Abstract
Background
Neuroticism is a strong predictor for a variety of social and behavioral outcomes, but the etiology is still unknown. Our study aims to provide a comprehensive investigation of causal effects of serum metabolome phenotypes on risk of neuroticism using Mendelian randomization (MR) approaches.
Methods
Genetic associations with 486 metabolic traits were utilized as exposures, and data from a large genome-wide association study of neuroticism were selected as outcome. For MR analysis, we used the standard inverse-variance weighted (IVW) method for primary MR analysis and 3 additional MR methods (MR-Egger, weighted median, and MR pleiotropy residual sum and outlier) for sensitivity analyses.
Results
Our study identified 31 metabolites that might have causal effects on neuroticism. Of the 31 metabolites, uric acid and paraxanthine showed robustly significant association with neuroticism in all MR methods. Using single nucleotide polymorphisms as instrumental variables, a 1-SD increase in uric acid was associated with approximately 30% lower risk of neuroticism (OR: 0.77; 95% CI: 0.62–0.95; PIVW = 0.0145), whereas a 1-SD increase in paraxanthine was associated with a 7% higher risk of neuroticism (OR: 1.07; 95% CI: 1.01–1.12; PIVW = .0145).
Discussion
Our study suggested an increased level of uric acid was associated with lower risk of neuroticism, whereas paraxanthine showed the contrary effect. Our study provided novel insight by combining metabolomics with genomics to help understand the pathogenesis of neuroticism.
Keywords: Neuroticism, genetically determined metabolite, Mendelian randomization, uric acid, paraxanthine
Significance Statement.
This is the first Mendelian randomization (MR) study, to our knowledge, to assess the causal relationship between serum metabolites and neuroticism. MR study could avoid confounding, reverse causation, and various biases compared with traditional observational study. Using genetic variants as instrumental variables, our results showed that urate had protective effects on neuroticism while paraxanthine had adverse effects on neuroticism. We also found some significant metabolic pathways that may be involved in neuroticism. By combining metabolomics with genomics, we provide new insight in the causal relationship between serum metabolites and neuroticism, which may contribute to the understanding of pathogenesis and potential therapeutic targets for neuroticism.
Introduction
Neuroticism is a human personality trait characterized by a tendency to respond with negative emotions to threat, frustration, or loss (Lahey, 2009). Individuals with neuroticism often believe that the world is dangerous and one’s ability is not enough to cope with challenging events (Barlow et al., 2014). Neuroticism can also predict a variety of social and behavioral outcomes, especially common psychiatric symptoms, including anxiety, depressive, social phobia, and substance use (Ormel et al., 2013b, 2013a). Genetic factors contribute substantially to the pathogenesis of neuroticism, accounting for 40% to 60% of the variance of this trait (Clark et al., 1994; Hettema et al., 2005; Fullerton, 2006). Recent genome-wide association studies (GWASs) have yielded substantial findings for improving our understanding of the etiological mechanism of neuroticism (Okbay et al., 2016; Luciano et al., 2018; Nagel et al., 2018). However, functional annotations towards biological processes are still lacking.
Substantial effort has been expended to understand the biological mechanisms through which GWAS-identified risk variants act on target diseases, including the introduction of expression quantitative trait loci, methylation quantitative trait loci, and protein quantitative trait loci (Westra et al., 2013; Consortium et al., 2017; McRae et al., 2018; Yao et al., 2018). Recent studies have proposed metabolic traits as functional intermediates to reveal the relevant biological processes of disease-related genetic variants (Gieger et al., 2008; Suhre et al., 2011). Metabolites are a group of small molecules widely distributed throughout the cells, tissues, and fluids of human bodies (Quinones and Kaddurah-Daouk, 2009). Alterations in the concentration of metabolites can serve as useful biomarkers and provide early evidence for the diagnosis and prognosis of pathologies (Assfalg et al., 2008; Holmes et al., 2008). Especially in recent years, the rapidly evolving field of metabolomics is providing a systematic readout of the metabolic state of an individual. GWASs with non-targeted metabolomics have been conducted and the concept of “genetically determined metabotypes (GDMs)” proposed to further our understanding of the relationships and interactions between genetic variations and environmental triggers of disease. GDMs serve as useful intermediate phenotypes to explain the causal roles of genetic variants in human diseases.
