Significance
Precise genome engineering allows researchers to modify gene function, tag endogenous proteins, or model human disease mutations. Here, we adapt prime editing, a new CRISPR-based technology that uses reverse transcription to write precise changes into a target genomic location, for the model organism Drosophila melanogaster. We created and optimized genetic tools to edit three genes (ebony, white, and forked) in cultured cells and in vivo. Importantly, we demonstrate efficient germ-line transmission of a precise edit in ebony. As Drosophila is the first nonmammalian animal to be tested using this method, this study demonstrates the potential wide impact and translatability of prime editing in other animal species.
Keywords: prime editing, Drosophila, genome engineering, pegRNA, CRISPR
Abstract
Precise genome editing is a valuable tool to study gene function in model organisms. Prime editing, a precise editing system developed in mammalian cells, does not require double-strand breaks or donor DNA and has low off-target effects. Here, we applied prime editing for the model organism Drosophila melanogaster and developed conditions for optimal editing. By expressing prime editing components in cultured cells or somatic cells of transgenic flies, we precisely introduce premature stop codons in three classical visible marker genes, ebony, white, and forked. Furthermore, by restricting editing to germ cells, we demonstrate efficient germ-line transmission of a precise edit in ebony to 36% of progeny. Our results suggest that prime editing is a useful system in Drosophila to study gene function, such as engineering precise point mutations, deletions, or epitope tags.
For several decades, genome editing has been a vital and versatile tool to study and modify gene function in model organisms. For example, targeted gene deletions or point mutations can be used to disrupt gene function, create gain-of-function alleles, model human disease mutations, or study complex traits (1). Furthermore, insertions can be used for gene tagging to detect or manipulate endogenous proteins (2). The advent of CRISPR has revolutionized genome editing in animal, plant, and microbe species (1). Researchers working with the model organism Drosophila melanogaster have utilized multiple CRISPR tools to study gene function, including Cas9, Cas12, and Cas13 (3–9). Drosophila is an important model because of its easy genetic manipulation, rich genomic resources, and usefulness to study human disease and disease transmission by insect vectors (10–12). Therefore, new CRISPR-based tools are likely to be functional and have wide-ranging impact on genome editing in this organism.
Prime editing is a recently developed CRISPR-based tool to engineer precise edits in the genome (13). Unlike precise editing using Cas9 and homology-directed repair (HDR), prime editing does not induce double-strand breaks and does not require a DNA template containing the edit. In addition, this method has low off-target effects (13, 14). Prime editing consists of two components, 1) a single guide RNA (sgRNA) with a 3′ extension encoding the edit, referred to as a prime editing guide RNA (pegRNA), and 2) a nickase mutant of Cas9 (nCas9H840A) fused with an engineered Moloney murine leukemia virus (M-MLV) reverse-transcriptase (RT) enzyme, referred to as prime editor 2 (PE2). The pegRNA–PE2 complex induces a nick at the target site and reverse transcribes the edit from the pegRNA into the genome via the RT domain. Like Cas9/HDR, prime editing allows many types of precise edits, such as single-base changes, deletions, or insertions.
Whereas prime editing was originally developed in human cells (13), it has been quickly adopted in other organisms including mice (13, 15, 16) and plants (17–25). Prime editing has been used to help correct disease mutations (13, 26), introduce herbicide-resistant alleles (18, 19, 21, 24), alter plant morphology (21), and model human disease mutations in organoids (16, 27). Here, we develop reagents and optimize conditions to conduct prime editing in an insect species, Drosophila.
Results
Prime Editing in Cultured S2R+ Cells.
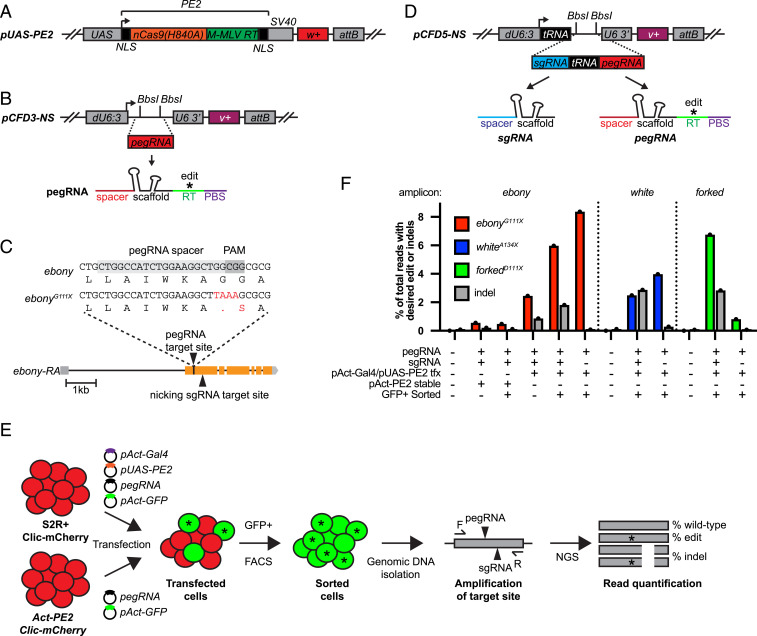
To initially test prime editing in Drosophila, we expressed prime editing components in Drosophila-derived cultured S2R+ cells by transfection. To achieve this, we constitutively expressed PE2 using two alternative expression plasmids. pAct-PE2 expresses PE2 under the Drosophila Actin5c promoter (SI Appendix, Fig. S1A), and pUAS-PE2 (Fig. 1A) expresses PE2 when used in combination with pAct-Gal4 (abbreviated as pAct-Gal4/pUAS-PE2). The latter expression plasmid was used to test high levels of PE2 expression due to signal amplification of the Gal4/UAS system (28). In addition, to express pegRNAs in cells, we constructed an empty expression vector (pCFD3-NS) that lacks the sgRNA scaffold sequence (NS, no scaffold) (Fig. 1B), which is a modified version of the sgRNA expression plasmid pCFD3 (29). A pegRNA cloned into this backbone is written as pCFD3-PE-geneedit.
Fig. 1.
Prime editing in cultured S2R+ cells. (A) Diagram of PE2 expression plasmid pUAS-PE2. attB, phiC31 recombination site; NLS, nuclear localization sequence; PBS, primer-binding site; SV40, 3′ untranslated region; UAS, upstream activating sequence; w+, white+ rescue transgene. (B) Diagram of pCFD3-NS pegRNA expression plasmid. BbsI sites indicate cloning site for pegRNA encoding sequence. dU6:3, U6 promoter; U6 3′, U6 downstream region; v+, vermillion+ rescue transgene. (C) ebony genomic region showing target site and edit (ebonyG111X). (D) Dual sgRNA and pegRNA expression plasmid pCFD5-NS. tRNA, D. melanogaster and O.s. Gly tRNA sequence. (E) Schematic of S2R+ prime editing experiment. (F) Approximate quantification of precise editing and indels from S2R+ transfection experiments by amplicon sequencing. tfx, transfection.
First, we designed a pegRNA to insert a 23-bp barcode (BC) sequence into the ebony gene (SI Appendix, File S1). This strategy was chosen to enable sensitive detection of insertion events by PCR. Four days after transfection of PE2 (pAct-PE2 or pAct-Gal4/pUAS-PE2) and pCFD3-PE-ebony23bpBC into S2R+ cells, genomic DNA was collected and insertion-specific primers were used to amplify the putative insertion (SI Appendix, Fig. S1B). Gel images and Sanger sequencing of PCR products confirmed the presence of the ebony23bpBC insertion using either pAct-PE2 or pAct-Gal4/pUAS-PE2 (SI Appendix, Fig. S1 C and D). To approximate the insertion rate, we performed amplicon sequencing of the target region from transfected cells. Transfections using pAct-Gal4/pUAS-PE2 resulted in an insertion efficiency of 0.42%, whereas transfections using pAct-PE2 were substantially lower (0.006%) (SI Appendix, Fig. S1E). Although our editing efficiencies were lower than reported in mammalian cells with an equivalent-sized insertion (13), these initial results demonstrated that prime editing was possible in Drosophila S2R+ cells.
