Significance
Genetic heterogeneity is a significant driver of antibiotic resistance in bacteria. Understanding copy number (CN) heterogeneity is important because minority subclones with increased CN can drive resistance during antibiotic exposure, but revert and escape detection during clinical susceptibility testing. Despite its clinical relevance, CN variation has eluded quantification at single-molecule resolution. Here, we report nanopore sequencing of arylomycin-resistant mutants carrying tandem repeats ranging in size from 4.8 to 50.0 kb and encompassing the arylomycin target gene lepB. Reads spanning individual repeat arrays show vast differences in CN, underscoring the importance of amplifications in driving the emergence of genetic heterogeneity. This is a direct observation of cell-to-cell CN differences in an antibiotic-resistant bacterial population.
Keywords: antibiotic resistance, heterogeneity, amplification, optimized arylomycins
Abstract
Tandem gene amplification is a frequent and dynamic source of antibiotic resistance in bacteria. Ongoing expansions and contractions of repeat arrays during population growth are expected to manifest as cell-to-cell differences in copy number (CN). As a result, a clonal bacterial culture could comprise subpopulations of cells with different levels of antibiotic sensitivity that result from variable gene dosage. Despite the high potential for misclassification of heterogenous cell populations as either antibiotic-susceptible or fully resistant in clinical settings, and the concomitant risk of inappropriate treatment, CN distribution among cells has defied analysis. Here, we use the MinION single-molecule nanopore sequencer to uncover CN heterogeneity in clonal populations of Escherichia coli and Acinetobacter baumannii grown from single cells isolated while selecting for resistance to an optimized arylomycin, a member of a recently discovered class of Gram-negative antibiotic. We found that gene amplification of the arylomycin target, bacterial type I signal peptidase LepB, is a mechanism of unstable arylomycin resistance and demonstrate in E. coli that amplification instability is independent of RecA. This instability drives the emergence of a nonuniform distribution of lepB CN among cells with a range of 1 to at least 50 copies of lepB identified in a single clonal population. In sum, this remarkable heterogeneity, and the evolutionary plasticity it fuels, illustrates how gene amplification can enable bacterial populations to respond rapidly to novel antibiotics. This study establishes a rationale for further nanopore-sequencing studies of heterogeneous cell populations to uncover CN variability at single-molecule resolution.
Antibiotic resistance is a serious threat to human health. Classically, emergence of resistance during antibiotic therapy has been attributed to expansion of preexisting clones that confer resistance via stable DNA mutations. Under antibiotic selection, resistant cells continue to divide, while their susceptible counterparts do not. If selection continues, resistant cells become predominant, forming a homogeneous population that is genetically stable when selection is removed. More recently, a growing appreciation has emerged for the contribution of unstable resistance determinants. In this case, expansion of a resistant clone in the absence of antibiotics can generate a heterogenous population of susceptible and resistant cells (1, 2). This exacerbates the potential for unexplained treatment failure because resistant subpopulations are available for enrichment during antibiotic exposure, but can revert and escape detection once selection is removed. An understanding of the genetic heterogeneity underpinning these dynamics is needed to inform durable therapeutic strategies.
Gene amplification is a common mechanistic explanation for unstable resistance phenotypes, both in vitro and in clinical isolates (3–7). Amplification instability is driven by the stochasticities associated with the frequent formation and loss of the amplifications themselves, selection, and the costs that maintenance of amplified DNA impose on fitness (8, 9). We expect the complex interplay among these factors to manifest as cell-to-cell differences in copy number (CN) that emerge during propagation of amplification-containing clones, but this has not been explicitly investigated at the level of individual DNA molecules.
With continuing advances in long-read, single-molecule sequencing technologies, we are now able to interrogate tandem amplifications at single-molecule resolution. The MinION (Oxford Nanopore Technologies [ONT]) nanopore sequencer can produce ultralong reads (reads of length ≥100 kb sum to at least half the total sequenced bases) capable of spanning entire tandem repeat arrays (10), and because nanopore sequencing directly detects the input molecule without the need for PCR amplification, differences in CN among individual array-spanning reads reflect CN variation as it exists in the underlying population. Here, we present MinION sequencing of mutants resistant to a recently discovered arylomycin derivative, G0775 (11). We show that tandem amplification of the target gene lepB is a mechanism of unstable arylomycin resistance and demonstrate that the observed instability is independent of RecA activity. This instability fosters genetic diversity via generation of vast cell-to-cell differences in lepB CN. We discuss how genetic heterogeneity could complicate the assessment of resistance in clinical settings.
