We describe how genes repressed by FLOWERING LOCUS C contribute to the regulatory network that controls flowering in response to environmental cues and developmental age in annual and perennial Brassicaceae.
Keywords: Floral transition, FLOWERING LOCUS C, FT, MADS-domain, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1, vernalization
Abstract
Responses to environmental cues synchronize reproduction of higher plants to the changing seasons. The genetic basis of these responses has been intensively studied in the Brassicaceae. The MADS-domain transcription factor FLOWERING LOCUS C (FLC) plays a central role in the regulatory network that controls flowering of Arabidopsis thaliana in response to seasonal cues. FLC blocks flowering until its transcription is stably repressed by extended exposure to low temperatures in autumn or winter and, therefore, FLC activity is assumed to limit flowering to spring. Recent reviews describe the complex epigenetic mechanisms responsible for FLC repression in cold. We focus on the gene regulatory networks controlled by FLC and how they influence floral transition. Genome-wide approaches determined the in vivo target genes of FLC and identified those whose transcription changes during vernalization or in flc mutants. We describe how studying FLC targets such as FLOWERING LOCUS T, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15, and TARGET OF FLC AND SVP 1 can explain different flowering behaviours in response to vernalization and other environmental cues, and help define mechanisms by which FLC represses gene transcription. Elucidating the gene regulatory networks controlled by FLC provides access to the developmental and physiological mechanisms that regulate floral transition.
Introduction
Many plant developmental programmes are responsive to environmental cues. This is particularly evident in the characteristic seasonal patterns of flowering. These patterns have adaptive value in synchronizing reproduction with appropriate environmental conditions, maximizing the number of progeny produced, and thereby contributing to fitness in natural environments or yield in agriculture. In temperate climates, winter temperatures (vernalization) and daylength (photoperiod) provide two major floral induction cues (Andrés and Coupland, 2012). How these environmental signals regulate flowering has been studied extensively in the Brassicaceae, and, in this family, the MADS-domain transcription factor FLOWERING LOCUS C (FLC) plays a central role in conferring a response to vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999). The transcriptional and post-transcriptional regulation of FLC and how these contribute to environmental responses have been reviewed in detail (Whittaker and Dean, 2017; Costa and Dean, 2019), but the downstream functions of FLC and how these provide further environmental and developmental check points on floral transition have received less attention. We discuss recent progress in defining the functions of FLC, how these integrate responses to daylength, temperature, and developmental age, and how they can confer developmental traits associated with divergence of annual and perennial life history.
FLC blocks floral transition by binding directly to genes encoding activators of flowering and repressing their transcription. During vernalization, FLC mRNA levels decrease and are stably repressed after vernalization, allowing target genes to be transcribed and flowering to occur (Michaels and Amasino, 1999; Sheldon et al., 1999). The mechanisms by which FLC expression is stably repressed by cold have been recently reviewed (Whittaker and Dean, 2017; Costa and Dean, 2019), and a detailed description of these is outside the scope of this article. However, in general, three aspects of FLC regulation are critical in considering its effect on downstream targets. First, FLC repression occurs progressively during exposure to cold, and extended exposure of several weeks that is typical of winter is required for stable repression (Michaels and Amasino, 1999; Sheldon et al., 1999). Exposure to cold for shorter periods can lead to partial repression of FLC and an incomplete flowering response (Coustham et al., 2012; Lazaro et al., 2018). Furthermore, the progressive repression of FLC across a tissue is due to a cell-autonomous mechanism in which FLC is fully, stably repressed in some cells but not at all in others (Angel et al., 2011). The consequence of this progressive cell-autonomous repression of FLC activity for target gene activity probably varies for different genes and tissues. Secondly, stable FLC repression occurs through accumulation of modified histone 3 at the FLC gene. As the plant is exposed to cold, transcription of FLC is repressed and then trimethylation on lysine 27 of histone 3 (H3K27me3) accumulates at a nucleation point near the transcriptional start site, followed by spreading of the modification across FLC after return to warm temperatures (Finnegan and Dennis, 2007; Swiezewski et al., 2009; Yang et al., 2014). In Arabidopsis, the H3K27me3 mark persists on FLC after cold exposure has ended, resulting in the stable repression of FLC expression after cold exposure, although the duration of cold required for stable repression varies among accessions (Coustham et al., 2012). In other Brassicaceae species, transcription of FLC orthologues is reactivated after vernalization (R. Wang et al., 2009). These two contrasting patterns of FLC regulation have important consequences for the roles of downstream flowering genes and pathways (Hyun et al., 2019). Thirdly, similar to most MADS-domain transcription factors (de Folter et al., 2005), FLC binds DNA as heterodimers with other members of the family (Li et al., 2008; Gu et al., 2013) and, therefore, in considering its regulation of specific targets, it is important to assess the specificity of MADS-domain complexes that include FLC and the availability of partner proteins that might influence FLC function (Mateos et al., 2015).
Identification of FLC targets in leaves and apices
FLC is expressed broadly and therefore can potentially regulate targets in a wide range of tissues, including leaves, shoot apices, and root tips (Michaels and Amasino, 1999; Sheldon et al., 2002; Bastow et al., 2004). The significance of FLC activity in different tissues was supported by misexpression studies, which demonstrated that FLC can repress flowering when expressed in the phloem companion cells or the shoot meristem (Searle et al., 2006). Furthermore, two major direct target genes that were identified early during the elucidation of Arabidopsis flowering pathways, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), are expressed in different tissues. FT encodes a RAF kinase inhibitor-like protein and is expressed in the vascular tissue of leaves, specifically in phloem companion cells (Kardailsky et al., 1999; Kobayashi et al., 1999; Chen et al., 2018). It encodes a systemic flowering signal whose transcription is induced by inductive long-day photoperiods, and moves to the shoot apex, where it interacts with the bZIP transcription factor FD (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Abe et al., 2019). FLC represses FT transcription by binding to FT chromatin, and specifically to the first intron of FT (Helliwell et al., 2006; Searle et al., 2006). In this way, FLC blocks expression of a critical component of the photoperiodic induction pathway. The second FLC target is SOC1 (Hepworth et al., 2002; Helliwell et al., 2006; Searle et al., 2006), which is expressed in leaves and the shoot apex (Samach et al., 2000; Michaels et al., 2005) and is one of the first genes induced by the FT signal at the shoot apex (Searle et al., 2006). SOC1 encodes a MADS-domain transcription factor that regulates several genes involved in floral transition at the shoot apex (Samach et al., 2000; Immink et al., 2012), and also contributes to floral transition in non-inductive short days (Moon et al., 2003), when the FT pathway is not active. Thus, by repressing SOC1, FLC blocks expression of an early acting gene at the shoot meristem that contributes to several flowering pathways.