Next, we designed a pegRNA to introduce a premature stop codon in ebony (ebonyG111X) (Fig. 1C). In addition, we designed an sgRNA for PE2 to nick the nonedited DNA strand, since this approach, known as the prime editor 3 (PE3) system, can bias mismatch repair and boost editing efficiencies in mammalian cells (13, 30, 31). To simultaneously coexpress a pegRNA and sgRNA, we constructed an empty dual-expression vector called pCFD5-NS (Fig. 1D). This vector uses transfer RNA (tRNA) processing to produce both pegRNA and sgRNA, and is a modified version of the multiplex sgRNA expression plasmid pCFD5 (32). A pegRNA/sgRNA pair cloned into this dual-expression backbone is written as pCFD5-PE3-geneedit.
After transfecting S2R+ cells with pCFD5-PE3-ebonyG111X, pAct-Gal4/pUAS-PE2, and pAct-GFP, we isolated green fluorescent protein-positive (GFP+) cells using fluorescence-activated cell sorting (FACS) and performed amplicon sequencing from their genomic DNA (Fig. 1E). Under these conditions, the precise editing efficiency of ebony was 6.0% (Fig. 1F). Furthermore, we found that editing efficiency was ∼2.5× lower without FACS enrichment and, unexpectedly, ∼12× lower using a stable PE2 cell line (Act-PE2) (Fig. 1 E and F). Like in mammalian cells (13), the PE3 system caused a low percentage of insertions and deletions (indels) (0.86%) (Fig. 1F). Finally, we compared editing efficiency using only a pegRNA (pCFD3-PE-ebonyG111X). Unexpectedly, editing efficiency was slightly higher (8.4%) without a nicking sgRNA (Fig. 1F). As expected, excluding the nicking sgRNA reduced the frequency of indels to background levels.
To test prime editing at other genomic sites, we designed pegRNAs to introduce premature stop codons into white and forked (whiteA134X and forkedD111X), along with sgRNAs to nick on the nonedited strand (SI Appendix, Fig. S1F). Editing efficiencies using pegRNA + sgRNA were roughly similar to ebony, producing 2.5 and 6.7% precise editing of white and forked, respectively (Fig. 1F). In addition, results with pegRNA only showed 4.0 and 0.8% precise editing of white and forked, respectively. Therefore, unlike ebony and white, forked editing efficiency was substantially improved by including a nicking sgRNA. In conclusion, using optimized prime editing conditions, we demonstrate precise editing efficiencies in S2R+ cells of ∼4 to 8%.
Prime Editing in Fly Somatic Cells.
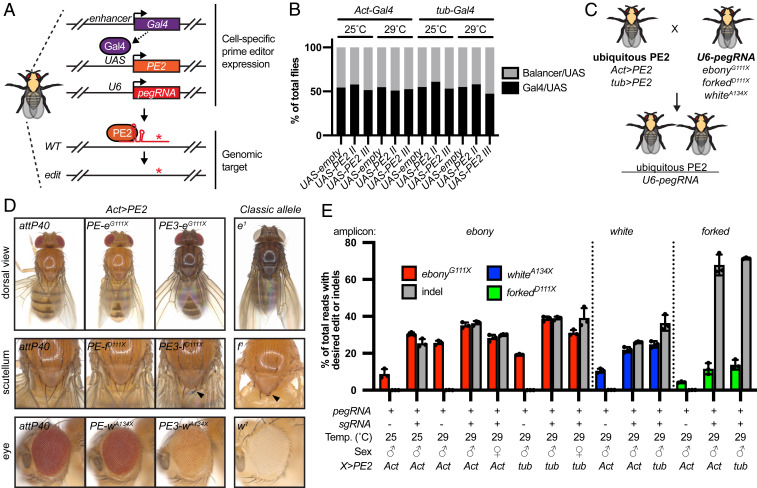
To test prime editing in vivo, we performed crosses between PE2- and pegRNA-expressing transgenic flies. This strategy has been used with Cas9 (5) and Cas12a (6) to edit somatic and germ cells, and it is generally associated with higher editing efficiencies than embryo injection. To express PE2 in vivo, we generated UAS-PE2 transgenic flies, which express PE2 when crossed with a Gal4 driver line (Fig. 2A). In addition, we generated transgenic flies expressing pegRNAs to introduce premature stop codons into ebony, white, and forked. These genes/edits were chosen to enable easy identification of mutant flies with body phenotypes. Transgenic pegRNA flies were created using the same plasmids validated in S2R+ cells (pCFD3-PE-geneedit and pCFD5-PE3-geneedit).
Fig. 2.
Prime editing in somatic cells. (A) Schematic of transgenic expression of prime editing components in flies and editing at an endogenous locus. Enhancer-specific Gal4 directs the tissue-specific expression of PE2. WT, wild type. (B) Quantification of adult fly viability after ubiquitous PE2 expression during all developmental stages and raised at either 25 or 29 °C. Act-Gal4/CyO or tub-Gal4/TM3 were crossed with UAS-PE2 (ChrII), UAS-PE2 (ChrIII), or UAS-empty (negative control), and the percentage of progeny with or without the balancer was calculated. Number of flies scored from left to right: 748, 687, 655, 157, 267, 202, 294, 413, 226, 131, 277, 238. (C) Schematic of genetic crosses between ubiquitous PE2 and pegRNA transgenic flies. (D) Images of adult flies with somatic editing using Act>PE2. Views of the dorsal side of whole adults (Top), scutellum (Middle), and eye (Bottom). Negative control is attP40 and positive control are classical loss-of-function alleles (Right). Females are shown for editing of ebony and forked, and males are shown for white editing. e1: (w1;; TM3,e1/TM6b,e1); f1: (y1, w1, f1). (E) Approximate quantification of precise somatic editing and indel percentage in adult flies by amplicon sequencing. Error bars show mean with SD. n = 3 adult flies.
Many groups have reported toxicity in Drosophila from expression of Cas9 (33–35) and Cas13 (7). To test for toxicity from PE2 expression, we crossed UAS-PE2 to two ubiquitous Gal4 drivers (Act-Gal4 and tub-Gal4) and analyzed the resulting progeny (abbreviated as Act>PE2 and tub>PE2). Act>PE2 and tub>PE2 larvae, pupae, and adults were morphologically normal. Furthermore, the observed number of Act>PE2 and tub>PE2 adult progeny was similar to negative control crosses when raised at 25 or 29 °C and when using two different UAS-PE2 transgenes (Fig. 2B). Finally, Act>PE2 and tub>PE2 flies were fertile and could be propagated as a stock. Therefore, ubiquitous expression of PE2 does not result in obvious toxicity in flies.
Next, we crossed Act>PE2 or tub>PE2 to transgenic pegRNA lines and analyzed progeny for evidence of editing in somatic cells (Fig. 2C). Crosses involving expression of pegRNA only (pCFD3-PE-geneedit) resulted in progeny that were wild-type in appearance (Fig. 2D and SI Appendix, Fig. S2). In contrast, somatic editing using pegRNA + sgRNA (pCFD5-PE3-geneedit) resulted in progeny with mutant phenotypes similar to classical alleles (Fig. 2D and SI Appendix, Fig. S2). In all cases, mutant phenotypes were slightly more severe at 29 compared with 25 °C. To approximate the type and frequency of DNA changes at target sites, we performed amplicon sequencing from single adult fly genomic DNA. For ebony, forked, and white, precise editing efficiency using Act>PE2 was highest with pegRNA + sgRNA, resulting in 35.2, 11.6, and 21.9% reads, respectively, with the intended edit (Fig. 2E). Comparable results were obtained using tub>PE2 (Fig. 2E). In addition, editing of ebony using Act>PE2 was higher at 29 than 25 °C but slightly lower in females compared with males (Fig. 2E). The PE3 system led to a significant percentage of indels at the target site, with an exceptionally high percentage for forked (67.9%). Since both the precise edit and frameshift indels would cause loss of gene function, our sequencing results explain the strong mutant phenotypes when using the PE3 system in somatic cells.
