A CRISPR-based system enables safer multimodal in vivo reporter gene cell tracking.
Abstract
Imaging reporter genes provides longitudinal information on the biodistribution, growth, and survival of engineered cells in vivo. A translational bottleneck to using reporter genes is the necessity to engineer cells with randomly integrating vectors. Here, we built homology-independent targeted integration (HITI) CRISPR-Cas9 minicircle donors for precise safe harbor-targeted knock-in of fluorescence, bioluminescence, and MRI (Oatp1a1) reporter genes. Our results showed greater knock-in efficiency using HITI vectors compared to homology-directed repair vectors. HITI clones demonstrated functional fluorescence and bioluminescence reporter activity as well as significant Oatp1a1-mediated uptake of the clinically approved MRI agent gadolinium ethoxybenzyl diethylenetriamine pentaacetic acid. Contrast-enhanced MRI improved the conspicuity of both subcutaneous and metastatic Oatp1a1-expressing tumors before they became palpable or even readily visible on precontrast images. Our work demonstrates the first CRISPR-Cas9 HITI system for knock-in of large DNA donor constructs at a safe harbor locus, enabling multimodal longitudinal in vivo imaging of cells.
INTRODUCTION
Molecular-genetic imaging with reporter genes permits the in vivo visualization and tracking of engineered cells and thus allows one to track the biodistribution, persistence, viability, and, in some cases, activation state of such cells (1, 2). Several reporter genes currently exist for visualizing engineered cells using preclinical optical fluorescence imaging (FLI) and bioluminescence imaging (BLI) (3–5) as well as those for clinical modalities such as magnetic resonance imaging (MRI), positron emission tomography (PET), and photoacoustic imaging (6–8). These noninvasive cell tracking tools are invaluable for understanding mechanisms of disease progression and the evaluation of treatments in preclinical animal models. Important examples in cancer research include the tracking of therapeutic stem cells (9–11); tracking immune cell migration, cancer progression, and metastasis (12–14); and evaluating tumor response to novel anticancer therapeutics (15, 16). More recently, the use of reporter genes to track therapeutic cells has been translated into the clinic. In this case, cytotoxic T cells were engineered to express a chimeric antigen receptor to target glioma cells, as well as a herpes simplex virus type 1 thymidine kinase (HSV1-TK) dual reporter-suicide gene (that selectively uptakes the PET tracer [18F]FHBG) to track the localization and viability of the injected therapeutic cells in glioma patients (17, 18).
Although reporter genes have great potential for therapeutic cell tracking, their functionality is best used when the genes are stably integrated into the desired cell’s genome, allowing reporter gene expression throughout the lifetime of the cell and in any subsequent daughter cells. Retroviral vectors, such as those derived from HIV lentiviruses, have generally been used for transgene integration due to their high transfection efficiency, large transgene capacity, and their ability to transduce a variety of dividing and nondividing cell types. However, the low acceptance of using reporter genes for tracking cell-based therapies may, in part, be due to the increased risk of random or quasi-random insertional mutagenesis when transgenes are delivered using viral vectors (19). In previous clinical trials involving children with X-linked severe combined immunodeficiency, a Moloney murine leukemia virus–based γ-retrovirus vector expressing the interleukin-2 receptor γ-chain (γc) complementary DNA successfully restored immunity in most patients. However, 5 of the 20 patients also developed leukemia, of which one child died, as a result of insertional mutagenesis and transactivation of proto-oncogenes (20–22). An alternative to viral-based engineering is to use nonviral transposase-based systems such as the Sleeping Beauty or piggyBac transposon systems (23, 24). Transposase systems can integrate large expression cassettes into mammalian cells, but lack specificity, tending to integrate at multiple sites within the genome almost randomly, or with preference for transcriptional start sites and long terminal repeat elements. For future cell-based therapies, it is therefore highly desirable to edit cells with reporter genes in a safe and site-specific manner. The application of such editing tools would allow longitudinal cell tracking to confirm that the cells are performing their intended role and to detect any ectopic growths or misplaced targeting at the earliest time point. This will ultimately give the clinician greater control and confidence in the outcomes of the targeted therapy.
Genomic safe harbors can incorporate exogenous pieces of DNA and permit their predictable function but do not cause alterations to the host genome or pose a risk to the host cell or organism (25). Several studies have successfully used genome editing tools such as zinc finger nucleases (ZFNs) and transcription activator–like effector nucleases (TALENs) to incorporate reporter genes at the adeno-associated virus integration site 1 (AAVS1) safe harbor locus, with no detrimental effects (26–28). Although ZFNs and TALENs have shown great promise as targeted DNA editors, they are time consuming, expensive, and challenging to engineer as unique nuclease sequences must be generated for every genomic target. Alternatively, clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR-Cas9), which was developed by several groups in 2013 (29–32), allows quicker, cheaper, and easier-to-design human genome editing. CRISPR-Cas9 uses short guide RNAs [gRNAs; ~20 base pairs (bp) in length] to direct the Cas9 endonuclease to a specific genomic locus and induce a double-stranded DNA break. Both the Cas9 enzyme and gRNA sequences can be encoded in a single plasmid and, when cotransfected with a donor DNA plasmid, can lead to higher homology-directed repair (HDR) knock-in efficiency than previous editing tools (33). We have previously described the first CRISPR-Cas9 system for AAVS1 integration of donor constructs containing an antibiotic resistance selection gene and both fluorescence (tdTomato) and bioluminescence (Firefly luciferase) reporter genes (34). We were able to confirm the correct and stable integration of donor DNA at the AAVS1 site and functional reporter gene expression in vivo. However, some of the limitations of our study include (i) the low editing efficiency (~3.8%) of human embryonic kidney (HEK)–293T cells; (ii) the use of large CRISPR-Cas9 and donor DNA plasmids that contained bacterial and antibiotic resistance genes, which limit transfection efficiency and would have associated safety concerns for clinical translation; and (iii) the lack of a translationally relevant reporter gene. In this study, we aimed to address these limitations by improving the efficiency and clinical safety of reporter gene integration at the AAVS1 safe harbor site and included a translationally relevant reporter gene.
We posited that the low editing efficiency of our first system was due, in part, to reduced transfection and knock-in efficiency, which is common with larger DNA plasmids, and the use of the HDR repair pathway for integration, which is intrinsically inefficient and not readily accessible to nondividing cells (35). In contrast to HDR-mediated DNA repair, the nonhomologous end joining (NHEJ) pathway is active in both proliferating and nonproliferating cells and is generally considered more efficient than HDR in mammalian cells (36). Recent studies have shown that by designing a CRISPR-Cas9 system that includes the same gRNA cut site in the donor vector as the genomic target site, the NHEJ repair pathway will more efficiently lead to transgene integration in zebrafish (37) and mammalian cells (38, 39). Suzuki et al. (40) refer to this mechanism as homology-independent targeted integration (HITI), which is expected to lead to increased insertion in the forward rather than the reverse direction, as intact gRNA target sequences will be preserved in the latter. Therefore, we postulated that HITI will increase the efficiency of reporter gene integration at the AAVS1 site (Fig. 1A) compared to HDR. To address the problem of size and bacterial/antibiotic resistance genes in plasmids, our group and Suzuki et al. (40) previously designed minicircles (MCs) to express genes of interest (41, 42). First described by Darquet et al. (43), MCs lack the bacterial backbone and antibiotic resistance genes that would otherwise compromise biosafety and clinical translation. In addition, the removal of the prokaryotic backbone also greatly reduces the size of the vector, thus improving transfection efficiency or providing space for the inclusion of other transgenes. To that end, we aimed to improve on our previous work by including a translationally relevant reporter gene in a multimodality imaging HITI MC donor. We determined that the rat organic-anion-transporting polypeptide 1A1 (Oatp1a1) gene was an ideal candidate. Oatp1a1 is a positive contrast MRI reporter gene due to its ability to uptake a clinically approved, liver-specific paramagnetic contrast agent called gadolinium ethoxybenzyl diethylenetriamine pentaacetic acid (Gd-EOB-DTPA; Primovist/Eovist) (44). We have previously shown that Oatp1a1 is a sensitive, quantitative, MRI reporter for three-dimensional (3D) cancer cell distribution in vivo (45). The purpose of this study was to develop HITI MC donor vectors for CRISPR-Cas9 editing of cells at the AAVS1 locus with three reporter genes to allow multimodality, longitudinal in vivo monitoring of their fate following transplantation.
Fig. 1. HITI experimental design.
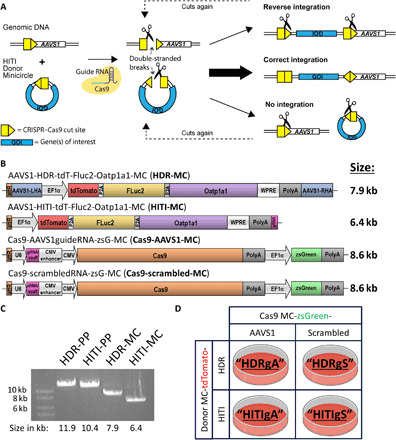
(A) HITI minicircles (MCs) contain a Cas9 cut site identical to that at the AAVS1 safe harbor locus. Both genomic and MC DNA are cut in the presence of a gRNA and Cas9. Genes of interest (GOI) are only stably integrated into the genome when inserted in the correct orientation; otherwise, the Cas9 cut sites are preserved, which increases the likelihood of continuous Cas9 cutting. (B) Trimodality HDR, HITI, and Cas9 MC constructs designed for this study. (C) Restriction digest agarose gel of parental plasmids (PPs) and MCs and indicated band sizes. (D) Transfection regimen for combinations of donor and Cas9 MCs and simplified abbreviations for each condition.
