Abstract
Viruses, despite their great abundance and significance in biological systems, remain largely mysterious. Indeed, the vast majority of the perhaps hundreds of millions of viral species on the planet remain undiscovered. Additionally, many viruses deposited in central databases like GenBank and RefSeq are littered with genes annotated as ‘hypothetical protein’ or the equivalent. Cenote-Taker 2, a virus discovery and annotation tool available on command line and with a graphical user interface with free high-performance computation access, utilizes highly sensitive models of hallmark virus genes to discover familiar or divergent viral sequences from user-input contigs. Additionally, Cenote-Taker 2 uses a flexible set of modules to automatically annotate the sequence features of contigs, providing more gene information than comparable tools. The outputs include readable and interactive genome maps, virome summary tables, and files that can be directly submitted to GenBank. We expect Cenote-Taker 2 to facilitate virus discovery, annotation, and expansion of the known virome.
Keywords: virus discovery, virus annotation, genome annotation, viral metagenomics, research tool, prophage
1. Introduction
Virus hunters have a challenging signal-to-noise problem to consider. For example, animals and bacteria share homologous genes with more amino acid identity and alignment fraction than even the most-conserved genes in some virus families (e.g. GenBank sequences: polyomavirus Large T antigen [NP_043127 vs. YP_009110677] and 50S ribosomal protein L14/60S ribosomal protein L23 [CUU95522 vs. NP_000969]). Further, there are no universal genes found in all viral genomes that could be used to probe complex datasets for viruses, whereas cellular genomes can be detected through polymerase chain reaction targeting ribosomal genes and alignment of sequences to other single-copy marker genes (Parks et al. 2018). Finally, at least hundreds of millions of virus species are likely to exist on Earth (Koonin et al. 2020), but sequences for only tens of thousands of virus species are deposited in the central GenBank virus database and high-quality genomes exist for approximately 10,000 virus species in the authoritative RefSeq database (Brister et al. 2015). Sequence space thus covers at, at best, 0.0001 per cent of the virosphere. To address these challenges, Cenote-Taker 2 is presented as a flexible tool to detect and annotate highly divergent virus sequences and facilitate deposition of these records into the central GenBank repository.
Several tools have previously been developed to detect virus sequences in complex datasets. Strategies include detection of hallmark genes conserved within known virus families (but absent in cellular genomes) (Roux et al. 2015; Starikova et al. 2020), detection of short nucleotide sequences believed to be enriched in viruses (Ren et al. 2020) (or other machine learning approaches; Amgarten et al. 2018; Zheng et al. 2019), or the ratio of genes common to virus genomes versus genes common to non-viral sequences (Paez-Espino et al. 2017). Each of these tools has pitfalls that can lead to false-positives or false-negatives and some tools are limited by minimum sequence length or are only geared to detect a limited range of virus families.
Beyond discovery and detection, de novo annotation of contigs representing viruses presents a number of challenges. To list a few, determination of genome topology, accurate calling of open reading frames (ORFs), estimation of the virus-chromosome junction in integrated proviruses, resolution of taxonomy, and, especially, accurate annotation of highly divergent homologs of known genes all present technical hurdles (Roux et al. 2019a). An even deeper problem is the misannotation of some existing GenBank entries. One random example is accession number YP_009506243, which is annotated as a densovirus virion structural protein despite the fact that it is clearly a bidnavirus type B DNA Polymerase. The error has been propagated into more recently deposited bidnavirus sequences (e.g. AWB14612, QJI53745). Relatedly, viral genes and genomes are often misidentified as host sequences (Krupovic et al. 2014). For example a mitovirus replication protein (ABK28172) is annotated as an Arabidopsis thaliana protein of ‘unknown’ function.
This article presents version 2.0 of our Cenote-Taker pipeline, which was originally geared toward elementary annotation of viruses with circular DNA genomes (Tisza et al. 2020). Cenote-Taker 2 is a more flexible tool that enables the discovery and annotation of all virus classes with DNA or RNA genomes, starting from genomic, metagenomic, transcriptomic, and metatransciptomic assemblies. It is available for use on Linux terminal and as a graphical user interface with free compute cluster usage on CyVerse (Devisetty et al. 2016) (https://de.cyverse.org/de/). The wiki (https://github.com/mtisza1/Cenote-Taker2/wiki#use-case-suggestions) contains a section on suggested parameters for different data types. Cenote-Taker 2 outpaces other currently available annotation tools, providing information for a higher percentage of genes with a higher degree of accuracy, especially for virus hallmark genes, and producing human-editable genome maps that can be opened in any number of genome viewers. Additionally, Cenote-Taker 2 performs better for discovery of viral sequences in complex datasets, with lower false-positive and false-negative rates than comparable tools.
2. Methods
2.1 Cenote-Taker 2 code
Cenote-Taker 2 was written in Bash, Perl, and Python. All scripts can be accessed on GitHub. In-depth discussion of use-cases and considerations can be found on the wiki. Installation uses Conda to manage packages (Gruning et al. 2018). BLAST and Hmmer databases developed for this tool can be found on Zenodo (https://zenodo.org/record/4031657).
2.2 Annotations of challenging viral genomes
Viral genomes with highly divergent ORFs were analyzed with Cenote-Taker 2 using default settings except “–enforce_start_codon False.”. Since VIGA default settings are highly stringent, several custom options were used to improve annotation: ‘–diamondevalue 1e-04 –diamondidthr 25 –hmmeridthr 25 –blastidthr 25’. Genome maps were visualized with MacVector 16.
2.3 Virus discovery comparison
Reads from each sequencing run were trimmed with Fastp (Chen et al. 2018), assembled with Megahit (Li et al. 2016) (default settings), and scaffolded with SOAPdenovo2 (Luo et al. 2012). Cenote-Taker 2 hallmark gene HMM (Hmmer) database (version from 21 April 2020) was used with viral hits having one or more detected hallmark genes. The Cenote-Taker 2 script requires a E value of 1e−8 as a minimum threshold for virion structural genes and 1e−15 for replication genes. VirSorter was used with ‘virome’ settings and categories 1, 2, 4, and 5 were kept. DeepVirFinder was used with the default training set and P value threshold of 0.005. Non-targeted pipeline was used with default settings. Comparisons were run on 23 April 2020.