Prime Editing in the Fly Germ Line.
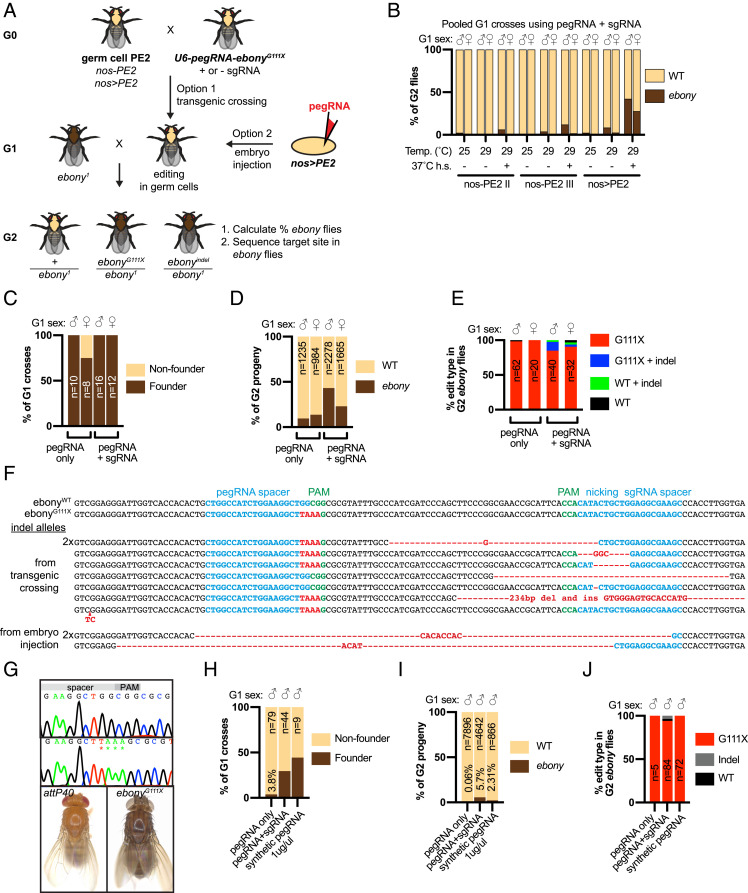
Adapting prime editing to the germ line could enable the creation and propagation of edited fly stocks. To accomplish this, we generated transgenic flies with PE2 under the control of the germ cell-specific nanos (nos) promoter, either as a single transgene (nos-PE2) (SI Appendix, Fig. S3A) or by combination of nos-Gal4 with UAS-PE2 (nos>PE2). To test for evidence of germ-line editing, we first crossed nos-PE2 or nos>PE2 to pCFD5-PE3-ebonyG111X (pegRNA + sgRNA) to generate G1 progeny with editing components expressed in germ cells (Fig. 3A). Next, pools of 10 G1 males or females were crossed with ebony1 and the transmission rate was calculated as the percentage of mutant ebony G2 progeny (ebonymut/ebony1). To optimize editing conditions, we compared transmission via the male versus female germ line and tested raising G1 animals at three different temperatures (25, 29, or 29 °C with 37 °C heat shocks). We observed the highest transmission rate from the male germ line using nos>PE2 and raising G1 animals at 29 °C with 37 °C heat shocks, where 217/514 (42.2%) of G2 progeny were phenotypically ebony (Fig. 3B). Single male G1 crosses using these optimized conditions produced similar results to pooled G1 crosses, with 16/16 (100%) of G1 males yielding a total of 982/2,278 (43.1%) ebony progeny (Fig. 3C, Table 1, and SI Appendix, Fig. S3B). Germ-line transmission of ebony was lower in females (Fig. 3 B–D and SI Appendix, Fig. S3 B and C) and for both sexes when using pegRNA only (pCFD3-PE-ebonyG111X) (Fig. 3 C and D and SI Appendix, Fig. S3C).
Fig. 3.
Prime editing in the germ line. (A) Schematic of genetic crosses to express PE2 and ebonyG111X pegRNA in germ cells and detect transmission of mutations in ebony. (B–E) Quantification of ebony transmission and edit type using transgenic crossing. pegRNA only: pCFD3-PE-ebonyG111X; pegRNA + sgRNA: pCFD5-PE3-ebonyG111X. Sex of G1 parents and sample size are indicated on the graph unless otherwise noted. (B) Quantification of ebony transmission from the germ line of G1 parents to G2 progeny, expressed as the percent of G2 flies with dark cuticle pigmentation (phenotypically ebony). For each condition (temperature raised, PE2 genotype), 10 G1 flies were crossed as a combined pool. The number of G2 flies analyzed was (left to right) 453, 518, 574, 413, 702, 405, 514, 454, 514, 405, 376, 493, 557, 492, 510, 562, 471, 481. (C) Quantification of single G1 flies that transmit at least one ebony progeny. (D) Quantification of G2 ebony progeny transmitted from single G1 crosses in C. (E) Quantification of sequenced edit types in individual G2 flies from single G1 crosses. (F) Sequence structure at the ebony target site, showing wild type, the intended G111X edit, indel alleles, pegRNA and sgRNA spacer (blue), PAM (green), and changes to wild-type sequence (red). (G) Sequence chromatograms (Top) and images (Bottom) of wild-type and ebonyG111X homozygous adult flies. (H–J) Quantification of ebony transmission and edit type using embryo injection of plasmid or synthetic pegRNA. pegRNA only: pCFD3-PE-ebonyG111X; pegRNA + sgRNA: pCFD5-PE3-ebonyG111X. Data from plasmid injections were combined for all concentrations tested. (H) Quantification of single G1 flies that transmit at least one ebony progeny. Data from synthetic pegRNA injection are shown using 1 μg/μL. (I) Quantification of G2 ebony progeny transmitted from single G1 crosses in H. (J) Quantification of sequenced edit types in individual G2 flies from single G1 crosses. Data for synthetic pegRNA were combined for all concentrations tested.
Table 1.
Quantification of ebony germ-line transmission by crossing nos>PE2 with pegRNA transgenes
| G0 cross | G1 parent sex | No. G1 crosses* | Founders, no. (%)† | Total G2 ebony progeny, no. (%) | ebony progeny with G111X edit, no. (%)‡ | Estimated% ebonyG111X progeny§ |
| nos>PE2 x pCFD3-PE-ebonyG111X (pegRNA only) | Male | 10 | 10/10 (100) | 118/1,117 (10.6) | 61/62 (98.4) | 10.6 |
| Female | 8 | 6/8 (75.0) | 135/849 (15.9) | 20/20 (100) | 15.9 | |
| nos>PE2 x pCFD5-PE3-ebonyG111X (sgRNA + pegRNA) | Male | 16 | 16/16 (100) | 982/2,278 (43.1) | 34/40 (85.0) | 36.6 |
| Female | 12 | 12/12 (100) | 382/1,665 (22.9) | 29/32 (90.6) | 20.7 | |
| nos>PE2 II x pCFD5-PE3-ebonyG111X (sgRNA + pegRNA) | Male | 9 | 9/9 (100) | 227/731 (31.1) | N.D. | N.D. |
| nos>PE2 III x pCFD5-PE3-ebonyG111X (sgRNA + pegRNA) | Male | 8 | 8/8 (100) | 243/732 (33.2) | N.D. | N.D. |
N.D., not determined.
