Significance
Although miRNAs are emerging as important regulators of diverse physiological and pathological processes, our knowledge of their potential role in regulation of circadian rhythms is still limited. We deployed a cell-based genome-wide screening approach and successfully identified mature miRNAs as cell-autonomous circadian modulators. We then specifically focused on the miR-183/96/182 cluster among the candidate miRNA hits and revealed their circadian function both in vitro and in vivo. This study provides resources for further understanding the role of miRNAs in the circadian network. It also highlights the importance of miRNAs as a genome-wide layer of circadian clock regulation.
Keywords: circadian rhythms, miRNA, genome-wide screen, miR-183/96/182 cluster
Abstract
The regulatory mechanisms of circadian rhythms have been studied primarily at the level of the transcription–translation feedback loops of protein-coding genes. Regulatory modules involving noncoding RNAs are less thoroughly understood. In particular, emerging evidence has revealed the important role of microRNAs (miRNAs) in maintaining the robustness of the circadian system. To identify miRNAs that have the potential to modulate circadian rhythms, we conducted a genome-wide miRNA screen using U2OS luciferase reporter cells. Among 989 miRNAs in the library, 120 changed the period length in a dose-dependent manner. We further validated the circadian regulatory function of an miRNA cluster, miR-183/96/182, both in vitro and in vivo. We found that all three members of this miRNA cluster can modulate circadian rhythms. Particularly, miR-96 directly targeted a core circadian clock gene, PER2. The knockout of the miR-183/96/182 cluster in mice showed tissue-specific effects on circadian parameters and altered circadian rhythms at the behavioral level. This study identified a large number of miRNAs, including the miR-183/96/182 cluster, as circadian modulators. We provide a resource for further understanding the role of miRNAs in the circadian network and highlight the importance of miRNAs as a genome-wide layer of circadian clock regulation.
Most organisms on Earth have internal timing systems that optimally adapt and respond to 24-h light–dark cycles. In mammals, the endogenous timing systems function and synchronize at multiple levels, from molecular to neural circuits (1). At the molecular level, the circadian clock is generated by a cell-autonomous regulatory feedback loop, where gene activators, such as BMAL1 and CLOCK, turn on the expression of gene repressors, such as PER and CRY, which in turn repress the expression and transcriptional activity of the activators (2, 3). These simplified steady-state messenger RNA (mRNA)/protein rhythms actually undergo complicated dynamic regulation by many other processes including RNA splicing, polyadenylation, nuclear export, RNA degradation, and microRNA (miRNA) regulation (4, 5).
miRNAs are a class of small noncoding RNAs. Genes encoding miRNAs can be found in the genome either as single genes or as clusters; i.e., miRNAs are localized in the same genomic region (usually less than 10 kb), transcribed in the same orientation, and highly coexpressed (6–8). These genes are first transcribed in the nucleus as a single primary transcript of miRNAs (pri-miRNAs) and are then cleaved to ∼70-nucleotide (nt) precursor miRNA (premiRNA) hairpins followed by further maturation in the cytoplasm. The biologically active mature miRNAs are single stranded and ∼22 nt in length (9, 10). In mammals, mature miRNAs usually bind to sites in the 3′ untranslated region (3′ UTR) of their target mRNAs through base-pairing of the seed region, mainly at position 2–7 from the 5′ end of the miRNA. This base-pairing is generally considered the minimal requirement to engage a target mRNA (10). Beyond the seed region, the binding between the whole mature miRNA sequence and the target mRNA is not perfectly complementary. These imperfect bindings interfere with the stability or translation of mRNAs (11, 12). They result in a complex network where a single miRNA can concurrently regulate a large number of target genes, and one gene can be regulated by multiple miRNAs (13–15). The regulations between miRNAs and target genes are highly time and tissue specific (16–18) and play important roles in diverse physiological and pathophysiological processes (19–23).
Although miRNAs are emerging as major regulators of diverse physiological and pathological processes, particularly in cancer (12), the knowledge of their roles in circadian rhythms is still limited to a few genes. To address this, several studies, including some genome-wide profiling experiments, focused on identifying miRNAs expressed in a circadian manner in various tissues and across different species, including mouse, Drosophila, and Arabidopsis (24–27). The focus of these studies was clock-controlled miRNAs, but the observed rhythmic expression alone of these miRNAs is neither necessary nor sufficient for them to be considered as regulators of circadian rhythms. Most of these studies did not determine how the oscillation of the miRNAs is relevant to specific physiological and behavioral rhythms. Instead, more convincing evidence for the regulatory role of miRNAs in clock function came from a genetic study in mice, where global knockout of Dicer, the enzyme responsible for cleaving premiRNA to generate mature miRNAs, significantly shortened the circadian period (28). Several other studies have also investigated specific miRNAs that regulate the expression of core clock components; for example, miR-142 regulates Bmal1, miR-17 regulates Clock, and miR-192/194 cluster regulates the Per gene family (29–32). These studies together suggest a pervasive regulation of the clock by miRNAs that requires further functional and mechanistic understanding.
In 2007, Xu et al. (27) identified a sensory organ-specific miRNA cluster of miR-183, miR-96, and miR-182, named the miR-183/96/182 cluster. Members of the miR-183/96/182 cluster are located within a 4-kb genomic region on mouse chromosome 6qA3, and this genomic arrangement is highly conserved in mammals (27). Some speculation exists regarding a circadian function for the miR-183/96/182 cluster or its members for several reasons. First, the gene expression of the miR-183/96/182 cluster oscillates in mouse retina in a circadian manner (27). Second, the predicted targets of the miR-183/96/182 cluster include some circadian rhythm relevant genes, and Adenylyl cyclase type 6 (Adcy6), a clock-controlled gene that modulates melatonin synthesis, is expressed in antiphase with the miR-183/96/182 cluster (27). Third, in humans, a polymorphism in premiR-182 was found to be associated with a marked reduction in the expression of the circadian gene CLOCK (33). However, to date, there has been no direct evidence to assign specific roles of the miR-183/96/182 cluster in the modulation of circadian rhythms.
In the present study, we used a cell-based genome-wide screening approach to systematically identify the mature miRNAs that can potentially alter circadian rhythms. We show here that our cell-based miRNA screening successfully identified cell-autonomous circadian modulators. We specifically focused on the miR-183/96/182 cluster among the candidate miRNA hits and revealed their circadian function both in vitro and in vivo.