Results and Discussion
Amplification of lepB Is a Mechanism of Arylomycin Resistance.
In previous work (11), we characterized the activity of G0775, an optimized arylomycin analog with potent activity against the essential bacterial type I signal peptidase LepB and no preexisting resistance in clinical isolates. In that study, we explored the genetic basis of resistance in mutants of Escherichia coli isolated in the presence of inhibitory concentrations of G0775 using Illumina short-read whole-genome sequencing (WGS) data. We identified single-nucleotide variants and short indels, but did not explore other types of genomic aberrations, including tandem amplifications. Of 272 mutants selected in the presence of G0775, the vast majority (94%) contained single-nucleotide polymorphisms (SNPs) in lepB that resulted in amino acid substitutions within the arylomycin binding pocket. However, one mutant exhibited a 16-fold shift in G0775 susceptibility (minimum inhibitory concentration [MIC] = 4; wild-type [WT] MIC = 0.25), but showed no evidence of point mutations. A screen for structural variants using short-read Illumina data identified a 16-fold increase in sequencing coverage corresponding to amplification of an ∼4.8-kb region containing lepB (Fig. 1A). Identification of read pairs consistent with the sequence junction that would result from a simple head-to-tail arrangement of the region confirmed that the amplification exists in direct tandem configuration. These data suggest that the observed increase in lepB dosage produces LepB protein levels high enough to overcome inhibition by the G0775 available in culture and are consistent with the previous observation that experimental overexpression of lepB in uropathogenic E. coli CFT073 decreases susceptibility to G0775 by eightfold (LepBHigh MIC = 1; WT MIC = 0.125) (11). Resistance caused by amplification-mediated overproduction of target molecules is not unique to arylomycins and has been observed for numerous classes of antibiotics (8, 12, 13)
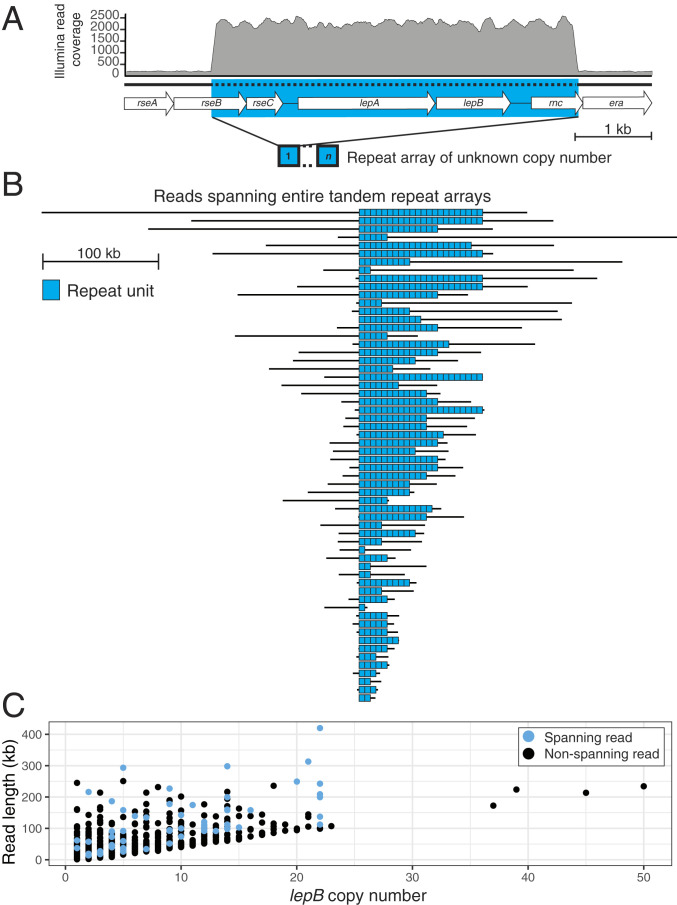
Fig. 1.
Variable CN of an ∼4.8-kb tandem repeat containing bacterial signal peptidase gene lepB. (A) E. coli ATCC strain 25922 gene map, with depth of mapped Illumina reads shown. The blue rectangle indicates the tandem repeat unit. (B) Sixty MinION reads that traverse the entire tandem array, with black horizontal lines representing reads and line length proportional to read length. Individual copies of the ∼4.8-kb repeat unit are show as blue rectangles. (C) Number of lepB copies per MinION read, for reads that traverse or partially overlap with the tandem array.
lepB Amplification Mutants Exhibit Extreme CN Heterogeneity.