A broader understanding of FLC targets came from the genome-wide approach of ChIP sequencing (ChIP-seq). Applying this method, Deng et al. (2011) identified 505 target genes, including FT and SOC1. Plant MADS-domain proteins such as FLC bind to a specific CArG box DNA motif with the consensus sequence CC(A/T)6GG (Schwarz-Sommer et al., 1992; Huang et al., 1993). Consistent with this, the majority of genome-wide binding sites in promoter targets of FLC contained at least one consensus CArG-box motif CCAAAAAT(G/A)G with an AAA extension at the 3' end (Deng et al., 2011). A later repetition of this experiment Mateos et al. (2015) identified 340 target genes with a 40% overlap with those identified by Deng et al. (2011), including many of the key flowering-related genes. The list of FLC targets included those active in pathways throughout development, although many were implicated in flowering (Table 1), including TEMPRANILLO1 (Castillejo and Pelaz, 2008) and those encoding the AP2-domain transcription factors SCHLAFMÜTZE and TARGET OF EAT3 (Aukerman and Sakai, 2003; Schmid et al., 2003), the SQUAMOSA BINDING PROTEIN LIKE (SPL) family member SPL15, which contributes to progression from the juvenile to the adult phase as well as floral transition (Schwarz et al., 2008; Hyun et al., 2016), and many genes involved in stress responses to cold, and light and hormone response pathways [jasmonic acid (JA), gibberellin (GA), ethylene, and auxin]. Additional targets include four genes involved in the circadian clock (REVEILLE 2, FIONA 1, LATE ELONGATED HYPOCOTYL, and CONSTANS-LIKE1), which might be relevant for the observation that FLC influences circadian period length (Salathia et al., 2006). Comparison of the ChIP-seq list with genome-wide expression data comparing FLC and flc genotypes indicated that ~90% of the direct targets that were differentially expressed in flc mutants were increased in expression, supporting the idea that FLC acts mainly as a repressor of transcription (Mateos et al., 2015).
Table 1.
Validated direct gene targets of FLC/PEP1 involved in flowering time.
| Gene | Target | Reference |
|---|---|---|
| FLC | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 | Helliwell et al. (2006) |
| Hepworth et al. (2002) | ||
| Searle et al. (2006) | ||
| FLC | FT | Deng et al. (2011) |
| Searle et al. (2006) | ||
| FLC | TARGET OF FLC AND SVP 1 | Richter et al. (2019) |
| FLC/PEP1 | SEPALLATA3 | Deng et al. (2011) |
| Mateos et al., 2017 | ||
| FLC | SQUAMOSA PROMOTER-BINDING-LIKE PROTEIN 3 | Deng et al. (2011) |
| FLC/PEP1 | SQUAMOSA PROMOTER-BINDING-LIKE PROTEIN 15 | Deng et al. (2011) |
| Mateos et al. (2017) | ||
| PEP1 | SQUAMOSA PROMOTER-BINDING-LIKE PROTEIN 8 | Mateos et al. (2017) |
| FLC/PEP1 | SHORT VEGETATIVE PHASE | Deng et al. (2011) |
| Mateos et al., 2017 | ||
| FLC | AGAMOUS-LIKE 16 | Deng et al. (2011) |
| FLC | TEMPRANILLO 1 | Deng et al. (2011) |
| PEP1 | TARGET OF EAT 2 | Mateos et al. (2017) |
| FLC | TARGET OF EAT 3 | Deng et al. (2011) |
| FLC | SCHLAFMÜTZE | Deng et al. (2011) |
| PEP1 | GIBBERELLIN 2-BETA-DIOXYGENASE 8 | Mateos et al. (2017) |
| PEP1 | GIBBERELLIN 3-BETA-DIOXYGENASE 2 | Mateos et al. (2017) |
| PEP1 | GIBBERELLIN RECEPTOR GID1B | Mateos et al. (2017) |
Helliwell et al. (2006) showed that FLC is part of a high molecular weight complex (600–800 kDa), suggesting that it might function as a tetramer and interact with other classes of proteins involved in transcriptional regulation, as described for other plant MADS-domain proteins (Theissen and Saedler, 2001; Smaczniak et al., 2012). The SHORT VEGETATIVE PHASE (SVP) MADS-domain protein is another repressor of flowering in Arabidopsis (Hartmann et al., 2000) and, similarly to FLC, decreases gradually in expression during floral transition (Gregis et al., 2009; Jang et al., 2009). The SVP and FLC proteins interact in vivo, and negatively regulate SOC1 transcription by combinatorially binding to adjacent CArG-binding sites in the SOC1 promoter (Li et al., 2008). There is also a striking overlap between FLC and SVP target genes genome-wide (Tao et al., 2012; Gregis et al., 2013; Mateos et al., 2015). In direct comparisons, FLC and SVP bound to 183 common genes, often to the same genomic regions within those genes; however, some genes were exclusively regulated by binding of one of the proteins (Mateos et al., 2015). Furthermore, a notable difference was that FLC regulated twice as many genes in leaves as SVP, and SVP had a more significant effect on gene expression in the apex than FLC. Mutation of SVP did not completely suppress the delay in flowering caused by FLC, suggesting that FLC might interact with other proteins or act alone. In agreement with this conclusion, FLC also physically interacts with the related floral repressor proteins MADS AFFECTING FLOWERING 3 (MAF3), MAF4, and FLOWERING LOCUS M (FLM) (de Folter et al., 2005; Gu et al., 2013; Posé et al., 2013), and these interactions are required for full floral repression by FLC (Gu et al., 2013). Furthermore, in addition to the CArG-box motif found at peaks surrounding FLC-binding sites in the ChIP-seq data, the G-box motif, CACGTG (Deng et al., 2011; Mateos et al., 2015), was enriched, suggesting possible combinatorial interactions at target genes between FLC and unrelated transcription factors.