Results
Genome-wide miRNA Screen.
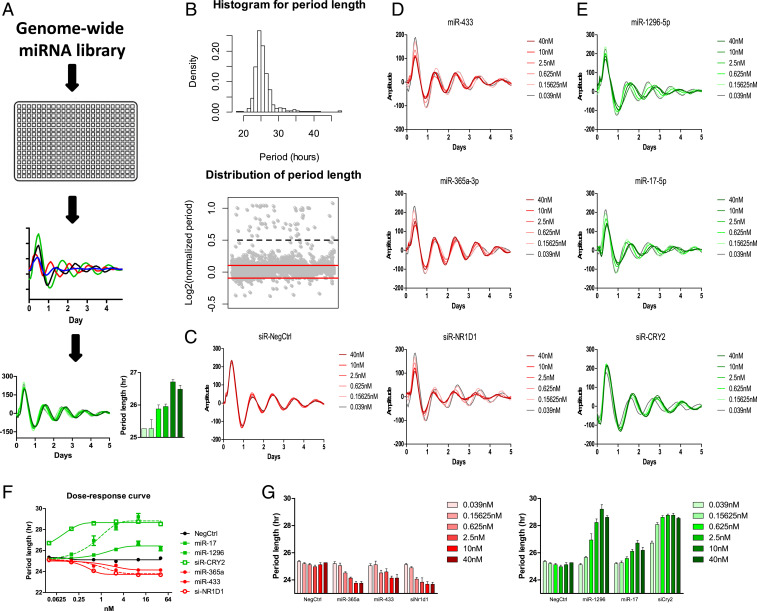
To identify miRNAs that have the potential to modulate circadian rhythms, we first adopted a mammalian cell-based assay to screen a library of miRNAs (Fig. 1A). We used U2OS cells which contain either the Bmal1-dLuc or Per2-dLuc gene to report circadian oscillations (34). We screened a Qiagen miRNA library containing 989 synthesized human miRNA mimics, which are chemically synthesized small-RNA molecules imitating mature miRNAs. An equal amount of these miRNA mimics was prespotted in each well of 384-well plates individually and then reverse-transfected into cells. By integrating the screening results from both Bmal1-dLuc and Per2-dLuc reporter cell lines, we found 113 long-period hits (11%) and 7 short-period hits (1%) from both sets of screens (Fig. 1B and SI Appendix, Table S1). In addition, we observed many hits that only show low amplitude rather than period change. As the low-amplitude phenotypes were most likely caused by noncircadian related cell fitness issues rather than circadian clock deficiency, these miRNA hits were not included in the follow-up studies.
Fig. 1.
A cell-based genome-wide screen for miRNA modifiers of circadian rhythms. (A) A schematic diagram of the genome-wide miRNA screen. Bmal1-dLuc and Per2-dLuc reporter cells were transfected with miRNAs in 384-well plates. Bioluminescence was recorded and analyzed to obtain circadian parameters and select candidate hits. Validation was performed by measuring the dose-dependent effects of some candidate miRNAs on circadian phenotypes. (B) Distribution of circadian period lengths of the screen. The histogram shows a tail toward the long period. Gray dots in the bottom panel represent normalized period lengths. The period length of each well was divided by the negative control and indicated in Log2 space. The cutoff was −0.1 and +0.1 (red lines). Values above 0.5 (black dash line) were considered outliers. (C) Representative bioluminescence profiles for dose-dependent phenotypic validation of negative control, short-period positive control (siNR1D1), and long-period positive control (siCRY2). (D) Short-period candidate hits (miR-433, miR-365a-3p) and (E) long-period candidate hits (miR-1296-5p, miR-17). (F) The dose–response curve of the indicated candidate miRNAs and control RNAs. (G) The period length parameters of the indicated candidate miRNAs and control RNAs. Per2-dLuc U2OS cells were transfected with the indicated amount of miRNAs (six 4-fold dilution series from 0.039 to 40 nM/well). Data represent the mean ± SD (n = 4).
Based on sequence similarity in the seed regions (35), miRNAs can be classified into families, which are usually evolutionarily related but do not strictly share common ancestry (10). The similar seed regions between members in the same family, in theory, lead to similar functions. Among the 120 candidate hits, 65 (54%) miRNAs belong to 35 families that have more than two members in humans (e.g., multimember family), and the rest of the hits are in families that currently contain only one member, themselves (SI Appendix, Table S2). Some of these identified families were highly enriched in candidate hits. For example, eight miRNAs from family let-7 and four miRNAs from family miR-17 were identified and showed a circadian phenotype in the screen. These might be attributed to similar seed regions shared by members of the miRNA family.
The other way to group miRNAs is based on their genomic locations because miRNAs tend to colocalize as clusters (6). Using all human miRNA data from miRBase (35), miRNAs are clustered if two neighboring miRNAs are on the same strand and within 10 kb, and in human, ∼20% of all miRNAs are clustered (36). In our screen, 54 out of the 120 miRNA hits identified were mapped to 35 clusters (SI Appendix, Table S3). This number accounts for 45% of all miRNA hits identified in the screen, which is higher than the overall ratio in all human miRNAs. Although miRNA clusters are defined by physical distance rather than the sequence similarity, we noticed that some clusters were enriched in candidate hits. For instance, all three members in cluster let-7a-1/let-7f-1/let-7d and all three members in cluster miR-183/miR-96/miR-182 were identified by the screen. Based on the seed sequence similarity, we further divided the 35 identified clusters into three different categories (36): 7 homo-seed clusters (miRNA members having identical seed sequences), 19 hetero-seed clusters (miRNA members having different seed sequences), and 9 homo-hetero-seed clusters (a combination of the former two classes). We found that homo-seed clusters have a significantly higher percentage of candidate miRNA enrichment than that of hetero-seed clusters and homo-hetero-seed clusters (SI Appendix, Fig. S1).