Single male or female parents were crossed with w;;TM3,e1/TM6b,e1 and only those resulting in at least 100 progeny were counted.
Founders are defined as parents that produced at least one ebony progeny.
Correct edit is defined as a 1-kb region flanking the target site that contains the G111X allele with no other indels or mutations.
Defined as the % ebony progeny x % ebony progeny with the G111X edit.
To determine if the ebonyG111X edit was transmitted via the germ line, we sequenced the target site in ebony G2 progeny from single fly G1 crosses. Using pCFD5-PE3-ebonyG111X (pegRNA + sgRNA), most G2 ebony progeny from nine independent G1 crosses had the intended G111X edit with no other changes (63/72 [87.6%]) (Fig. 3E, Table 1, and SI Appendix, Fig. S3D). Of the nine G2 ebony progeny with an unintended change, two had an indel, six had the G111X edit and an indel, and one had no changes within a 1-kb region surrounding the target site (Fig. 3 E and F and SI Appendix, Fig. S3D). Furthermore, the frequency of nonprecise edits in G2 progeny appeared similar between individual G1 crosses and between male and female G1 crosses (Fig. 3E and SI Appendix, Fig. S3D). The frequency of correct edits was even higher when using pCFD3-PE-ebonyG111X (pegRNA only), where 81/82 (98.8%) ebony progeny from 11 independent G1 crosses had the intended edit (Fig. 3E, Table 1, and SI Appendix, Fig. S3E). Finally, to demonstrate that the G111X allele produces a loss-of-function phenotype, we generated homozygous ebonyG111X flies, which were viable, fertile, and exhibited dark body pigment (Fig. 3G).
Another method to induce heritable genomic changes in Drosophila is by embryo injection (5). To test this method with prime editing, we introduced pegRNA encoding ebonyG111X into fertilized nos>PE2 embryos by injecting plasmid DNA (pCFD3-PE3-ebonyG111X, pCFD5-PE3-ebonyG111X) or synthesized pegRNA (Fig. 3A). To detect edit transmission, single injected G1 adult males were crossed with ebony1 females and the percentage of mutant ebony (ebonymut/ebony1) flies in their G2 progeny was calculated. For all three injection types, we identified founder G1 crosses that gave rise to at least one ebony fly (Fig. 3H, Table 2, and SI Appendix, Fig. S4A). Injection of pCFD5-PE3-ebonyG111X (pegRNA + sgRNA) gave the highest rate of transmission, where 264/4,642 (5.7%) of G2 progeny were ebony (Fig. 3I and Table 2). Sequencing individual ebony G2 progeny from pCFD5-PE3-ebonyG111X injections revealed that 79/84 (94%) contained the correct edit (Fig. 3J, Table 2, and SI Appendix, Fig. S4B), three had an indel that disrupted the target site (Fig. 3F), and two had no changes. Remarkably, all G2 ebony flies resulting from injection of pCFD3-PE-ebonyG111X or synthesized pegRNA had the correct edit (Fig. 3J, Table 2, and SI Appendix, Fig. S4B). While there were no obvious differences in editing efficiency or edit type using a range of plasmid concentrations (200 to 1,000 ng/µL), germ-line transmission was highest using 1 µg/µL synthetic pegRNA (Table 2 and SI Appendix, Fig. S4A). In summary, our observations demonstrate that prime editing is functional in the fly germ line by transgenic crossing or embryo injection.
Table 2.
Quantification of ebony germ-line transmission by injection into nos>PE2 embryos
| Reagent injected | Concentration, ng/μL | No. embryos injected | No. fertile G1 crosses* | Founders, no. (%)† | Total G2 ebony progeny, no. (%) | No. ebony progeny with G111X edit‡ | Estimated % ebonyG111X progeny§ |
| Plasmid pCFD3-PE-ebonyG111X (pegRNA only) | 1,000 | 50 | 14/16 | 1/14 (7.1) | 1/1,449 (0.07) | 1/1 | 0.07 |
| 800 | 50 | 22/24 | 1/22 (4.5) | 1/2,180 (0.05) | 1/1 | 0.05 | |
| 600 | 50 | 16/18 | 0/16 (0) | 0/1,600 (0) | N/A | N/A | |
| 400 | 50 | 18/21 | 1/18 (5.6) | 3/1,767 (0.17) | 3/3 | 0.17 | |
| 200 | 50 | 9/10 | 0/9 (0) | 0/900 (0) | N/A | N/A | |
| Plasmid pCFD5-PE3-ebonyG111X (sgRNA + pegRNA) | 1,000 | 50 | 10/17 | 1/10 (10) | 45/1,039 (4.33) | 8/10 | 3.5 |
| 800 | 50 | 5/9 | 1/5 (20) | 33/517 (6.38) | 10/10 | 6.38 | |
| 600 | 50 | 7/16 | 2/7 (28.6) | 35/722 (4.85) | 19/19 | 4.85 | |
| 400 | 50 | 15/21 | 6/15 (40) | 119/1,594 (7.47) | 28/30 | 7.0 | |
| 200 | 50 | 7/8 | 3/7 (42.9) | 32/770 (4.16) | 14/15 | 3.9 | |
| Synthetic RNA ebonyG111X pegRNA | 1,000 | 50 | 9/11 | 4/9 (44.4) | 20/866 (2.31) | 19/19 | 2.31 |
| 500 | 50 | 14/15 | 3/14 (21.1) | 22/1,620 (1.36) | 21/21 | 1.36 | |
| 250 | 50 | 12/14 | 3/12 (25) | 15/1,515 (0.99) | 15/15 | 0.99 | |
| 125 | 50 | 18/18 | 3/18 (16.7) | 13/1,994 (0.65) | 13/13 | 0.65 | |
| 62.5 | 50 | 18/19 | 1/18 (5.6) | 4/1,985 (0.20) | 4/4 | 0.20 | |
| 31.5 | 50 | 14/16 | 0/14 (0) | 0/1,659 (0) | N/A | 0 |
N/A, not applicable.
Single injected males were crossed with w;;TM3,e1/TM6b,e1 females and only those resulting in at least 100 progeny were counted.
Founders are defined as parents that produced at least one ebony progeny.
Correct edit is defined as a 1-kb region flanking the target site that contains the G111X allele with no other indels or mutations.
Defined as the % ebony progeny x % ebony progeny with the G111X edit.
For germ-line editing experiments described so far, we used a nos>PE2 fly strain where the nos-Gal4 and UAS-PE2 transgenes are located on different chromosomes. To facilitate the removal of prime editing component transgenes from genomic edits, we constructed fly stocks with nos-Gal4 and UAS-PE2 transgenes recombined onto the same chromosome, one version on chromosome 2 (nos>PE2 II) and one on chromosome 3 (nos>PE2 III). Germ-line editing using these two strains was assessed by crossing with pCFD5-PE3-ebonyG111X (pegRNA + sgRNA). Single male G1 progeny were crossed with ebony1 females and we counted the number of ebony G2 progeny. One hundred percent of G1 males transmitted at least one G2 ebony progeny (Table 1 and SI Appendix, Fig. S5A), and 31 and 33% of G2 progeny were ebony using nos>PE2 II and nos>PE2 III, respectively (Table 1 and SI Appendix, Fig. S5 B and C).
Discussion
Currently, precise genome editing in Drosophila is performed by CRISPR-Cas9 and HDR (5). HDR enables a wide variety of edits, yet is a relatively low efficiency process, and a number of unintended side effects have been documented, such as off-target mutations (36), imprecise integration of the donor DNA (37), or genome rearrangement (38). In addition, HDR is not as useful for tissue-specific editing because HDR events only occur in dividing cells. Furthermore, molecular cloning of donor constructs can be technically challenging and time-consuming.
