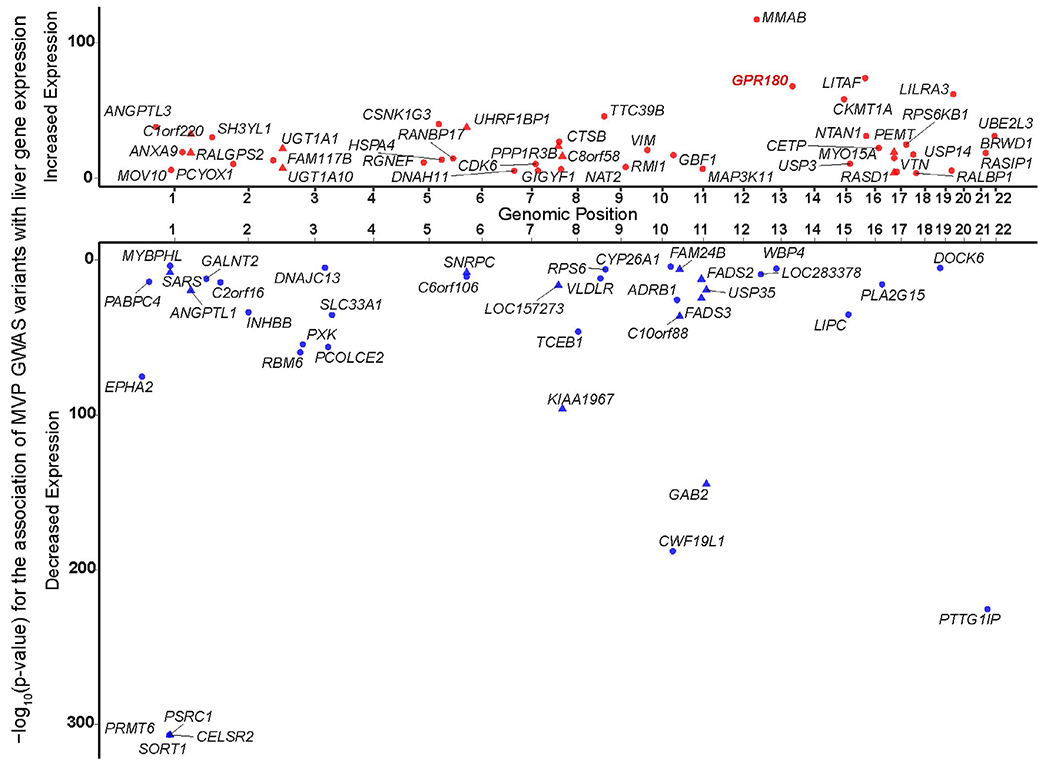
Lipid GWAS (genome-wide association study) loci that have a colocalized liver cis-eQTL (expression quantitative trait locus). Symbols represent the lead variant from the MVP (Million Veteran Program) GWAS20 colocalized with an eQTL for the named gene. The x-axis shows chromosomal positions of the lead GWAS variants, and the y-axis shows the −log10 P values of the GWAS variant’s association with gene expression levels in liver. Increased (red) or decreased (blue) expression corresponds to the direction of association of the risk allele with the MVP GWAS lipid trait, where risk is defined as increased total cholesterol, LDL (low density lipoprotein), or triglycerides, or decreased HDL (high density lipoprotein). Triangles: GWAS variants that were colocalized with a cis-eQTL for more than one gene. Circles: variants that were colocalized with a single gene. Only the associations with false discovery rate < 0.01 are shown; full results can be found in Table S6.
Translational evidence of cis-eQTLs for metabolic traits: the example of GPR180
GPR180, one of the 84 genes identified in the colocalization analysis of MVP GWAS variants, codes for a G protein-coupled receptor with an unknown endogenous ligand that has been proposed to play a role in vascular remodeling.21 However, its role in lipid metabolism is unknown. The minor allele of rs2298058 was associated with increased triglyceride levels in the MVP GWAS and colocalized with rs9561643 (r2 = 0.95, Table S6), the most significant cis-eQTL (Q value = 4.60 E-69, T statistic = 18.01) for GPR180 (Figure 3c), which associated with increased expression (Figure S3C, Figure S6). In our liver analysis, rs2298058 was also associated with increased GPR180 expression (Q value = 4.07 E-65, T statistic = 17.50). Development of treatments aimed at reducing GPR180 levels could represent a therapeutic target for the reduction of triglycerides in patients.