RESULTS
CRISPR-Cas9 engineering of multiple human cell types with trimodal reporter gene MCs
In this study, we designed our trimodal reporter gene system in MC constructs to reduce the size and immunogenicity of our donor DNA and to remove antibiotic resistance genes. To compare the efficiency of HDR versus HITI editing at the AAVS1 site, we designed two donor and two Cas9-expressing MCs, as shown in Fig. 1B. The HDR and HITI constructs were engineered to express tdTomato (tdT), firefly luciferase (Fluc2), and rat organic anion transporting polypeptide 1a1 (Oatp1a1) genes under the control of an EF1α promoter and 2A self-cleaving peptide system (Fig. 1B). The HDR and HITI parental plasmids (PPs) initially measured 11.9 and 10.4 kb in size, which were then reduced to 7.9 and 6.4 kb when recombined into MCs, respectively, as confirmed by agarose gel electrophoresis (Fig. 1C). The HDR-MC was flanked by left and right AAVS1 homologous arms either side of the AAVS1 genomic cut site, whereas the HITI donor contained the same CRISPR-Cas9 cut site as the AAVS1 genomic site (fig. S1). In this instance, if the MC DNA inserted in the correct orientation at the AAVS1 site, the CRISPR-Cas9 cut sites would be lost and the trimodal reporter genes would be stably integrated into the genome (fig. S1). The Cas9-expressing MCs were designed to contain the necessary RNA scaffolding and gRNA sequences targeting the AAVS1 site or a scrambled gRNA control, alongside a zsGreen (zsG) fluorescent reporter gene (Fig. 1B). Both the pCas9-AAVS1-MC and pCas9-scrambled-MC constructs measured 12.5 kb in PP form and 8.6 kb in MC form (Fig. 1B).
Our first objective was to determine the correct integration of our donor MCs in three human cell lines: HEK-293T, HeLa, and PC3 cells. All three were cotransfected with the HDR-MC or HITI-MC together with either the Cas9-AAVS1-MC or Cas9-scrambled-MC (as outlined in Fig. 1D) and grown for 48 hours. The cells were then fluorescence-activated cell (FAC) sorted for tdT+/zsG+ cells to purify cells that were successfully cotransfected, and tdT fluorescence was then tracked every 7 days using flow cytometry (fig. S2, A and B). In two separate experimental groups, the cells were then resorted 14 or 21 days later for tdT+/zsG− cells to ensure that the cell populations had not randomly integrated the Cas9-zsG MCs into the genome (fig. S2C). Both PC3 experimental groups were resorted 14 days after the initial sort (and not 21 days later) due to lower transfection rates. However, resorting the cells 14 or 21 days later had a negligible effect on tdT+ cell populations across the time points. For almost all cell types, there was a higher percentage of tdT+ fluorescence cells at end point in the HITI-AAVS1 groups (pink shading, fig. S2C), suggesting better or more stable integration compared to HDR-AAVS1 groups. For the 293T and HeLa cell groups, specifically, the difference was at least two to three times greater for HITI-transfected cells versus HDR. The only exception was the PC3 #1 group, which had a higher incidence of HDR-AAVS1 tdT+ cells and was likely a result of poor transfection efficiency of the HITI construct for that group.
MCP integration and BLI analysis
We next performed junctional polymerase chain reaction (PCR) analysis on extracted DNA samples to determine whether the tdT+ mixed cell populations (MCPs) had correctly incorporated the trimodal donor MCs into the AAVS1 site in the right orientation (fig. S3A). A correct integration band (1.4 kb) was detected for all HITI-guideAAVS1 (HITIgA)–engineered cells (very low transfection efficiency for PC3 cells may explain why the integration band was weak) as well as a correct integration band (1.3 kb) for HDR-guideAAVS1 (HDRgA) cells for 293T and HeLa MCPs. There were no integration bands for the control naïve cells or cells engineered with scrambled gRNA (HITI/HDRgS). Next, we performed in vitro BLI experiments to determine whether the integrated reporter gene was functioning in the MCPs. Varying numbers of each cell type were imaged with BLI after addition of d-luciferin to visualize FLuc2 expression (fig. S3B). In all cell types, there was a positive correlation between BLI signal and cell number (fig. S3B). There was a consistently higher signal seen in the HITIgA cell populations compared to HDRgA, with approximately three times higher average radiance for the 293T and HeLa HITIgA MCPs and almost six times greater for PC3 HITIgA cells at a concentration of 1 × 105 cells (fig. S3B, right).
HITI is more efficient than HDR in clonal populations
Next, we used clonal cell isolation to determine whether HITI or HDR was more efficient at correctly integrating our large donor MCs at the AAVS1 site. Single-cell tdT+ clones were isolated from the 293T and PC3 MCPs into 96-well plates during a third FAC sort (FACS). We decided to use the 293T cells as a proof-of-principle cell line and the PC3 cells as a relevant prostate cancer model cell line; hence, the HeLa cells were not included in studies from this point onward. PCR integration checks were performed on the 293T and PC3 clonal populations to determine the efficiency of HITI- versus HDR-mediated reporter gene integration at the AAVS1 site (Fig. 2, A and B). The number of 293T clonal populations with correct integration was 11.8% (4 of 34) for HDRgA-engineered cells and 36.1% (13 of 36) for HITIgA clones (Fig. 2, A and B). PC3 cells grew fewer colonies but showed zero integration at the AAVS1 site for tdT+ HDR-engineered cells (0 of 14), whereas 10.5% (2 of 19) of the HITI-engineered colonies had correct reporter gene integration, indicating that HITI was more efficient in both cell types.
Fig. 2. Junctional PCR integration checks for 293T and PC3 clonal cell populations.
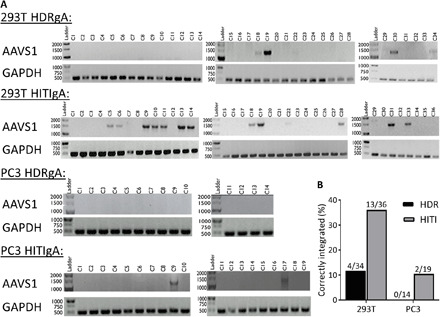
(A) PCR integration checks at the AAVS1 site. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as a DNA loading control. (B) Quantification shows a higher number of positive integration clones for HITI-engineered cells compared to HDR for both 293T and PC3 cell lines.
In vitro reporter gene imaging
Next, we expanded single 293T and PC3 HITIgA clonal cells (with correct integration bands) for further in vitro reporter gene functionality testing. First, we confirmed tdT fluorescence for both the 293T-HITI (Fig. 3A) and PC3-HITI (Fig. 3G) clones via fluorescence microscopy. Next, we confirmed a positive correlation between BLI signal and increasing cell numbers for 293T (Fig. 3, B and C; r2 = 0.9718) and PC3 (Fig. 3, H and I; r2 = 0.9897) cells. BLI signal measured to at least 10 passages showed stable FLuc2 expression over time for both clonal cell lines (Fig. 3, D and J). To test for Oatp1a1 functionality, 293T naïve, 293-HITI, PC3 naïve, and PC3-HITI cells were incubated with or without Gd-EOB-DTPA (6.4 mM) in normal medium for 90 min, washed thoroughly, pelleted, and inserted into an agarose phantom. Inversion recovery MRI was performed at 3 T, and spin-lattice relaxation rate (R1) maps were generated for 293T (Fig. 3E) and PC3 (Fig. 3K) cell populations. Neither the naïve 293T/PC3 nor untreated 293T-HITI and PC3-HITI cell populations exhibited any change in R1 rates (Fig. 3, F and L). Only HITI clones expressing Oatp1a1 had significantly increased R1 rates after Gd-EOB-DTPA incubation, with ~10-fold increase for 293T-HITI cells (7.952 ± 0.87 s−1) compared with naïve, treated controls (0.806 ± 0.038 s−1; n = 3, P < 0.001; Fig. 3F) and ~5-fold increase for PC3-HITI cells (3.426 ± 0.217 s−1) compared with naïve, treated controls (0.6402 ± 0.045 s−1; n = 3, P < 0.001; Fig. 3L).
Fig. 3. In vitro FLI, BLI, and MRI characterization.
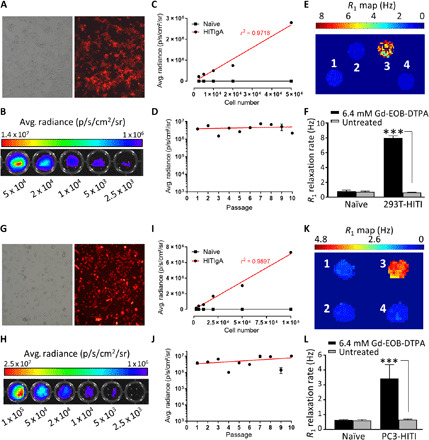
(A to F) Represents 293T-HITI and (G to L) represents PC3-HITI clonal cells, respectively. (A and G) Brightfield and tdT fluorescence. (B and H) BLI intensity maps related to cell number. (C and I) Quantification of BLI signal to cell number. (D and J) BLI signal over successive passages. (E and K) Spin-lattice relaxation maps of representative phantoms containing pellets of cells untreated or treated with 6.4 mM Gd-EOB-DTPA, as follows: 1, naïve, treated; 2, naïve, untreated; 3, HITI treated; 4, HITI untreated. (F and L) Quantification of spin-lattice relaxation rates. Means ± SE, n = 3; *** P < 0.001.