We then randomly selected some candidate miRNA hits and conducted a dose-dependent assay for further validation. We tested these miRNAs at six different doses (fourfold dilution series; from 40 to 0.039 nM). All of them showed dose-dependent effects on circadian parameters (Fig. 1 C–E). The dose–response curve of miRNA dose-dependent effects showed that the phenotype reaches a plateau at about 10 nM, which was the dosage used in our miRNA screen (Fig. 1 F and G). In addition to miRNA, small-interfering RNA (siRNA) is another class of small RNAs (synthetic double-stranded RNA, ∼20–25 base pairs in length) that is central to RNA interference (37). Despite differences in origins, siRNAs and miRNAs share similarities in terms of downstream cellular machinery (37). We, therefore, compared the dose dependency of miRNAs with that of siRNAs. We found that the potency of the miRNAs was generally lower than that of the siRNAs in our dose-dependent assay (Fig. 1 D–G). For example, the CRY2 siRNA could significantly shorten the circadian period length of U2OS-reporter cells at a concentration lower than 0.02 nM, but the miRNAs typically did not show a circadian phenotype in reporter cells at a concentration lower than 0.5 nM.
The miR-183/96/182 Cluster Was Important for Maintaining Circadian Period Length and Amplitude in Human Cells.
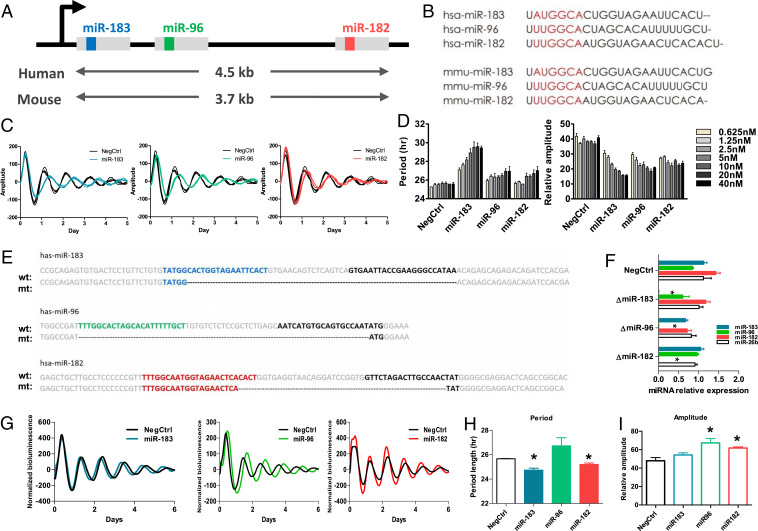
The miR-183/96/182 cluster drew our attention because all members from this cluster were identified by the screen. This cluster is conserved in both human and mouse and has members sharing similar, though not identical, seed regions (Fig. 2 A and B). This miRNA cluster was suggested to be involved in circadian rhythms, given that the predicted targets include some circadian relevant genes (33, 38). However, no direct evidence has validated the role of this miRNA cluster in circadian rhythm modulation. In our study, all members from this cluster showed a longer-period and lower-amplitude phenotype after being overexpressed in reporter cells, in a dose-dependent manner (Fig. 2 C and D).
Fig. 2.
The cluster of miR-183/96/182 altered the circadian period length and amplitude at the cellular level. (A) Schematic diagram showing the miR-183/96/182 cluster on the chromosome. Members of the miR-183/96/182 cluster locate on the same chromosome and are cotranscribed through the same promoter. The gray blocks represent the primary miRNA. The inside color boxes (blue, green, and red) represent the sequences for miR-183-5p, miR-96-5p, and miR-182-5p, respectively. (B) The sequences of mature miRNA for members of the miR-183/96/182 cluster in human and mouse. The red region represents the seed region of each miRNA, which exhibits high similarity. (C) Representative bioluminescence profiles of U2OS Per2-dLuc reporter cells which were transfected with 10 nM synthetic miRNA mimics of each member of the miR-183/96/182. (D) The dose-dependent effects of members of the miR-183-/96/182 cluster on period length. U2OS Per2-dLuc reporter cells were transfected with the indicated amount of miRNAs (five 2-fold dilution series from 2.5 to 40 nM/well). The data represent the mean ± SD (n = 4). (E) Knockout of each member of the miR-183/96/182 cluster using a CRISPR-Cas9 approach. For each miRNA, the “wt” sequence represents the sequence of wild-type primary miRNA, and the “mt” sequence represents the mutant sequence of primary miRNA after deletion. The colored sequences (blue, green, and red) represent the 5′ miRNA, and the black sequences represent the 3′ miRNA. Deletions are marked by dashes. (F) The mature miRNA expression levels detected by qPCR confirmed the complete deletion of each miRNA. (G) Representative bioluminescence profiles for each CRISPR miRNA deletion cell line. (H) Period lengths of CRISPR miRNA deletion cell lines. (I) Amplitude of CRISPR miRNA deletion cell lines. Data represent the mean ± SEM (n = 3), *P < 0.05.
To confirm that the observed circadian phenotypes were not due to ectopic effects generated by the massive transfection of synthetic miRNAs, we generated miRNA knockout reporter cell lines using a CRISPR-Cas9 approach. Because of their very short length and noncoding properties, there is very limited space in miRNAs for identifying protospacer adjacent motif (PAM) sequences. Small indels induced by CRISPR-Cas9 do not cause a frameshift in the noncoding gene and may be tolerable during the imperfect complementary binding between miRNAs and their targets. We therefore chose to use paired CRISPR RNAs (crRNAs) to individually excise a genomic sequence encoding each member of the miR-183/96/182 cluster (Fig. 2E), resulting in a complete loss of expression of the mature miRNAs (Fig. 2F). The knockout of miR-183 or miR-182 shortened the circadian period length (Fig. 2 G and H), and the knockout of miR-96 or miR-182 increased the amplitude (Fig. 2I). These results were opposite from the observed effects when the miRNA mimics were transfected in the cells. These results indicate an intrinsic effect of the miR-183/96/182 cluster on circadian rhythms.
Circadian Behaviors Were Altered in miR-183/96/182 Cluster–Deficient Mice.