Prime editing has the potential to address some of these limitations. PE2 uses a nickase mutant of Cas9 (H840A) that induces single-strand breaks, which are known to decrease undesired genome changes and increase HDR:indel ratios (13, 39). In addition, prime editing does not require cell division and functions in postmitotic cultured cells (13). pegRNAs contain both targeting sequence and edit template and are simple to generate, thus facilitating multiple editing experiments in parallel. Furthermore, transgenic pegRNAs enable temporal and spatial control of precise editing, similar to transgenic sgRNAs used for CRISPR-Cas9 knockout (6, 29, 35, 40, 41). One important caveat is that prime editing is currently limited to small (<100-bp) edits that are identified by molecular assays (e.g., PCR).
Precise editing efficiencies in S2R+ cells were ∼4× lower than in mammalian cells, and nicking sgRNAs (PE3 system) did not always increase efficiency. It is not clear if this is due to biological differences (e.g., DNA repair pathways) or technical differences (e.g., transfection method, promoter use, temperature) between these two culture systems. Further optimization of prime editing will likely improve its efficiency in cultured Drosophila cells. Regardless, our results suggest that prime editing can be used as a tool to generate edited S2R+ cell lines.
Ubiquitous PE2 and pegRNA expression in whole animals led to editing efficiencies of 10 to 40% for ebony, white, and forked. Although nicking sgRNAs led to higher editing frequencies, they also caused frequent indels (26 to 68%), which presumably contributed to the robust loss-of-function phenotypes we observed. Conversely, single pegRNAs did not cause obvious mutant phenotypes despite evidence of precise editing (4 to 26%). Therefore, unlike existing transgenic crossing techniques for somatic knockout (6, 29, 32, 34, 35, 40), we were unable to introduce a precise edit in the majority of cells in the fly using ubiquitous expression of prime editing components. Nevertheless, some applications may be compatible with our reported somatic editing efficiencies, such as edits that drive tumorigenesis or mosaic protein tagging.
By restricting expression of PE2 to germ cells, we demonstrated transmission of a precise edit (ebonyG111X) from founder flies to progeny. Transmission rates were higher using transgenic crossing compared with embryo injection, similar to observations with Cas9 (5). For example, transgenic expression of pegRNA + sgRNA in the male germ line gave transmission rates (% ebony progeny × % ebony progeny with correct edit) of ∼36%, compared with 5.3% using embryo injection. These rates are comparable to using HDR to make similarly sized edits in injected embryos (29, 42–44) and facilitates molecular screening of a small number of progeny. However, generating transgenic pegRNA fly lines takes ∼1 mo, and thus delays germ-line editing experiments compared with embryo injection. Furthermore, commercial synthetic pegRNA can be used for embryo injections, which obviates any plasmid cloning. Interestingly, synthetic pegRNA outperformed pegRNA-only plasmid (pCFD3-PE3-ebonyG111X), perhaps due to the chemical modifications that increase RNA stability. Finally, we edited viable body marker genes to facilitate phenotypic analysis, but it will be important to determine the generality of this method by editing additional genes, especially essential genes.
High levels of germ cell PE2 expression via the Gal4/UAS system resulted in higher germ-line transmission rates, perhaps because PE2 is limiting for efficient prime editing, similar to base editors (45). In addition, our observation that higher temperatures increase transmission rates may be due to boosted Gal4/UAS expression (46) and/or PE2 M-MLV RT enzyme activity (47). To avoid causing stress to injected embryos, we did not test raising them at temperatures greater than 25 °C. Further manipulating this temperature sensitivity could be useful to optimize germ-line editing in Drosophila.
Prime editing efficiency can be boosted in mammalian cell lines by including a nicking sgRNA (PE3 system) (13), but recent evidence in injected mouse embryos suggests that indels caused by the double nick occur at undesirable rates, higher than the desired edit (48). Indeed, we observe similar high indel rates in fly somatic cells, in particular with editing of forked. Interestingly, using the PE3 system in germ cells, only 6 to 15% of ebony progeny contained an indel or other nonprecise edit. Perhaps germ cells express different DNA repair components that lead to higher-fidelity repair of double-nicking events. Some ebonymut/ebony1 G2 flies had no obvious changes by sequencing, but this could be due to a large deletion, as has been observed in mouse embryos (48). Therefore, while the PE3 system in its current form may be less desirable in fly somatic cells, it is still useful in the germ line.
Currently, designing an effective pegRNA for precise editing is less straightforward than for sgRNAs. We deliberately selected pegRNA spacer sequences based on previously validated sgRNAs (Methods), but this might have led to better than average editing efficiency. The recent introduction of software tools has made pegRNA design easier (49–52). When possible, we recommend testing editing efficiency in cultured cells before proceeding in vivo. While amplicon sequencing produces high-quality quantitative data, there are faster and cheaper molecular assays such as dinucleotide signature capture (53) or tracking of indels by decomposition (54).
In summary, we have developed genetic tools to express prime editing components in Drosophila, and optimized conditions for efficient editing in cultured cells and in vivo. By designing/cloning a pegRNA and optional sgRNA, Drosophila researchers can generate a wide variety of precise genome modifications such as point mutations, epitope tag insertions, or deletions. Furthermore, the ability to use prime editing in the fly germ line makes it useful to create custom fly strains for gene function analysis. Since CRISPR-based tools are continually engineered for optimal efficiency or new functions, it is likely that future variant prime editor systems will improve this method in Drosophila. Finally, the tools and optimized conditions we developed for prime editing in Drosophila may be useful in other insect species, such as to develop new methods of disease vector control.
Methods
pegRNA and sgRNA Design.
pegRNA spacer sequences were selected based on previously validated sgRNA target sites for ebony (55), white (41), and forked (32). Thirteen base pairs were used for the pegRNA primer-binding site (PBS). For the reverse transcribed (RT) template, we used either a 34-bp (ebony23bpBC) or 18-bp (ebonyG111X, whiteA134X, forkedD111X) region. In all of our pegRNA designs, the pegRNA protospacer adjacent motif (PAM) is disrupted by the edit. Nicking sgRNAs were designed to nick the DNA strand opposite the pegRNA-nicked strand within 40 to 90 bp of the pegRNA nick (ebonyG111X: +57; whiteA134X: +70; forkedD111X: +57). See SI Appendix, File S1 for pegRNA and sgRNA sequences. See SI Appendix, File S3 for additional pegRNA and sgRNA design parameters.
Plasmid Cloning.
Plasmid DNAs were constructed and propagated using standard protocols as follows. PCR fragments were amplified using Phusion polymerase (New England Biolabs; M0530). Plasmids were digested with restriction enzymes at 37 °C for 2 to 16 h. Linearized plasmid and PCR fragments were gel purified using QIAquick columns (28115; Qiagen). Inserts and backbones were assembled using Gibson assembly (New England Biolabs; E2611) or T4 ligation (New England Biolabs; M0202). Gateway-compatible expression and entry vectors were recombined using LR Clonase II (Thermo Fisher Scientific; 11791020). Chemically competent TOP10 Escherichia coli (Invitrogen; C404010) were transformed with plasmids containing either ampicillin or kanamycin resistance genes and selected on lysogeny broth (LB)-agar plates with 100 µg/mL ampicillin or 50 µg/mL kanamycin. ccdB-resistant chemically competent E. coli (Invitrogen; A10460) were transformed with plasmids containing a Gateway cassette (ccdB, Chlor.R.) and selected on LB-agar plates with 100 µg/mL ampicillin and colonies were grown with 100 µg/mL ampicillin and 20 µg/mL chloramphenicol. Plasmid DNA was isolated from bacterial cultures using a QIAprep Spin Miniprep Kit (Qiagen; 27104) and Sanger sequenced at the Dana-Farber/Harvard Cancer Center DNA Resource Core or Genewiz. Oligo and double-strand DNA (dsDNA) sequences are listed in SI Appendix, File S2.