Oatp1a1 sensitivity
The MR detection limit of Oatp1a1-expressing 293T and PC3 HITI cell clones was investigated by varying the ratio of naïve:HITI cells in MR phantoms and in vivo with subcutaneous PC3-HITI cell injections. In all instances, MRI and BLI were performed on the same day of the experimental setup. For MR phantoms, all cells (except a naïve-no Gd control, “naïve-NG”) were incubated with Gd-EOB-DTPA (5.2 mM, 60 min), thoroughly washed, and then combined so that each 20 × 106 cell sample would contain a defined number of Oatp1a1-expressing HITI cells and naïve cells (expressed as percent-HITI of the total cell population; see Fig. 4 and fig. S4). The cell pellets were then placed into an agarose phantom, and inversion recovery MRI was performed at 3 T. Spin-lattice relaxation rate (R1) maps were generated for both PC3 (Fig. 4A) and 293T (fig. S4A) cells. Note that the phantoms in Fig. 4A were rearranged in a linear format for figure presentation only and at two different scales—one on a full scale of R1 values and the second on a saturated scale (maximum 2.0 Hz as the highest concentration) to show visual differences at the lower concentrations. The change in sensitivity with increasing HITI cell numbers can be clearly seen in stacked histograms showing pixel-by-pixel R1 counts for each PC3 (Fig. 4B) and 293T (fig. S4B) cell pellet. For both cell types, a significant increase in the average R1 values, compared to naïve controls, was evident when only 10% of the cell pellet contained Oatp1a1-expressing HITI cells (Fig. 4C for PC3 and fig. S4C for 293T).
Fig. 4. PC3-HITI MRI and BLI sensitivity.
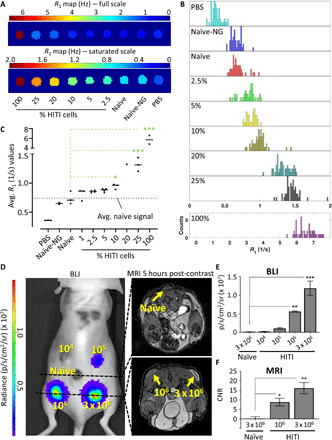
(A) Representative spin-lattice relaxation map of a phantom containing cell pellets of PC3 naïve and/or combinations of HITI cells treated with 5.2 mM Gd-EOB-DTPA for 90 min. Two scales are shown of the same phantom to convey the sensitivity at lower HITI concentrations. PBS, no cell control; Naïve-NG, naïve cells without Gd-EOB-DTPA. (B) Stacked histograms of all pixel-by-pixel R1 counts for each phantom sample shown in (A). The 100%-HITI sample is shown on a separate scale. (C) Quantification of all R1 rates calculated from three phantoms, except for the 20% sample, which was n = 1. The dotted line represents the average of the naïve samples that were treated with Gd-EOB-DTPA. (D) A total of 3 × 106 PC3 cells were injected subcutaneously into five locations on the back of nude mice, with increasing concentrations of HITI-engineered cells, as indicated in yellow. After precontrast imaging, the mice were injected with Gd-EOB-DTPA and imaged 5 hours later using MRI and BLI. Representative transverse images are shown. (E and F) Quantification of BLI [average radiance (photons s−1 cm−2 sr−1)] and MRI (CNR) signals from ROIs drawn around the injection sites. Note, 104 and 105 PC3-HITI injections lacked enough contrast to measure CNR values and are not shown. Means ± SE, n = 3 mice. *P < 0.05, **P < 0.01, and ***P < 0.001.
We next injected various combinations of PC3-naïve and PC3-HITI cells at five sites subcutaneously on the backs of nude mice to analyze Oatp1-expressing cell detection sensitivity in vivo (Fig. 4, D and E, and fig. S5). A total of 3 × 106 cells were injected per site with the following number of PC3-HITI cells: 0 (naïve cell only control), 104, 105, 106, 3 × 106 (PC3-HITI cell only control). Naïve cells were included with HITI cells so that each injection contained a total of 3 × 106 per site. BLI signal intensity increased as PC3-HITI cell numbers increased (representative mouse shown in Fig. 4D), with 106 and 3 × 106 HITI injections showing significant signal increase above naïve background controls (Fig. 4E). Transverse MR images from the same mouse showed positive contrast at both the 106 and 3 × 106 HITI injection sites 5 hours after Gd-EOB-DTPA injection (Fig. 4F). Similar to the BLI data, these sites also exhibited significantly higher contrast-to-noise ratios (CNRs) than naïve controls (Fig. 4F). The 104 and 105 PC3-HITI injections were difficult to visualize on MRI as the phosphate-buffered saline (PBS) of these injections spread out over the ~6 hours after injection and had no discernible positive contrast, so they could not be measured. These data were consistent across all three mice (see fig. S5) and showed that the very minimum number of Oatp1a1-expressing cells we could detect with Gd-EOB-DTPA–based MR contrast was 106 cells in a 50-μl subcutaneous injection volume.
Effect of Gd-EOB-DTPA on HITI-engineered Oatp1a1-expressing cells
Trypan blue exclusion assays and cell counts were performed to determine whether the uptake of Gd-EOB-DTPA into Oatp1a1-expressing 293T and PC3 cells affected cell viability and growth over time. Cells were plated (day 0) and incubated with 5.2 mM Gd-EOB-DTPA contrast agent or saline the next (day 1) for 90 min. There was no significant difference in viability or growth rate between 293T-naïve and 293T-HITI cells or between control (saline) or Gd-EOB-DTPA–treated cell groups (fig. S6A). In contrast, the PC3-HITI clone exhibited a slower growth rate compared to the mixed population of naïve cells (fig. S6B). Exposure to Gd-EOB-DTPA, however, did not affect the viability or growth of either cell type compared to their no-Gd (saline) controls (fig. S6B). These data indicate that Gd-EOB-DTPA uptake had no negative effect on the cells. However, the single PC3-HITI clone we obtained was slower growing compared to the mixed cell naïve population.
PC3-HITI Oatp1a1 tumor models for MRI detection
As a proof of principle that our HITI-engineered cells could show Gd-EOB-DTPA–induced positive MRI contrast in subcutaneous tumors, we injected 293T-naïve and 293T-HITI cells on either flank of a nude mouse (fig. S7). For both cell types, the large masses were visible on precontrast images and showed noticeable positive contrast 20 min after Gd-EOB-DTPA injection. However, 5 hours after contrast, the naïve tumor had returned to precontrast background levels, whereas the HITI tumor had very prominent positive contrast that also showed heterogeneity within the tumor mass (fig. S7). This heterogeneity, in contrast, likely reflects areas of viable (enhancing) and nonviable (non-enhancing) Oatp1-expressing cells within the tumors, as we have reported previously (45).
Moving into a more relevant cancer model, we next injected PC3-naïve and PC3-HITI clonal cells subcutaneously on either flank of nude mice and followed BLI and MRI signal changes over time (Fig. 5). At only 11 days after injection, before the tumors were visible or palpable, clear positive contrast was observed for HITI-engineered cells 5 hours after Gd-EOB-DTPA injection, whereas naïve tumors were undistinguishable (Fig. 5A and fig. S8). The same mice were then imaged at day 46, where the naïve tumor was visible due to pooled Gd-EOB-DTPA at 20 min after contrast injection. In a similar fashion to 5 hours after contrast, only the HITI-engineered cells retained the Gd agent and showed bright, positive contrast (Fig. 5B and fig. S9). Contrast-to-noise measurements, tumor size, and estimated cell numbers for PC3-HITI tumors all increased from days 11 to 46 (Fig. 5, C to E). These data suggest that the Oatp1a1 MRI reporter can detect tumor burden at stages where the tumors are not visible or palpable, and tumor growth can be tracked longitudinally with Gd-EOB-DTPA–enhanced MRI.
Fig. 5. Longitudinal in vivo MRI of subcutaneous PC3-HITI cells.
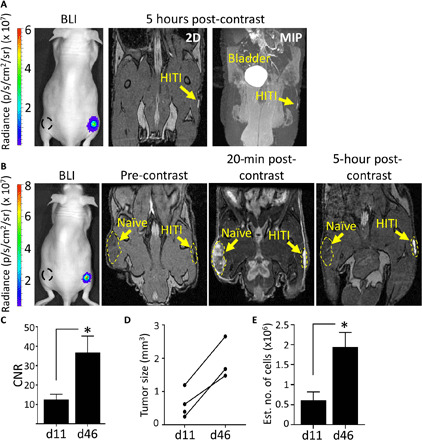
Mice were injected subcutaneously with 1 × 106 naïve and PC3-HITI cells on the left and right flanks, respectively. BLI signal was present on right flank only. Naïve tumor locations are denoted by black dashed line. (A) Day 11 after PC3 injection. 2D and maximum intensity projection (MIP) images acquired 5 hours after Gd-EOB-DTPA injection. Naïve tumors were undistinguishable at this stage. (B) The same mouse was reimaged at day 46. Pre-, 20-min post-, and 5-hour post-contrast images were obtained. (C) CNRs of PC3-HITI tumors 5 hours after contrast showed significant increase from days 11 to 46. (D and E) Increase in PC3-HITI tumor volume (D) and estimated cell numbers (E) from days 11 to 46. Means ± SE, n = 4 (day 11) and n = 3 (day 46). *P < 0.05.