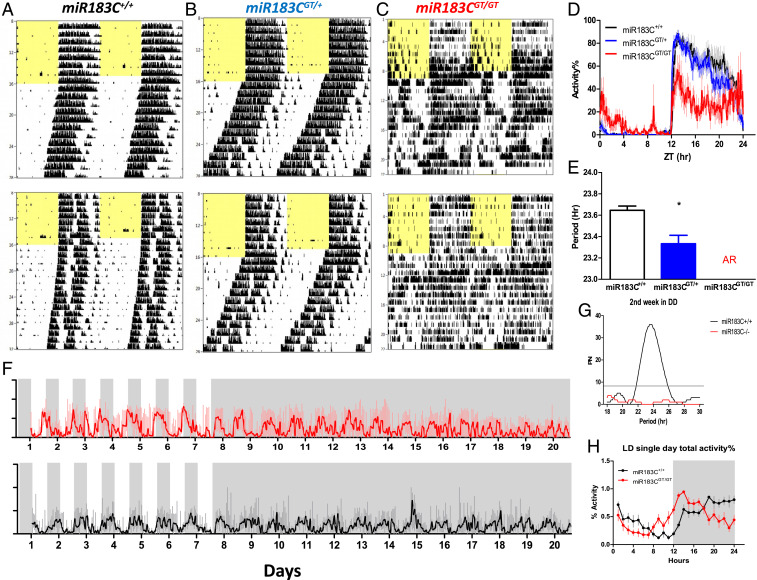
Next, we analyzed the circadian behaviors of miR-183/96/182 cluster–deficient mice in running-wheel cages. The miR-183/96/182 cluster gene-trap (miR-183CGT/GT) mouse has the expression of the miR-183/96/182 cluster inactivated throughout the whole body (38). Compared with wild-type littermate control mice, heterozygous miR-183CGT/+ mice had shorter free-running periods in constant darkness, and homozygous miR-183CGT/GT mice became arrhythmic after release into constant darkness (Fig. 3 A–C). Voluntary wheel-running activity is a subset of the total general activity. Access to running wheels may artificially affect the activity patterns. We noticed that the wheel-running activity level was extremely low in miR-183CGT/GT mice (SI Appendix, Fig. S2A), although the miR-183CGT/GT mice were hyperactive compared with wild-type mice (SI Appendix, Fig. S2B). We found that the miR-183CGT/GT mice had circling behaviors, which might be caused by vestibular dysfunction (39). The discrepancy between wheel-running activity and total general activity in miR-183CGT/GT mice suggested reduced access to running wheels. The reduced wheel-running activity might flatten the diurnal rhythmicity (40), causing a false-positive scoring of arrhythmicity. To preclude such a possibility, we compared the nonwheel beam-break total activities between miR-183C+/+ and miR-183CGT/GT mice. In the first week of constant darkness, both miR-183C+/+ and miR-183CGT/GT mice had similar period lengths (SI Appendix, Fig. S2C). About 1 wk after release into constant darkness, compared with wild-type controls, the miR-183CGT/GT mice gradually lost their rhythmicity in general total activity (Fig. 3 F and G). Furthermore, the miR-183CGT/GT mice also exhibited a phase advance compared with wild-type mice under entrained conditions (Fig. 3H). It has been found that the deficiency of the miR-183/96/182 cluster can cause retinal degeneration (38, 41–43). As intrinsically photosensitive retinal ganglion cells (ipRGCs) in the retina are the main conduit of light information to the master circadian clock, the suprachiasmatic nucleus (SCN) (44, 45), we asked if the circadian disruption in the miR-183CGT/GT mice may be caused by dysfunction of light entrainment. We found that the sensitivity of ipRGCs, tested using the pupillary light reflex (PLR), was not significantly different between miR-183C+/+ and miR-183CGT/GT mice and therefore excluded this possibility (SI Appendix, Fig. S3). Together, these results validated a functional role of the miR-183/96/182 cluster in circadian regulation at the behavioral level.
Fig. 3.
Circadian behaviors were altered in the miR-183/96/182 cluster–deficient mice. (A–C) Representative wheel-running locomotor activity profiles of homozygous wild-type, heterozygous, and homozygous gene-trap mice (miR-183C+/+, n = 14; miR-183CGT/+, n = 22; miR-183CGT/GT, n = 10). Animals were maintained on LD12:12 entrained conditions for the first week, indicated by the yellow filled and open area in the records, and then were released to constant darkness (DD) for 3 wk to measure free-running periods. (D) Diurnal wheel-running activity profiles under the entrained condition. Wheel-running activities were normalized to the highest activity level of each animal and then averaged across 7 d prior to DD. (E) Free-running periods under constant darkness. The free-running period of heterozygous mice was significantly shorter than that of wild-type mice (t test: *P = 0.004). The miR-183CGT/GT mice became arrhythmic (AR) after release into DD. (F) Beam-break total activities of homozygous wild-type and gene-trap mice (miR-183C+/+, n = 4; miR-183CGT/GT, n = 4). Animals were maintained on LD12:12 for the first 7 d, indicated by the gray and white shading, and then released to DD for 14 d. The red and black curves denote the mean activities of mutant mice and wild-type mice, respectively. The light red and light gray vertical lines denote the SD of the mean activities of mutant mice and wild-type mice, respectively. (G) Circadian periodicity of the beam-break total activity DD condition was detected by Lomb–Scargle periodogram. Activity of miR-183CGT/GT in the second week of DD did not pass the threshold for period detection. PN on y-axis stands for normalized power. (H) Diurnal beam-break total activity profiles under the entrained condition. Activities were normalized to the highest activity level of each animal and then averaged across 7 d prior to DD. Data represent the mean ± SD.
The miR-183/96/182 Cluster Altered Circadian Rhythms in a Tissue-Specific Way.