Mendelian randomization (MR) is a novel technique that uses genetic variants as instrumental variables to assess causal inferences between risk factors and clinical outcomes of interest (Choi et al., 2019). The fundamental principle of the MR study design is that genetic variants, which indicate the biological effects or level of effects of a modifiable exposure, should be causally associated with exposure-related disease risk. This approach is largely independent of the biases inherent in observational studies, given that genetic variation is unlikely to be affected by environmental factors (Pierce et al., 2011). During the past years, the explosion in publicly available GWAS summary data has made the MR popular in inferring causality, especially the 2-sample MR design that utilizes just 1 pair of GWAS summary statistics (Zheng et al., 2017). Based on the 2-sample MR approach, we are able to link GDMs with neuroticism using GWAS estimates on both metabolic phenotypes and neuroticism. The aims of the present study were to: (1) assess the causal relationships between genetically determined levels of serum metabolites and risk of neuroticism, (2) investigate the genetic variants that might have important roles in determining the variation of corresponding metabolites and risk of neuroticism, and (3) identify potential metabolic pathways that may be involved with the pathogenesis of neuroticism.
Methods
Genetically Determined Metabolites
Data for genetic associations with serum metabolites were downloaded from the Metabolomics GWAS server (http://metabolomics.helmholtz-muenchen.de/gwas/). Briefly, Shin et al. (Shin et al., 2014) conducted the most comprehensive genome-wide association estimates with high-throughput metabolic profiling as phenotypes and developed the atlas of genetic influences on human blood metabolites. The study sample was composed of 7824 adult individuals from 2 European population studies. All participants gave written informed consent, and the study was approved by local ethics committees. Metabolic profiling was performed based on non-targeted mass spectrometry analysis on human fasting serum. After merging the datasets of the 2 studies, a subset of 486 metabolites was available for GWAS analysis. Of the 486 total metabolites, 309 were classified as known, implying that their chemical identity had been determined according to Metabolon’s spectra library (Metabolon, Inc., Morrisville, NC). The 309 known metabolites were further assigned to 8 biochemical classes (amino acids, peptides, lipids, cofactors and vitamins, carbohydrates, energy, nucleotides, and xenobiotics), as described in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The genotyping and imputation steps for the 2 cohorts were described in detail in previous studies (Suhre et al., 2011; Shin et al., 2014). Finally, approximately 2.1 million single nucleotide polymorphisms (SNPs) present in both cohorts were tested in the primary association meta-analysis.
Selection of Instrumental Variables
The principle of MR relies on the basic assumption of valid instrumental variables; this particularly requires the extracted SNPs to be (1) significantly related to exposure, (2) independent of any confounder, and (3) associated with the outcome directly through the exposure. To determine the instrumental variables for the 486 metabolites, we first extracted genetic variants with association thresholds at P < 1 × 10−5 to ensure they conferred substantial variance for the corresponding metabolites. Second, we identified independent variants using a clumping procedure implemented in PLINK1.9 (http://www.cog-genomics.org/plink2/), setting a linkage-disequilibrium threshold of r2 < 0.1 with 500 kb in the European 1000 Genomes Project Phase 3 reference panel. We next assessed whether these instrumental variables were strong enough to predict metabolite levels by 2 parameters: the explained variance (adjusted R2) and F statistic. The adjusted R2 was estimated using the “gtx” package in R software (https://www.r-project.org/), and the F statistic was calculated based on the formula F = R2(n–1–k)/(1–R2)k, where n is the sample size and k represents the number of genetic variants (Brion et al., 2013). A threshold of F > 10 was typically recommended for MR analyses (Burgess et al., 2013).
GWAS of Neuroticism
We obtained the GWAS summary statistics for neuroticism from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). Briefly, Okbay et al. (Okbay et al., 2016) conducted the large GWAS meta-analysis on 170 910 individuals from 59 cohorts. Neuroticism was measured according to a 12-item version of the Eysenck Personality Inventory Neuroticism scale. Genotyping was performed separately for each cohort using different versions of Affymetrix arrays. Quality-control procedures were conducted using the EasyQC software following standard protocols (Winkler et al., 2014). Association analysis was carried out with a logistic regression model correcting for age, gender, and study-specific covariates. All participants provided written informed consent, and ethical approvals were obtained for each cohorts. Detailed information for cohort descriptions, quality-control filters, and imputation procedures can be found in the previously published study. Finally, a total of 6 524 442 SNPs were included in the GWAS summary statistics.
MR Analyses
We used the inverse-variance weighted (IVW) method as the primary MR analysis to determine the causal relationships between genetically determined levels of serum metabolites and neuroticism. Briefly, the IVW method provided a consistent estimate of the causal effect of the exposure when each variant satisfied the assumption of valid instrumental variable (Burgess et al., 2013). We then used additional methods to provide sensitivity analysis for the robustness of the MR estimates. MR-Egger was a useful sensitivity analysis that provided consistent estimates with invalid instruments (Bowden et al., 2015). The MR-Egger approach was able to detect certain violations of the standard instrumental variable assumptions and provide an effect estimate not subject to these violations. The weighted median method was another approach that provided consistent estimates even when up to 50% of the variants were deemed invalid instruments (Bowden et al., 2016). MR-PRESSO (MR pleiotropy residual sum and outlier) was another newly developed MR approach that could detect and correct for horizontal pleiotropic outliers to provide a corrected estimate (Verbanck et al., 2018). Furthermore, we conducted the MR-PRESSO Global test to evaluate whether horizontal pleiotropy existed. All analyses were performed using the MendelianRandomization and MR-PRESSO packages in R software version 3.4.4. P < .05 was considered statistically significant.