pCFD3-NS (Addgene 149545; DGRC 1528).
pCFD3 (Addgene; 49410) (29) was digested with BbsI (Fermentas; ER1011) and XbaI (New England Biolabs; R0145), which remove the sgRNA scaffold and Drosophila U6 downstream region, and the backbone was purified using a QIAquick column (28115; Qiagen). A gBlock (IDT) containing two BbsI sites and the U6 downstream region was inserted into a digested pCFD3 backbone by Gibson assembly.
pCFD5-NS (Addgene 149546; DGRC 1529).
pCFD5 (Addgene; 73914) (32) was digested with BbsI (Fermentas; ER1011) and XbaI (New England Biolabs R0145), which remove the sgRNA scaffold, Oryza sativa (O.s.) Gly tRNA, sgRNA scaffold, and U6 downstream region. The backbone was purified using a QIAquick column (28115; Qiagen). A gBlock (IDT) containing two BbsI sites and the U6 downstream region was inserted into the digested pCFD5 backbone by Gibson assembly. The D. melanogaster Gly tRNA sequence remains 5′ to the first BbsI site.
pEntr_PE2 (Addgene 149548; DGRC 1526).
PE2 coding sequence was PCR amplified from pCMV-PE2 (Addgene; 132775). pEntr backbone was PCR amplified from pEntr_D-TOPO (Invitrogen; K240020). PE2 coding sequence was cloned into the pEntr backbone by Gibson assembly.
pNos-PE2-attB (Addgene 149549; DGRC 1525).
PE2 coding sequence was PCR amplified from pCMV-PE2 (Addgene; 132775) and gel purified. pNos-Cas9-attB (56) was digested with XbaI/AvrII (New England Biolabs; R0145, R0174) to remove Cas9 sequences and the backbone fragment was gel purified. PE2 coding sequence was inserted into digested pNos-attB by Gibson assembly.
pAct-GW-HygroR (Addgene 149610; DGRC 1524).
Act5c promoter was amplified from pAWF (Murphy laboratory; https://emb.carnegiescience.edu/drosophila-gateway-vector-collection) and gel purified. The backbone was PCR amplified from pMK33-GW (Ram Viswanatha, Harvard Medical School, Boston, MA), using primers that exclude the Metallothionein promoter, and gel purified. The Act5c fragment was inserted into the pMK33-GW backbone by Gibson assembly.
pUAS-PE2-attB (Addgene 149550; DGRC 1527) and pAct-PE2-HygroR (Addgene; 149552) were generated by Gateway reactions between pEntr_PE2 and pWalium10-roe (57) or pAct-GW-HygroR, respectively.
To clone the pCFD3-PE-ebony23bpBC expression plasmid, oligos encoding the spacer, scaffold, and extension were inserted into pCFD3-NS by ligation. Briefly, pCFD3-NS was digested with BbsI and purified on a QIAquick column. Top and bottom oligo pairs encoding either the spacer, scaffold, or extension sequence (SI Appendix, File S2) were designed such that they had overlapping sticky ends with each other and digested pCFD3-NS. Oligo pairs were separately annealed and all were ligated into digested pCFD3-NS using T4 ligase (New England Biolabs; M0202). See SI Appendix, File S3 for detailed cloning protocols.
To clone pCFD3-PE-ebonyG111X, pCFD3-PE-whiteA134X, and pCFD3-PE-forkedD111X, gBlock (IDT) dsDNA fragments encoding the entire pegRNA were inserted into pCFD3-NS by Gibson assembly. Briefly, pCFD3-NS was digested with BbsI and purified on a QIAquick column. gBlock fragments were designed such that the pegRNA sequence was flanked by sequences homologous to digested pCFD3-NS (SI Appendix, File S2). For each gene target, a gBlock was inserted into digested pCFD3-NS by Gibson assembly. See SI Appendix, File S3 for detailed cloning protocols.
To clone pCFD5-PE3-ebonyG111X, pCFD5-PE3-whiteA134X, and pCFD5-PE3-forkedD111X, two overlapping gBlock (IDT) dsDNA fragments encoding the pegRNA and nicking sgRNA were inserted into pCFD5-NS by Gibson assembly. Briefly, pCFD5-NS was digested with BbsI and purified on a QIAquick column. gBlock 1 encoded the sgRNA sequence flanked by a sequence homologous to pCFD5-NS and a partial sequence encoding the O.s. Gly tRNA, and gBlock 2 encoded the pegRNA flanked by the O.s. Gly tRNA and a sequence homologous to pCFD5-NS (SI Appendix, File S2). For each gene target, gBlocks 1 and 2 were inserted together into digested pCFD5-NS by Gibson assembly. See SI Appendix, File S3 for detailed cloning protocols.
Cell Culture.
Drosophila S2R+ cells were cultured at 25 °C using Schneider’s media (21720-024; Thermo Fisher Scientific) with 10% fetal bovine serum (A3912; Sigma) and 50 U/mL penicillin-streptomycin (15070-063; Thermo Fisher Scientific). S2R+ cells were transfected using Effectene (301427; Qiagen) following the manufacturer’s instructions. For all S2R+ cell-culture experiments, we used the PT5 cell line that expresses clic-mCherry (58).
The stably expressing PE2 cell line was generated by transfecting pAct-PE2-HygroR into S2R+ cells. S2R+ cells were transfected in a 6-well dish at a concentration of 1.8 × 106 cells per milliliter (2 mL total volume). Twenty-four hours after transfection, 200 µg/mL hygromycin B (Calbiochem; 400051-1MU) was added to the media. Five days after transfection, cells were resuspended and transferred to a T75 flask with fresh media containing 200 µg/mL hygromycin B. One week later, cells were resuspended, centrifuged at 100 × g for 10 min, and resuspended in 3 mL fresh media containing 200 µg/mL hygromycin B. Resuspended cells were transferred serially into each well of a 6-well plate as a dilution series. Visible colonies were resuspended and expanded after ∼3 wk.
Plasmids were transfected into S2R+ or Act-PE2/S2R+ cells. Briefly, S2R+ or Act-PE2/S2R+ cells were seeded at 600,000 cells per well of a 24-well plate and transfected with a total of 200 ng plasmid DNA. S2R+ cells were transfected with pAct-Gal4 (Y. Hiromi, National Institute of Genetics, Mishima, Japan), pUAS-PE2, pegRNA plasmid, and pAct-GFP (also known as pLib6.6; Ram Viswanatha, Harvard Medical School) at a 3:3:3:1 ratio. Act-PE2/S2R+ cells were transfected with pegRNA plasmid and pAct-GFP at a 3:1 ratio. To increase the chances that GFP+ cells contained prime editing plasmids, we transfected less pAct-GFP plasmid relative to the other cotransfected plasmids.
Four days after transfection, GFP+ cells were isolated by FACS. Cells were first resuspended in culture media and pipetted into a cell-straining FACS tube (352235; Corning) to break up the cell clump. Fifty thousand cells with GFP fluorescence in the 60 to 80th percentile of fluorescence intensity were sorted on an Aria 561 instrument into a single well of a 96-well plate and incubated at 25 °C for 24 h.
Five days after transfection, genomic DNA was isolated from sorted and nonsorted cells using QuickExtract reagent (Lucigen; QE09050). In addition, genomic DNA was isolated from nontransfected S2R+ cells as a negative control. Briefly, culture media were removed and replaced with the same volume of QuickExtract reagent. The solution was resuspended by pipetting, transferred to a PCR strip tube, and incubated at 65 °C for 15 min and then at 98 °C for 2 min.
Fly Culture and Crosses.