In addition to A. thaliana, FLC orthologues repress flowering and contribute to vernalization response in many other species from different Brassicaceae clades, including Arabidopsis lyrata (Kemi et al., 2013), Arabis alpina (R. Wang et al., 2009; Albani et al., 2012), Arabidopsis halleri (Aikawa et al., 2010; Nishio et al., 2016), Arabidopsis arenosa (Baduel et al., 2016), Capsella rubella (Guo et al., 2012), Cardamine hirsuta (Cartolano et al., 2015), Brassica rapa (Schranz et al., 2002), and Boechera stricta (Lee et al., 2018). However, the relatedness of their target genes in these different species has not been extensively studied. Comparative ChIP-seq analysis of FLC in A. thaliana and its orthologue PERPETUAL FLOWERING 1 (PEP1) in A. alpina identified a conserved group of 39 target genes, and these included genes with important functions in floral transition, such as SPL15, SOC1, and SEP3 (Mateos et al., 2017). However, overall, only 14% of binding sites were conserved. In addition, FLC and PEP1 also regulate common pathways related to GA and cold response, but mostly by binding to different genes, and many of these binding sites were specific to the A. alpina lineage, suggesting that the regulation of these pathways has been evolutionarily recruited independently following speciation and might represent convergent evolution (Mateos et al., 2017; Tilmes et al., 2019). The involvement of FLC in regulating GA responses was also supported by demonstration of an in vivo interaction between the C-terminus of the MADS domain of FLC and the N-terminal leucine heptad repeat I (LHRI) domain of DELLA proteins such as RGA in Arabidopsis (Li et al., 2016). These DELLA proteins are important mediators of GA signalling that are degraded in the presence of GA (Schwechheimer, 2012). Because DELLA proteins are believed not to bind DNA directly, their association with flowering time genes might in part be mediated by FLC (Li et al., 2016).
The FLC target SPL15 is regulated by miR156 to confer age-dependent responses to vernalization
One of the targets of FLC, SPL15, represents a convergence point of flowering pathways regulated by vernalization, the age of the plant, and GA. In Arabidopsis, SPL15 and a further 10 members of the SPL family of transcription factors are negatively regulated at the post-transcriptional level by miRNA156 (miR156) and closely related miR157 (Rhoades et al., 2002; Guo et al., 2008). These miRNAs are expressed at high levels in the cotyledons and leaves produced early in shoot development, and progressively decrease in abundance in leaves formed later on the shoot (Wu and Poethig, 2006; Yang et al., 2013; Yu et al., 2013). During vegetative development, these miRNAs repress the transition from the juvenile to adult phase, so that their progressive reduction during shoot development allows the acquisition of adult traits such as abaxial trichomes on leaves (Wu et al., 2009). The abundance of miR156 also falls in shoot apices, where it controls floral transition (J.W. Wang et al., 2009; Bergonzi et al., 2013). Overexpression of MIR156f in the shoot apical meristem (SAM) delayed floral transition (J.W. Wang et al., 2009). Several SPL transcription factors that contain miR156 target sequences in their cognate mRNA are expressed in the SAM, including SPL3, SPL4, SPL5, SPL9, and SPL15 (Cardon et al., 1997; J.W. Wang et al., 2009; Yamaguchi et al., 2009; Hyun et al., 2016). Among these, loss-of-function mutant alleles of SPL15 delay flowering under short days (SDs) (Hyun et al., 2016; Xu et al., 2016). Furthermore, mutations in the miR156 recognition sequence of SPL15 (rSPL15) increase the abundance of SPL15 and confer early flowering (Hyun et al., 2016). In wild-type plants, SPL15 accumulates in the SAM under SDs prior to floral induction, and rSPL15 accumulates earlier (Hyun et al., 2016), correlating with earlier flowering. Taken together, these results indicate that the FLC target SPL15 promotes flowering under SDs and its ability to promote flowering is repressed in younger plants by miR156 (Fig. 1A). In contrast to its late-flowering phenotype under SDs, the spl15 mutant is not late flowering under long days (LDs). This conditionality of the phenotype dependent on daylength was proposed to be due to the photoperiodic flowering pathway bypassing the requirement for SPL15 under LDs. This model was confirmed genetically by combining the ft and twin sister of ft (tsf) mutations, which block the photoperiodic pathway, with the spl15 mutant (Hyun et al., 2019). The triple mutant spl15 ft tsf flowered much later than ft tsf under LDs. Thus, spl15 exhibits a conditional effect on flowering time, only being required under conditions in which the photoperiodic pathway is not active.
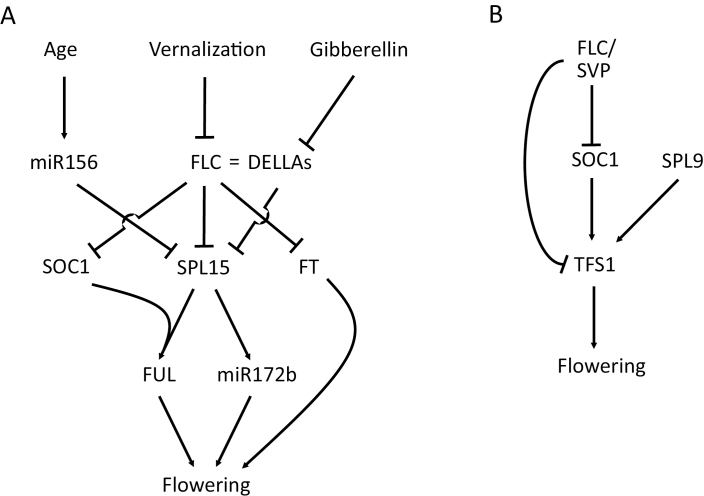
Fig. 1.
Schematic representation of the gene regulatory network controlled by FLC. (A) FLC is a master regulator of flowering that integrates cues from several flowering pathways in Arabidopsis. FLC expression is repressed by the cold-induced vernalization pathway. FLC directly represses the florigen-encoding gene FT and SQUAMOSA PROMOTER BINDING-LIKE PROTEIN 15 (SPL15). SPL15 is additionally post-translationally regulated by the gibberellin pathway via DELLA proteins and at the post-transcriptional level by the ageing pathway via miR156. The MADS-domain protein SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) is encoded by another direct target of FLC, and cooperates with SPL15 to activate target genes such as FRUITFULL and MIR172B. (B) A type II coherent feed-forward loop regulates expression of the positive floral regulator TARGET OF FLC AND SVP1 (TFS1). FLC and SVP directly repress TFS1 transcription and that of its positive activator SOC1. Activation of TFS1 expression in the inflorescence meristem also involves SPL9.