Metabolic Pathway Analysis
Metabolic pathway analysis was conducted using the web-based MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/) (Chong et al., 2018). We analyzed the metabolite sets from 2 libraries in our study, including the Small Molecule Pathway Database and the KEGG Database (Frolkis et al., 2010; Kanehisa et al., 2012). All metabolites that passed the suggestive threshold of association by IVW (PIVW < .05) were included in the metabolic pathway analysis.
Results
Strength of the Instrumental Variables
We performed a 2-sample MR analysis to assess the causal effects of human blood metabolites on neuroticism using a pair of GWAS summary statistics. The instrumental variables generated were composed of 3–675 SNPs and could explain 0.8%–83.5% of the variance of their corresponding metabolites. The minimum F statistic for indicating the strength of these instrumental variables was 20.33, meaning that all instrumental variables for the 486 metabolites were sufficiently effective (F statistic >10) for MR analysis.
Effects of Genetically Determined Metabolites on Neuroticism
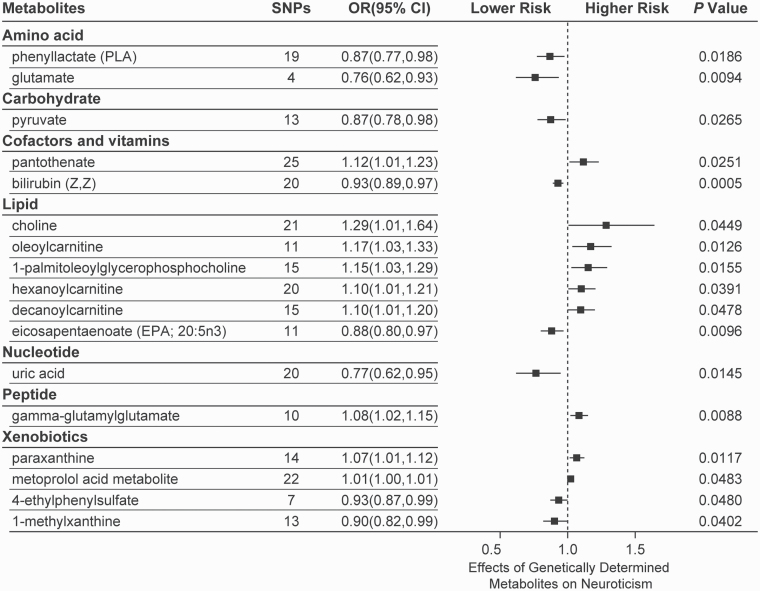
The IVW identified 17 known metabolites and 14 unknown metabolites that were significantly associated with neuroticism (Figure 1; supplementary Table 1). Of the 17 known metabolites, bilirubin (Z,Z) showed the most significant association with neuroticism (PIVW = .0005). Using 20 SNPs as proxy predictors, a 1-SD increase in the level of bilirubin (Z,Z) was associated with an approximately 10% lower risk of neuroticism (odds ratio [OR]: 0.93; 95% confidence interval [CI]: 0.89–0.97). We also identified 7 other metabolites that were associated with lower risk of neuroticism, including phenyllactate (OR: 0.87; 95% CI: 0.77–0.98; PIVW = .0186), glutamate (OR: 0.76; 95% CI: 0.62–0.93; PIVW = .0094), pyruvate (OR: 0.87; 95% CI: 0.78–0.98; PIVW = .0256), eicosapentaenoate (20:5n3) (OR: 0.88; 95% CI: 0.80–0.97; PIVW = .0096), uric acid (OR: 0.77; 95% CI: 0.62–0.95; PIVW = .0145), 4-ethylphenylsulfate (OR: 0.93; 95% CI: 0.87–0.99; PIVW = .0480), and 1-methylxanthine (OR: 0.90; 95% CI: 0.82–0.99; PIVW = .0402). The other 9 metabolites were associated with increased risk of neuroticism, including pantothenate (OR: 1.12; 95% CI: 1.01–1.23; PIVW = .0251), choline (OR: 1.29; 95% CI: 1.01–1.64; PIVW = .0449), oleoylcarnitine (OR: 1.17; 95% CI: 1.03–1.33; PIVW = .0126), 1-palmitoleoylglycerophosphocholine (OR: 1.15; 95% CI: 1.03–1.29; PIVW = .0155), hexanoylcarnitine (OR: 1.10; 95% CI: 1.01–1.21; PIVW = .0391), decanoylcarnitine (OR: 1.10; 95% CI: 1.01–1.20; PIVW = .0478), gamma-glutamylglutamate (OR: 1.08; 95% CI: 1.02–1.15; PIVW = .0088), paraxanthine (OR: 1.07; 95% CI: 1.01–1.12; PIVW = .0117), and metoprolol acid metabolite (OR: 1.01; 95% CI: 1.00–1.01; PIVW = .0483).