DISCUSSION
This is the largest liver eQTL study reported to date. The increased statistical power resulting from inclusion of 1,183 individuals resulted in the ability to detect novel eQTLs, providing greater understanding of variation of gene expression and its genetic regulation. By performing a combined analysis, 1.4–15.5-fold more cis-eQTLs were identified when compared with the individual data sets alone. A recently reported analysis including 588 livers22 mapped liver cis-eQTLs for variants with a minor allele frequency (MAF) ≥ 0.05 and focused on loci for age-related macular degeneration. Our analysis has focused on three important liver-related phenotypes: drug response, metabolic traits, and sex dimorphism in liver-related traits. The implications of obtaining a new, comprehensive map of liver eQTLs are vast, due to the central role of the liver in homeostasis, and this large analysis of liver eQTLs provides a resource to make new discoveries pertaining to the genetic basis of liver-related traits.
Canonical pathway analysis demonstrated significant enrichment of cis-eQTLs associated with genes in pathways highly relevant in liver phase I-II metabolic processes, including cytochrome P450 enzymes, hepatic transporters, uridine 5′-diphospho-glucuronosyl-transferases, and glutathione S-transferases. There was an enrichment of cis-eQTLs in pathways associated with detoxification of reactive intermediates of oxidative stress, which is involved in diseases such as atherosclerosis,23 and in genes associated with retinoic × receptor down regulation in liver acute phase response to inflammation, a key feature of metabolic disorders such as dyslipidemia and diabetes.24
It has been reported that GWAS loci associated with complex traits are more likely to be eQTLs.25 Analysis of liver cis-eQTLs in loci associated with any phenotype in the GWAS catalog demonstrated a more than threefold enrichment. Similar enrichment was seen when GWAS loci were restricted to drug response traits. When enrichment of cis-eQTLs associated with the expression of 1,496 drug response genes was investigated in GWAS loci of drug response, there was an 11-fold enrichment of the most highly significant cis-eQTLs (P value < 1 E-50). This supports the notion that noncoding variation is particularly relevant to interindividual variation in drug response.
Investigation of eQTLs of drug response, using preliminary results obtained from this analysis, have identified new genetic variants affecting clinically relevant response in patients. We have demonstrated a link between rs8192675, a cis-eQTL regulating expression of the glucose transporter gene SLC2A2, and metformin efficacy26 and identified ATM as a target gene for variants within an enhancer region associated with metformin efficacy.27
Here, we report the identification of novel genetic variants that might identify patients at risk of severe 5-FU toxicity. Despite 5-FU being among the most commonly prescribed chemotherapeutic agents, up to 34% of patients treated with 5-FU and other fluoropyrimidines develop severe toxicity. Up to 85% of administered 5-FU is metabolized by DPD, making DPD a crucial detoxifying enzyme. Our analysis identified a haplotype block of eQTLs which associated with decreased DPYD expression. The most significant cis-eQTL in this haplotype block, rs59353118, is in high LD with variants associated with decreased DPD activity in Europeans, rs72728438 (r2 = 0.97),13 and toxicity of capecitabine (a 5-FU prodrug), rs12022243 (r2 = 0.94).14 A variant in high LD with rs59353118 and rs72728438 (r2 > 0.97), rs72728443, is located in an open chromatin region enriched for histone modifications associated with enhancer activity (Table S7). A p53 binding motif (Figure 3a) also spans rs72728443. Altered binding of the DPYD repressor, p53, to the variant allele of rs72728443 suggests a mechanism by which cis-eQTLs identified in this analysis might alter liver DPYD expression, leading to toxic effects of fluoropyrimidines. After 30 years of research on DPYD genetics and 5-FU, new genetic variants of the risk of 5-FU severe toxicity should be discovered to improve the limited predictive power of DPYD genetic testing.12
Zhang et al.5 have reported that 3.7% of genes in human liver demonstrate sex-biased expression, while Dimas et al.28 found that ~ 12%–15% of eQTLs expressed in lymphoblastoid cell lines are sex biased. Sex-biased differences in energy storage and metabolism have been shown to result in variability in response to pharmacologic agents as well as the onset and manifestation of diseases, including dyslipidemia, diabetes, and CAD. Therefore, a better understanding of the mechanisms involved in sex-biased differential regulation of gene expression in human liver could be of substantial biological and clinical importance. Plasma levels of LDL-C have been shown to be sex-biased, with concentrations higher in men than women, and regulated, in part, by PCSK9.15 Liver overexpression of PCSK9 leads to increased liver and plasma PCSK9 levels, reduced liver LDL receptor levels, and reduced plasma LDL-C clearance.29 PCSK9 has also been associated with the severity of coronary atherosclerosis resulting from the inability to achieve optimal reductions in plasma LDL levels.30 The PCSK9 inhibitors alirocumab and evolocumab are recently approved monoclonal antibodies that target and inactivate PCSK9. A sex-biased response to PCSK9 antibody therapy has been observed, with men experiencing greater reductions in plasma LDL-C levels than women.7 Our analysis has identified sex-biased cis-eQTLs associated with increased liver PCSK9 expression in men. Specifically, rs114525994 is predicted to affect binding of transcription factors, while rs12145732 is associated with chromatin marks indicating an active enhancer region and is predicted to alter binding motifs of forkhead box protein A (FOXA) transcription factors (Table S8). Variants at FOXA binding sites have been shown to decrease binding of both FOXA transcription factors and estrogen receptor alpha to their targets in human liver.19 Liver cis-eQTLs that alter FOXA binding sites could modulate the effect of sex hormones on gene expression, suggesting a mechanism by which sex-biased liver cis-eQTLs might differentially regulate liver expression of PCSK9.