The functional importance of SPL15 and miR156 in vernalization response downstream of FLC first became evident from analysis of the perennial Brassicaceae species A. alpina and Cardamine flexuosa (Bergonzi et al., 2013; Zhou et al., 2013). These plants do not flower if exposed to vernalization soon after germination, but do when exposed to vernalization when 4–5 weeks old. This dependency on the age of the plant can be defined as the acquisition of competence to flower in response to vernalization. It cannot be explained by age-dependent reduction in FLC mRNA, because exposure of young plants to vernalization caused repression of FLC transcription (Bergonzi et al., 2013; Zhou et al., 2013). However, because the ability to respond to vernalization correlated with miR156 reaching trough levels, the progressive reduction in miR156 level in the shoot apex was proposed to confer age-dependent responses to vernalization. Consistent with this, transgenic plants that expressed MIR156f from the viral 35S promoter never flowered in response to vernalization, and a reduction in miR156 activity by expressing a miR156 mimic that sequesters miR156 enabled younger plants to flower in response to vernalization (Bergonzi et al., 2013; Zhou et al., 2013). This effect of miR156 in A. alpina was later shown to depend upon the orthologue of SPL15 (AaSPL15) (Hyun et al., 2019). Loss-of-function mutations in AaSPL15 prevented flowering in response to vernalization, as observed for MIR156f-overexpressing plants; furthermore, rSPL15 plants acquired competence to respond to vernalization earlier than the wild type. Notably, age was measured independently in the SAM and each axillary meristem, suggesting that the mechanism of miR156 down-regulation by age is regulated autonomously in different meristems of the same plant (Park et al., 2017; Hyun et al., 2019). Therefore, an age-dependent vernalization response can be explained by repression of SPL15 transcription by FLC and SPL15 post-transcriptional repression by miR156 in individual meristems, so that this double lock on SPL15 expression is only relieved when FLC expression is reduced during vernalization and miR156 levels decrease in older meristems (Fig. 2).
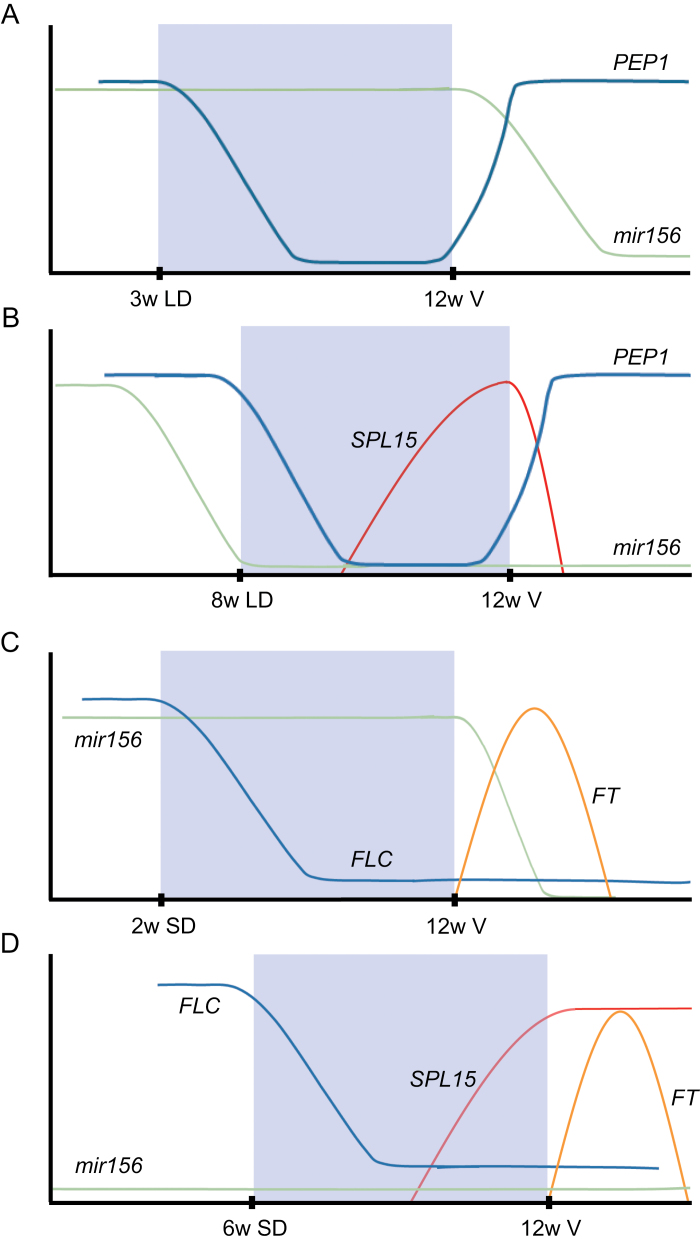
Fig. 2.
FLC and its orthologues in other species regulate flowering in response to vernalization by controlling two major floral-promoting pathways involving SPL15 and FT. In perennial Arabis alpina plants exposed to vernalization, FT expression is repressed by the FLC orthologue PEP1 both before and after vernalization, when PEP1 is reactivated by warm conditions (A and B). Therefore, flowering is dependent on the miR156/SPL15 module. When young meristems are vernalized, miR156 levels are high and repress floral transition by maintaining low SPL15 levels, so no flowering occurs (A). Only older meristems can flower during the vernalization period when the level of miR156 has fallen and SPL15 mRNA can increase during vernalization (B). In the winter-annual Arabidopsis thaliana, FLC is transcriptionally repressed during vernalization in short days in the cold and remains stably repressed in subsequent warm temperatures (C and D). This allows plants to flower by activation of FT transcription after cold treatment via the photoperiodic pathway irrespective of age, even when miRNA156 levels are high in young plants that are vernalized (C). Stable repression of FLC transcription in annuals therefore allows plants to flower independently of age through the FT pathway after vernalization, thus bypassing dependency on the miRNA156/SPL15 module. The graphs (A–D) depict the relative level of FLC, PEP1, SPL15, and FT mRNAs, and miRNA156 on the y-axis. V, vernalization; w, weeks; SD. short days; LD, long days. The blue box represents a vernalization treatment.
Finally, SPL15 is negatively regulated at the post-translational level by GA. SPL15 interacts with the DELLA protein RGA in yeast and in planta, and reduction in GA levels in the shoot meristem increases the abundance of RGA and the amount of the RGA–SPL15 complex (Hyun et al., 2016). The interaction between RGA and SPL15 was proposed to reduce SPL15 activity, because lowering GA levels at the shoot apex by expression of a GA catabolic enzyme reduced the ability of rSPL15 to promote early flowering. DELLA proteins also interact with SPL9 (Yu et al., 2012; Yamaguchi et al., 2014), suggesting that the regulation of the activity of SPL transcription factors by GA during reproductive development is more prevalent (Zhang et al., 2007). It is relevant that FLC and its orthologue in A. alpina, PEP1, directly bind to several genes that encode enzymes required for GA biosynthesis or for GA signalling (Deng et al., 2011; Mateos et al., 2015; Tilmes et al., 2019). The transcription of some of these genes also increases during vernalization (Tilmes et al., 2019). This observation suggests that FLC ensures that SPL15 is inactive prior to vernalization by effectively regulating its expression via a feed-forward loop: first, FLC binds directly to the SPL15 promoter to repress its transcription; and, secondly, FLC reduces GA biosynthesis, which increases the level of DELLA proteins that inhibit SPL15 activity. SPL15 expression is then activated transcriptionally and post-translationally during vernalization as FLC transcription is repressed. Because GA levels increase at about the time of flowering in apices of plants grown under SDs (Eriksson et al., 2006), this post-translational regulation of SPL15 probably also contributes to its appropriate temporal activation under non-inductive conditions.