Figure 1.
Mendelian randomization associations between serum metabolites and neuroticism based on inverse-variance weighted (IVW) method.
Sensitivity Analysis
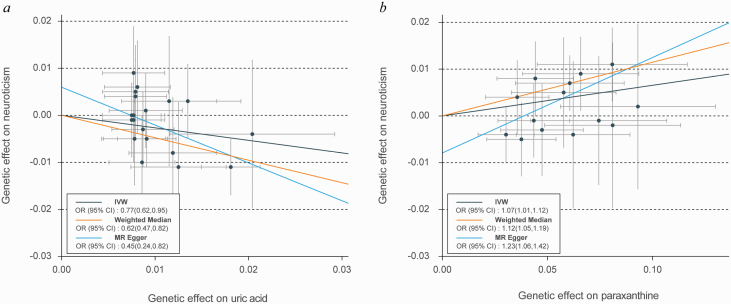
Although the IVW approach is highly effective for inferring causality of an exposure for a complex disease outcome, it is known to be vulnerable to weak instrument bias. We conducted sensitivity analyses to overcome these biases. Table 1 showed the sensitivity analysis results for the IVW identified metabolites. Two metabolites showed robust causal relationships with neuroticism when sensitivity analyses were applied, which were uric acid (PMR-Egger = .0091; Pweighted-median = .0006 and PMR-PRESSO = .0244) and paraxanthine (PMR-Egger = .0073; Pweighted-median = .0004 and PMR-PRESSO = .0256), respectively, and no evidence of horizontal pleiotropy was observed for either metabolite (PGlobal test = .0573 for uric acid and PGlobal test = .1376 for paraxanthine). Using 20 SNPs as predictors, a 1-SD increase in uric acid was associated with an approximately 30% lower risk of neuroticism (OR: 0.77; 95% CI: 0.62–0.95; Figure 2A), whereas a 1-SD increase in paraxanthine was associated with a 7% higher risk of neuroticism (OR: 1.07; 95% CI: 1.01–1.12; Figure 2B).
Table 1.
Sensitivity Analysis of Causal Associations Between IVW Identified Metabolites and Neuroticism
| Metabolites | MR-Egger | Weighted median | MR-PRESSO | Global test | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) |
P value | OR (95% CI) |
P value | OR (95% CI) |
P value | RSS | P value | |
| Amino acid | ||||||||
| Phenyllactate | 0.98 (0.70,1.37) | .9082 | 0.87 (0.74,1.01) | .0756 | 0.87 (0.77,0.98) | .0301 | 24.42 | .2602 |
| Glutamate | 0.01 (0.00,540.44) | .3870 | 0.73 (0.55,0.96) | .0268 | 0.76 (0.64,0.90) | .0514 | 3.59 | .5940 |
| Carbohydrate | ||||||||
| Pyruvate | 0.70 (0.48,1.04) | .0752 | 0.81 (0.69,0.94) | .0070 | 0.87 (0.78,0.98) | .0466 | 17.50 | .2715 |
| Cofactors and vitamins | ||||||||
| Pantothenate | 1.20 (0.91,1.59) | .2002 | 1.13 (0.98,1.30) | .1057 | 1.12 (1.03,1.22) | .0185 | 19.94 | .7968 |
| Bilirubin (Z,Z) | 0.93 (0.81,1.07) | .2975 | 0.94 (0.89,0.99) | .0450 | 0.93 (0.89,0.97) | .0025 | 26.18 | .2416 |
| Lipid | ||||||||
| Choline | 0.88 (0.32,2.45) | .8120 | 1.34 (0.95,1.89) | .1010 | 1.29 (1.03,1.60) | .0344 | 17.55 | .7241 |
| Oleoylcarnitine | 1.68 (0.71,3.94) | .2369 | 1.19 (1.01,1.41) | .0375 | 1.17 (1.06,1.30) | .0129 | 8.14 | .7654 |
| 1-Palmitoleoylglycerophosphocholine | 1.28 (0.88,1.85) | .1917 | 1.19 (1.02,1.40) | .0292 | 1.15 (1.04,1.28) | .0210 | 13.86 | .6112 |
| Hexanoylcarnitine | 1.06 (0.79,1.42) | .7129 | 1.15 (1.02,1.30) | .0223 | 1.10 (1.01,1.21) | .0530 | 26.67 | .2009 |
| Decanoylcarnitine | 1.32 (0.83,2.11) | .2433 | 1.18 (1.04,1.33) | .0121 | 1.10 (1.01,1.20) | .0678 | 18.48 | .3151 |
| Eicosapentaenoate (20:5n3) | 0.80 (0.65,1.01) | .0518 | 0.89 (0.77,1.02) | .0842 | 0.88 (0.80,0.97) | .0244 | 12.79 | .4204 |
| Nucleotide | ||||||||
| Uric acid | 0.45 (0.24,0.82) | .0091 | 0.62 (0.47,0.82) | .0006 | 0.77 (0.62,0.95) | .0244 | 33.25 | .0573 |
| Peptide | ||||||||
| Gamma-glutamylglutamate | 1.07 (0.91,1.25) | .4155 | 1.09 (0.99,1.19) | .0685 | 1.08 (1.02,1.15) | .0278 | 11.23 | .4629 |
| Xenobiotics | ||||||||
| Paraxanthine | 1.23 (1.06,1.42) | .0073 | 1.12 (1.05,1.19) | .0004 | 1.07 (1.01,1.12) | .0256 | 22.18 | .1376 |