Last, PC3-HITI cells were injected via the tail vein into immunocompromised NOD scid gamma (NSG) and nude mice to investigate the ability of Oatp1a1 as a reporter gene for visualizing metastases (Fig. 6 and fig. S10). Using BLI as a guide, we were able to detect metastatic tumors in the head of NSG (Fig. 6) and nude (fig. S10) mice, 84 and 114 days after injection, respectively. Tumors were undistinguishable in precontrast MR images for the NSG mouse. However, 5 hours after Gd-EOB-DTPA injection, a cluster of small, enhanced PC3-HITI tumors were clearly visible (yellow arrows, Fig. 6), which equated to an overall tumor size of 2.05 mm3 and, assuming that a 1-cm3 tumor contains around 1 × 109 cells, contained around 2.1 × 106 cells. Similarly, in the nude mouse model, a larger, more defined tumor showed strong enhancement 5 hours after contrast injection (fig. S10), which measured 31.42 mm3 in size and contained ~3.1 × 107 cells. These data indicate the usefulness of this reporter gene for detecting metastatic burden with contrast-enhanced MRI.
Fig. 6. In vivo BLI and MRI detection of PC3-HITI metastases.
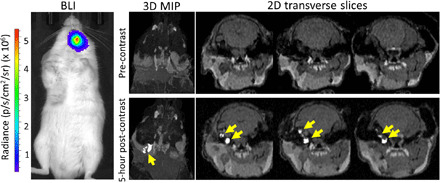
BLI signal was evident in the head region of an NSG mouse 83 days after tail vein injection of PC3-HITI cells. (Top) The tumor was not evident in precontrast 3D MIP or 2D transverse T1-weighted MRI images. (Bottom) However, 5 hours after Gd-EOB-DTPA injection, there were clear clusters of enhanced PC3-HITI tumors that were easily discernible from surrounding tissue (yellow arrows).
DISCUSSION
As personalized medicine and CRISPR-editing become a reality in the clinic, there is a greater need to (i) improve the efficiency, efficacy, and safety of genetically engineered cell therapies, and (ii) improve our understanding of disease progression and treatment response in preclinical models of disease. Reporter gene-based imaging allows us to track the location, viability, growth, and efficacy of such treatments, and in preclinical models of cancer progression and treatment. In this study, we have developed a non–viral vector–based engineering system for large DNA multimodality reporter gene integration into the AAVS1 safe harbor genomic locus. To improve safety further, we used MCs as the DNA vector of choice, which eliminates bacterial DNA contamination and antibiotic resistance genes. In addition, we showed that using the NHEJ repair pathway with HITI could improve DNA editing efficiency in human cells compared to the more commonly used HDR pathway. Last, building off our previous work (34), we have engineered a trimodality reporter gene construct that contains a clinically relevant MRI reporter, Oatp1a1, in addition to fluorescent and bioluminescent genes, which enabled cell sorting and noninvasive BLI/MRI of engineered cells in a preclinical cancer model.
One of the major limitations of engineering cells with large, multimodality reporter gene DNA plasmids is the reduced efficiency of both transfection and gene editing with increasing construct/insert size (46–48). In addition, the presence of bacterial and antibiotic resistance genes in PPs has the potential to exert immunological responses and raises safety concerns. To circumvent these issues, we designed our study to use MCs, which remove the bacterial backbone from PPs and thus reduce the size of the DNA donor constructs. Using MCs instead of PPs allowed us to remove ~4 kb of unwanted DNA from our HDR construct, with a further reduction of ~1.5 kb for the HITI MC when the homologous arm sequences were replaced with a 20-bp gRNA sequence (saving a total of ~5.5 kb). These large-scale reductions thus provided us with room to upgrade our dual-modality tdTomato and FLuc2 reporter gene construct we previously reported (34) to a trimodality reporter gene construct with the addition of the Oatp1a1 MRI reporter (44, 45). To improve safety and translatability, we also removed the puromycin resistance gene to reduce the MC size by a further 600 bp and used FACS of tdTomato-positive cells to obtain mixed and clonal cell populations instead of antibiotic selection. Our final step for improving safety was to design our system to target a “safe harbor” locus in genomic DNA. Several of these loci have now been reported in the literature (49) and are described as sites where inserted genetic elements can function as intended, without causing alterations that would pose a risk to the host cell or organism (25). For this study, we targeted the AAVS1 site found within the human protein phosphatase 1 regulatory subunit 12C (PP1R12C) gene, as this has been one of the best characterized, to date. No known side effects are associated with disrupting the PP1R12C gene; however, it has been reported that mechanisms such as DNA methylation can silence transgenes targeted to this genomic region (50). Because our studies rely on stable reporter gene expression over time for accurate cell detection and proliferation, we investigated whether reporter gene expression in our AAVS1-engineered 293T-HITI and PC3-HITI cell populations changed over time. We found that BLI signal was stable to at least 10 passages and tdT fluorescence was expressed in both cell lines, indicating consistent transgene expression.
We have shown here that HITI-based CRISPR-Cas9 cell engineering is more efficient than the more commonly used HDR method for integrating large DNA donor constructs. Targeted transgene integration is typically achieved using homologous arms and the HDR pathway; however, this mechanism is highly inefficient and not usually active in nondividing cells (35). Our previous study showed that only 3.8% of selected cells were correctly edited using the HDR mechanism (34). In contrast, the HITI method that uses the NHEJ pathway is active in all stages of the cell cycle and in quiescent cells (38) and thus has been used to improve editing efficiency. Using the method described by Suzuki et al. (40), our engineered 293T and PC3 clonal cell populations did have greater DNA integration at the AAVS1 site compared with HDR (36 and 12% for HITI versus 10.5 and 0% for HDR, respectively). However, the NHEJ repair pathway is error prone and often leads to insertions and deletions (indels) at the DNA junctions. Consequently, this mechanism is often taken advantage of to produce DNA disruptions, gene silencing, and knockouts. These issues would need to be considered if using the HITI method for correctional DNA editing and promoter-less vector integration, because these require specific DNA sequences, either upstream or downstream, to be preserved. In this case, we engineered cells with the only requirements being that the transgene inserts into the AAVS1 site (confirmed with junctional PCR) and that the reporter genes are consistently expressed (confirmed with imaging). Therefore, indels at either the 5′ or 3′ junction would likely have a negligible impact on our experiments.
Although we confirmed correct transgene integration at the AAVS1 site in our study, we cannot rule out integration at other off-target sites in HITI- and HDR-engineered populations. Several 293T and PC3 single-cell clonal populations expressed the tdTomato fluorescence reporter gene but did not show integration bands for the AAVS1 site. Evidence suggests that CRISPR-Cas9 is not 100% accurate and off-target effects have been reported as a common problem associated with CRISPR (51, 52). Thus, it is likely that some MCs integrated into off-target Cas9 cut sites in clones where the correct AAVS1 integration bands were absent or that the MCs inserted into the AAVS1 site in the wrong direction. Although HITI is designed to minimize integration in the wrong orientation, the error-prone NHEJ repair mechanism of blunt-ended DNA breaks could lead to indels at the CRISPR-Cas9 cut site boundaries, which could then disrupt the ability of Cas9 to recognize and recut those sites. Although our preliminary data suggest that stable integration in the wrong direction with HITI was absent in our sorted MCPs, there remains a possibility of indel formation using Cas9 HITI. This could be reduced in future studies by adopting a similar method to that recently reported by Li and colleagues (53), where 5-bp overhangs created by Cas12a could lead to more precisely edited genomes, in a process coined microhomology-dependent TI (MITI) (53). Independently of CRISPR, MCs, like plasmids, can also randomly integrate into the genome of cells, albeit at very low rates. Future work will need to analyze the rate of off-target integrations and possible indel disruptions at the CRISPR-Cas9 cut sites using techniques, such as next-generation sequencing, to determine the full safety profile of HITI at safe harbor loci. To improve targeting specificity, studies have shown that high-fidelity Cas9 enzymes in ribonucleoprotein complexes (RNPs), instead of Cas9 DNA vectors, improve on-target activity while reducing off-target editing (54, 55). In combination with RNPs, AAVs are now commonly used as DNA donors for CRISPR experiments due to their high transduction capabilities in hard-to-transfect cell lines, their low risk of random integration, and reduced immunogenic response. However, AAVs are still limited by their loading capacity of ~4.5 kb, which would be a problem for large, multimodality imaging vectors as presented here, but conceivable for future studies where only one imaging reporter gene is required. With these emerging technologies, it is likely that CRISPR gene editing will become highly specific and thus safer in the near future.