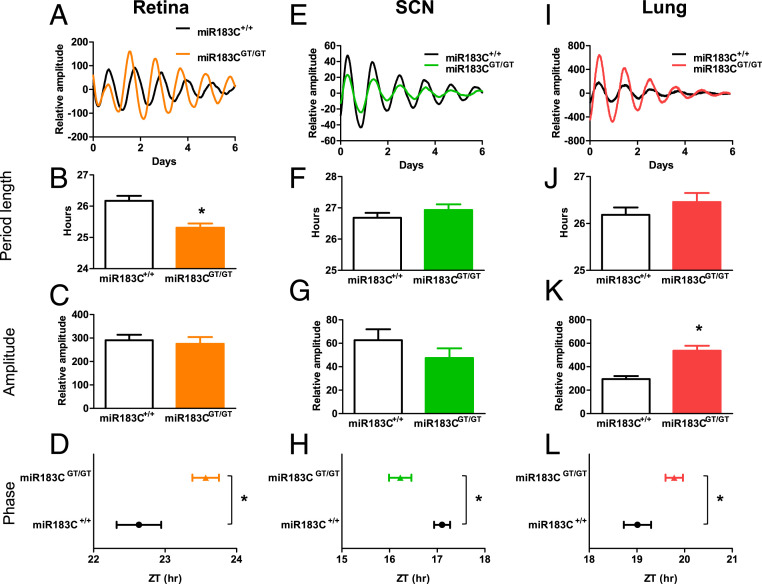
The expression of miRNAs is highly tissue specific (16–18). The miR-183/96/182 cluster has very high expression in many sensory organs and is considered to be a neuronal-specific miRNA cluster (39, 46). To delineate the circadian function of the miR-183/96/182 cluster at a tissue level, we generated a miR-183/96/182 cluster luciferase reporter mouse line by crossing a miR-183CGT/GT mouse with a Per2::Luc reporter knock-in mouse (47). The Per2::Luc reporter knock-in mouse has a luciferase-coding sequence inserted before the endogenous Per2 stop codon, thus fused with an intact endogenous Per2-3′ UTR. Mice expressing PER2::LUC fusion protein have indistinguishable behavioral and molecular characteristics compared with wild-type mice (47, 48). We compared ex vivo circadian bioluminescence rhythms between wild-type and mutant mice in three selected tissues, which have detectable expression of the miR-183/96/182 cluster: the retina which has high expression level of the cluster, the lung tissue which contains peripheral clock function, and the SCN which contains the master clock in the brain. Interestingly, the three tissues showed different circadian phenotypes (Fig. 4). The inactivation of the miR-183/96/182 cluster shortened the circadian period in the retina (Fig. 4 A and B) but did not change period lengths in the SCN (Fig. 4 E and F) and the lung tissue (Fig. 4 I and J). The phases of all tissues tested were significantly changed by the inactivation of the miR-183/96/182 cluster, but in different directions. Retina and lung had a phase delay (Fig. 4 D–L), whereas SCN had a phase advance (Fig. 4H), which was consistent with the phase advance in general activity (Fig. 3H). This indicates that the effects of the miR-183/96/182 cluster on circadian rhythms were tissue specific. Additionally, the amplitude of the PER2::LUC fusion protein rhythm was significantly higher in the mutant lung tissue than the wild-type lung tissue (Fig. 4K).
Fig. 4.
The circadian phenotype of the miR-183/96/182 cluster was tissue-specific. The miR-183/96/182 cluster altered the circadian period length, amplitude, and phase in the retina (A–D), SCN (E–H), and lung (I–L) from Per2::Luc mice carrying homozygous wild-type or homozygous gene-trap alleles (miR-183C+/+, n = 14∼23; miR-183CGT/GT, n = 12∼19). Animals were maintained under an LD12:12 entrained condition before dissection. Tissue dissection was performed 2–3 h before lights off, and the tissue was immediately cultured for recording for 1 wk. Representative bioluminescence recording profiles of miR-183C+/+ and miR-183CGT/GT are on the Top (A, E, and I). Circadian period length (B, F, and J) and relative amplitude (C, G, and K) were calculated by the linear model fit (damped sin) method. The phase of each tissue (D, H, and L) was represented by the time of peak luminescence on the first day of constant conditions (first day in Lumicycle), *P < 0.05. Data represent the mean ± SD.
PER2 Is a Direct Target of miRNA-96.
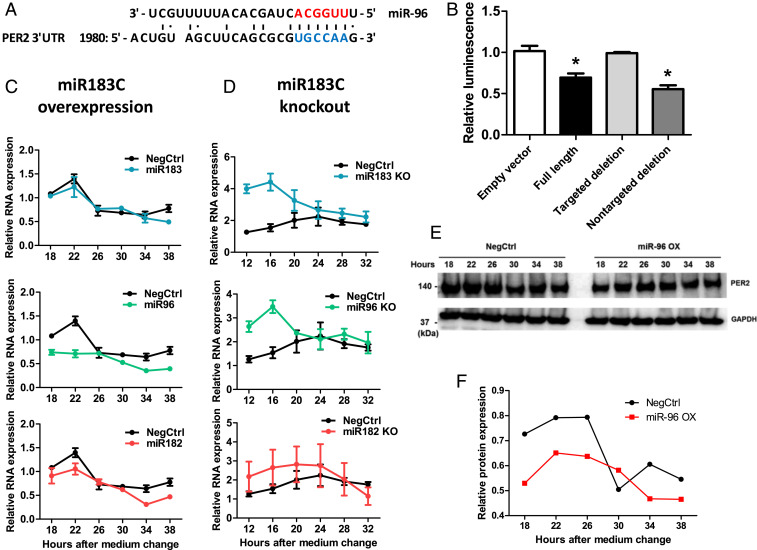
We further investigated how the miR-183/96/182 cluster contributed to circadian clock regulation and whether there were any core circadian genes among their direct targets. We first used two independent computational methods, DIANA-microT-CDS and miRanda (14, 49, 50), to predict the potential targets for members of the miR-183/96/182 cluster. Among the canonical circadian genes, PER2 and CLOCK were predicted to be direct targets by both methods (SI Appendix, Table S4), so we focused on these two genes. Both methods identified the same potential binding region on PER2-3′ UTR for miR-96 (Fig. 5A). We then validated the direct bindings between miR-96 and PER2-3′ UTR by performing a luciferase reporter assay (Fig. 5B). Consistently, the overexpression of miR-96 decreased while the knockout of miR-96 increased PER2 mRNA levels (Fig. 5 C and D). The overexpression of miR-96 also decreased PER2 at the protein level (Fig. 5 E and F). Although CLOCK was predicted by both methods to be a target for miR-96, this was not validated by the 3′ UTR luciferase reporter assay (SI Appendix, Fig. S4). On the other hand, CLOCK was computationally predicted to be a direct target of miR-182. This was validated through a 3′ UTR luciferase reporter assay from other studies (33, 51), as well as in our present study (SI Appendix, Fig. S5 A and B). The overexpression of miR-182, however, did not repress the expression of CLOCK at either mRNA or protein levels (SI Appendix, Fig. S5 C and D). Due to the complexity of circadian networks, although CLOCK could be directly targeted by the miR-182, the down-regulation effect might be compensated by other factors.
Fig. 5.