Changes in levels of metabolites are important biomarkers for many diseases, and integration of liver eQTLs with metabolic traits has the potential to identify novel genetic loci associated with perturbations in metabolic homeostasis. Dysregulation of blood lipid levels has been associated with metabolic disorders, including type 2 diabetes, obesity, and metabolic syndrome. This analysis identified 23 novel cis-eQTLs that colocalized with GWAS variants associated with blood lipid traits, suggesting that alterations in liver gene expression may provide an explanation for the effect of these variants on blood lipid levels. For example, rs9561643 is a cis-eQTL that increased expression of GPR180 and colocalized with rs2298058, a GWAS variant associated with higher plasma triglycerides. Bioinformatic analyses of these two variants and variants in strong LD (r2 > 0.8) indicated the strongest evidence for functional effects for rs2298058 and rs9561643. Both rs2298058 and rs9561643 are predicted to be in regions of open chromatin and associated with chromatin marks for active regulatory regions. Activating transcription factor 3 (ATF3) and nuclear respiratory factor 1 (NRF1) have been shown to bind in the region of rs2298058 in liver cells (Table S9). Decreased expression of ATF3 has been shown to increase serum triglyceride levels,31 while binding of both homodimers of ATF3 and heterodimers of ATF3 with NRF1 have been shown to repress gene transcription.32 These results suggest that cis-eQTLs that decrease the binding of either ATF3 or NRF1 may lead to increased GPR180 expression and serum triglyceride levels. A GWAS variant in GPR180 has been associated with the circulating mass of lipoprotein-associated phospholipase A2, a proinflammatory enzyme that binds to LDL-C.33 When this analysis was adjusted for baseline LDL-C, HDL-C, and triglycerides, however, the association was no longer significant, suggesting a link between GPR180 and lipid levels. To date, there are no definitive studies linking GPR180 function and lipid metabolism. GPR180 represents a druggable target and the development of GPR180 antagonists could represent an effective therapeutic intervention for hypertriglyceridemia. G-coupled protein receptors have been commonly used as therapeutic targets, particularly in the treatment of obesity and dyslipidemia,34 and using this liver analysis, we have identified GPR180 as a potential new target of drug development.
A limitation of our study is that tissue samples for each data set were collected using different sample collection and storage protocols, and patient populations differed in health status and exposure to clinical interventions prior to tissue collection. We have controlled for this variability by adjusting our expression analysis for hidden biological and technical variation that might affect gene expression. We have also utilized a fixed effect model which has been shown to increase power of detection in the presence of heterogeneity among data sets. This statistical approach allows for greater discovery of eQTLs. However, the cis-eQTLs with the strongest signals were those that were common across data sets, indicating the robustness of these signals to heterogeneity among the data sets. Since these common cis-eQTLs were also utilized for the pathway analysis, the results of this analysis are also refractory to the heterogeneity among data sets.
This study was performed in individuals of European ancestry. Care should be taken when applying results for eQTLs generated in Europeans to other ethnic populations, in particular when MAFs or haplotype structure differ markedly. Our study poses the first basis for testing the most significant cis-eQTLs in samples and data sets of subjects with different ethnicity.
In conclusion, as a result of the increased statistical power resulting from the large sample size of 1,183 individuals, this analysis has identified novel cis-eQTLs associated with interindividual variability in drug response and metabolic profiles, including sex-biased differences in risk and severity of disease and response to drug therapy. This comprehensive liver eQTL analysis represents an invaluable foundational resource to expand our biological knowledge of liver-related diseases and can serve as a guide to the discovery of genetic markers and novel targets of drug development. Among the many possible applications of this data set to drive future investigations, our study can inform the biological basis of previously annotated large GWAS. These discoveries can be used to identify and guide development of new drug targets. They can also enrich existing or new genotyping platforms with new noncoding, regulatory genetic biomarkers. We also envision a role for these results in improving transcriptome-wide association studies.