Contigs uniquely called by Cenote-Taker 2 were determined to either have hits in the virion structural or viral genome-packaging gene HMM set and/or in the virus genome replication-associated gene HMM set. Putative viral contigs called uniquely by other sources were annotated with Cenote-Taker, using RPS-BLAST with the CDD database (1e−4 e value cutoff) and HHsearch (80% probability cutoff) with CDD, Pfam, and PDB. All annotated genes were scanned for names of viral replication or virion structural genes and domains.
Venn diagrams were prepared with InteractiVenn (Heberle et al. 2015).
2.4 Comparison of prophage pruning module to real prophage data
Reads and genomes were downloaded from NCBI. Reads from virions containing induced prophages were trimmed with Fastp (Chen et al. 2018) and aligned to their respective reference genomes with Bowtie2 (Langmead and Salzberg 2012). Edges of prophage read coverage were determined by manual inspection. Bacterial reference genomes were run through Cenote-Taker 2 with default settings and ‘–prune_prophage True --virus_domain_db virion’. Genome tracks were visualized with Integrative Genomics Viewer (Thorvaldsdottir et al. 2013).
This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov).
3. Results
3.1 Cenote-Taker 2 process overview
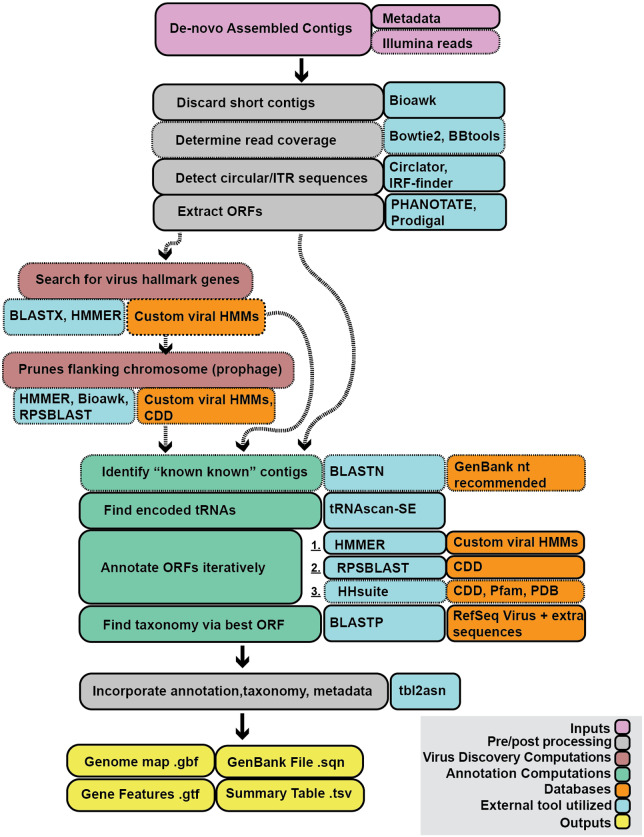
A basic run of Cenote-Taker 2 requires only a file of contigs from any biological source and a file with metadata that enables submission of annotated sequences to GenBank. A number of optional settings allow users to customize the pipeline. In-depth discussion of the options can be found at the Cenote-Taker 2 GitHub repo (https://github.com/mtisza1/Cenote-Taker2) and wiki (https://github.com/mtisza1/Cenote-Taker2/wiki). Figure 1 provides a visual of the Cenote-Taker 2 workflow. First, Cenote-Taker 2 analyzes contigs above a user-determined length and detects contigs with inverted or direct terminal repeats. Contigs with direct terminal repeats are circularized and rotated to a position where no ORFs overlap the wrap-point. Since de novo assemblies from short reads are not able to distinguish between circular DNA molecules and molecules with long direct terminal repeats, some virus genomes with long direct terminal repeats may be falsely circularized in this step. Another step uses Illumina read data to calculate the average depth of coverage for each contig. Provision of Illumina reads and coverage calculation is optional. All input contigs are then scanned for the presence of a curated set of hallmark genes specific to known virus families. To detect virus sequences in complex datasets, only contigs containing the minimum user-determined number of virus hallmark genes are moved forward for further analysis. For users who have indicated that their input contigs are pre-filtered to only contain viral contigs, all contigs are kept and annotated. Therefore, Cenote-Taker 2 can be used simply as an annotation tool, if desired.
Figure 1.
Schematic of Cenote-Taker 2 Processes. Visual representation of Cenote-Taker 2 virome analysis. Boxes with dashed lines and dashed arrows represent optional inputs or processes. The tool can be broken down into four partitions: preprocessing, virus discovery, virus annotation, and postprocessing. The preprocessing partition formats contigs, assesses their topology, and extracts ORFs based on bacteriophage/eukaryotic virus typing. The virus discovery partition (entirely optional) detects and quantifies any virus hallmark genes encoded by the contig, then the pruning module assigns prophage/chromosome borders. The annotation partition compares each putatively viral contig with a GenBank-formatted nucleotide database (optional), detects tRNA sequences, annotates ORFs using a three-tiered process, and determines family-level taxonomy based on an informative hallmark gene. The postprocessing step formats all the information into a genome map, gene feature file, and GenBank submission file for each contig, and a virome summary table is made for the entire run.
Many virus genomes are integrated into host chromosomes. In datasets likely to containing cellular chromosomes, a single contig might thus contain a virus sequence flanked on one or both sides by a cellular sequence. Users can choose to allow Cenote-Taker 2 to prune flanking cellular sequences and generate a genome map for the viral portion of the contig. Then, an optional ‘known knowns’ module step queries a nucleotide database, such as GenBank nt, with BLASTN (Altschul et al. 1990) and marks contigs with at least 90 per cent average nucleotide identity to existing database entries.
Next, candidate tRNA genes are detected and annotated (Lowe and Chan 2016). A tentative taxonomy of each contig is then inferred using BLASTX against a custom database containing Refseq virus and plasmid sequences from GenBank. This taxonomy is used to determine the best ORF-caller (PHANOTATE for putative bacteriophage (McNair et al. 2019), Prodigal for other viruses (Hyatt et al. 2010)). ORFs are then functionally annotated based on validated datasets using tools for detection of remote homologs (i.e. hmmscan (Potter et al. 2018), RPS-BLAST (Marchler-Bauer et al. 2017), then HHblits/HHsearch (Meier and Soding 2015)). In these steps, only carefully curated databases (CDD, PFam, PDB, Cenote-Taker 2 hallmark database) are queried in order to avoid propagation of mis-annotated sequences in databases such as GenBank nr. For each sequence, a hallmark gene sequence is queried against a reference database of viral proteins using BLASTP. All annotation, taxonomy information, and metadata are combined to generate several outputs. Each contig is represented as an interactive genome map file (.gbf), a gene feature file (.gtf), and a file that can be used for GenBank submission (.sqn). Finally, key information on all annotated contigs is provided in a single virome summary table (.tsv).