We engineered cells with a multimodality reporter gene construct to enable us to go from single cell, optical imaging methods (FLI) to higher-sensitivity whole-animal planar imaging (BLI) and superior 3D high-resolution tomographic imaging (MRI) in animals. This offers several advantages. First, fluorescently activated cell sorting of tdTomato-expressing cells eliminates the need for an antibiotic resistance selection gene, which constitutes a safety risk and has been associated with structural plasmid instabilities (56). Second, the firefly luciferase gene (FLuc2), in combination with its substrate d-luciferin, allows us to directly visualize engineered cells in vivo using BLI. Inclusion of bioluminescent genes in preclinical cancer models is a relatively inexpensive and valuable tool that also allows one to track cell migration and cell seeding in metastatic cancer models, assess cell viability, and follow cell/tumor growth longitudinally (14). A limitation of BLI is that it is restricted to small animal models of disease. However, it is useful for determining sites of cell arrest/seeding/growth and thus can be used in conjunction with other reporter genes as a guide for determining when and where to perform relatively more expensive, higher-resolution clinical imaging, such as MRI (13). To build off our previous dual FLI-BLI study (34), we decided to include the MRI reporter gene, Oatp1a1, as a translationally relevant and sensitive reporter gene to complete our trimodality construct for HITI-based CRISPR engineering. First described by Patrick et al. (44), Oatp1a1 selectively, but reversibly, uptakes the clinically approved Gd3+ contrast agent Gd-EOB-DTPA and thus provides positive contrast in T1-weighted MR images. The authors concluded, therefore, that Oatp1a1-engineered cells and tumors should be easier to detect than the negative contrast generated by T2 agents, such as superparamagnetic iron oxide (SPIO) and ferromagnetic agents (44, 57). In addition, engineering cells with integrated Oatp1a1 expression means that MR images can be obtained longitudinally to track cell migration and growth, and signal intensity can be directly correlated with cell viability. Last, we and others have found that Oatp1a1 also enhances the uptake of d-luciferin for BLI (45, 58) and the fluorescent dye indocyanine green (using the human ortholog OATP1B3) for both fluorescent (59) and photoacoustic imaging (60), which gives an added advantage of using Oatp1 for multimodality imaging. Because we, and others, have now shown that the human OATP1B3 gene also functions as a useful fluorescent, photoacoustic, and MRI reporter gene, in vivo (59, 60), future studies will focus on exchanging Oatp1a1 for the more translationally favorable OATP1B3 ortholog.
In this study, we set out to determine the smallest number of Oatp1a1-expressing HITI-engineered cells that we could detect using MRI phantoms and in vivo injections (with BLI as a guide). Our in vitro MRI phantom experiments showed significant increases in R1 rates when 10% of the cell pellet, and thus only 10% of each voxel, contained Oatp1a1-expressing cells. For in vivo sensitivity measurements, we were able to reliably detect 1 million cells in a 50-μl volume. The cell and contrast injections and imaging were all performed on the same day so that the cell numbers would not vary with migration, cell death, or proliferation. Similar experiments have been performed for PET reporter genes. However, there are some issues with this experimental setup as we prepared cells in PBS rather than within a matrix such as Matrigel so as not to impede diffusion of the contrast agent. These were merely subcutaneous injections of a known number of cells in a known volume of PBS, not tumors. Without time to develop blood vessels, as you would with tumors, it is likely that some of the contrast agent could not diffuse into the injection site fully. This may explain why the contrast enhancement appears as a ring around the cell injection sites. We also believe that this problem likely contributed to the lack of signal enhancement in the 104 and 105 injection sites, which without a matrix to hold them in place also had the problem of cell spreading over a larger surface area. In summary, there are no easy ways to measure the sensitivity of exact cell numbers in vivo, especially with a reporter gene system that relies on an injected substrate; however, the combination of in vitro and in vivo experiments give us a better idea of the minimum number of cell numbers we can confidently detect.
When expressing reporter genes in cells, it is important to know whether the protein and/or its substrates affect the normal function or viability of the cell. In our study, we noticed that the Oatp1a1-expressing PC3-HITI cells grew slower compared to the mixed cell naïve population in vitro and in vivo, whereas their 293T-HITI counterpart showed no difference compared to naïve cells. Although unfortunate for comparative purposes, we believe that the slow growth rate of the PC3-HITI cells was simply due to heterogeneity in clonal populations—especially when comparing to mixed naïve populations—and not due to expression of Oatp1a1. Our data also support this because the 293T-HITI cells were not affected. In addition, we and others have already shown that there is no impact of Oatp1a1 on cell growth in several different cell types (44, 45). Ideally, several correctly engineered clones would be combined for biological and imaging experiments. We were limited in this case by obtaining just one usable PC3-HITI clone. However, the clone still grew primary and metastatic tumors, which we could sensitively detect with BLI and MRI. In addition, the uptake of Gd-EOB-DTPA did not have any detrimental impact on cell viability or growth, indicating that Oatp1 is a viable option for reporter gene imaging and cell tracking.
The improved safety profile and expression of multimodal reporter genes proposed here could have several uses in cell engineering, or at least help answer several concerns with in vivo cell therapies. For example, the U.S. Food and Drug Administration have listed potential safety concerns related to unproven stem cell therapies (61), including (i) the ability of cells to move from placement sites and change into inappropriate cell types or multiply, (ii) failure of cells to work as expected, and (iii) the growth of tumors. In addition, the long-term safety profiles of cells engineered with randomly integrating viruses still require further investigation and optimization. These are concerns that could be addressed by targeting nonviral DNA vectors, such as MCs, to specific safe harbor loci, such as AAVS1, and reducing the use of integrating viruses. Incorporating reporter genes for clinical grade imaging will also help improve patient safety by allowing one to track cellular therapies in vivo [such as for stem cells or cancer-homing theranostic cells (62)]. Clinicians could then determine whether the therapeutic cells are localizing to the correct anatomical feature, such as a solid tumor (18), or to determine their persistence and viability for short- and long-term treatment strategies. Future work will focus on evaluating our system in stem cells and other clinically relevant cell types. Translation will also need to consider building donor vectors that lack optical reporter genes and use other selection methods (e.g., magnetic sorting). It is easily feasible to switch out genes from our trimodality construct for other imaging purposes, such as replacing FLuc2 with a PET reporter gene for dual PET-MR imaging. Suicide switch genes could also be incorporated to further improve safety by killing the engineered cells in cases where they become oncogenic (63), for example. These tools not only are useful for clinical cell-based therapies but also are extremely useful in preclinical studies for investigating cancer progression/aggression, metastatic burden, and treatment strategies. Avoiding the use of random-integrating viruses and targeted editing should also help reduce off-target effects of gene editing that may alter the normal characteristics of the cell type being studied.
CONCLUSION
Our work demonstrates the first CRISPR-Cas9 HITI MC system for safe harbor integration of a large donor construct encoding three reporter genes for multimodal longitudinal imaging of cells in vivo. We have shown that inclusion of the translationally relevant MR reporter gene, Oatp1a1, can enable localization and tracking of small primary and metastatic tumors that are not readily detectable visually or in precontrast MR images. This work lays the foundation for an effective and safer nonviral genome editing tool for noninvasive reporter gene tracking of multiple cell types in vivo.
MATERIALS AND METHODS
Constructs
Construct designs are shown in Fig. 1B. The pCas9-AAVS1guideRNA-zsG-MC (Cas9-AAVS1-MC) and pCas9-scrambledRNA-zsG-MC (Cas9-scrambled-MC) PPs originated from pCas-Guide-AAVS1 and pCas-Guide-Scrambled plasmids purchased from Origene (MD, USA). The Cas9 enzyme and gRNA sequences were cloned between attB and attP recombination sites in an MC bacterial backbone containing a ZsGreen (zsG) fluorescence reporter driven by the elongation factor 1-α promoter (hEF1α). The AAVS1-HDR-tdT-Fluc2-Oatp1a1-MC (HDR-MC) construct was derived from an HDR vector lacking the Oatp1a1 gene as we described previously (34). This plasmid is driven by the hEF1α promoter and expresses tdTomato (tdT), firefly luciferase (Fluc2), and organic anion transporting polypeptide 1a1 (Oatp1a1) using a self-cleaving 2a peptide system. For improved expression, the plasmids also contain the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) followed by the human growth hormone polyadenylation signal (hGH polyA). The HDR plasmid contains the left and right homologous arms (RHA: 527 bp, LHA: 481 bp) that are complementary to the region flanking the AAVS1 cut site; the homologous arms were obtained from the pAAVS1-puroDNR plasmid from Origene (MA, USA). The Oatp1a1 gene was added through PCR amplification from a previously made vector we constructed using PGK_Straw_E2A_Oatp1a1 (a gift from K. Brindle’s laboratory; University of Cambridge). Using the HDR-MC PP as the template, we generated the pAAVS1-HITI-tdT-Fluc2-Oatp1a1-MC (HITI-MC) PP using the In-Fusion Cloning Kit from Clontech (Takara Bio, CA, USA). Using restriction enzyme digestion, we extracted the bacterial backbone and MC recombination sites and then extracted the three reporter genes (without the homologous arms)—tdT, Fluc2, and Oatp1a1—from the HDR-MC construct using PCR. However, for HITI functionality, we designed our primers to also include a 23-bp extension (5′-GTTAATGTGGCTCTGGTTCTGGG-3′) downstream of the polyA sequence, which incorporates the same cut site and protospacer adjacent motif (PAM) sequence for our AAVS1 gRNA, which allows Cas9 cutting of both the MC and genomic DNA.