FT and SPL15 are genetically redundant FLC targets that determine whether flowering occurs during or after cold exposure
Expression of FLC prior to vernalization blocks floral induction by directly repressing the transcription of a set of flowering genes that are described above, whereas the transcriptional repression of FLC by cold during vernalization allows these targets to be expressed and floral induction to proceed. In A. thaliana, floral induction is usually considered to occur after vernalization, when plants in which FLC transcription is repressed are exposed to LDs that mimic spring conditions and allow activation of the photoperiodic pathway (Coustham et al., 2012) (Fig. 2). During the vernalization period, when plants are exposed to cold, the photoperiodic pathway is not active, because vernalization typically occurs under SDs or low light, analogous to winter conditions. Thus, stable repression of FLC transcription renders A. thaliana competent to flower through the photoperiodic pathway after vernalization. The mobile florigen protein FT is a component of the photoperiodic pathway and is transcribed only in LDs following vernalization. Furthermore, as FT transcription is directly repressed by FLC, the photoperiodic flowering response after vernalization depends upon stable repression of FLC transcription by cold. Thus, A. thaliana, and other species in which FLC transcription is stably repressed by vernalization, can undergo floral induction after vernalization through this FT-based route. As described above, because FT can also bypass the requirement for the miR156/SPL15 module, stable repression of FLC transcription allows floral induction to occur after vernalization, independently of the age of the plant. Genetic support for this model came recently from comparing the vernalization response of A. thaliana FRI FLC ft tsf and FRI FLC plants (Hyun et al., 2019). Young FRI FLC ft tsf plants, in which miR156 levels were still high, flowered much later after vernalization than FRI FLC plants, whereas older FRI FLC ft tsf plants, in which miR156 levels had declined, flowered at the same time as FRI FLC when exposed to vernalization.
In contrast, transcription of FLC orthologues in perennial Brassicaceae species, such as PEP1 of A. alpina, is reactivated after vernalization (R. Wang et al., 2009; Baduel et al., 2016; Kiefer et al., 2017) and, therefore, PEP1 represses transcription of FT when A. alpina is exposed to LDs after vernalization (Hyun et al., 2019) (Fig. 2). Consequently, flowering in A. alpina is initiated during cold exposure when PEP1 transcription is repressed. Because the photoperiodic pathway is effectively blocked by PEP1 before and after vernalization, these plants are dependent upon the miR156/SPL15 module for floral induction during vernalization, which explains why they can only respond to vernalization when meristems reach the age at which miR156 levels have fallen. The importance of reactivation of PEP1 after vernalization in conferring an age-dependent vernalization response in A. alpina was recently supported by genetic data. Arabis alpina pep1 mutants carrying a stably repressed allele of FLC from annual Arabis montbretiana could respond to vernalization as young plants, as previously shown for A. thaliana (Hyun et al., 2019).
Thus, by negatively regulating both SPL15 and FT, FLC and its orthologues control both major pathways that promote flowering in response to vernalization. Stable repression of FLC transcription in annuals allows plants to flower independently of age through the FT pathway after vernalization, whereas reactivation of FLC orthologues in perennials blocks FT transcription, which forces these plants to flower through the age-dependent SPL15 pathway. Recently, winter varieties of Brassica napus that flower in response to vernalization were found to produce flower buds in late autumn and open flowers in spring (O’ Neill et al., 2019). In these plants, floral induction occurs under vernalizing conditions in autumn when the photoperiodic pathway is not expected to be active, and therefore might initiate flowering via the miR156/SPL15 pathway. This possibility extends the previously noted similarities between flowering of B. napus crops and perennials such as A. alpina (O’ Neill et al., 2019), and is significant for breeding for flowering time in winter crops.
A regulatory module involving MADS-domain and B3-type transcription factors controlled by FLC at the shoot meristem
A further direct target of FLC that was recently shown to contribute to a feed-forward loop in the shoot meristem is TARGET OF FLC AND SVP 1 (TFS1), which encodes a B3-type transcription factor that promotes flowering (Richter et al., 2019). TFS1 was identified by mining genome-wide ChIP-seq and RNA-seq data as a gene specifically expressed in shoot apices and directly bound by FLC and its interacting partner SVP (Mateos et al., 2015; Richter et al., 2019). The gene is expressed on the flanks of the shoot meristem specifically during and after floral transition, and tfs1 mutants are late flowering. In FRI FLC plants, the mRNA level of TFS1 increases during vernalization as FLC expression falls. Transcriptional activation of TFS1 in this precise temporal and spatial pattern involves SPL9, the paralogue of SPL15, and the MADS-domain transcription factor SOC1 (Richter et al., 2019) (Fig. 1B). SOC1 and SPL9 physically interact and both bind directly to different sites within the TFS1 promoter, suggesting that their interaction might induce looping at the locus that is required to activate TFS1 transcription. SOC1 is not only a well-established target of FLC, whose transcription is repressed by direct binding of FLC to its promoter and activated during vernalization when FLC transcription is repressed (Hepworth et al., 2002; Searle et al., 2006; Deng et al., 2011), but is also one of the earliest acting transcription factors during floral transition (Samach et al., 2000; Immink et al., 2012). Thus, FLC represses TFS1 transcription both by direct binding to its promoter and by repressing transcription of its major direct upstream regulator SOC1, creating a coherent type-2 feed-forward loop (Alon, 2007). Such a loop might delay the expression of TFS1 rather than inducing a more rapid response to FLC down-regulation during vernalization (Mangan and Alon, 2003).
Mechanism by which FLC represses transcription of targets
The predominant genome-wide function of FLC is as a repressor of transcription (Mateos et al., 2015), but the mechanism by which this occurs has not been elucidated in detail, although it probably involves protein partners. FLC is a type II MADS-domain protein that possesses multiple domains that can potentially interact with other proteins. These include the keratin-like domain, also known as the K-box, the intervening domain, the MADS-domain that binds DNA, and the C-terminal domain that can stabilize protein complexes and regulate transcription (Kaufmann et al., 2005). Thus, the target specificity of FLC and efficacy of its repression function is potentially conferred by its interaction partners, or the cofactors of the MADS-domain proteins with which it heterodimerizes (Li et al., 2008; Gu et al., 2013).