3.2 Generation of hidden Markov models for virus hallmark genes
Amino acid sequences from public virus databases, RefSeq and GenBank, were downloaded in batches based on family-level taxonomy. Sequences were dereplicated at 70 per cent Identity with CD-HIT (Fu et al. 2012), then these representative sequences were clustered using EFI-EST (Gerlt et al. 2015) (pairwise E value cutoff <1e−10). Clusters were visualized in Cytoscape (Su et al. 2014) and multi-lobed clusters were manually divided (removing interstitial sequences) or discarded. Clusters were then further pruned with MCL cluster (Morris et al. 2011). Each cluster of three or more proteins was aligned using MAFFT (Katoh and Standley 2013) with default settings. The resulting multiple sequence alignments (MSAs) were used as queries for HHsearch structural prediction and distant homology detection searches against PDB, CDD, and Pfam (80% probability cutoff). MSAs without confident alignment to any models in this search were again used as queries for HHblits against UniProt (80% probability cutoff). Each MSA with a hit in either search was named based on the HHsearch/HHblits top hit and used to generate a hidden Markov model (HMM) using Hmmer. All HMMs were kept for further consideration if the name corresponded to a possible viral hallmark gene (e.g. major capsid protein (MCP)). All Putative Hallmark HMMs were tested for specificity with a two-step validation by first querying against a negative control database, namely, human proteins from RefSeq, using Hmmer (hmmscan, 1e−6 E value cutoff). Second, protein sequences from a variety of human and environmental metagenome-derived contigs were queried against the database of the remaining HMMs using Hmmer and any proteins with hits to the database were then cross-queried using HHsearch against PDB, CDD, and Pfam. If these proteins had HHsearch hits to models in these databases that were qualitatively different from the identity of the putative Hallmark HMM, the Hallmark HMM was discarded. To acquire hallmark genes not represented by GenBank or RefSeq database, genomes from the human gut virome database (Gregory, 2019) and virome assemblies from seawater (Beaulaurier et al. 2020) were translated, and amino acid sequences were processed as above. Finally, HMMs from pVOGs (Grazziotin et al. 2017) and Pfam (El-Gebali et al. 2019) were considered and validated in the same manner. Some replication-related Hallmark HMMs were later removed because they were similar to genes typically found on plasmids or conjugative transposons. Virion structural, virion processing, and virion packaging gene HMMs are used by Cenote-Taker 2 with a cutoff of 1e−8 and genome replication gene HMMs are used with a cutoff of 1e−15.
3.3 Cross-comparison of currently available virus annotation modules
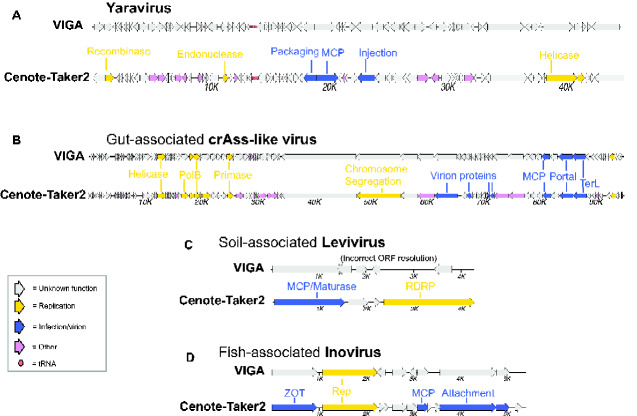
At present, VIGA (González-Tortuero et al. 2018) is the only publicly available genome annotation tool specifically designed for viruses. To compare VIGA’s genome annotation function to Cenote Taker 2, we arbitrarily chose four ‘challenging’ viral genomes as case studies (Fig. 2). For the a newly described amoeba-tropic DNA virus named Yaravirus (Boratto et al. 2020) (Fig. 2A), only Cenote-Taker 2 could discern an annotation for any genes, with the MCP, packaging ATPase, and replicative helicase all being recognizable. For a crAss-like phage assembled from a human gut metagenome dataset (Fig. 2B), Cenote-Taker 2 again annotates more genes than VIGA. For a soil-associated levivirus (Starr et al. 2019), only Cenote-Taker 2 could identify the capsid/maturase gene and the RNA-dependent RNA polymerase (RDRP). In this case, the levivirus RDRP gene lacked a stop codon and this prevented VIGA, but not Cenote-Taker 2, from calling the ORF correctly. For a fish-associated inovirus, only Cenote-Taker 2 was able to identify the packaging ATPase (ZOT), MCP, and attachment protein. For the most important functional annotations for each genome, supporting evidence is shown from HHpred and DELTA-BLAST (Supplementary Fig. S1). Direct genome map outputs from VIGA and Cenote-Taker 2 are available for each genome (Supplementary Files S1–S8).
Figure 2.
Comparison of genome maps from VIGA and Cenote-Taker 2. Cenote-Taker 2 and VIGA were run with optimized options (see Methods). (A) Yaravirus (accession MT293574) is a newly reported midsize DNA virus found in amoebae. (B) crAss-like viruses are tailed phages. The species shown here was assembled from a human gut metagenome SRA dataset SRR6128032. (C) Leviviridae sp. isolate H1_Bulk_28_FD_scaffold_59 (accession MN033558) is a levivirus genome identified in a soil metatranscriptome. (D) Inoviridae sp. isolate ctba29 (accession MH616818) is an inovirus found in a haddock virome dataset.