| Metoprolol acid metabolite | 1.01 (0.99,1.02) | .0718 | 1.01 (0.99,1.01) | .1977 | 1.01 (1.00,1.01) | .0516 | 20.83 | .5946 |
| 4-Ethylphenylsulfate | 1.02 (0.83,1.25) | .8765 | 0.91 (0.83,0.99) | .0386 | 0.93 (0.88,0.99) | .0677 | 6.49 | .5966 |
| 1-Methylxanthine | 1.08 (0.53,2.20) | .8410 | 0.91 (0.80,1.04) | .1697 | 0.90 (0.84,0.97) | .0187 | 8.04 | .8662 |
Abbreviations: CI, confidence interval; IVW, inverse-variance weighted; MR-PRESSO, MR pleiotropy residual sum and outlier; OR, odds ratio; RSS, residual sum of squares.
Figure 2.
Genetic associations of uric acid and paraxanthine with neuroticism. (A) Genetic effect of uric acid on neuroticism. (B) Genetic effect of paraxanthine on neuroticism. Each of the single nucleotide polymorphisms (SNPs) associated with metabolite are represented by a black dot with the error bar depicting the SE of its association with metabolite (horizontal) and neuroticism (vertical). The slopes of each line represent the causal association for each method.
Genetic Determinants
We also investigated the leading SNPs that played important roles in inferring the causal relationships between the identified metabolites and neuroticism. Among the 20 SNPs composing the instrumental variable of uric acid, rs17246501 showed the most significant association (P = 2.35 × 10–30) with uric acid (supplementary Table 2). Notably, the rs17246501 also showed a significant association signal (P = .0016) with neuroticism. Of the 14 SNPs for paraxanthine, rs3768372 (P = .0103), rs10791723 (P = .0120), and rs4371296 (P = .0499) were also significantly associated with neuroticism (supplementary Table 3). These SNPs might provide valuable information serving as diagnostic or therapeutic targets for neuroticism.
Metabolic Pathway Analysis
Our study identified 7 significant metabolic pathways that were involved in the pathogenesis of neuroticism (Table 2). The most significant metabolic pathway was the alanine, aspartate, and glutamate metabolism (P = .0041) from the KEGG database. Two neuroticism-related metabolites, pyruvate and glutamate, were involved in the metabolic pathway of alanine, aspartate, and glutamate metabolism. We also identified 5 other metabolic pathways from the KEGG database, including pantothenate and CoA biosynthesis (P = .0052), butanoate metabolism (P = .0111), glycine, serine and threonine metabolism (P =.0158), arginine and proline metabolism (P = .0385), and D-glutamine and D-glutamate metabolism (P = .0449). There was also a significant metabolic pathway identified from the Small Molecule Pathway Database, which was the caffeine metabolism (P = .0295).
Table 2.
Significant Metabolic Pathways Involved in the Pathogenesis of Neuroticism
| Metabolic pathway | Metabolites involved |
P value | Database |
|---|---|---|---|
| Alanine, aspartate, and glutamate metabolism | Pyruvate; glutamate | .0041 | KEGG |
| Pantothenate and CoA biosynthesis | Pyruvate; pantothenate | .0052 | KEGG |
| Butanoate metabolism | Pyruvate; glutamate | .0111 | KEGG |
| Glycine, serine, and threonine metabolism | Pyruvate; choline | .0158 | KEGG |
| Caffeine metabolism | Paraxanthine; 1-methylxanthine | .0295 | SMPDB |
| Arginine and proline metabolism | Pyruvate; glutamate | .0385 | KEGG |
| D-Glutamine and D-glutamate metabolism | Glutamate | .0449 | KEGG |
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; SMPDB, Small Molecule Pathway Database.