Several proteins that interact with FLC are implicated in chromatin modifications: FLC physically interacts with EMBRYONIC FLOWER 1 (EMF1) and is required for FLC-mediated FT repression (Wang et al., 2014). EMF1, LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), and the H3K4me3 demethylase JMJ14/PKDM7B form a polycomb group protein complex termed EMF1c, which is a polycomb repressive complex1 (PRC1)-like complex that represses FT transcription (Wang et al., 2014). EMF1 is also associated with many genomic sites, including FT chromatin, which are marked with H3K27me3 deposited by PRC2 (Kim et al., 2012). This involvement of FLC with polycomb repressive complexes suggests that PcG-mediated gene silencing contributes to repression of gene expression by FLC. This was supported for the direct target of FLC and SVP, TFS1, which was found to possess a high level of repressive H3K27me3 marks at sites that were also occupied by LHP1 and were dependent on FLC and SVP (Richter et al., 2019). The level of H3K4me3 is enriched at the chromatin of actively transcribed genes, and the level of this mark was low at the TFS1 locus in the presence of FLC and SVP, and was enriched in single and double mutants of flc and svp (Richter et al., 2019). Therefore, the presence of repressive and permissive chromatin modifications at TFS1 is consistent with the known antagonistic dynamics of chromatin marks during floral transition (Engelhorn et al., 2017).
Transcription of TFS1 is repressed by binding of FLC and SVP 3' to the TFS1 stop codon and their interaction with a PRC complex and LHP1 that associate with the gene body of TFS1, suggesting that the resulting complex forms a chromosomal loop. The formation of this ‘locked’ loop is dependent on CURLY LEAF (CLF), FLC, or SVP (Richter et al., 2019). Therefore, the transcriptional repression of TFS1 by FLC and SVP consists of two component processes: the formation of a chromatin loop that requires FLC; and SVP binding at the 3' end of the gene and high levels of H3K27me3 within the gene body. SOC1 also activates TFS1 transcription by binding to the 3' end of TFS1 and reducing SVP recruitment to the same region (Richter et al., 2019). It remains unclear how general the mechanisms of action of FLC on FT and TFS1 are for other targets. However, based on TFS1 regulation, antagonistic chromatin remodelling via FLC and other MADS-domain transcription factors appears to represent the central mechanism that defines the spatiotemporal expression of downstream targets.
Quantitative effects of FLC on target genes and its consequences for floral transition and inflorescence development
The level of FLC mRNA varies tremendously among Arabidopsis accessions and is correlated with flowering time (Michaels et al., 2003; Lempe et al., 2005; Shindo et al., 2005; Sasaki et al., 2018). In a genome-wide expression analysis of 132 accessions, a subset of 38 genes whose expression was correlated with flowering time included FLC and three downstream targets, FT, SOC1, and SPL15 (Sasaki et al., 2018). Thus, increasing levels of FLC mRNA can lead to later flowering phenotypes, presumably by reducing the expression of known target genes. Variation in the FLC expression level is largely due to allelic variation either at FLC or at the upstream FRIGIDA (FRI) locus. Natural allelic variation in FRI has been analysed for >1000 Arabidopsis accessions (Zhang and Jimenez-Gomez, 2020), and has been estimated to account for ~70% of diversity in flowering time (Shindo et al., 2005). FRI is recruited to the FLC locus as part of a large protein complex containing chromatin modifiers to increase FLC transcription (Li et al., 2018). Many early-flowering summer-annual Arabidopsis accessions arose from winter-annual progenitors by the loss of FRI function (Johanson et al., 2000). Other early-flowering accessions possess FLC alleles that are expressed weakly (Michaels et al., 2003) or in which the duration of vernalization required for stable repression varies (Coustham et al., 2012) due to cis-acting variation. Among 173 natural Swedish accessions, enormous variation for flowering time at growth temperatures of 10°C and 16°C was detected, and genome-wide association analysis identified a peak on the promoter of the FLC gene (Sasaki et al., 2015). Variation in the expression of FLC orthologues is probably also important in determining flowering time in other Brassicaceae species but, as many are allopolyploids containing several copies of different FLC orthologues, the combined effects of these on flowering time and vernalization response can be complex (Schiessl et al., 2019; Takada et al., 2019).
In addition to affecting flowering time in the absence of vernalization treatment, allelic heterogeneity at the FLC locus can determine the duration or temperature of vernalization required to induce flowering. Accessions differ in the duration of vernalization required for stable repression of FLC, and therefore for activation of the FT target gene after vernalization (Coustham et al., 2012). The coding sequence of FLC is remarkably conserved among Arabidopsis accessions (Li et al., 2014) and most polymorphisms at the FLC locus are associated with cis-acting variation in FLC non-coding regions. Some of these, for example in the lov-1 accession, are near the transcriptional start site, close to the site at which the H3K27me3 mark first increases during vernalization (Coustham et al., 2012). The lov-1 plants require exposure to cold for longer than is necessary for full vernalization of other A. thaliana accessions and, after shorter vernalization periods, FLC expression in lov-1 is reactivated in individual cells in subsequent warm conditions (Coustham et al., 2012), a feature that is characteristic of perennial Brassicaceae species. A combination of cis-acting single nucleotide polymorphisms (SNPs) at lov-1 FLC quantitatively mediates instability of FLC repression by disrupting stable long-term chromatin silencing of the locus, probably via histone modification feedback (Qüesta et al., 2020). Another study tested the responses of 47 Arabidopsis accessions containing the major haplotypes of FLC to short vernalization treatments of 4 weeks, and detected variation in the ability to stably repress FLC transcription (Li et al., 2014). Variation of these haplotypes in this response was again associated with non-coding sequence variation presumably linked to the histone modifications at FLC related to stable repression (Li et al., 2014). Differences in FLC expression patterns due to variations in chromatin silencing between annual and perennial Brassicaceae depend on cis-polymorphisms in non-coding FLC sequences (Kiefer et al., 2017), and might therefore represent a general mechanism for life history evolution. Thus, accessions vary in the duration of vernalization required to stably repress FLC, and allow the promotion of flowering through activation of FT, and presumably other targets. Furthermore, the duration of cold treatment required for full vernalization is likely to be more complex in the fluctuating conditions that prevail during natural growth (Burghardt et al., 2016). In addition to affecting floral transition, partial vernalization treatments can affect the extent of inflorescence development. This was most clearly demonstrated in A. alpina, where incomplete vernalization led to reactivation of the FLC orthologue PEP1 in the inflorescence, causing floral reversion and reduced inflorescence size (Lazaro et al., 2018). Similarly, in A. thaliana, high levels of FLC expression caused by allelic variation at the upstream regulator ENHANCER of AG-4 2 (HUA2) can also cause floral reversion phenotypes in the inflorescence and vegetative rosettes to form within the inflorescence (Wang et al., 2007). These phenotypes, which are probably caused by repression of FLC targets during inflorescence development, may also persist after incomplete vernalization.