3.4 Comparison of virus discovery module
Cenote-Taker 2 was compared with three leading virus discovery tools, each with its own method for detecting viral sequences. Like Cenote-Taker 2, VirSorter (Roux et al. 2015) uses a virus hallmark gene detection approach. One limitation is that it is only designed to detect bacteriophages. DeepVirFinder (Ren et al. 2020) uses a machine learning approach to find short nucleotide motifs common in viral sequences. An additional pipeline, non-targeted (Paez-Espino et al. 2017) (used for ‘Uncovering Earth’s Virome’ (Paez-Espino et al. 2016)), compares predicted protein sequences encoded by a contig to a curated set of known viral and cellular proteins. A limitation of non-targeted is that it only considers contigs greater than 5 kb, while the other tools have no strict minimum length. The main categories of complex datasets that might be searched for new viruses are as follows: assembled contigs derived from DNA samples enriched for viral sequences (DNA virome), RNA samples enriched for viral sequences (RNA virome), DNA from unenriched samples (genomes and metagenomes), or RNA from unenriched samples (transcriptomes and metatranscriptomes). An additional parameter to consider is the fact that ssDNA viruses may require a second-strand synthesis step for DNA samples, such as multiple displacement amplification (MDA). DNA subject to MDA also becomes selectively enriched for circular viral genomes through rolling circle amplification (RCA) effects (Gu et al. 2018). Examples of each category of dataset were assembled and scaffolded (see Methods), and contigs >1,000 nucleotides were analyzed with the four virus discovery pipelines. Cenote-Taker 2 outperformed all other discovery tools for finding contigs with genes encoding for virion components (i.e. ‘structural’) or replication genes for each type of dataset (Figs 3 and 4).
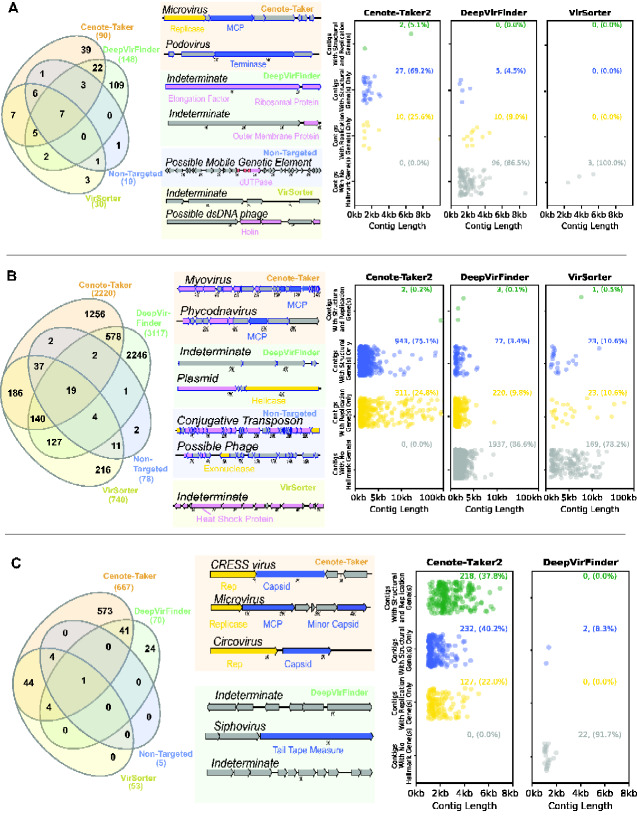
Figure 3.
Comparison of virus discovery tools for DNA datasets. Contigs >1,000 nucleotides were analyzed using four virus detection/discovery pipelines. (A) A dataset for human stool enriched for nuclease-resistant DNA in virus-sized particles (SRR6128021). Left panel: Venn diagram displaying the overlap of contigs identified as viruses by the various pipelines. Middle panel: maps showing representative examples of unique calls from each pipeline. Left panel: scatter plots display the length distribution for calls unique to each software package. Rows of plots show contigs that contain both virion structural genes and DNA or RNA replication-associated genes, only one of the two categories of gene, or neither. (B) Similar display of a dataset for Amazon River water samples analyzed with shotgun total DNA sequencing (ERR2338392). (C) Similar display of a dataset for wastewater enriched for nuclease-resistant DNA in virus-sized particles amplified using MDA/RCA (SRR3580070).
For a DNA virome dataset (i.e., virus-like particle enrichment with nuclease digestion, followed by DNA sequencing) from human gut (Shkoporov et al. 2018), DeepVirFinder had the most total virus calls (149), of which 109 were unique to DeepVirFinder (Fig. 3A). However, when unique calls were analyzed in more detail by functionally annotating genes with RPS-BLAST and HHpred, most contigs uniquely called by DeepVirFinder were found to lack any virus hallmark genes, making these calls ambiguous. Cenote-Taker 2 also had many unique calls (39) and, by definition, all uniquely called contigs encoded a virion structural/packaging or replicative viral hallmark gene (Fig. 3A), implying that Cenote-Taker 2 has higher specificity for viruses. A similar trend can be seen for a large metagenome (dsDNA) assembly from Amazon river water (Santos Junior et al. 2019) (Fig. 3B). For the metagenome dataset DeepVirFinder again had the most calls, but most of the unique calls were ambiguous upon closer inspection and some were false-positives.
A different pattern was seen for a waste water sample from which DNA was subjected to second-strand synthesis through MDA/RCA (Pearson et al. 2016) (Fig. 3C). For the amplified DNA dataset, Cenote-Taker 2 detected more total calls (5.2× all other tools combined) and more unique calls (23.8× all other tools combined), all of which have at least one type of hallmark gene. Single-stranded DNA viruses are highly abundant members of many microbial communities (Roux et al. 2019b; Malki et al. 2020; Tisza et al. 2020) making Cenote-Taker 2's discovery module a particular advance for researchers interested in these viruses.