Flies were maintained on standard fly food at 25 or at 29 °C when noted. Fly stocks were obtained from individual laboratories or the Bloomington Drosophila Stock Center (BDSC) (indicated as BL). Stocks used in this study are as follows: yw (N.P. laboratory), yw; Sp hs-hid/CyO (derived from BL7757), yw;; TM3,Sb/TM6, Tb (N.P. laboratory), ywf (BL1493), yv nos-phiC31int; attP40 (BL25709), yv nos-phiC31int;; attP2 (BL25710), yw; tub-Gal4 (BL5138), yw; Act-Gal4 (BL4414), yw; nos-Gal4 (BL4442), nos-Gal4 (BL4937), UAS-emptyVK37 (Bellen laboratory, Baylor College of Medicine, Houston, TX).
Transgenic flies generated in this study (submitted to the BDSC) are as follows:
yw; UAS-PE2,w+ attP40 (BL90971)
yw;; UAS-PE2,w+ attP2 (BL90968)
yv; pCFD3-PE-ebonyG111X,v+ attP40 (BL90969)
yv; pCFD3-PE-whiteA134X,v+ attP40 (BL90973)
yv; pCFD3-PE-forkedD111X,v+ attP40 (BL90970)
yv; pCFD5-PE3-ebonyG111X,v+ attP40 (BL90975)
yv; pCFD5-PE3-whiteA134X,v+ attP40 (BL90976)
yv; pCFD5-PE3-forkedD111X,v+ attP40 (BL90978)
yscv; nos-PE2,v+ attP40
yv;; nos-PE2,v+ attP2
Fly stocks with multiple transgenes (submitted to the BDSC) are as follows:
w; Act-Gal4/CyO; UAS-PE2,w+ attP2 (BL90977) (abbreviated as Act>PE2)
w; UAS-PE2,w+ attP40; Tub-Gal4/TM6b (BL90974) (abbreviated as tub>PE2)
w; nos-Gal4; UAS-PE2,w+ attP2 (BL90972) (abbreviated as nos>PE2)
w; nos-Gal4, UAS-PE2,w+ attP40 (BL91349) (abbreviated as nos>PE2 II)
w;; nos-Gal4, UAS-PE2,w+ attP2 (BL91350) (abbreviated as nos>PE2 II)
Transgenic flies were generated by phiC31 integration of attB-containing plasmids into either attP40 or attP2 landing sites. Briefly, plasmid DNA was purified twice on QIAquick columns and eluted in injection buffer (100 µM NaPO4, 5 mM KCl) at a concentration of 200 ng/µL. Plasmid DNA was injected into ∼50 fertilized embryos (yv nos-phiC31int; attP40 or yv nos-phiC31int;; attP2) and resulting progeny were outcrossed to screen for transgenic founder progeny. nos-PE2 and pegRNA insertions were isolated by screening for vermillion+ eye color. UAS-PE2 insertions were isolated by screening for white+ eye color.
For PE2 toxicity experiments, Act-Gal4/CyO or tub-Gal4/TM3-Sb was crossed with either UAS-empty (ChrII), UAS-PE2 (ChrII), or UAS-PE2 (ChrIII) and progeny were raised at either 25 or 29 °C starting at egg deposition. The frequency of PE2-expressing progeny was determined by counting the number of adult nonbalancer progeny and dividing by the total number of flies (no. nonbalancer/no. nonbalancer + no. balancer).
For somatic editing experiments, Act>PE2 or tub>PE2 flies were crossed with pegRNA flies and adult PE2/pegRNA progeny were analyzed for mutant phenotypes.
For germ-line editing experiments involving transgenic crossing, nos-PE2 or nos>PE2 flies were crossed with pCFD5-PE3-eG111X flies and G1 progeny were crossed with TM3,e1/TM6b,e1. To screen different germ cell PE2 genotypes and temperature conditions, G1 crosses were performed as pools of 10 PE2/pegRNA males or females. G1 crosses were performed as single PE2/pegRNA male or female crosses for optimal conditions (nos>PE2, 29 °C + heat shock; h.s.). The phenotypes of G2 progeny were scored as either wild-type or ebony (dark cuticle pigment) on a fly-dissecting scope. To heat shock G1 larvae, we incubated larvae at 37 °C for 1 h in five separate treatments after egg deposition: 24, 48, 72, 96, and 120 h.
For germ-line editing experiments involving embryo injection of either plasmid DNA or synthetic pegRNA, nos>PE2 adult flies were used to lay fertilized eggs on collection plates. Fertilized eggs were injected at the Harvard Medical School Transgenic RNAi Project (TRiP) facility using standard procedures. Injected embryos were raised at 25 °C throughout all developmental stages and resulting single adult males were crossed with TM3,e1/TM6b,e1 females. The phenotypes of progeny were scored the same as by using transgenic crossing. Synthetic pegRNA was synthesized by Agilent (custom pegRNA service) with 3× 2′-O-methyl 3′-phosphorothioate at 3′ and 5′ ends and diluted in H2O. pegRNA sequence: mC*mU*mG*GCCAUCUGGAAGGCUGGGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCGGCAAAUACGCGCUUUAAGCCUUCCAGAUGGmU*mU*mU*U.
Focal stack images of adult flies were obtained using a Zeiss Axio Zoom V16 fluorescence microscope and merged using Helicon Focus 7. Images were then processed using Adobe Photoshop CS6.
Fly genomic DNA was isolated by grinding a single fly in 50 µL squishing buffer (10 mM Tris⋅Cl, pH 8.2, 1 mM ethylenediaminetetraacetate, 25 mM NaCl) with 200 µg/mL proteinase K (3115879001; Roche) and incubating at 37 °C for 30 min and 95 °C for 2 min. For somatic editing experiments, genomic DNA was collected from adult male flies unless otherwise noted. For germ-line editing experiments, genomic DNA was collected from both male and female G2 adult flies. For Sanger sequencing experiments, Taq PCR was used to amplify the target site and purified fragments were sequenced at Genewiz.
Amplicon Sequencing.
Genomic edit sites were amplified by PCR to yield amplicons for next-generation sequencing (NGS). Briefly, 1 µL of S2R+ or fly genomic DNA was used in a PCR using Q5 High-Fidelity DNA polymerase (New England Biolabs; M0491L). Primer pairs (SI Appendix, File S2) were designed to yield amplicons ∼200 to 280 bp in size with the intended editing site located within 100 bp of either the forward or reverse primer. PCR fragments were purified using QIAquick columns (28115; Qiagen) and submitted to the Massachusetts General Hospital Center for Computational and Integrative Biology DNA Core (CRISPR sequencing) or Genewiz (Amplicon-EZ).
NGS reads were analyzed using CRISPResso2 (version 2.0.38) (59). To calculate the percent of reads with the precise edit, we used the following parameters: “–prime_editing_pegRNA_spacer_seq,” “–prime_editing_pegRNA_extension_seq,” “–prime_editing_pegRNA_scaffold_sequence,” “–ignore_substitutions,” and “–discard_indel_reads.” The precise editing frequency was calculated from “CRISPResso_quantification_of_editing_frequency.txt” for the “prime-edited” amplicon, by dividing the no. reads found under these headers - "unmodified"/"reads aligned all amplicons." To determine the percent of reads with indels, we ran CRISPResso2 with standard settings and the “–ignore_substitutions” parameter. The indel frequency was calculated from “CRISPResso_quantification_of_editing_frequency.txt,” as the no. modified/no. reads_aligned.
For S2R+ and fly experiments involving the edits ebonyG111X, whiteA134X, and forkedD111X, we specified a quantification window (“-qwc”) that encompasses the region between the pegRNA and nicking sgRNA (spanning the −6 position relative to the pegRNA PAM to the −6 position relative to the sgRNA PAM) (ebony: 96 to 158; forked: 97 to 159; white: 112 to 187).