The stable repression of FLC by vernalization occurs cell-autonomously via an ‘all or nothing’ bistable state (Angel et al., 2011). Therefore, incomplete vernalization would be expected to result in tissues in which FLC is completely switched off in some cells, but is still expressed at pre-vernalization levels in others (Angel et al., 2011). How this would affect the expression of target genes in different tissues is unknown. However, at least for SOC1, a gradual increase that was the reciprocal of the effect on FLC transcription was observed during vernalization (Sheldon et al., 2006), suggesting that SOC1 may be activated in a cell-autonomous manner, although regulation of SOC1 is complicated by an FLC-independent acute induction by cold (Sheldon et al., 2006). Activation of FT transcription in a cell-autonomous manner may be sufficient to activate floral induction, because the FT protein induces flowering cell non-autonomously (Corbesier et al., 2007). However, for targets encoding transcription factors such as SOC1, SPL15, SEP3, or TFS1, it is unclear that they could confer inflorescence and floral meristem identity cell non-autonomously. Thus, partial vernalization treatments would be expected to have complex and unpredictable effects on inflorescence development.
Perspectives
Only a subset of FLC target genes has been incorporated into gene regulatory networks that control floral transition. Notably, those studied in detail encode transcription factors (e.g. SOC1, SPL15, and TFS1) or components of transcriptional complexes (e.g. FT), whereas genes encoding other classes of protein, including enzymes of unknown function or involved in phytohormone biosynthesis, remain to be studied in detail. Established target genes of FLC, such as FT and SPL15, define key pathways that regulate the floral transition in different environments or physiological contexts. These pathways intersect with the patterns of transcriptional repression of FLC or its orthologues in particular species to confer different flowering behaviours. For example, the unstable repression of the FLC orthologue PEP1 in A. alpina forces this species to flower through the SPL15 pathway, which is also controlled by miR156, generating an age-based check point on vernalization response (Hyun et al., 2019) (Fig. 2). Other such check points may exist and explain how, in some contexts, FLC repression and floral induction can occur during vernalization and in autumn, but that flowers and inflorescences only fully develop in spring (Kemi et al., 2019; O’ Neill et al., 2019). Determining the spatial and temporal patterns of expression of further FLC targets and the regulatory networks they control during vernalization may contribute to defining other check points at which flowering can be environmentally regulated downstream of FLC. Comparative approaches among species are likely to be essential to understand the full role of FLC in flowering processes (Mateos et al., 2017; Hyun et al., 2019; Kemi et al., 2019; O’ Neill et al., 2019), as the flowering behaviour of A. thaliana is highly derived and dominated by the FT pathway. Similarly, further studies under natural conditions will define how the networks controlled by FLC contribute to seasonal patterns in ecologically relevant conditions (Burghardt et al., 2016; O’ Neill et al., 2019).
Acknowledgements
The laboratory of GC is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2048/1 Project ID: 390686111) and is supported by a core grant from the Max Planck Society.
References
- Abe M, Kosaka S, Shibuta M, Nagata K, Uemura T, Nakano A, Kaya H. 2019. Transient activity of the florigen complex during the floral transition in Arabidopsis thaliana. Development 146, dev171504. [DOI] [PubMed] [Google Scholar]
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. 2010. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proceedings of the National Academy of Sciences, USA 107, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani MC, Castaings L, Wötzel S, Mateos JL, Wunder J, Wang R, Reymond M, Coupland G. 2012. PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genetics 8, e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U 2007. Network motifs: theory and experimental approaches. Nature Reviews. Genetics 8, 450–461. [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nature Reviews. Genetics 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Arnold B, Weisman CM, Hunter B, Bomblies K. 2016. Habitat-associated life history and stress-tolerance variation in Arabidopsis arenosa. Plant Physiology 171, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. 2004. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordström KJ, Wang R, Schneeberger K, Moerland PD, Coupland G. 2013. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340, 1094–1097. [DOI] [PubMed] [Google Scholar]
- Burghardt LT, Runcie DE, Wilczek AM, Cooper MD, Roe JL, Welch SM, Schmitt J. 2016. Fluctuating, warm temperatures decrease the effect of a key floral repressor on flowering time in Arabidopsis thaliana. New Phytologist 210, 564–576. [DOI] [PubMed] [Google Scholar]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P. 1997. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. The Plant Journal 12, 367–377. [DOI] [PubMed] [Google Scholar]
- Cartolano M, Pieper B, Lempe J, Tattersall A, Huijser P, Tresch A, Darrah PR, Hay A, Tsiantis M. 2015. Heterochrony underpins natural variation in Cardamine hirsuta leaf form. Proceedings of the National Academy of Sciences, USA 112, 10539–10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. 2008. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Current Biology 18, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Chen Q, Payyavula RS, Chen L, Zhang J, Zhang C, Turgeon R. 2018. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proceedings of the National Academy of Sciences, USA 115, 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Costa S, Dean C. 2019. Storing memories: the distinct phases of Polycomb-mediated silencing of Arabidopsis FLC. Biochemical Society Transactions 47, 1187–1196. [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587. [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, et al. 2005. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. The Plant Cell 17, 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. 2011. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J, Blanvillain R, Kröner C, Parrinello H, Rohmer M, Pose D, Ott F, Schmid M, Carles CC. 2017. Dynamics of H3K4me3 chromatin marks prevails over H3K27me3 for gene regulation during flower morphogenesis in Arabidopsis thaliana. Epigenomes 1, 8–40. [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O. 2006. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. The Plant Cell 18, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. 2007. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Current Biology 17, 1978–1983. [DOI] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, et al. 2013. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biology 14, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. The Plant Journal 60, 626–637. [DOI] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y. 2013. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nature Communications 4, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. 2008. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418, 1–8. [DOI] [PubMed] [Google Scholar]
- Guo YL, Todesco M, Hagmann J, Das S, Weigel D. 2012. Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics 192, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. The Plant Journal 21, 351–360. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 46, 183–192. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. 2002. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. The EMBO Journal 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. 1993. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Research 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. 2016. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Developmental Cell 37, 254–266. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G. 2019. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363, 409–412. [DOI] [PubMed] [Google Scholar]
- Immink RG, Posé D, Ferrario S, et al. 2012. Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiology 160, 433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology 17, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G. 2009. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. The Plant Journal 60, 614–625. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. 2005. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. [DOI] [PubMed] [Google Scholar]