MATERIALS AND METHODS
Data sets
This analysis utilized deidentified genotype and gene expression data, including data from the database of Genotypes and Phenotypes (dbGaP) and Gene Expression Omnibus (GEO) repositories, and has been determined to be nonhuman subject research by the University of North Carolina at Chapel Hill Institutional Review Board (study number 10–2253). Our analysis included four human liver data sets of genotype and gene expression microarray data (see Table 1, for demographics, details of platforms used, and GEO accession numbers). Schroder et al. (data set 1) profiled 149 samples from normal noncancerous liver tissue resected from patients with liver cancer.35 All tissue samples were examined by a pathologist, and only histologically noncancerous tissues were used for analysis. Innocenti et al. (data set 2) profiled 205 normal (nondiseased) postmortem liver samples from organ donors.2 Schadt et al. (data set 3) profiled 427 postmortem and surgical resection liver samples from organ donors,36 and similar to data set 2, tissue samples came from donor livers that were not used for whole organ transplants or from tissue that remained following a partial graft into a smaller recipient. Greenawalt et al. profiled 960 liver samples (data set 4) collected at the time of Roux-en-Y gastric bypass surgery.37
Cis-eQTL analysis
To test for associations between genotype and gene expression, an additive (codominant) linear model was employed in the Matrix eQTL software package (http://www.bios.unc.edu/research/genomic_software/Matrix_eQTL/).38 A 1 megabase window flanking the transcriptional start/stop sites was used to identify cis-eQTLs. For each data set, minor allele dosage, filtered to exclude variants with MAF < 0.01, was used to examine genotype association with rank inverse quantile normalized gene expression. Covariates included sex and age, the first 1 (data sets 1–3) or 5 (data set 4) principal components from genetic ancestry analysis, and 15–35 hidden factors identified using PEER (https://www.sanger.ac.uk/science/tools/PEER).39
Following identification of cis-eQTLs in each individual data set, cis-eQTLs identified in at least two data sets were included in the combined analysis. The T statistics from the additive linear model for each cis-eQTL within each data set were used to generate a meta-T-statistic as follows:
For each data set i in this equation the T statistic generated by Matrix eQTL (ti) is weighted by wi, and the sum of these weighted T statistics is calculated. The rationale for the use of these weights follows a principle that the variance of regression coefficients is inversely proportional to sample size. The true accuracy of each platform is unknown, and expression on the platforms is only measured in a relative sense. As the collective sample size was large, this meta-T-statistic was assumed to be normally distributed under the null and was used as a measure of the effect size of eQTLs and also to calculate the associated P value. Cis-eQTLs with a false discovery rate Q value < 0.05 were considered statistically significant.
Cis-eQTLs in genes of drug response
A list of 1,496 genes of drug response was compiled from the Pharmacogenomics Knowledge Base (PharmGKB) (http://www.pharmgkb.org/), a comprehensive database that curates information about the impact of genetic variation on drug response; the PharmaADME Working group list of absorption, distribution, metabolism, and excretion genes; the US Food and Drug Administration (FDA) Pharmacogenomics Biomarkers, the Nuclear Receptor Signaling Atlas (NURSA) Consortium; the DrugBank catalog (https://www.drugbank.ca/), a comprehensive database containing information on drug targets; and the literature35,40 (Table S1). This list was used to filter eQTLs for association with genes of drug response.
Identification of sex-biased cis-eQTLs
The interaction model of Matrix eQTL was used to test for sex-biased eQTLs. This model tests for equality of effect sizes between males and females by adding a genotype-by-sex interaction term to the linear regression analysis. Following determination of sex-biased cis-eQTLs within each data set, the resulting T statistics for each cis-eQTL from each data set were used to generate a meta-T-statistic as described above. False discovery rate Q values < 0.05 were considered statistically significant.