PER2 was a direct target of miR-96. (A) Sequence alignment between miR-96 and its putative binding sites (blue letters) in the PER2-3′ UTR. Red letters indicate canonical seed nucleotides. (B) HEK-293T cells were transfected with a luciferase reporter plasmid containing one of the following sequences: 1) no 3′ UTR; 2) a full-length wild-type PER2-3′ UTR; 3) a full-length PER2-3′ UTR with a targeted deletion at the predicted binding site of miR-96 (1992_2002 del: gcgcgTGCCAA; uppercase is the potential seed-binding site of miR-96); 4) a full-length PER2-3′ UTR with a deletion in a sequence (1712_1734 del: ttcataaacacaagaacacttta) predicted not to be bound by miR-96, *P < 0.05. (C) PER2 mRNA levels in U2OS cells that transfected with miRNA mimics of each member of the miR-183/96/182 cluster. RNA was sampled 18 h after medium change, every 4 h across 1 d. The mRNA levels were determined by qPCR (n = 3). (D) PER2 mRNA levels in miR-183/96/182 cluster knockout cells. RNA from U2OS cells with the deletion of each member of the miR-183/96/182 cluster was harvested 12 h after medium change, every 4 h across 1 d. The mRNA levels were determined by qPCR (n = 3). Data represent the mean ± SEM (E) U2OS cells were transfected with either control or miR-96 mimics, and PER2 protein levels were analyzed by Western blot 48 h posttransfection. (F) Quantification of PER2 protein band intensity compared with the negative control on Western blot.
Given the significant phenotypes in circadian rhythms in miR-183/96/182 cluster mutants, we further investigated the expression of some core circadian genes from different mouse tissues as shown in SI Appendix, Fig. S6. Under light-entrained conditions, the gene expression levels of core circadian genes were not changed in all three tissues tested. The peak expression of Clock, Per1, and Per2 in mutants was slightly delayed compared with wild types, except for Per2 that was advanced in the SCN. Under conditions of constant darkness, however, the expression levels of the core circadian genes in the eyes of mutant mice were all significantly lower than those of the wild-type mice. The expression peaks of Bmal1, Clock, and Per1 in SCN of mutant mice were advanced compared with those of wild types.
Discussion
For more than a decade, miRNAs have been found to play important roles in many physiological processes and disease conditions. Although a few miRNAs have been identified as circadian modulators, knowledge of the genome-wide picture of miRNA modulation of circadian rhythms is still limited. Previous studies usually started from either miRNA expression profiling or in silico miRNA target prediction to narrow down specific candidate miRNAs before functionally validating the role of the miRNA as a circadian modulator. In this phenotype-driven study, we started from a genome-wide cell-based functional screen and identified miRNAs that modulate circadian rhythms. The majority of the hits lengthened the period, but a very small number of hits were found to shorten the period, which is similar to what was observed in genome-wide siRNA studies (52). Approximately half of the candidate hits could be assigned to an miRNA family or a cluster. Multiple members from a family were likely to be coidentified from the screen, and homo-seed clusters had a higher percentage of candidate hits, suggesting that the conserved seed sequences were important determinants of functions. However, there were still members from the same family that did not show the same phenotypes. This suggests that differences in secondary structures (53) or in sequences beyond the seed regions still exist in members of the same family, which might be the reason for identifying only a portion in any particular family.
Between the threshold level and saturation level of miRNA concentration, these candidate miRNAs affect the circadian parameters in a dose-dependent manner, but not as efficiently as siRNAs. This may be due to the partial complementary binding between the mammalian miRNA seed regions and their mRNA targets, whereas siRNAs have a perfect binding sequence complementarity to their targets. Ideally, 100 molecules of an siRNA may specifically target 100 molecules of one circadian gene mRNA. However, 100 molecules of an miRNA may target mRNAs of 10 different genes with an average of 10 molecules of each gene. If only one mRNA affects circadian rhythms among the 10 different miRNA target mRNAs, the effect of miRNA on circadian rhythms would be greatly diluted. The relatively weaker repression effects of miRNAs compared to that of siRNAs may have their biological implication in the cell. Like a buffer solution requires a mixture of a weak acid and its conjugate base or vice versa, the cellular transcriptome may require a mixture of moderate affinity miRNAs and their conjugate targets for stabilization.
Our screen also identified some miRNA hits that have been previously reported to play a role in circadian rhythms in other studies, which validates our screen as an effective strategy to identify miRNAs as circadian clock regulators. For example, five out of six of the members in the miR-17 cluster were identified from our screen, and miR-17 was reported to participate in circadian period regulation by directly targeting the Clock gene (32). In our screen, we found that the transfection of miR-17 mimics lengthened the circadian period in U2OS reporter cells. Other candidate miRNA hits identified from our screen need further validation for their intrinsic function in circadian modulation. Of course, our screen could not identify all miRNA circadian modulators. One reason is that the miRNA library used in this screen only covers a portion of the currently known miRNA space. A larger-size library would be expected to identify more miRNA hits. The phenotype of a given miRNA depends greatly on the cellular context. Using different reporter cell lines may harvest distinct miRNA hits, given that miRNAs are highly cell-type specific.
Our study focused on the miR-183/96/182 cluster, which has been predicted to be involved in circadian rhythms because of previously observed circadian expression patterns, and the predicted targets include certain core circadian genes or circadian rhythm relevant genes (27, 33, 38). Previous reports link the miR-183/96/182 cluster to circadian rhythms because Adcy6 is predicted to be a target of miR-182 and miR-96, and Adcy6 regulates arylalkylamine–N-acetyltransferase (Aanat), an enzyme in the melatonin synthesis pathway (27). Melatonin is a hormone produced primarily in the pineal gland and is implicated in the regulation of circadian rhythms in most animals (54). However, most laboratory mouse strains are melatonin deficient (55–58). Therefore, it is less likely that the miR-183/96/182 cluster affects the circadian rhythms in melatonin-deficient mice through modulating the melatonin pathway, although Adcy6 was experimentally validated to be a target of the miR-182 and miR-96 (27). So far, there have been no experiments that validate the function of the miR-183/96/182 cluster as circadian modulators or dissect the circadian role of each member. In the present study, we first determined that each member in this cluster could change the circadian rhythms in human cells through transient transfection of these miRNA mimics. We then confirmed that these circadian effects were endogenous and cell autonomous by knocking them out in human cells. We found that each member in the miR-183/96/182 cluster exhibits different circadian phenotypes in human cells after being knocked out. Because the seed regions of each member of the miR-183/96/182 cluster are similar but not identical, they have both common and unique targets. Thus, they can show cooperative or opposing effects on biological processes, implying the complexity of this miRNA cluster in mediating gene regulation. We found that PER2 is one of the direct targets of miR-96. Per2 has been experimentally validated to be directly targeted by multiple miRNAs, such as miR-24 and miR-30a in the other studies (28, 48). All these miRNAs together with the miR-96 identified from our study regulate the Per2 expression by targeting its 3′ UTR. However, the validation of CLOCK as a direct target of miR-182 was not conclusive. The expression of endogenous CLOCK was not affected by miR-182, even though the 3′ UTR luciferase reporter assay supported the effects. This is probably because the endogenous CLOCK expression is affected by other factors in a complex network context that is not limited to miR-182.