Discussion
Our study performed a 2-sample MR analysis to provide an unbiased estimate of the causal relationships between genetically determined levels of serum metabolites and risk of neuroticism. To the best of our knowledge, this was the first MR study that linked metabolic phenotyping with genomics to assess causal relationships between human serum metabolites and neuroticism. Using SNPs as proxies, our study found that an increased level of uric acid was associated with lower risk of neuroticism, while an increased level of paraxanthine was associated with higher risk of neuroticism. Our study also reported several other metabolites that might contribute to the development of neuroticism.
Uric acid is the end product of adenosine metabolism in humans. Lower uric acid has long been recognized as a risk factor for neurological and psychiatric disorders, such as Parkinson’s disease, multiple sclerosis, and major depressive and anxiety disorders (Weisskopf et al., 2007; Moccia et al., 2015; Black et al., 2018). Ostojic et al. (Simeunovic Ostojic and Maas, 2018) suggested that uric acid could be a biomarker of mood dysfunction, personality traits, and behavioral patterns. Bartoli et al. (Bartoli et al., 2018b) reported that lower uric acid serum levels were associated with psychological distress and suicidal ideation severity. In addition, a recent meta-analysis further highlighted an inverse association between serum uric acid levels and depression (Bartoli et al., 2018a). The neurotic personality usually incorporated the tendency to experience frequent, intense negative emotions and was strongly associated with psychiatric symptoms. Our study reported that lower serum uric acid levels were associated with higher risk of neuroticism, which supported the findings from previous studies. In addition, rs17246501 (corresponding to the SLC2A9 gene) was a genetic predictor for uric acid, which also showed significant association with neuroticism. Interestingly, a previous cross-sectional analysis reported that serum uric acid and SLC2A9 variant were strongly associated with depressive and anxiety disorders (Lyngdoh et al., 2013). Though further evidence is needed, the identified SNPs might serve as important targets for revealing the pathogenesis and therapies of neuroticism.
Paraxanthine is another metabolite that showed causal association with neuroticism. Notably, parexanthine also shows close interconnection with adenosine. Indeed, paraxanthine is a non-selective antagonist of adenosine A1 and A2A receptors that play important roles in caffeine metabolism. A previous study has reported that heavy caffeine ingestion might cause or exacerbate anxiety and might be associated with depression and increased use of antianxiety drugs. Caffeine might also exacerbate anxiety and panic in panic disorder patients (Clementz and Dailey, 1988). The effect of caffeine on anxiety was also shown in several subsequent studies (Childs and de Wit, 2006; Nardi et al., 2009; Trapp et al., 2014; Yudko and McNiece, 2014; Botton et al., 2017). Furthermore, coffee and caffeine consumption had also been reported to have antidepressant effects (Yudko and McNiece, 2014; Wang et al., 2016). As a whole, our study showed consistent findings with previous studies, supporting the possible role of the purinergic system in neuroticism-related disorders, such as anxiety and depression (Cheffer et al., 2018; Bartoli et al., 2020).
Our study also reported several metabolic pathways that might be involved in the pathogenesis of neuroticism, such as the alanine, aspartate, and glutamate metabolism pathway. Glutamate is a common excitatory and inhibitory neurotransmitter in the human brain and has been proved to play a major role in the pathogenesis of mental disorders (Hashimoto et al., 2013; Hasler et al., 2019). Our study also suggested that pantothenate and CoA biosynthesis was associated with risk of neuroticism. Pantothenate and CoA biosynthesis was also reported to be associated with risk of major depression and other affective disorders (Chu et al., 2009; Hammerschlag et al., 2017). Although additional evidence is needed, these metabolic pathways might provide valuable information to help understand the underlying biological mechanisms in the pathogenesis of neuroticism.
Our study has several limitations. First, the strength of the instrumental variables relied on the sample size of GWASs, so more data should be collected to improve the veracity of the generated GDMs. Second, though MR proved to be a powerful approach to assess the causal associations between serum metabolites and neuroticism, the results require further verification based on experimental data. Third, genetic architectures played an essential role in determining the causal relationships between metabolites and neuroticism. However, further work should be done to determine the role of genetic variants in affecting variations of metabolites in neuroticism.
Conclusions
In conclusion, our study suggested an increased level of uric acid was associated with lower risk of neuroticism, whereas paraxanthine showed the contrary effect. Our MR study also identified multiple metabolic pathways and genetic risk variants that might have causal effects on neuroticism. Our study provided novel insight by combining metabolomics with genomics to help understand the pathogenesis of neuroticism.
Supplementary Materials
Supplementary data are available at International Journal of Neuropsychopharmacology (IJNPPY) online.
Supplementary Table 1. Unknown metabolites identified by the Mendelian randomization estimates.