- Kemi U, Leinonen PH, Savolainen O, Kuittinen H. 2019. Inflorescence shoot elongation, but not flower primordia formation, is photoperiodically regulated in Arabidopsis lyrata. Annals of Botany 124, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemi U, Niittyvuopio A, Toivainen T, Pasanen A, Quilot-Turion B, Holm K, Lagercrantz U, Savolainen O, Kuittinen H. 2013. Role of vernalization and of duplicated FLOWERING LOCUS C in the perennial Arabidopsis lyrata. New Phytologist 197, 323–335. [DOI] [PubMed] [Google Scholar]
- Kiefer C, Severing E, Karl R, Bergonzi S, Koch M, Tresch A, Coupland G. 2017. Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Molecular Ecology 26, 3437–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. 2012. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genetics 8, e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Lazaro A, Obeng-Hinneh E, Albani MC. 2018. Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiology 176, 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Hsieh JW, Schranz ME, Mitchell-Olds T. 2018. The functional change and deletion of FLC homologs contribute to the evolution of rapid flowering in Boechera stricta. Frontiers in Plant Science 9, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genetics 1, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. 2008. A repressor complex governs the integration of flowering signals in Arabidopsis. Developmental Cell 15, 110–120. [DOI] [PubMed] [Google Scholar]
- Li M, An F, Li W, Ma M, Feng Y, Zhang X, Guo H. 2016. DELLA proteins interact with FLC to repress flowering transition. Journal of Integrative Plant Biology 58, 642–655. [DOI] [PubMed] [Google Scholar]
- Li P, Filiault D, Box MS, et al. 2014. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes & Development 28, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang D, He Y. 2018. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nature Plants 4, 836–846. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences, USA 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, Fornara F, Schneeberger K, Krajewski P, Coupland G. 2015. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biology 16, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G. 2017. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proceedings of the National Academy of Sciences, USA 114, E11037–E11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proceedings of the National Academy of Sciences, USA 100, 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. 2005. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiology 137, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. 2003. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. The Plant Journal 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Nishio H, Buzas DM, Nagano AJ, Suzuki Y, Sugano S, Ito M, Morinaga S, Kudoh H. 2016. From the laboratory to the field: assaying histone methylation at FLOWERING LOCUS C in naturally growing Arabidopsis halleri. Genes & Genetic Systems 91, 15–26. [DOI] [PubMed] [Google Scholar]
- O’Neill CM, Lu X, Calderwood A, Tudor EH, Robinson P, Wells R, Morris R, Penfield S. 2019. Vernalization and floral transition in autumn drive winter annual life history in oilseed rape. Current Biology 29, 4300–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim H, Lee I. 2017. Comparative analysis of molecular and physiological traits between perennial Arabis alpina Pajares and annual Arabidopsis thaliana Sy-0. Scientific Reports 7, 13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. 2013. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. [DOI] [PubMed] [Google Scholar]
- Qüesta JI, Antoniou-Kourounioti RL, Rosa S, Li P, Duncan S, Whittaker C, Howard M, Dean C. 2020. Noncoding SNPs influence a distinct phase of Polycomb silencing to destabilize long-term epigenetic memory at Arabidopsis FLC. Genes & Development 34, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G. 2019. Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genetics 15, e1008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathia N, Davis SJ, Lynn JR, Michaels SD, Amasino RM, Millar AJ. 2006. FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biology 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sasaki E, Frommlet F, Nordborg M. 2018. GWAS with heterogeneous data: estimating the fraction of phenotypic variation mediated by gene expression data. G3 8, 3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Zhang P, Atwell S, Meng D, Nordborg M. 2015. ‘Missing’ G × E variation controls flowering time in Arabidopsis thaliana. PLoS Genetics 11, e1005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl SV, Quezada-Martinez D, Tebartz E, Snowdon RJ, Qian L. 2019. The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Scientific Reports 9, 14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. 2003. Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC. 2002. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. 2008. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Molecular Biology 67, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. 1992. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. The EMBO Journal 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C 2012. Gibberellin signaling in plants—the extended version. Frontiers in Plant Science 2, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. The Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. The Plant Cell 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Finnegan EJ, Dennis ES, Peacock WJ. 2006. Quantitative effects of vernalization on FLC and SOC1 expression. The Plant Journal 45, 871–883. [DOI] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiology 138, 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, et al. 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences, USA 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Takada S, Akter A, Itabashi E, et al. 2019. The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Scientific Reports 9, 13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. The Plant Journal 70, 549–561. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. 2001. Plant biology. Floral quartets. Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- Tilmes V, Mateos JL, Madrid E, Vincent C, Severing E, Carrera E, López-Díaz I, Coupland G. 2019. Gibberellins act downstream of Arabis PERPETUAL FLOWERING1 to accelerate floral induction during vernalization. Plant Physiology 180, 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V. 2007. HUA2 caused natural variation in shoot morphology of A. thaliana. Current Biology 17, 1513–1519. [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu X, Yuan W, Schmitz RJ, He Y. 2014. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Developmental Cell 28, 727–736. [DOI] [PubMed] [Google Scholar]
- Whittaker C, Dean C. 2017. The FLC locus: a platform for discoveries in epigenetics and adaptation. Annual Review of Cell and Developmental Biology 33, 555–575. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ML, Hu TQ, Zhao JF, Park MY, Earley KW, Wu G, Yang L, Poethig RS. 2016. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics 12, e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. 2009. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Developmental Cell 17, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D. 2014. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344, 638–641. [DOI] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C. 2014. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Current Biology 24, 1793–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS. 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2, e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW. 2013. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2, e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. 2012. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. The Plant Cell 24, 3320–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jimenez-Gomez JM. 2020. Functional analysis of FRIGIDA using naturally occurring variation in Arabidopsis thaliana. The Plant Journal (in press). doi: 10.1111/tpj.14716 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schwarz S, Saedler H, Huijser P. 2007. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Molecular Biology 63, 429–439. [DOI] [PubMed] [Google Scholar]
- Zhou CM, Zhang TQ, Wang X, Yu S, Lian H, Tang H, Feng ZY, Zozomova-Lihová J, Wang JW. 2013. Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340, 1097–1100. [DOI] [PubMed] [Google Scholar]