Colocalization analysis of cis-eQTLs and GWAS variants of blood metabolite levels
To investigate whether liver gene regulation might influence blood metabolite levels, the colocalization of cis-eQTLs with GWAS loci for blood metabolites reported by Shin et al.41 was investigated. PLINK (http://zzz.bwh.harvard.edu/plink/download.shtml) was used to estimate the LD between the metabolite GWAS variants reported by Shin et al.41 and, for any variants with LD r2 > 0.8, one of the variants was pruned from the analysis. Colocalization was defined as metabolite GWAS variants in strong LD (r2 > 0.8 in European samples from the 1,000 Genomes Project Phase 3) with the most significant liver cis-eQTL for the same gene. A similar analysis was performed to determine the colocalization of liver cis-eQTLs and blood lipid trait (triglycerides, HDL-C, LDL-C, and total cholesterol) variants reported by the Million Veteran Program (MVP).20 The MVP lipid GWAS included European, black, and Hispanic participants and, in order to assess the maximum number of lipid-associated variants, our colocalization analysis included GWAS lipid variants that were significant in any population. Using the bioinformatics tool swiss (github.com/statgen/swiss), representative MVP GWAS variants associated with one or more lipid traits and present in the 1,000 Genomes Project Phase 3 were selected and clumped such that the pairwise LD of the representative MVP GWAS variants have r2 < 0.8 with all other representative MVP GWAS variants. Colocalization analysis between MVP GWAS variants and liver cis-eQTLs was then performed using European samples from the 1,000 Genomes Project Phase 3 as the reference panel for determination of LD between GWAS variants and cis-eQTLs.
Supplementary Material
Table S7. Bioinformatic analysis of DPYD cis-eQTLs. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs59353118 in Europeans from the 1,000 Genomes Project and the region is relative to the DPYD gene. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChiP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. DNase 1, Deoxyribonuclease 1; eQTLs, expression quantitative trait loci; NA, data not available in relevant cell lines.
Table S8. Bioinformatic analysis of two sex-biased cis-eQTLs associated with PCSK9 expression. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs114525994 in Europeans from the 1,000 Genomes Project. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChiP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. DNase 1, Deoxyribonuclease 1; eQTLs, expression quantitative trait loci; LD, linkage disequilibrium.
Table S9. Bioinformatic analysis of the lead MVP GWAS variant for plasma triglyceride levels and colocalized cis-eQTL in GPR180 and variants in LD r2 > 0.8. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs2298058 in Europeans from the 1,000 Genomes Project. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChIP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. SNPnexus noncoding variation scoring predicts functional impact of noncoding variants using 8 noncoding variant scoring algorithms. Variants were ranked based on these scores (1 = most likely to be functional/deleterious, 10 = least likely to be functional/deleterious). DNase 1, Deoxyribonuclease 1; eQTL, expression quantitative trait locus; GWAS, genome-wide association study; LD, linkage disequilibrium; MVP, Million Veteran Program.
Table S10. Positive controls in genes of drug response. Cis-eQTL signals previously associated with gene expression in the human liver were reproduced in our study. eQTLs, expression quantitative trait loci.
Table S11. Ingenuity Pathway Analysis (IPA) of 3,165 genes containing at least one significant cis-eQTL. eQTL, expression quantitative trait locus.
Table S1. List of 1,496 genes of drug response compiled from PharmGKB, PharmADME, FDA Pharmacogenomics Biomarkers, the Nuclear Receptor Signaling Atlas Consortium, the DrugBank Catalog, and the literature.
Table S2 Summary of cis-eQTLs in 1,496 genes of drug response in each data set and the combined analysis. eQTLs, expression quantitative trait loci.
Table S3. Cis-eQTLs with the lowest P value for each of the 300 drug response genes with the most significant eQTLs. eQTLs, expression quantitative trait loci.
Table S4. Summary of sex-biased cis-eQTLs in autosomal genes in each data set and the combined analysis. eQTLs, expression quantitative trait loci.
Table S5. Sex-biased cis-eQTLs with the lowest P value for each of the 300 genes with the most significant eQTLs. eQTLs, expression quantitative trait loci.
Figure S1. Sample quality control.
Figure S2. Genotype microarray preprocessing and quality control.
Figure S3. Genotype versus mRNA (messenger RNA) expression plots for (A) rs59353118 and DPYD, (B) rs12145732 and PCSK9, and (C) rs9561643 and GPR180.
Figure S4. Schematic showing the association of rs59353118 with decreased DPYD expression and increased fluoropyrimidine toxicity. The direction of the effect is based on the underlined allele. rs59353118 colocalizes with rs12022243 which was associated with increased fluoropyrimidine toxicity in a candidate gene study.
Figure S5. Schematic showing the association of rs114525994 with increased PCSK9 expression in males but not females. Increased PCSK9 expression is associated with increased plasma LDL-C (low density lipoprotein cholesterol) levels and may help explain a greater reduction in LDL-C levels observed in some men following PCSK9 inhibitor treatment. The direction of the effect is based on the underlined allele.
Figure S6. Schematic showing the association of rs9561643 with increased GPR180 expression and increased plasma triglyceride levels. The direction of the effect is based on the underlined allele. rs9561643 colocalized with rs2298058 which was associated with increased plasma triglyceride levels in a GWAS (genome-wide association study) of lipid traits.