Supplementary Table 2. Genetic predictors of uric acid and their association with neuroticism.
Supplementary Table 3. Genetic predictors of paraxanthine and their association with neuroticism.
Acknowledgments
We thank the High-Performance Computing Cluster of the First Affiliated Hospital of Xi’an Jiaotong University for data processing.
This work was supported by the National Natural Science Foundation Award of China (grant no. 81771471).
Statement of Interest
None.
Data Availability Statement
Data for GWAS scans with human serum metabolites are freely accessible through the Metabolomics GWAS Server (http://metabolomics.helmholtz-muenchen.de/gwas/). GWAS summary statistics for neuroticism can be freely downloaded from the GWAS Catalog website (https://www.ebi.ac.uk/gwas/).
References
- Assfalg M, Bertini I, Colangiuli D, Luchinat C, Schäfer H, Schütz B, Spraul M (2008) Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci U S A 105:1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DH, Ellard KK, Sauer-Zavala S, Bullis JR, Carl JR (2014) The origins of neuroticism. Perspect Psychol Sci 9:481–496. [DOI] [PubMed] [Google Scholar]
- Bartoli F, Trotta G, Crocamo C, Malerba MR, Clerici M, Carrà G (2018a) Antioxidant uric acid in treated and untreated subjects with major depressive disorder: a meta-analysis and meta-regression. Eur Arch Psychiatry Clin Neurosci 268:119–127. [DOI] [PubMed] [Google Scholar]
- Bartoli F, Crocamo C, Bava M, Castagna G, Di Brita C, Riboldi I, Trotta G, Verrengia E, Clerici M, Carrà G (2018b) Testing the association of serum uric acid levels with behavioral and clinical characteristics in subjects with major affective disorders: a cross-sectional study. Psychiatry Res 269:118–123. [DOI] [PubMed] [Google Scholar]
- Bartoli F, Burnstock G, Crocamo C, Carra G (2020) Purinergic signaling and related biomarkers in depression. Brain Sci 10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black CN, Bot M, Scheffer PG, Snieder H, Penninx BWJH (2018) Uric acid in major depressive and anxiety disorders. J Affect Disord 225:684–690. [DOI] [PubMed] [Google Scholar]
- Botton PH, Pochmann D, Rocha AS, Nunes F, Almeida AS, Marques DM, Porciúncula LO (2017) Aged mice receiving caffeine since adulthood show distinct patterns of anxiety-related behavior. Physiol Behav 170:47–53. [DOI] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Haycock PC, Burgess S (2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion MJ, Shakhbazov K, Visscher PM (2013) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42:1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheffer A, Castillo ARG, Corrêa-Velloso J, Gonçalves MCB, Naaldijk Y, Nascimento IC, Burnstock G, Ulrich H (2018) Purinergic system in psychiatric diseases. Mol Psychiatry 23:94–106. [DOI] [PubMed] [Google Scholar]
- Childs E, de Wit H (2006) Subjective, behavioral, and physiological effects of acute caffeine in light, nondependent caffeine users. Psychopharmacology (Berl) 185:514–523. [DOI] [PubMed] [Google Scholar]
- Choi KW, Chen CY, Stein MB, Klimentidis YC, Wang MJ, Koenen KC, Smoller JW; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2019) Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample Mendelian randomization study. JAMA Psychiatry 76:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TT, Liu Y, Kemether E (2009) Thalamic transcriptome screening in three psychiatric states. J Hum Genet 54:665–675. [DOI] [PubMed] [Google Scholar]
- Clark LA, Watson D, Mineka S (1994) Temperament, personality, and the mood and anxiety disorders. J Abnorm Psychol 103:103–116. [PubMed] [Google Scholar]
- Clementz GL, Dailey JW (1988) Psychotropic effects of caffeine. Am Fam Physician 37:167–172. [PubMed] [Google Scholar]
- GTEx Consortium, et al. (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, Xia J, Liang Y, Shrivastava S, Wishart DS (2010) SMPDB: the small molecule pathway database. Nucleic Acids Res 38:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton J. (2006) New approaches to the genetic analysis of neuroticism and anxiety. Behav Genet 36:147–161. [DOI] [PubMed] [Google Scholar]
- Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, Illig T, Suhre K (2008) Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 4:e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschlag AR, et al. (2017) Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet 49:1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Malchow B, Falkai P, Schmitt A (2013) Glutamate modulators as potential therapeutic drugs in schizophrenia and affective disorders. Eur Arch Psychiatry Clin Neurosci 263:367–377. [DOI] [PubMed] [Google Scholar]