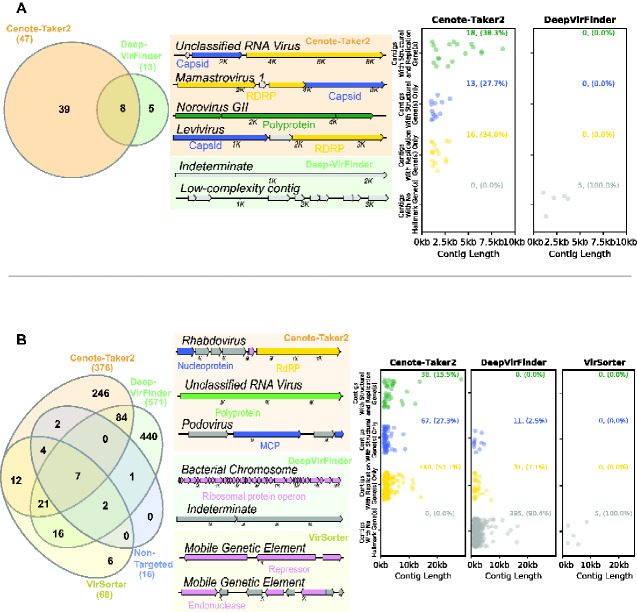
Cenote-Taker 2 also detected more RNA viruses from a sewage RNA virome dataset (virus-like particle enrichment with RNA sequencing) than DeepVirFinder (Fig. 4A). Furthermore, nearly all the unique calls from DeepVirFinder are low complexity sequences or only contain unrecognizable ORFs. VirSorter and non-targeted did not detect any viruses in this dataset, consistent with the fact that they were not designed to detect RNA viruses.
Figure 4.
Comparison of virus discovery tools for RNA datasets. Contigs >1,000 nucleotides were analyzed using four virus detection/discovery pipelines. (A) A dataset for sewage analyzed with viral particle capture and RNA sequencing (ERR3201762). Left panel: only Cenote-Taker 2 and DeepVirFinder had any virus calls. Middle panel: maps of representative examples of contigs which each pipeline uniquely called as viral. Right panel: Contig attribute chart showing only contigs called uniquely by Cenote-Taker 2 and DeepVirFinder. (B) A dataset for stool from a Tasmanian Devil with whole metatranscriptome RNA sequencing (SRR8048119). Left panel: comparison of the overlap of contigs the various pipelines designated as viral. Middle panel: maps of representative examples of contigs which each pipeline uniquely called as viral. Right panel: contig attribute chart showing only contigs called uniquely by Cenote-Taker 2, DeepVirFinder and VirSorter.
Metatranscriptome samples are perhaps the most complex category of dataset because they are expected to contain RNA virus genomes alongside transcripts from DNA viruses or other mobile genetic elements. Cenote-Taker 2 detected both RNA viruses and DNA virus transcripts in a metatranscriptome dataset for Tasmanian devil stool samples (Fig. 4B) (Chong et al. 2019).
Both Cenote-Taker 2 and VirSorter employ a virus hallmark gene approach. Cenote-Taker 2 has hallmark genes for both eukaryotic viruses and phages, whereas VirSorter is putatively only for phage detection, so we wanted to know if the excess calls from Cenote-Taker 2 were populated mostly with eukaryotic virus sequences. Looking at taxonomy of contigs called by Cenote-Taker2 but not VirSorter, it is clear that only a small percentage are eukaryotic viruses, with the exception of the MDA dataset in which Cenote-Taker 2 called dozens of (putatively eukaryotic) CRESS virus sequences (Supplementary Table S1).
DeepVirFinder detected more unique calls for 3/5 datasets but most contigs lacked hallmark genes, so random subsets of hallmark gene-negative contigs from DeepVirFinder were pulled for manual inspection. Gene content (using HHPred and DELTA-BLAST methods) and sequence similarity to known taxa (BLAST) was analyzed for these contigs. A minority of contigs had mobile genetic element-like genes (e.g. integrase), and one contig contained a gene with strong similarity to an Enterobacteria phage GEC 3S integrase (Supplementary Table S2). Other hallmark gene-negative DeepVirFinder calls appear to represent sequences of bacterial or unknown origin. Although it is conceivable that some manually unidentifiable DeepVirFinder calls might represent virus families that await formal discovery, Cenote-Taker 2 shows better overall sensitivity and specificity for detecting contigs that are verifiably viral by manual inspection in all types of dataset.
3.5 Prophage pruning module
When the Cenote-Taker 2 prophage pruning module is selected, linear contigs are assigned ORF calls via Prodigal, then ORFs are iteratively searched with 1, HMMSCAN of the custom virus hallmark gene database; 2, HMMSCAN of the custom common virus gene database; and 3, RPS-BLAST of CDD. Each gene is then considered to be 1, a virus hallmark gene; 2, a common viral gene (hit in the custom ‘common’ (but not hallmark) virus gene database or hit in CDD of a domain found in 10 or more RefSeq Caudovirales genomes or hit in CDD with ‘PHA0’ prefix); 3, a common host gene (all other CDD hits); or 4, an unknown gene (no hits in any of these databases). Based on the coordinates of the ORFs and their categorization, each nucleotide position in the contig is scored as likely virus or likely host. Bases within virus hallmark or common viral genes are scored as 10. Bases within unknown genes, which are more common in viruses, are scored as 5. Bases in intergenic regions are scored as 0 and bases within known bacterial genes are scored as −3. The sum of 5 kb windows tiled every 50 bases is calculated, then scores are smoothed based on the scores of adjacent windows. Contig segments with one or more consecutive windows with a positive score are resolved, and segments containing virus hallmark genes are designated as viruses or virus fragments.
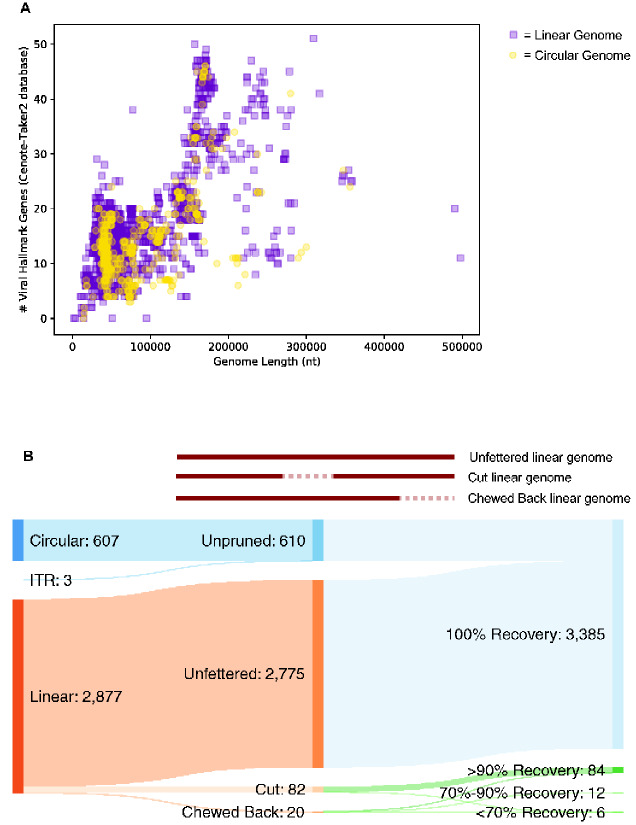
To show that Cenote-Taker 2 can identify virus genomes from order Caudovirales, which commonly occur as prophages and encode a variable assemblage of accessory genes, all 3,493 putatively complete Caudovirales genomes were downloaded from virus RefSeq. Each sequence was fed to Cenote-Taker 2 with the prophage pruning module on. First, 99.8 per cent (3487/3493) of genomes were identified as having at least one viral hallmark gene in the Cenote-Taker 2 database (Fig. 5A), with almost all also having several hallmark genes. Three of the six hallmark-negative sequences (NC_042064, NC_042059, NC_042564) were incomplete genome fragments. One (NC_002670) was a phage satellite (Chopin et al. 2001). One (NC_023591) was a mobile genetic element with many conjugative genes but no genes annotated as virion structural or packaging genes, suggesting that it is non-viral. The last (NC_029050) was a sequence that had almost no callable ORFs and is perhaps a degraded prophage relic.