Fastq files containing amplicon reads are available from the National Center for Biotechnology Information (NCBI) BioProject (https://www.ncbi.nlm.nih.gov/bioproject/), accession no. PRJNA655492.
Supplementary Material
Acknowledgments
We thank Rich Binari for general lab assistance and help with fly genetics (particularly during the COVID-19 shutdown), the TRiP and Drosophila RNAi Screening Center for help generating transgenic flies, Ram Viswanatha for sharing plasmids and general discussions, Gillian Millburn for discussions on pegRNA transgene nomenclature, Cathryn King for general lab assistance, Cooper Cavers for help isolating transgenic flies, Jorden Rabasco for help with molecular cloning, Andrew Anzalone for advice with synthetic pegRNAs, and Ben Ewen-Campen, Jonathan Zirin, and Thai LaGraff for comments on the manuscript and help with fly stocks. J.A.B. was supported by the Damon Runyon Foundation and a “Training Grant in Genetics” T32 Ruth Kirschstein-National Research Service Award institutional research training grant funded through the NIH/National Institute of General Medical Sciences. This work was also supported by NIH Grants R24OD01984, R24OD030002, and P41GM132087. N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021996118/-/DCSupplemental.
Data Availability.
The raw sequence read data reported in this article have been deposited in the NCBI BioProject (accession no. PRJNA655492).
References
- 1.Pickar-Oliver A., Gersbach C. A., The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandemoortele G., Eyckerman S., Gevaert K., Pick a tag and explore the functions of your pet protein. Trends Biotechnol. 37, 1078–1090 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Venken K. J., et al. , Genome engineering: Drosophila melanogaster and beyond. Wiley Interdiscip. Rev. Dev. Biol. 5, 233–267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korona D., Koestler S. A., Russell S., Engineering the Drosophila genome for developmental biology. J. Dev. Biol. 5, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bier E., Harrison M. M., O’Connor-Giles K. M., Wildonger J., Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Port F., Starostecka M., Boutros M., Multiplexed conditional genome editing with Cas12a in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 117, 22890–22899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchman A. B., et al. , Programmable RNA targeting using CasRx in flies. CRISPR J. 3, 164–176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh N., Depner N., Larson R., King-Jones K., A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 21, 279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viswanatha R., Zaffagni M., Zirin J., Perrimon N., Kadener S., CRISPR-Cas13 mediated knock down in Drosophila cultured cells. bioRxiv: 10.1101/2020.11.01.364166 (1 November 2020). [DOI] [Google Scholar]
- 10.Hales K. G., Korey C. A., Larracuente A. M., Roberts D. M., Genetics on the fly: A primer on the Drosophila model system. Genetics 201, 815–842 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ugur B., Chen K., Bellen H. J., Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9, 235–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raban R. R., Marshall J. M., Akbari O. S., Progress towards engineering gene drives for population control. J. Exp. Biol. 223 (suppl. 1), jeb208181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anzalone A. V., et al. , Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D. Y., Moon S. B., Ko J. H., Kim Y. S., Kim D., Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 48, 10576–10589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sürün D., et al. , Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes (Basel) 11, 511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., et al. , Efficient generation of mouse models with the prime editing system. Cell Discov. 6, 27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Li J., Chen J., Yan L., Xia L., Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13, 671–674 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Xu W., et al. , Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 13, 675–678 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Hua K., Jiang Y., Tao X., Zhu J. K., Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 18, 2167–2169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Q., et al. , Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Butt H., et al. , Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 18, 2370–2372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X., et al. , Plant prime editors enable precise gene editing in rice cells. Mol. Plant 13, 667–670 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Veillet F., et al. , Prime editing is achievable in the tetraploid potato, but needs improvement. bioRxiv: 10.1101/2020.06.18.159111 (18 June 2020). [DOI]
- 24.Jiang Y.-Y., et al. , Prime editing efficiently generates W542L and S621I double mutations in two ALS genes of maize. Genome Biol. 21, 257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., et al. , Spelling changes and fluorescent tagging with prime editing vectors for plants. bioRxiv: 10.1101/2020.07.16.206276 (17 July 2020). [DOI] [PMC free article] [PubMed]
- 26.Rousseau J., et al. , Specific mutations in genes responsible for Alzheimer and for Duchenne muscular dystrophy introduced by base editing and PRIME editing. bioRxiv: 10.1101/2020.07.31.230565 (1 August 2020). [DOI]
- 27.Schene I. F., et al. , Prime editing for functional repair in patient-derived disease models. Nat. Commun. 11, 5352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty U., Alani E., Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Res. 16, fow071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modrich P., Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 281, 30305–30309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huynh N., Zeng J., Liu W., King-Jones K., A Drosophila CRISPR/Cas9 toolkit for conditionally manipulating gene expression in the prothoracic gland as a test case for polytene tissues. G3 (Bethesda) 8, 3593–3605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Port F., et al. , A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 9, e53865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poe A. R., et al. , Robust CRISPR/Cas9-mediated tissue-specific mutagenesis reveals gene redundancy and perdurance in Drosophila. Genetics 211, 459–472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll D., Staying on target with CRISPR-Cas. Nat. Biotechnol. 31, 807–809 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Skryabin B. V., et al. , Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9-mediated genome editing events. Sci. Adv. 6, eaax2941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ledford H., CRISPR gene editing in human embryos wreaks chromosomal mayhem. Nature 583, 17–18 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Maizels N., Davis L., Initiation of homologous recombination at DNA nicks. Nucleic Acids Res. 46, 6962–6973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meltzer H., et al. , Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat. Commun. 10, 2113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo S., Ueda R., Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gratz S. J., et al. , Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi T., Sloutskin A., Kalifa R., Juven-Gershon T., Gerlitz O., Efficient in vivo introduction of point mutations using ssODN and a co-CRISPR approach. Biol. Proced. Online 22, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge D. T., Tipping C., Brodsky M. H., Zamore P. D., Rapid screening for CRISPR-directed editing of the Drosophila genome using white coconversion. G3 (Bethesda) 6, 3197–3206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koblan L. W., et al. , Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duffy J. B., GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 34, 1–15 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Baranauskas A., et al. , Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng. Des. Sel. 25, 657–668 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Aida T., et al. , Prime editing primarily induces undesired outcomes in mice. bioRxiv: 10.1101/2020.08.06.239723 (6 August 2020). [DOI]
- 49.Bhagwat A. M., et al. , multicrispr: gRNA design for prime editing and parallel targeting of thousands of targets. Life Sci. Alliance 3, e202000757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H. K., et al. , Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol., 10.1038/s41587-020-0677-y (2020). [DOI] [PubMed] [Google Scholar]
- 51.Chow R. D., Chen J. S., Shen J., Chen S., A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng., 10.1038/s41551-020-00622-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu J. Y., et al. , PrimeDesign software for rapid and simplified design of prime editing guide RNAs. bioRxiv: 10.1101/2020.05.04.077750 (4 May 2020). [DOI] [PMC free article] [PubMed]
- 53.Billon P., et al. , Detection of marker-free precision genome editing and genetic variation through the capture of genomic signatures. Cell Rep. 30, 3280–3295.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sentmanat M. F., Peters S. T., Florian C. P., Connelly J. P., Pruett-Miller S. M., A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8, 888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Port F., Muschalik N., Bullock S. L., Systematic evaluation of Drosophila CRISPR tools reveals safe and robust alternatives to autonomous gene drives in basic research. G3 (Bethesda) 5, 1493–1502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren X., et al. , Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. U.S.A. 110, 19012–19017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perkins L. A., et al. , The Transgenic RNAi Project at Harvard Medical School: Resources and validation. Genetics 201, 843–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neumüller R. A., et al. , Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics 190, 931–940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clement K., et al. , CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence read data reported in this article have been deposited in the NCBI BioProject (accession no. PRJNA655492).