MC production
ZYCY10P3S2T Escherichia coli (System Biosciences, Palo Alto, CA, USA) were transformed with the original PPs of all four constructs (HDR-MC or HITI-MC, Cas9-AAVS1-MC, and Cas9-scrambled-MC), and viable colonies were selected using kanamycin plates. Colonies were picked 24 hours after transformation and grown in 6 ml of lysogeny broth (LB) with kanamycin for 6 hours at 37°C, followed by growth in terrific broth (TB) for 12 hours at 37°C. To induce expression of the phiC31 integrase for MC production via attB and attP recombination, 100 ml of LB broth together with 100 μl of 20% arabinose induction solution (System Biosciences, Palo Alto, CA, USA) and 4 ml of 1 N NaOH was added to the culture and grown for 5.5 hours at 30°C. An endotoxin-free maxi kit (Qiagen, Valencia, CA, USA) was used to purify both PP and MC. Following purification of the MC products, PP contamination was removed using the Plasmid Safe ATP-dependent DNase Kit (Epicentre, WI, USA), and the products were cleaned and concentrated using the Clean & Concentrator-25 Kit (Zymo Research, CA, USA).
Cell culture and transfection
HEK-293T cells and human adenocarcinoma HeLa cells (both from the American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium (Wisent Bioproducts, Québec, Canada) supplemented with 10% fetal bovine serum (FBS; Wisent Bioproducts, Québec, Canada) and 1× antibiotic-antimycotic (Thermo Fisher Scientific, Waltham, MA, USA). Human grade 4 adenocarcinoma PC3 cells were a gift from H. Leong (Western University, ON, Canada) and were grown in RPMI (Wisent Bioproducts, Québec, Canada) supplemented with 5% FBS and 1× antibiotic-antimycotic. Cells were transfected with the linear polyethylenimine transfection agent jetPEI (Polyplus-transfection, Illkirch, France), according to the manufacturer’s instructions. Briefly, cells were grown in six-well plates until 80 to 90% confluency and cotransfected with 1 μg each of Cas9-AAVS1-MC or Cas9-Scrambled-MC together with 1 μg of the donor MC constructs: HDR-MC or HITI-MC, for a total DNA mass of 2 μg. The DNA was prepared in 150 mM NaCl and complexed with 4 μl of jetPEI reagent per well.
FACS and flow analysis
All FACS and flow cytometry was performed at the London Regional Flow Cytometry Facility (Robarts Research Institute, London, Canada). Forty-eight hours after transfection, the population of cells displaying both red (tdTomato) and green (zsGreen) fluorescence were sorted using a BD FACSAria III cell sorter (BD Biosciences, San Jose, CA, USA). At selected time points following FACS, the cells were analyzed for tdTomato fluorescence using a FACSCanto flow cytometer (BD Biosciences, San Jose, CA, USA). Either 14 or 21 days after the initial sort, the cells were again sorted on the FACSAria III to purify tdTomato-positive cells only (referred to as the pooled population). In this regard, our protocol aimed to sort cells that had incorporated the MC inserts (based on tdTomato fluorescence) into the genome and excluded any cells that had randomly integrated Cas9 MC DNA (zsGreen). At the same time as the second (tdTomato) sort, individual cells were plated into wells of a 96-well plate to enable single-cell colonies to be grown and expanded (referred to as clonal cell populations).
Genomic DNA extractions and AAVS1 integration analysis
Extraction of genomic DNA from the pooled population of cells was performed using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. DNA quality and concentrations were measured on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Extraction of genomic DNA from clonal populations was performed as we described previously (34). Briefly, cell pellets were resuspended in a QuickExtract DNA extraction solution (Lucigen, Middleton, WI, USA), incubated at 65°C for 10 min, vortexed, and incubated at 98°C for 5 min. The DNA was then directly used for PCR or stored at −20°C. To check for integration at the AAVS1 site, two primers were designed to amplify the 3′ junction between the donor cassette and the AAVS1 site outside of the homologous arm region. The forward primer was uniquely complementary to the polyA tail in the MC cassette (5′-CCTGGAAGTTGCCACTCCAG-3′) and the reverse primer to the AAVS1 site (5′-AAGGCAGCCTGGTAGACAGG-3′). A 1.3-kb PCR product was produced if the MC-HDR was correctly integrated at the AAVS1 site, and a 1.7-kb PCR product was produced if MC-HITI was correctly integrated. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were designed as DNA loading controls and to confirm successful DNA extractions: forward 5′-TTGCCCTCAACGACCACTTT-3′ and reverse 5′-GTCCCTCCCCAGCAAGAATG-3′ and yielded a PCR product of 502 bp. Agarose gel electrophoresis with 1% agarose gels and RedSafe (FroggaBio, ON, Canada) was used to separate and visualize PCR products.
In vitro fluorescence and BLI
The pooled and clonal cell populations were evaluated for tdTomato fluorescence expression on an EVOS FL auto 2 microscope (Thermo Fisher Scientific, Waltham, MA, USA). For BLI experiments, varying cell numbers were plated in triplicate into black walled 96-well plates. d-Luciferin (0.1 mg/ml; PerkinElmer, Waltham, MA, USA) was added to each well, and images were rapidly collected on the IVIS Lumina XRMS In Vivo Imaging System (PerkinElmer) equipped with a cooled charge-coupled device (CCD) camera. Average radiance values in photons s−1 cm−2 sr−1 were measured from regions of interest drawn around each well using LivingImage software (PerkinElmer).
In vitro MRI
Naïve and Oatp1a1-expressing cell clones were seeded in 15-cm tissue culture dishes and grown to confluency. Cells were incubated with medium containing 5.2 mM Gd-EOB-DTPA or with medium containing an equivalent volume of PBS for 90 min at 37°C and 5% CO2. Cells were then washed three times with PBS, trypsinized, and pelleted (20 × 106 cells per pellet) in 0.2-ml Eppendorf tubes. For sensitivity experiments, various numbers of naïve:HITI cells were combined in Eppendorf tubes, mixed well, and then pelleted. The tubes were placed into a 1% agarose phantom mold, and MRI was performed on a 3-T GE clinical MRI scanner with an eight-channel head RF coil (General Electric Healthcare Discovery MR750 3.0 T, Milwaukee, WI, USA). A fast spin echo inversion recovery (FSE-IR) pulse sequence was used with the following parameters: matrix size, 256 × 256; repetition time (TR), 5000 ms; echo time (TE), 16.3 ms; echo train length (ETL), 4; number of excitations (NEX), 1; receiver bandwidth (rBW), 25 kHz; inversion times (TIs), 25, 50, 100, 200, 350, 500, 750, 1000, 1500, 2000, 2500, and 3000 ms; in-plane resolution, 0.27 mm × 0.27 mm; slice thickness, 2.0 mm; scan time, 5 min and 25 s per TI. Spin-lattice relaxation rates (R1) were determined by nonlinear least-squares fitting (MATLAB, MathWorks, Natick, MA, USA) of the following equation to the signal intensity across the series of TIs on a pixel by pixel basis
Here, S, Mss, and Mi represent the acquired signal, the longitudinal magnetization in steady-state equilibrium, and the initial longitudinal magnetization acquired after the inversion pulse, respectively. T1 is the spin-lattice relaxation time such that and TI is the inversion time.
Animal models
All animal protocols were approved by the University Council on Animal Care at the University of Western Ontario (protocol #2015-058) and follow the Canadian Council on Animal Care (CCAC) and Ontario Ministry of Agricultural, Food and Rural Affairs (OMAFRA) guidelines. Crl:NU-Foxn1nu (nude) male mice (Charles River Laboratories, Wilmington, MA, USA; N = 3 to 5) aged 6 to 8 weeks were used for subcutaneous and metastatic tumor model injections, and NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ (NSG) immunodeficient male mice (obtained from the Humanized Mouse and Xenotransplantation Facility at the Robarts Research Institute, University of Western Ontario, London, Canada; N = 3) were used for experimental metastasis models (intravenous cell injections).
In vivo BLI
BLI was performed on the same IVIS Lumina XRMS system described for in vitro imaging. Mice were anesthetized with 2% isoflurane in 100% oxygen using a nose cone attached to an activated carbon charcoal filter for passive scavenging and kept warm on a heated stage. Anesthetized mice received a 100-μl intraperitoneal injection of d-luciferin (30 mg/ml), and BLI images were acquired with automatic exposure times until the peak BLI signal was obtained (up to 40 min). Regions of interest were manually drawn using LivingImage software to measure average radiance (photons s−1 cm−2 sr−1). The peak average radiance was used for quantification for each mouse.