Figure S7. Canonical pathway analysis of cis-eQTLs. Genes whose expression was associated with a cis-eQTL using a P Simes’ threshold of 1 E-20 were analyzed for enrichment in canonical pathways using Ingenuity Pathway Analysis (Qiagen Bioinformatics, Germantown, MD). Pathways which showed significant enrichment in liver eQTLs (Q value < 0.05) are shown. Numbers in bold indicate the total number of genes in the canonical pathway. eQTLs, expression quantitative trait loci.
Figure S8. Enrichment of liver cis-eQTLs in traits from the NHGRI-EBI Catalog of Published GWAS. All GWAS (genome-wide association study) loci identified in populations of European descent were downloaded from the NHGRI-EBI Catalog of Published GWAS and variants in high LD (r2 > 0.8) were identified using European individuals from the 1,000 Genomes Project Phase 3 data as a reference population. Liver cis-eQTLs that colocalized with any GWAS locus (GWAS) or GWAS loci associated with drug response as a phenotype (GWAS Drug Response) were divided into bins based on P value and the ratio of the number of colocalized cis-eQTLs vs. all cis-eQTLs identified in the combined analysis in each bin was plotted. Similarly, colocalization of liver cis-eQTLs identified in drug response genes with GWAS loci of drug response phenotypes was determined and plotted (Drug Response eQTL in GWAS Drug Response). The red line indicates a ratio = 1 (i.e., no enrichment). eQTLs, expression quantitative trait loci; LD, linkage disequilibrium.
Table S6. Colocalization of liver cis-eQTLs with lipid GWAS variants reported by the Million Veterans Program (MVP) (20). eQTLs, expression quantitative trait loci; GWAS, genome-wide association study.
Table S12. Colocalization of liver cis-eQTLs with blood metabolite GWAS variants reported by Shin et al. (41). eQTLs, expression quantitative trait loci.
Supplementary Methods and Results. Supplementary Methods and Results.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ In expression quantitative trait locus (eQTL) mapping, sample size greatly affects the probability of discovering novel eQTLs. This is the largest liver eQTL study reported to date, resulting in increased statistical power to detect novel eQTLs, thus providing a greater understanding of variation of liver gene expression and its genetic regulation.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This analysis has identified novel cis-eQTLs associated with interindividual variability in drug response, metabolic profiles, and sex-biased differences in risk and severity of disease and response to therapy.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ This comprehensive liver eQTL analysis represents an invaluable foundational resource to expand our biological knowledge of liver-related diseases and can serve as a guide to the discovery of genetic markers and novel targets of drug development.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Increased knowledge of genetic variants responsible for variability in liver gene expression, especially the identification of novel regulatory variants in genes of drug response, will provide the basis for translating genetic variations into clinically relevant tools.
Acknowledgments
FUNDING
This work was funded in part by a National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R21DK081157-01A2, to F.I.
Footnotes
SUPPORTING INFORMATION
Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
CONFLICT OF INTEREST/DISCLOSURE
All authors declared no competing interests for this work.
References
- 1.Wright FA et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet 46, 430–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Innocenti F et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 7, e1002078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glubb DM & Innocenti F Mechanisms of genetic regulation in gene expression: examples from drug metabolizing enzymes and transporters. Wiley Interdiscip. Rev. Syst. Biol. Med 3, 299–313 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Glubb DM, Dholakia N & Innocenti F Liver expression quantitative trait loci: a foundation for pharmacogenomic research. Front. Genet 3, 153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y et al. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS One 6, e23506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franconi F, Brunelleschi S, Steardo L & Cuomo V Gender differences in drug responses. Pharmacol. Res 55, 81–95 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Endocrinologic and Metabolic Drugs Advisory Committee. FDA Advisory Committee Briefing Document Praluent (alirocumab) (US FDA, Silver Spring, 2015). [Google Scholar]
- 8.Mombelli G et al. Gender-related lipid and/or lipoprotein responses to statins in subjects in primary and secondary prevention. J. Clin. Lipidol 9, 226–233 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Westra HM, Franke L From genome to function by studying eQTLs. Biochim. Biophys Acta 1842, 1896–1902 (2014). [DOI] [PubMed] [Google Scholar]
- 10.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher PM et al. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amstutz U et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther 103, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seck K et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clin. Cancer Res 11, 5886–5892 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Rosmarin D et al. A candidate gene study of capecitabine-related toxicity in colorectal cancer identifies new toxicity variants at DPYD and a putative role for ENOSF1 rather than TYMS. Gut 64, 111–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakoski SG, Lagace TA, Cohen JC, Horton JD & Hobbs HH Genetic and metabolic determinants of plasma PCSK9 levels. J. Clin. Endocrinol. Metab 94, 2537–2543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayne J et al. Plasma PCSK9 levels correlate with cholesterol in men but not in women. Biochem. Biophys. Res. Commun 361, 451–456 