Figure 5.
Cenote-Taker 2 detects and does not prune the vast majority of Caudovirales genomes. (A) Putatively complete Caudovirales genomes (3,493) were downloaded from NCBI RefSeq. Each genome is represented as a dot with its length on the x-axis and the number of genes called as viral hallmarks by Cenote-Taker 2 on the y-axis. (B) Top: schematic visualizing perturbations/lack thereof to genomes caused by Cenote-Taker 2 pruning module. Bottom: Sankey diagram of input/output of 3487 Caudovirales RefSeq genomes.
To investigate whether the Cenote-Taker 2 pruning module might truncate Caudovirales sequences, the length of each genome was analyzed before and after pruning (Fig. 5B). Of the 2,877 genomes eligible for pruning (610 of the 3,487 were recognized as circular or flanked by ITRs and therefore not eligible), 96.5 per cent (2775/2877) were kept intact by the pruning module. 2.9 per cent (82/2877) of genomes were ‘cut’ in the middle because the pruning module removed loci incorrectly determined to be non-viral (each case only had one cut region). Over 90 per cent of the original genome was kept after pruning in all but one cut genome. 0.7 per cent (20/2877) of genomes were ‘chewed back’ from one end. Seven of twenty had >90 per cent recovery, twelve of twenty had 70–90 per cent recovery, and five of twenty had <70 per cent recovery.
To test the accuracy of virus-chromosome junction calls in real prophage data, comparison of Cenote-Taker 2's pruning module to experimental data of excised, encapsidated prophage sequences was conducted. The Sequence Read Archive was searched for deep sequencing runs for bacterial isolates treated with prophage-inducing agents followed by enrichment and sequencing of nuclease-resistant virions. We found and analyzed three Bioprojects covering five bacterial isolates and six prophages (Supplementary Figs S2–S4). This analysis shows that the Cenote-Taker 2 pruning module makes approximately correct determinations of prophage/chromosome borders. The genetic distance between the Cenote-Taker 2 calls and the edge of the encapsidated phage reads ranges from about 2 kb to <100 bp.
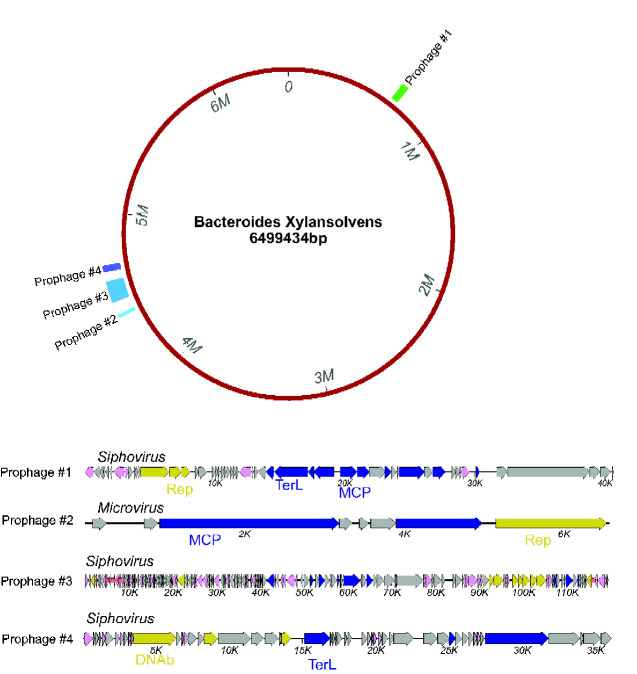
As a use case, the main chromosome of a Bacteroides xylanisolvens genome (genome assembly ASM654696v1) was analyzed with prophage pruning on. Prophage calls and virus genome maps are shown in Fig. 6, with three apparently full-length siphoviruses and one full-length microvirus prophage being detected.
Figure 6.
Cenote-Taker 2 analysis of Bacteroides xylanisolvens genome (GenBank assembly: ASM654696v1) with prophage pruning. The circular map represents the B. xylanisolvens genome annotated with coordinates of the prophage called with Cenote-Taker 2. The Cenote-Taker 2-generated map of each prophage is shown.
4. Discussion
We expect Cenote-Taker 2 will prove useful to scientists who wish to detect and annotate viruses, including divergent previously unknown virus species, in large and complex datasets. Cenote-Taker 2 empowers users both with the ability to easily discover viruses in complex datasets as well as the ability to quickly analyze candidate viruses through visualization of annotated genome maps in any available genome or plasmid map viewer. Further, combining discovery and annotation should dovetail nicely with other techniques to cluster viral sequences at the species level (Ondov et al. 2016; Gregory et al. 2019) or higher taxonomic levels (Bin Jang et al. 2019; Roux et al. 2019a), especially when visualizing pairwise comparisons of virus genomes within or between taxa (Sullivan et al. 2011). Another advantage of pairing virus discovery calls with genome map annotation is that it allows a user to more easily assess the veracity of a putative viral contig via gene content analysis of each contig. Furthermore, because Cenote-Taker 2 eases submission of annotated genomes to GenBank, even those who do not use Cenote-Taker 2 will indirectly benefit by having a larger, better-annotated, central sequence database.