In vivo MRI and quantification
All mouse MRI scans were performed with a custom-built gradient insert and a 3.5-cm-diameter birdcage RF coil (Morris Instruments, Ottawa, ON, Canada), as we described previously (45). Mice were kept anesthetized during the scan with 2% isoflurane administered via a nose cone attached to the coil. T1-weighted images were acquired using a 3D spoiled gradient recalled acquisition in steady-state pulse sequence using the following parameters: field of view, 50 mm; TR, 14.7 ms; TE, 3.3 ms; rBW, 31.25 MHz; matrix size, 250 × 250; flip angle, 60°; NEX, 3; 200-μm isotropic voxels; scan time, approximately 15 min per mouse. Precontrast images were acquired followed by administration of Gd-EOB-DTPA (1.67 mmol/kg) (Primovist; Bayer, Mississauga, ON, Canada) via the tail vein. Mice were then reimaged 20 min later for immediate postcontrast images, which provide positive contrast to many tissues, including the naïve tumors, as a result of Gd-EOB-DTPA pooling, and/or 5 hours later for Oatp1a1-specific uptake. This time point was determined to allow enough time for Gd-EOB-DTPA to be cleared, yet still provided strong positive contrast in Oatp1a1-expressing cells (44, 45). CNR and tumor size measurements were calculated from MR images using ITK-snap open source software (www.itksnap.org) (64). Tumors were manually segmented in three dimensions by tracing the tumor or control tissue (hind leg muscle) with polygon and paintbrush tools and pixel intensity recorded in every slice. The CNR of tumors was calculated by taking the signal intensity of the difference between tumor regions and muscle tissue divided by the SD of background signal
The number of cells in the tumors was estimated from the assumption that a tumor reaching a size of 1 cm3 is estimated to contain around 1 × 109 cells.
In vivo Oatp1a1-induced Gd-EOB-DTPA uptake MRI and BLI sensitivity
To evaluate the cellular detection sensitivity of Oatp1a1-expressing cells with Gd-EOB-DTPA–enhanced MRI, nude mice were injected with 50 μl of cell suspensions in PBS containing 3 × 106 total cells per injection at the following ratios: 3 × 106 naïve cells alone; 104 PC3-HITI + 2.99 × 106 naïve cells; 105 PC3-HITI + 2.9 × 106 naïve cells; 106 PC3-HITI + 2 × 106 naïve cells; and 3 × 106 PC3-HITI cells alone, subcutaneously in five locations on the back/flank region. Immediately after cell injections, Gd-EOB-DTPA (1.67 mmol/kg) was injected into the tail vein, and mice were imaged on a 3-T clinical grade MR scanner 5 hours later. This time point allows clearance of Gd-EOB-DTPA from the body yet provides sufficient time for the agent to penetrate the subcutaneous injections sites and accumulate in Oatp1a1-expressing cells. After MRI, mice were moved to the IVIS scanner and injected with 100 μl of d-luciferin (30 mg/ml) intraperitoneally and BLI was performed, as described earlier.
293T and PC3 tumor models
293T or PC3 naïve and HITI-engineered cells were injected subcutaneously (2.5 × 106 293Ts and 1 × 106 PC3s) on the left and right flanks of nude mice, respectively (293T, N = 2; PC3, N = 5). For experimental metastasis studies, 5 × 105 PC3 naïve or HITI-engineered cells were injected into the tail veins of NSG or nude mice (N = 3). Tumor growth was tracked on a weekly basis with BLI, as described above. MRI was performed on mice at various time points, as indicated in the results section. First, a precontrast scan was performed on all mice, followed immediately with injection of the Gd-EOB-DTPA contrast agent into the tail vein (1.67 mmol/kg). For some experiments, the mice were rescanned 15 to 20 min after contrast injection to show tumor and whole-body distribution of Gd-EOB-DTPA. In all instances, MRI scans were performed ~5 hours after contrast injection because Oatp1a1-expressing cells still retain Gd-EOB-DTPA and show strong positive contrast at this time point. This also allows enough time for washout of Gd-EOB-DTPA in most tissues and organs (except for the gastrointestinal tract and bladder where cleared Gd-EOB-DTPA accumulates before being excreted) (44).
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 7 (GraphPad Software Inc., CA, USA; www.graphpad.com) software. One-way analysis of variance (ANOVA) with Tukey’s multiple comparison test was used for in vitro and in vivo BLI and CNR data analysis. An unpaired one-tailed t test with Welch’s correction was used to analyze the increase in CNR/cell numbers for PC3-HITI day 11 versus day 46 tumors.
Supplementary Material
Acknowledgments
We thank K. Brindle (University of Cambridge) for providing the Oatp1a1 plasmid, Y. Xia for help with paper revisions, K. Chadwick for FACS and flow cytometry help, and D. Reese for MRI troubleshooting. Funding: This work was funded by a Canadian Institutes of Health Research (CIHR) project grant (grant #377071, J.A.R.), a Natural Sciences and Engineering Research Council (NSERC) discovery grant (grant #RGPIN-2016-05420, J.A.R.), and a University of Western Ontario Strategic Support for CIHR Success Seed grant (J.A.R.). Author contributions: J.A.R. designed the project. J.A.R., J.J.K., and M.S.-M. designed the experiments. J.J.K. directed the study and, with M.S.-M., carried out most of the experiments. N.N.N. developed the methods for Oatp1a1 MRI, and F.M.M. developed the MATLAB scripts for processing MRI phantom data. Y.C. helped perform MRI. M.M.E. processed and analyzed MRI data. A.M.H. helped develop the PPs. J.J.K. wrote the manuscript with help from M.S.-M., which J.A.R. reviewed and edited. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/4/eabc3791/DC1
REFERENCES AND NOTES
- 1.Brader P., Serganova I., Blasberg R. G., Noninvasive molecular imaging using reporter genes. J. Nucl. Med. 54, 167–172 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kircher M. F., Gambhir S. S., Grimm J., Noninvasive cell-tracking methods. Nat. Rev. Clin. Oncol. 8, 677–688 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Prescher J. A., Contag C. H., Guided by the light: Visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol. 14, 80–89 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Hong H., Yang Y., Cai W., Imaging gene expression in live cells and tissues. Cold Spring Harb. Protoc., pdb.top103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J. E., Kalimuthu S., Ahn B.-C., In vivo cell tracking with bioluminescence imaging. Nucl. Med. Mol. Imaging 49, 3–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M., Wang Y., Liu M., Lan X., Multimodality reporter gene imaging: Construction strategies and application. Theranostics 8, 2954–2973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilad A. A., Shapiro M. G., Molecular imaging in synthetic biology, and synthetic biology in molecular imaging. Mol. Imaging Biol. 19, 373–378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joo H. K., Chung J.-K., Molecular-genetic imaging based on reporter gene expression. J. Nucl. Med. 49, 164S–179S (2008). [DOI] [PubMed] [Google Scholar]
- 9.Reagan M. R., Kaplan D. L., Concise review: Mesenchymal stem cell tumor-homing: Detection methods in disease model systems. Stem Cells 29, 920–927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Cao F., De A., Cao Y., Contag C., Gambhir S. S., Wu J. C., Chen X., Trafficking mesenchymal stem cell engraftment and differentiation in tumor-bearing mice by bioluminescence imaging. Stem Cells 27, 1548–1558 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidd S., Spaeth E., Dembinski J. L., Dietrich M., Watson K., Klopp A., Battula V. L., Weil M., Andreeff M., Marini F. C., Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells 27, 2614–2623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton A. M., Parkins K. M., Murrell D. H., Ronald J. A., Foster P. J., Investigating the impact of a primary tumor on metastasis and dormancy using MRI: New insights into the mechanism of concomitant tumor resistance. Tomography 2, 79–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkins K. M., Dubois V. P., Hamilton A. M., Makela A. V., Ronald J. A., Foster P. J., Multimodality cellular and molecular imaging of concomitant tumour enhancement in a syngeneic mouse model of breast cancer metastasis. Sci. Rep. 8, 8930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkins K. M., Hamilton A. M., Makela A. V., Chen Y., Foster P. J., Ronald J. A., A multimodality imaging model to track viable breast cancer cells from single arrest to metastasis in the mouse brain. Sci. Rep. 6, 35889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandergaast R., Khongwichit S., Jiang H., De Grado T. R., Peng K.-W., Smith D. R., Russell S. J., Suksanpaisan L., Enhanced noninvasive imaging of oncology models using the NIS reporter gene and bioluminescence imaging. Cancer Gene Ther. 27, 179–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah K., Jacobs A., Breakefield X. O., Weissleder R., Molecular imaging of gene therapy for cancer. Gene Ther. 11, 1175–1187 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Yaghoubi S. S., Jensen M. C., Satyamurthy N., Budhiraja S., Paik D., Czernin J., Gambhir S. S., Noninvasive detection of therapeutic cytolytic T cells with 18F–FHBG PET in a patient with glioma. Nat. Clin. Pract. Oncol. 