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Croston GE, Milan LB, Marschke KB, Reichman M & Briggs MR Androgen receptor-mediated antagonism of estrogen-dependent low density lipoprotein receptor transcription in cultured hepatocytes. Endocrinology 138, 3779–3786 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Persson L, Henriksson P, Westerlund E, Hovatta O, Angelin B & Rudling M Endogenous estrogens lower plasma PCSK9 and LDL cholesterol but not Lp(a) or bile acid synthesis in women. Arterioscler. Thromb. Vasc. Biol 32, 810–814 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Tuteja G, Schug J & Kaestner KH Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 148, 72–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klarin D et al. Genetics of blood lipids among ~300,000 multiethnic participants of the Million Veteran Program. Nat. Genet 50, 1514–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukada S et al. Inhibition of experimental intimal thickening in mice lacking a novel G-protein-coupled receptor. Circulation 107, 313–319 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Strunz T et al. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci. Rep 8, 5865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimura J & Itoh K Role of Nrf2 in the pathogenesis of atherosclerosis. Free Radic. Biol. Med. 88, 221–232 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Plutzky J & Kelly DP The PPAR-RXR transcriptional complex in the vasculature. Circ. Res 108, 1002–1016 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME & Cox NJ Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou K et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet 48, 1055–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luizon MR et al. Genomic characterization of metformin hepatic response. PLoS Genet. 12, e1006449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimas AS et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 22, 2368–2375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagace TA et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest 116, 2995–3005 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhary R, Garg J, Shah N & Sumner A PCSK9 inhibitors: A new era of lipid lowering therapy. World J. Cardiol 9, 76–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou C-L et al. Role of activating transcription factor 3 in fructose-induced metabolic syndrome in mice. Hypertens. Res 41, 589–597 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Brown SL, Sekhar KR, Rachakonda G, Sasi S & Freeman ML Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2–related factor 2 (Nrf2)–regulated stress pathway. Cancer Res. 68, 364–368 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Chu AY et al. Genome-wide association study evaluating lipoprotein-associated phospholipase A2 mass and activity at baseline and after rosuvastatin. Therapy. Circ. Cardiovasc. Genet 5 676–685 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Kimple ME, Neuman JC, Linnemann AK & Casey PJ Inhibitory G proteins and their receptors: emerging therapeutic targets for obesity and diabetes. Exp. Mol. Med 46, e102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrüder A et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 13, 12–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schadt EE et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenawalt DM et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shabalin AA Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stegle O, Parts L, Durbin R & Winn J A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol 6, e1000770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhibber A et al. Genomic architecture of pharmacological efficacy and adverse events. Pharmacogenomics 15, 2025–2048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin SY et al. An atlas of genetic influences on human blood metabolites. Nat. Genet 46, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S7. Bioinformatic analysis of DPYD cis-eQTLs. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs59353118 in Europeans from the 1,000 Genomes Project and the region is relative to the DPYD gene. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChiP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. DNase 1, Deoxyribonuclease 1; eQTLs, expression quantitative trait loci; NA, data not available in relevant cell lines.
Table S8. Bioinformatic analysis of two sex-biased cis-eQTLs associated with PCSK9 expression. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs114525994 in Europeans from the 1,000 Genomes Project. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChiP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. DNase 1, Deoxyribonuclease 1; eQTLs, expression quantitative trait loci; LD, linkage disequilibrium.
Table S9. Bioinformatic analysis of the lead MVP GWAS variant for plasma triglyceride levels and colocalized cis-eQTL in GPR180 and variants in LD r2 > 0.8. The LD r2 refers to data obtained from the 1,000 Genomes Project and is in relation to rs2298058 in Europeans from the 1,000 Genomes Project. The RegulomeDB score represents the evidence that each variant functions in a regulatory role (1-strong evidence, 6-weak evidence). ENCODE data includes experimental information in HepG2 cells or HUVEC (human umbilical vein endothelial cells) for ChIP-seq and DNase 1 hypersensitivity, as well chromatin state and transcription factor binding motifs that were identified using a combination of computational and experimental data available in the Regulome DB and/or HaploReg v4 databases. SNPnexus noncoding variation scoring predicts functional impact of noncoding variants using 8 noncoding variant scoring algorithms. Variants were ranked based on these scores (1 = most likely to be functional/deleterious, 10 = least likely to be functional/deleterious). DNase 1, Deoxyribonuclease 1; eQTL, expression quantitative trait locus; GWAS, genome-wide association study; LD, linkage disequilibrium; MVP, Million Veteran Program.