The expression of miRNAs has been shown to be highly tissue specific (16–18). Consequently, the same miRNAs may behave differently in different tissues. This was observed in our study. Using Per2::Luc circadian reporter knock-in mice, we found that different tissues exhibited different circadian phenotypes. In miR-183/96/182 cluster–deficient Per2::Luc mice, the SCN showed phase advance, the lung had both phase delay and increased amplitude, and the retina showed a phase delay and shortened period compared with wild-type mice. These differences may be due to the tissue-specific gene expression of the miRNAs themselves, the tissue-specific gene network context, or the interactions between both. The SCN is believed to play an important role in synchronizing peripheral clocks throughout the body. However, our luciferase tissue culture study showed that the retina and lung explants behaved differently from the SCN in the miR-183/96/182 cluster–deficient mice. It was reported that the retinal circadian clock synchronizes directly to light–dark cycles, which is independent of the SCN phase (59). Our results from the retina seem to agree with this finding, showing distinct circadian patterns between the SCN and retina in mutants. However, it should be acknowledged that the in vitro tissue rhythms in the culture may not truly reflect the in vivo rhythmicity synchronization by SCN. Further in vivo luciferase imaging study in freely moving mice will be very helpful to answer these questions.
The miR-183/96/182 cluster is highly expressed in the nervous system, particularly in sensory organs such as the inner ear and retina (39, 60). Knocking out of members of the miR-183/96/182 cluster resulted in retinal degeneration (38, 41–43). The retina is not only a sensory organ but is also a self-sustained circadian clock (61). As the only source of photic input to the SCN in mammals, the retina has been proposed to contribute to the overall circadian organization (62). However, experiments from retinal dysfunction in rodents, generated either by a genetic approach or surgical approaches, had divergent observations. Most of them showed no effects on period length and phase, while some of them found phase change or shortened period (63). It has been demonstrated that the miR-183/96/182 cluster–deficient mice have severely degenerated retinas (38, 39). However, in our study the retina of the mutant still exhibited sustained Per2::Luc rhythmicity and normal pupillary light reflex mediated by melanopsin-containing retinal ganglion cells. The circadian mRNA profiling in the present study showed that the retina had dampened expression of core circadian genes in constant darkness but not in light-dark–entrained condition. All of these results in our study suggest that the circadian changes may not be due to a significant contribution from retinal degeneration.
Intriguingly, the miR-183/96/182 cluster–deficient mice showed a significant circadian behavioral abnormality. The homozygous mutants became arrhythmic in constant darkness conditions. Although we observed various circadian alterations at both the cellular and tissue levels in different types of miR-183/96/182 mutants, the cells and tissues still exhibited sustained rhythmicity. Behavior, however, does not necessarily reflect cell-autonomous clock phenotypes (64). The discrepancies between the circadian phenotypes at the behavioral and cellular levels may suggest that the miR-183/96/182 cluster not only regulates circadian rhythms in a cell-autonomous way but may also affect the circadian system downstream of the transcription–translation feedback loops or at a higher neuronal network level. For example, some circadian clock output genes critical to behaviors may be affected by the miR-183/96/182 cluster despite the intact central oscillator, or genes important for coding neuronal circuits downstream of the SCN may be affected so that the signaling between the SCN and the downstream brain regions or peripheral tissues are influenced. Thus, despite confirmation of the cell-autonomous function of the miR-183/96/182 cluster, their functions in circadian neuronal circuits, including coupling, should not be excluded.
In summary, our study revealed that the miR-183/96/182 cluster has functional impacts on circadian rhythms in mammals. They can modulate circadian systems either by direct targeting of the core clock machinery or through a more indirect mechanism that ultimately feeds into the circadian clock. Our genome-wide miRNA screen provides a valuable resource and insights into the layer control of the circadian networks by noncoding RNAs.
Materials and Methods
Animals.
All animal care and experimental procedures complied with University of Southern California guidelines for the care and use of animals and were approved by the University of Southern California Animal Care and Use Committee under protocol #20826.
The miR-183/96/182 cluster gene-trap mice (miR183C GT) were obtained as a gift from Changchun Xiao at Scripps Research. The miR-183CGT/GT; Per2Luc reporter mice were generated by crossing miR-183CGT/+ mice with Per2Luc knock-in mice. All mice were housed in a temperature- and humidity-controlled room with food and water ad libitum. For time-course tissue dissections, mice of each genotype at 2–3 mo of age were euthanized by isoflurane at each circadian time point, and the tissues were isolated, snap frozen, and stored at −80 °C until processing for RNA or protein analyses. For dissections of tissues during the dark phase, euthanasia of mice and dissections of eyes or retinas were performed under infrared illumination by a red lamp, and then the other tissues were dissected under the regular lights.
For other detailed materials and methods, please see SI Appendix, SI Materials and Methods. These methods include circadian assays in mice using wheel-running activity and total activity. Detailed methods of miRNA screen, CRISPR-Cas9 knockout of miRNAs in human cells, luciferase assays, RNA isolation, qRT-PCR, Western blotting, bioluminescence recording of tissues from Per2Luc reporter mice, and statistical analysis are also included.
Supplementary Material
Acknowledgments
We thank Dr. Chaungchun Xiao at Scripps Research for miR-183/96/182 cluster GT mice, and Dr. Amanda Roberts at Scripps Research Animal Models Core Facility for assistance with nonwheel behavioral assessment. We thank Dr. Tsuyoshi Hirota at Nagoya University for giving comments on the manuscript. We also thank our colleagues from the S.A.K. laboratory for discussion and suggestions. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant 5R01DK108087 (to S.A.K.).