Two known annotation challenges of viral coding regions that are not resolved with Cenote-Taker 2 are ribosomal frameshifting, which is documented in some RNA viruses and dsDNA bacteriophage, and intron-containing genes, which are common in eukaryotic viruses. Other non-canonical translated features could be missed, as well. We are not aware of current tools for automating the resolution of these features. Additionally, functional annotation is somewhat limited by only using databases with well-curated gene families, which precludes the annotation of newly characterized gene families not yet in gold standard databases. However, the custom database of over 3,000 HMMs of viral hallmark genes developed with Cenote-Taker 2 goes beyond Pfam, PDB, and CDD databases and mitigates some of these limitations.
Cenote-Taker 2 outperforms other currently available virus discovery pipelines for a variety of reasons. While both VirSorter and non-targeted employ HMMs of viral genes to some extent, it is likely that the models developed for Cenote-Taker 2 represent more of the diversity of viral hallmark genes. Further, since contigs are penalized by Non-Targeted if they contain common chromosomal genes, contigs representing a (pro)virus sequence flanked by a chromosomal sequence might be discarded instead of pruned. DeepVirFinder uses a fundamentally different approach, looking for nucleotide k-mers of different lengths to determine if a contig is a virus. Two reasons why this approach can fall short are as follows: 1, nucleotide sequence space may be unable to adequately capture the vast diversity of virus genomes; 2, DeepVirFinder was trained on ‘virome’ assemblies. Physical enrichment of virus-like particles is notoriously difficult (Zolfo 2019), so some training datasets may have been contaminated with host sequences. Moreover, it is known that some sequences, even in very clean virus-like particle preparations, are not viruses but mobile genetic elements that parasitize viral capsid machinery (Martinez-Rubio et al. 2017). Overall, because DeepVirFinder misses some contigs with virus hallmark genes while a small number DeepVirFinder calls are clearly derived from bacterial chromosomes, it is unclear what proportion of hallmark gene-negative contigs from this tools are likely to be virus ‘accessory regions’, virus sequences of previously undescribed types, other mobile genetic elements, or host chromosome sequences. Two new tools capable of virus discovery have come out very recently, only after we completed our analysis (Antipov et al. 2020; Kieft et al. 2020).
While there are likely new ‘types’ of yet-to-be discovered viruses encoding novel capsid and replication genes, Cenote-Taker 2 can readily be updated to include new hallmark gene models. For example, a new model was made for the highly derived RNA-dependent RNA polymerase gene of the proposed new family Quenyaviridae (Obbard et al. 2020).
Supplementary Material
Acknowledgements
The authors would like to acknowledge early users of Cenote-Taker 2, especially Leen Beller, Kema Malki, and Keir Balla, for their many helpful suggestions.
Funding
This research was funded by the Intramural Research Program of the NIH and NCI and NSF Award #1758974.
Data availability
GenBank submissions of ‘third-party annotations’ of virus genomes require a DOI number of the associated manuscript. The accession numbers of annotated genomes from this article will therefore be provided when this article gets a DOI number. In the meantime, see supplementary .gbf files.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
Contributor Information
Michael J Tisza, Lab of Cellular Oncology, NCI, NIH, Bethesda, MD 20892-4263, USA.
Anna K Belford, Lab of Cellular Oncology, NCI, NIH, Bethesda, MD 20892-4263, USA.
Guillermo Domínguez-Huerta, Department of Microbiology, Ohio State University, Columbus, OH, USA.
Benjamin Bolduc, Department of Microbiology, Ohio State University, Columbus, OH, USA.
Christopher B Buck, Lab of Cellular Oncology, NCI, NIH, Bethesda, MD 20892-4263, USA.
References
- Altschul S. F. et al. (1990) ‘ Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. [DOI] [PubMed] [Google Scholar]
- Amgarten D. et al. (2018) ‘ MARVEL, a Tool for Prediction of Bacteriophage Sequences in Metagenomic Bins’, Frontiers in Genetics, 9: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov D. et al. (2020) ‘ Metaviral SPAdes: Assembly of Viruses from Metagenomic Data’, Bioinformatics, 36: 4126–9. [DOI] [PubMed] [Google Scholar]
- Beaulaurier J. et al. (2020) ‘ Assembly-Free Single-Molecule Sequencing Recovers Complete Virus Genomes from Natural Microbial Communities’, Genome Research , 30: 437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Jang H. et al. (2019) ‘ Taxonomic Assignment of Uncultivated Prokaryotic Virus Genomes is Enabled by Gene-Sharing Networks’, Nature Biotechnology, 37: 632–9. [DOI] [PubMed] [Google Scholar]
- Boratto P. V. Met al. (2020) ‘ Yaravirus: A Novel 80-Nm Virus Infecting Acanthamoeba Castellanii’, Proceedings of the National Academy of Sciences, 117: 16579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister J. R. et al. (2015) ‘ NCBI Viral Genomes Resource’, Nucleic Acids Research, 43: D571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. F. et al. (2018) ‘ Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor’, Bioinformatics, 34: i884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong R. et al. (2019) ‘ Fecal Viral Diversity of Captive and Wild Tasmanian Devils Characterized Using Virion-Enriched Metagenomics and Metatranscriptomics’, Journal of Virology, 93: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A. et al. (2001) ‘ Analysis of Six Prophages in Lactococcus lactis IL1403: Different Genetic Structure of Temperate and Virulent Phage Populations’, Nucleic Acids Research, 29: 644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devisetty U. K. et al. (2016) ‘ Bringing Your Tools to CyVerse Discovery Environment Using Docker’, F1000Research, 5: 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S. et al. (2019) ‘ The Pfam Protein Families Database in 2019’, Nucleic Acids Research, 47: D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. et al. (2012) ‘ CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data’, Bioinformatics, 28: 3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A. et al. (2015) ‘ Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A Web Tool for Generating Protein Sequence Similarity Networks’, Biochimica et Biophysica Acta (Bba) - Proteins and Proteomics, 1854: 1019–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Tortuero E.,S. T. et al. (2018) ‘ VIGA: A Sensitive, Precise and Automatic de Novo VIral Genome Annotator’, bioRxiv 277509. [Google Scholar]
- Grazziotin A. L., Koonin E. V., Kristensen D. M. (2017) ‘ Prokaryotic Virus Orthologous Groups (pVOGs): A Resource for Comparative Genomics and Protein Family Annotation’, Nucleic Acids Research, 45: D491–D498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory A. C. (2019) ‘ The Human Gut Virome Database’, BioRxiv. [Google Scholar]