Table S10. Positive controls in genes of drug response. Cis-eQTL signals previously associated with gene expression in the human liver were reproduced in our study. eQTLs, expression quantitative trait loci.
Table S11. Ingenuity Pathway Analysis (IPA) of 3,165 genes containing at least one significant cis-eQTL. eQTL, expression quantitative trait locus.
Table S1. List of 1,496 genes of drug response compiled from PharmGKB, PharmADME, FDA Pharmacogenomics Biomarkers, the Nuclear Receptor Signaling Atlas Consortium, the DrugBank Catalog, and the literature.
Table S2 Summary of cis-eQTLs in 1,496 genes of drug response in each data set and the combined analysis. eQTLs, expression quantitative trait loci.
Table S3. Cis-eQTLs with the lowest P value for each of the 300 drug response genes with the most significant eQTLs. eQTLs, expression quantitative trait loci.
Table S4. Summary of sex-biased cis-eQTLs in autosomal genes in each data set and the combined analysis. eQTLs, expression quantitative trait loci.
Table S5. Sex-biased cis-eQTLs with the lowest P value for each of the 300 genes with the most significant eQTLs. eQTLs, expression quantitative trait loci.
Figure S1. Sample quality control.
Figure S2. Genotype microarray preprocessing and quality control.
Figure S3. Genotype versus mRNA (messenger RNA) expression plots for (A) rs59353118 and DPYD, (B) rs12145732 and PCSK9, and (C) rs9561643 and GPR180.
Figure S4. Schematic showing the association of rs59353118 with decreased DPYD expression and increased fluoropyrimidine toxicity. The direction of the effect is based on the underlined allele. rs59353118 colocalizes with rs12022243 which was associated with increased fluoropyrimidine toxicity in a candidate gene study.
Figure S5. Schematic showing the association of rs114525994 with increased PCSK9 expression in males but not females. Increased PCSK9 expression is associated with increased plasma LDL-C (low density lipoprotein cholesterol) levels and may help explain a greater reduction in LDL-C levels observed in some men following PCSK9 inhibitor treatment. The direction of the effect is based on the underlined allele.
Figure S6. Schematic showing the association of rs9561643 with increased GPR180 expression and increased plasma triglyceride levels. The direction of the effect is based on the underlined allele. rs9561643 colocalized with rs2298058 which was associated with increased plasma triglyceride levels in a GWAS (genome-wide association study) of lipid traits.
Figure S7. Canonical pathway analysis of cis-eQTLs. Genes whose expression was associated with a cis-eQTL using a P Simes’ threshold of 1 E-20 were analyzed for enrichment in canonical pathways using Ingenuity Pathway Analysis (Qiagen Bioinformatics, Germantown, MD). Pathways which showed significant enrichment in liver eQTLs (Q value < 0.05) are shown. Numbers in bold indicate the total number of genes in the canonical pathway. eQTLs, expression quantitative trait loci.
Figure S8. Enrichment of liver cis-eQTLs in traits from the NHGRI-EBI Catalog of Published GWAS. All GWAS (genome-wide association study) loci identified in populations of European descent were downloaded from the NHGRI-EBI Catalog of Published GWAS and variants in high LD (r2 > 0.8) were identified using European individuals from the 1,000 Genomes Project Phase 3 data as a reference population. Liver cis-eQTLs that colocalized with any GWAS locus (GWAS) or GWAS loci associated with drug response as a phenotype (GWAS Drug Response) were divided into bins based on P value and the ratio of the number of colocalized cis-eQTLs vs. all cis-eQTLs identified in the combined analysis in each bin was plotted. Similarly, colocalization of liver cis-eQTLs identified in drug response genes with GWAS loci of drug response phenotypes was determined and plotted (Drug Response eQTL in GWAS Drug Response). The red line indicates a ratio = 1 (i.e., no enrichment). eQTLs, expression quantitative trait loci; LD, linkage disequilibrium.
Table S6. Colocalization of liver cis-eQTLs with lipid GWAS variants reported by the Million Veterans Program (MVP) (20). eQTLs, expression quantitative trait loci; GWAS, genome-wide association study.
Table S12. Colocalization of liver cis-eQTLs with blood metabolite GWAS variants reported by Shin et al. (41). eQTLs, expression quantitative trait loci.
Supplementary Methods and Results. Supplementary Methods and Results.