6, 53–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keu K. V., Witney T. H., Yaghoubi S., Rosenberg J., Kurien A., Magnusson R., Williams J., Habte F., Wagner J. R., Forman S., Brown C., Allen-Auerbach M., Czernin J., Tang W., Jensen M. C., Badie B., Gambhir S. S., Reporter gene imaging of targeted T cell immunotherapy in recurrent glioma. Sci. Transl. Med. 9, eaag2196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milone M. C., O’Doherty U., Clinical use of lentiviral vectors. Leukemia 32, 1529–1541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacein-Bey-Abina S., Garrigue A., Wang G. P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K., Asnafi V., Intyre E. M., Cortivo L. D., Radford I., Brousse N., Sigaux F., Moshous D., Hauer J., Borkhardt A., Belohradsky B. H., Wintergerst U., Velez M. C., Leiva L., Sorensen R., Wulffraat N., Blanche S., Bushman F. D., Fischer A., Cavazzana-Calvo M., Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe S. J., Mansour M. R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M. H., Pike-Overzet K., Chatters S. J., de Ridder D., Gilmour K. C., Adams S., Thornhill S. I., Parsley K. L., Staal F. J. T., Gale R. E., Linch D. C., Bayford J., Brown L., Quaye M., Kinnon C., Ancliff P., Webb D. K., Schmidt M., von Kalle C., Gaspar H. B., Thrasher A. J., Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118, 3143–3150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M. P., Wulffraat N., Leboulch P., Lim A., Osborne C. S., Pawliuk R., Morillon E., Sorensen R., Forster A., Fraser P., Cohen J. I., de Saint Basile G., Alexander I., Wintergerst U., Frebourg T., Aurias A., Stoppa-Lyonnet D., Romana S., Radford-Weiss I., Gross F., Valensi F., Delabesse E., Macintyre E., Sigaux F., Soulier J., Leiva L. E., Wissler M., Prinz C., Rabbitts T. H., Le Deist F., Fischer A., Cavazzana-Calvo M., LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wilson M. H., Coates C. J., George A. L. Jr., PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Turchiano G., Latella M. C., Gogol-Döring A., Cattoglio C., Mavilio F., Izsvák Z., Ivics Z., Recchia A., Genomic analysis of Sleeping Beauty transposon integration in human somatic cells. PLOS ONE 9, e112712 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papapetrou E. P., Schambach A., Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 24, 678–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Zhang W. Y., Hu S., Lan F., Lee A. S., Huber B., Lisowski L., Liang P., Huang M., de Almeida P. E., Won J. H., Sun N., Robbins R. C., Kay M. A., Urnov F. D., Wu J. C., Genome editing of human embryonic stem cells and induced pluripotent stem cells with zinc finger nucleases for cellular imaging. Circ. Res. 111, 1494–1503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y., Liu C., Cerbini T., San H., Lin Y., Chen G., Rao M. S., Zou J., Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl. Med. 3, 821–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerbini T., Funahashi R., Luo Y., Liu C., Park K., Rao M., Malik N., Zou J., Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PLOS ONE 10, e0116032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho S. W., Kim S., Kim J. M., Kim J.-S., Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J., RNA-programmed genome editing in human cells. eLife 2013, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M., RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Q., Regan S. N., Xia Y., Oostrom L. A., Cowan C. A., Musunuru K., Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubois V. P., Zotova D., Parkins K. M., Swick C., Hamilton A. M., Kelly J. J., Ronald J. A., Safe harbor targeted CRISPR-Cas9 tools for molecular-genetic imaging of cells in living subjects. Cris. J. 1, 440–449 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Orthwein A., Noordermeer S. M., Wilson M. D., Landry S., Enchev R. I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G., Xia B., Peter M., Durocher D., A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieber M. R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auer T. O., Duroure K., De Cian A., Concordet J.-P., Del Bene F., Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki K., Izpisua Belmonte J. C., In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 63, 157–164 (2018). [DOI] [PubMed] [Google Scholar]
- 39.He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B., Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 44, e85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E. J., Hatanaka F., Yamamoto M., Araoka T., Li Z., Kurita M., Hishida T., Li M., Aizawa E., Guo S., Chen S., Goebl A., Soligalla R. D., Qu J., Jiang T., Fu X., Jafari M., Esteban C. R., Berggren W. T., Lajara J., Nuñez-Delicado E., Guillen P., Campistol J. M., Matsuzaki F., Liu G.-H., Magistretti P., Zhang K., Callaway E. M., Zhang K., Belmonte J. C. I., In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T. D., Chen Y., Ronald J. A., A novel approach for assessment of prostate cancer aggressiveness using survivin-driven tumour-activatable minicircles. Gene Ther. 26, 177–186 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Ronald J. A., Cusso L., Chuang H.-Y., Yan X., Dragulescu-Andrasi A., Gambhir S. S., Development and validation of non-integrative, self-limited, and replicating minicircles for safe reporter gene imaging of cell-based therapies. PLOS ONE 8, e73138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darquet A.-M., Cameron B., Wils P., Scherman D., Crouzet J., A new DNA vehicle for nonviral gene delivery: Supercoiled minicircle. Gene Ther. 4, 1341–1349 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Patrick P. S., Hammersley J., Loizou L., Kettunen M. I., Rodrigues T. B., Hu D.-E., Tee S.-S., Hesketh R., Lyons S. K., Soloviev D., Lewis D. Y., Aime S., Fulton S. M., Brindle K. M., Dual-modality gene reporter for in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 111, 415–420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyström N. N., Hamilton A. M., Xia W., Liu S., Scholl T. J., Ronald J. A., Longitudinal visualization of viable cancer cell intratumoral distribution in mouse models using Oatp1a1-enhanced magnetic resonance imaging. Invest. Radiol. 54, 302–311 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Paix A., Folkmann A., Goldman D. H., Kulaga H., Grzelak M. J., Rasoloson D., Paidemarry S., Green R., Reed R. R., Seydoux G., Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. U.S.A. 114, E10745–E10754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreiss P., Mailhe P., Scherman D., Pitard B., Cameron B., Rangara R., Aguerre-Charriol O., Airiau M., Crouzet J., Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 27, 3792–3798 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hornstein B. D., Roman D., Arévalo-Soliz L. M., Engevik M. A., Zechiedrich L., Effects of circular DNA length on transfection efficiency by electroporation into HeLa cells. PLOS ONE 11, e0167537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pellenz S., Phelps M., Tang W., Hovde B. T., Sinit R. B., Fu W., Li H., Chen E., Monnat R. J. Jr., New human chromosomal sites with “Safe Harbor” potential for targeted transgene insertion. Hum. Gene Ther. 30, 814–828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ordovás L., Boon R., Pistoni M., Chen Y., Wolfs E., Guo W., Sambathkumar R., Bobis-Wozowicz S., Helsen N., Vanhove J., Berckmans P., Cai Q., Vanuytsel K., Eggermont K., Vanslembrouck V., Schmidt B. Z., Raitano S., Van Den Bosch L., Nahmias Y., Cathomen T., Struys T., Verfaillie C. M., Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 5, 918–931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X.-H., Tee L. Y., Wang X.-G., Huang Q.-S., Yang S.-H., Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D. C., Wang X., Off-target genome editing: A new discipline of gene science and a new class of medicine. Cell Biol. Toxicol. 35, 179–183 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Li P., Zhang L., Li Z., Xu C., Du X., Wu S., Cas12a mediates efficient and precise endogenous gene tagging via MITI: Microhomology-dependent targeted integrations. Cell. Mol. Life Sci. 77, 3875–3884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vakulskas C. A., Dever D. P., Rettig G. R., Turk R., Jacobi A. M., Collingwood M. A., Bode N. M., McNeill M. S., Yan S., Camarena J., Lee C. M., Park S. H., Wiebking V., Bak R. O., Gomez-Ospina N., Pavel-Dinu M., Sun W., Bao G., Porteus M. H., Behlke M. A., A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J. S., Dagdas Y. S., Kleinstiver B. P., Welch M. M., Sousa A. A., Harrington L. B., Sternberg S. H., Joung J. K., Yildiz A., Doudna J. A., Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira P. H., Mairhofer J., Marker-free plasmids for biotechnological applications – implications and perspectives. Trends Biotechnol. 31, 539–547 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Xiao Y.-D., Paudel R., Liu J., Ma C., Zhang Z.-S., Zhou S.-K., MRI contrast agents: Classification and application (Review). Int. J. Mol. Med. 38, 1319–1326 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Patrick P. S., Lyons S. K., Rodrigues T. B., Brindle K. M., Oatp1 enhances bioluminescence by acting as a plasma membrane transporter for D-luciferin. Mol. Imaging Biol. 16, 626–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu M.-R., Liu H.-M., Lu C.-W., Shen W.-H., Lin I.-J., Liao L.-W., Huang Y.-Y., Shieh M.-J., Hsiao J.-K., Organic anion-transporting polypeptide 1B3 as a dual reporter gene for fluorescence and magnetic resonance imaging. FASEB J. 32, 1705–1715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nyström N. N., Yip L. C. M., Carson J. J. L., Scholl T. J., Ronald J. A., Development of a human photoacoustic imaging reporter gene using the clinical dye indocyanine green. Radiol. Imaging Cancer 1, e190035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marks P. W., Witten C. M., Califf R. M., Clarifying stem-cell therapy’s benefits and risks. N. Engl. J. Med. 376, 1007–1009 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Parkins K. M., Dubois V. P., Kelly J. J., Chen Y., Knier N. N., Foster P. J., Ronald J. A., Engineering circulating tumor cells as novel cancer theranostics. Theranostics 10, 7925–7937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivics Z., Self-destruct genetic switch to safeguard iPS cells. Mol. Ther. 23, 1417–1420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yushkevich P. A., Piven J., Hazlett H. C., Smith R. G., Ho S., Gee J. C., Gerig G., User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/4/eabc3791/DC1