- Gregory A. C. et al. (2019) ‘ Marine DNA Viral Macro- and Microdiversity from Pole to Pole’, Cell, 177: 1109–23 e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruning B., The Bioconda Team. et al. (2018) ‘ Bioconda: Sustainable and Comprehensive Software Distribution for the Life Sciences’, Nat Methods, 15: 475–6., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L. et al. (2018) ‘ Research Progress on Rolling Circle Amplification (RCA)-Based Biomedical Sensing’, Pharmaceuticals (Pharmaceuticals), 11: 35., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H. et al. (2015) ‘ InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams’, BMC Bioinformatics, 16: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D. et al. (2010) ‘ Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification’, BMC Bioinformatics, 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘ MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft K., Zhou Z., Anantharaman K. (2020) ‘ VIBRANT: automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences’, Microbiome, 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. et al. (2020) ‘ Global Organization and Proposed Megataxonomy of the Virus World’, Microbiology and Molecular Biology Reviews, 84: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Bamford D. H., Koonin E. V. (2014) ‘ Conservation of Major and Minor Jelly-Roll Capsid Proteins in Polinton (Maverick) Transposons Suggests That They Are Bona Fide Viruses’, Biology Direct, 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘ Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. (2016) ‘ MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices’, Methods, 102: 3–11. [DOI] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016) ‘ tRNAscan-SE on-Line: Integrating Search and Context for Analysis of Transfer RNA Genes’, Nucleic Acids Research, 44: W54–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R. et al. (2012) ‘ SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler’, Gigascience, 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K. et al. (2020) ‘ Prokaryotic and Viral Community Composition of Freshwater Springs in Florida, USA’, mBio, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. (2017) ‘ CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures’, Nucleic Acids Research, 45: D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rubio R. et al. (2017) ‘ Phage-Inducible Islands in the Gram-Positive Cocci’, The Isme Journal, 11: 1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNair K. et al. (2019) ‘ PHANOTATE: A Novel Approach to Gene Identification in Phage Genomes’, Bioinformatics, 35: 4537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A., Soding J. (2015) ‘ Automatic Prediction of Protein 3D Structures by Probabilistic Multi-Template Homology Modeling’, PLoS Computational Biology, 11: e1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. H. et al. (2011) ‘ clusterMaker: A Multi-Algorithm Clustering Plugin for Cytoscape’, BMC Bioinformatics, 12: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J. et al. (2020) ‘ A New Lineage of Segmented RNA Viruses Infecting Animals’, Virus Evolution, 6: vez061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B. D. et al. (2016) ‘ Mash: Fast Genome and Metagenome Distance Estimation Using MinHash’, Genome Biology, 17: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2016) ‘ Uncovering Earth's Virome’, Nature, 536: 425–30. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2017) ‘ Nontargeted Virus Sequence Discovery Pipeline and Virus Clustering for Metagenomic Data’, Nature Protocols, 12: 1673–82. [DOI] [PubMed] [Google Scholar]
- Parks D. H. et al. (2018) ‘ A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life’, Nature Biotechnology, 36: 996–1004. [DOI] [PubMed] [Google Scholar]
- Pearson V. M., Caudle S. B., Rokyta D. R. (2016) ‘ Viral Recombination Blurs Taxonomic Lines: Examination of Single-Stranded DNA Viruses in a Wastewater Treatment Plant’, PeerJ, 4: e2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. C. et al. (2018) ‘ HMMER Web Server: 2018 Update’, Nucleic Acids Research, 46: W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. et al. (2020) ‘ Identifying Viruses from Metagenomic Data Using Deep Learning’, Quantitative Biology, 8: 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. (2019a) ‘ Eloe-Fadrosh, E.A., 2019a. Minimum Information about an Uncultivated Virus Genome (MIUViG’, Nature Biotechnology, 37: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. (2015) ‘ VirSorter: Mining Viral Signal from Microbial Genomic Data’, PeerJ, 3: e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. (2019b) ‘ Cryptic Inoviruses Revealed as Pervasive in Bacteria and Archaea across Earth's Biomes’, Nature Microbiology, 4: 1895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Junior C. D. et al. (2019) ‘ Flood Season Microbiota from the Amazon Basin Lakes’, Microbiology Resource Announcements, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkoporov A. N. et al. (2018) ‘ Reproducible Protocols for Metagenomic Analysis of Human Faecal Phageomes’, Microbiome, 6: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starikova E. V. et al. (2020) ‘ Phigaro: High-Throughput Prophage Sequence Annotation’, Bioinformatics, 36: 3882–4. [DOI] [PubMed] [Google Scholar]
- Starr E. P. et al. (2019) ‘ Metatranscriptomic Reconstruction Reveals RNA Viruses with the Potential to Shape Carbon Cycling in Soil’, Proceedings of the National Academy of Sciences, 116: 25900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G. et al. (2014) ‘ Biological Network Exploration with Cytoscape 3’, Current Protocols in Bioinformatics, 47: 8.13.1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011) ‘ Easyfig: A Genome Comparison Visualizer’, Bioinformatics, 27: 1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P. (2013) ‘ Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration’, Briefings in Bioinformatics, 14: 178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza M. J. et al. (2020) ‘ Discovery of Several Thousand Highly Diverse Circular DNA Viruses’, eLife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T. et al. (2019) ‘ Mining, Analyzing, and Integrating Viral Signals from Metagenomic Data’, Microbiome, 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L. et al. (2018) ‘ A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core’, Journal of Molecular Biology, 430: 2237–43. [DOI] [PubMed] [Google Scholar]
- Zolfo M. P. F. et al. (2019) ‘ Detecting Contamination in Viromes Using ViromeQC’, Nature Biotechnology, 37: 1408–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBank submissions of ‘third-party annotations’ of virus genomes require a DOI number of the associated manuscript. The accession numbers of annotated genomes from this article will therefore be provided when this article gets a DOI number. In the meantime, see supplementary .gbf files.