- Hasler G, Buchmann A, Haynes M, Müller ST, Ghisleni C, Brechbühl S, Tuura R (2019) Association between prefrontal glutamine levels and neuroticism determined using proton magnetic resonance spectroscopy. Transl Psychiatry 9:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema JM, Prescott CA, Myers JM, Neale MC, Kendler KS (2005) The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry 62:182–189. [DOI] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK (2008) Metabolic phenotyping in health and disease. Cell 134:714–717. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey BB. (2009) Public health significance of neuroticism. Am Psychol 64:241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke TK, Shirali M, Harris SE, Marioni RE, Liewald DC, Fawns-Ritchie C, Adams MJ, Howard DM, Lewis CM, Gale CR, McIntosh AM, Deary IJ (2018) Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet 50:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngdoh T, Bochud M, Glaus J, Castelao E, Waeber G, Vollenweider P, Preisig M (2013) Associations of serum uric acid and SLC2A9 variant with depressive and anxiety disorders: a population-based study. PLoS One 8:e76336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, Marioni RE, Shah S, Yang J, Powell JE, Harris SE, Gibson J, Henders AK, Bowdler L, Painter JN, Murphy L, Martin NG, Starr JM, Wray NR, Deary IJ, Visscher PM, Montgomery GW (2018) Identification of 55,000 replicated DNA methylation QTL. Sci Rep 8:17605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia M, Lanzillo R, Costabile T, Russo C, Carotenuto A, Sasso G, Postiglione E, De Luca Picione C, Vastola M, Maniscalco GT, Palladino R, Brescia Morra V (2015) Uric acid in relapsing-remitting multiple sclerosis: a 2-year longitudinal study. J Neurol 262:961–967. [DOI] [PubMed] [Google Scholar]
- Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, Savage JE, Hammerschlag AR, Skene NG, Muñoz-Manchado AB, White T, Tiemeier H, Linnarsson S, Hjerling-Leffler J, Polderman TJC, Sullivan PF, van der Sluis S, Posthuma D; 23andMe Research Team (2018) Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 50:920–927. [DOI] [PubMed] [Google Scholar]
- Nardi AE, Lopes FL, Freire RC, Veras AB, Nascimento I, Valença AM, de-Melo-Neto VL, Soares-Filho GL, King AL, Araújo DM, Mezzasalma MA, Rassi A, Zin WA (2009) Panic disorder and social anxiety disorder subtypes in a caffeine challenge test. Psychiatry Res 169:149–153. [DOI] [PubMed] [Google Scholar]
- Okbay A, et al. ; LifeLines Cohort Study (2016) Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel J, Jeronimus BF, Kotov R, Riese H, Bos EH, Hankin B, Rosmalen JGM, Oldehinkel AJ (2013a) Neuroticism and common mental disorders: meaning and utility of a complex relationship. Clin Psychol Rev 33:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel J, Bastiaansen A, Riese H, Bos EH, Servaas M, Ellenbogen M, Rosmalen JG, Aleman A (2013b) The biological and psychological basis of neuroticism: current status and future directions. Neurosci Biobehav Rev 37:59–72. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40:740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones MP, Kaddurah-Daouk R (2009) Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis 35:165–176. [DOI] [PubMed] [Google Scholar]
- Shin SY, et al. ; Multiple Tissue Human Expression Resource (MuTHER) Consortium (2014) An atlas of genetic influences on human blood metabolites. Nat Genet 46:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeunovic Ostojic M, Maas J (2018) Anorexia nervosa and uric acid beyond gout: an idea worth researching. Int J Eat Disord 51:97–101. [DOI] [PubMed] [Google Scholar]
- Suhre K, et al. ; CARDIoGRAM (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp GS, Allen K, O’Sullivan TA, Robinson M, Jacoby P, Oddy WH (2014) Energy drink consumption is associated with anxiety in Australian young adult males. Depress Anxiety 31:420–428. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Chen CY, Neale B, Do R (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shen X, Wu Y, Zhang D (2016) Coffee and caffeine consumption and depression: a meta-analysis of observational studies. Aust N Z J Psychiatry 50:228–242. [DOI] [PubMed] [Google Scholar]
- Weisskopf MG, O’Reilly E, Chen H, Schwarzschild MA, Ascherio A (2007) Plasma urate and risk of Parkinson’s disease. Am J Epidemiol 166:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium (2014) Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9:1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, et al. (2018) Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 9:3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudko E, McNiece SI (2014) Relationship between coffee use and depression and anxiety in a population of adult polysubstance abusers. J Addict Med 8:438–442. [DOI] [PubMed] [Google Scholar]
- Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, Evans DM, Smith GD (2017) Recent developments in Mendelian randomization studies. Curr Epidemiol Rep 4:330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for GWAS scans with human serum metabolites are freely accessible through the Metabolomics GWAS Server (http://metabolomics.helmholtz-muenchen.de/gwas/). GWAS summary statistics for neuroticism can be freely downloaded from the GWAS Catalog website (https://www.ebi.ac.uk/gwas/).