Footnotes
Competing interest statement: S.A.K. and C.B.G. are coauthors on a 2017 Guidelines article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020454118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Partch C. L., Green C. B., Takahashi J. S., Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24, 90–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patke A., Young M. W., Axelrod S., Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Lim C., Allada R., Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 16, 1544–1550 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Kojima S., Green C. B., Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry 54, 124–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altuvia Y., et al. , Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 33, 2697–2706 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J., miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marco A., Ninova M., Ronshaugen M., Griffiths-Jones S., Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res. 41, 7745–7752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian M. R., Sonenberg N., Filipowicz W., Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Bartel D. P., Metazoan microRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H., Ingolia N. T., Weissman J. S., Bartel D. P., Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebert L. F. R., MacRae I. J., Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajewsky N., microRNA target predictions in animals. Nat. Genet. 38, S8–S13 (2006). [DOI] [PubMed] [Google Scholar]
- 14.John B., et al. , Human microRNA targets. PLoS Biol. 2, e363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B., Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Sood P., Krek A., Zavolan M., Macino G., Rajewsky N., Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. U.S.A. 103, 2746–2751 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagos-Quintana M., et al. , Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Ludwig N., et al. , Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang T.-C., Mendell J. T., microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 8, 215–239 (2007). [DOI] [PubMed] [Google Scholar]
- 20.O’Connell R. M., Rao D. S., Chaudhuri A. A., Baltimore D., Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Mendell J. T., Olson E. N., MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., Kwon E. J., Tsai L. H., MicroRNAs in learning, memory, and neurological diseases. Learn. Mem. 19, 359–368 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Kowdley K. V., MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics 10, 246–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H.-Y. M., et al. , microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatfield D., et al. , Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23, 1313–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima S., Gatfield D., Esau C. C., Green C. B., MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One 5, e11264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S., Witmer P. D., Lumayag S., Kovacs B., Valle D., MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282, 25053–25066 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Chen R., D’Alessandro M., Lee C., miRNAs are required for generating a time delay critical for the circadian oscillator. Curr. Biol. 23, 1959–1968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shende V. R., Neuendorff N., Earnest D. J., Role of miR-142-3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS One 8, e65300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan X., et al. , Clock-controlled mir-142-3p can target its activator, Bmal1. BMC Mol. Biol. 13, 27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagel R., Clijsters L., Agami R., The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 276, 5447–5455 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Gao Q., Zhou L., Yang S.-Y., Cao J.-M., A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci. Rep. 6, 30070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saus E., et al. , Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum. Mol. Genet. 19, 4017–4025 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Liu A. C., et al. , Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozomara A., Griffiths-Jones S., miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Luo J., Zhang H., Lu J., microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol. Biol. Evol. 33, 2232–2247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carthew R. W., Sontheimer E. J., Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lumayag S., et al. , Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 110, E507–E516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J., et al. , Maturation arrest in early postnatal sensory receptors by deletion of the miR-183/96/182 cluster in mouse. Proc. Natl. Acad. Sci. U.S.A. 114, E4271–E4280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leise T. L., et al. , Voluntary exercise can strengthen the circadian system in aged mice. Age (Dordr.) 35, 2137–2152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang L., et al. , miR-183/96 plays a pivotal regulatory role in mouse photoreceptor maturation and maintenance. Proc. Natl. Acad. Sci. U.S.A. 114, 6376–6381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busskamp V., et al. , miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 83, 586–600 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Zhu Q., et al. , Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 286, 31749–31760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Güler A. D., et al. , Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatori M., et al. , Inducible ablation of melanopsin-expressing retinal ganglion cells reveals their central role in non-image forming visual responses. PLoS One 3, e2451 (2008). Correction in: PLoS ONE4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krol J., et al. , Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Yoo S. H., et al. , PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo S. H., et al. , Period2 3′-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. U.S.A. 114, E8855–E8864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paraskevopoulou M. D., et al. , DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41, W169–W173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reczko M., Maragkakis M., Alexiou P., Grosse I., Hatzigeorgiou A. G., Functional microRNA targets in protein coding sequences. Bioinformatics 28, 771–776 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Ding X., et al. , The role of miR-182 in regulating pineal CLOCK expression after hypoxia-ischemia brain injury in neonatal rats. Neurosci. Lett. 591, 75–80 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Zhang E. E., et al. , A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139, 199–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belter A., et al. , Mature miRNAs form secondary structure, which suggests their function beyond RISC. PLoS One 9, e113848 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turek F. W., Gillette M. U., Melatonin, sleep, and circadian rhythms: Rationale for development of specific melatonin agonists. Sleep Med. 5, 523–532 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Kennaway D. J., Melatonin research in mice: A review. Chronobiol. Int. 36, 1167–1183 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Goto M., Oshima I., Tomita T., Ebihara S., Melatonin content of the pineal gland in different mouse strains. J. Pineal Res. 7, 195–204 (1989). [DOI] [PubMed] [Google Scholar]
- 57.Roseboom P. H., et al. , Natural melatonin ‘knockdown’ in C57BL/6J mice: Rare mechanism truncates serotonin N-acetyltransferase. Brain Res. Mol. Brain Res. 63, 189–197 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Kasahara T., Abe K., Mekada K., Yoshiki A., Kato T., Genetic variation of melatonin productivity in laboratory mice under domestication. Proc. Natl. Acad. Sci. U.S.A. 107, 6412–6417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buhr E. D., Van Gelder R. N., Local photic entrainment of the retinal circadian oscillator in the absence of rods, cones, and melanopsin. Proc. Natl. Acad. Sci. U.S.A. 111, 8625–8630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuhn S., et al. , miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc. Natl. Acad. Sci. U.S.A. 108, 2355–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruan G.-X., Gamble K. L., Risner M. L., Young L. A., McMahon D. G., Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators. PLoS One 7, e38985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tosini G., Pozdeyev N., Sakamoto K., Iuvone P. M., The circadian clock system in the mammalian retina. BioEssays 30, 624–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul K. N., Saafir T. B., Tosini G., The role of retinal photoreceptors in the regulation of circadian rhythms. Rev. Endocr. Metab. Disord. 10, 271–278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu A. C., et al. , Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.