Using read coverage as proxy for CN, the Illumina sequencing data show that the average number of lepB copies in the population of this mutant strain is 16. This could be indicative of a population in which each cell contains precisely 16 lepB copies, but more likely reflects a variable distribution of lepB CNs with a population average of 16. To distinguish between these possibilities, three replicates of the initial culture were sequenced with the MinION portable sequencer to generate ultralong sequencing reads (SI Appendix, Table S1). Reads were pooled, and the number of tandem repeated units on individual reads that spanned the entire tandem repeat array were counted (SI Appendix, Table S2). Overall, we generated 5.7 gigabases of sequencing data and identified 60 spanning reads (SI Appendix, Table S1). To avoid underestimates of the number of repeated units resulting from the complex error profiles of MinION reads, dot-plot analyses were used to infer repeat CN (SI Appendix, Figs. S1 and S2 and Materials and Methods). Among 60 spanning reads ranging in size from 13,359 to 419,870 bases, the number of lepB copies was nonuniformly distributed between 1 and 22, with a mean value of 10 (SD = 6) and a median of 9 (Fig. 1B). It is notable that the mean CNs of the population inferred from MinION data are lower than those inferred using Illumina data, 10 compared to 16. This could reflect amplification losses within the population resulting from the overnight subculture of the initial population in the absence of G0775 prior to MinION sequencing. However, a lack of reads long enough to span the highest CN repeat arrays present in the culture is also a likely contributing factor. The population-level dynamics of amplification gain and loss are discussed in Instability of lepB Amplifications Fuels CN Heterogeneity.
As a control, we also MinION-sequenced WT E. coli American Type Culture Collection (ATCC) strain 25922. Among 205 reads which span the region amplified in the mutant, all but one showed one copy of lepB as expected. The exception showed two copies resulting from an apparent inverted duplication (SI Appendix, Fig. S3). This is consistent with previous estimates of the frequency of amplifications of specified genes in unselected cultures, which can be as high as 1 in 100 (9), and underscores the utility of MinION sequencing for identifying variability arising from alterations at single-molecule resolution.
Among mutant reads that partially overlap with, but do not span, the entire repeat array, we identified a read 234,192 bases in length with at least 50 adjacent copies of the repeat (Fig. 1C and SI Appendix, Fig. S4). A further four nonspanning reads ranging in size from 106,871 to 223,798 bases supported a minimum number of tandem repeated units greater than the maximum of 22 indicated by the spanning reads. (Fig. 1C). The MinION data add direct evidence to the growing recognition that clonal bacterial population can exhibit substantial heterogeneity with respect to CN (3, 7) and are consistent with highly dynamic temporal expansion and/or contraction of the lepB repeat array (Fig. 1A) during clonal evolution.
CN heterogeneity in bacteria is of special interest because it is a likely explanation for isolates that exhibit phenotypic heterogeneity with respect to antibiotic resistance, so-called heteroresistance (3, 5–7). The underlying hypothesis is that variable gene dosages exist within such populations, something that has not been directly ascertained at the level of individual DNA molecules. This makes the observation of vastly distributed lepB CNs particularly notable. Although long-read sequencing of bacterial isolates is increasingly common, previous studies involving MinION sequencing of isolates containing gene amplifications have either not investigated CN at the level of individual reads (14) or have failed to produce reads long enough to span the repeat array under investigation (4). Because a 16-fold amplification involving an ∼4.8-kb unit is easily spanned by a 100-kb+ long read (4.8 kb × 16 = 76.8 kb), the G0775-resistant mutant with its inferred average lepB CN of 16 is particularly well-suited to exploring CN heterogeneity with nanopore sequencing. Nonetheless, the presence of a nonspanning read with 50 tandem repeated units suggests that we have not recovered the upper limit of CN range within the population. Even with ultralong reads, characterization of heterogeneity becomes less tractable the larger and/or higher-order the amplifications are, both of which can be substantial. A recent screen of DNA amplifications in 41 clinical isolates identified amplification units as large as 279.8 kb and amplification levels as least as high as 69.9-fold (6).
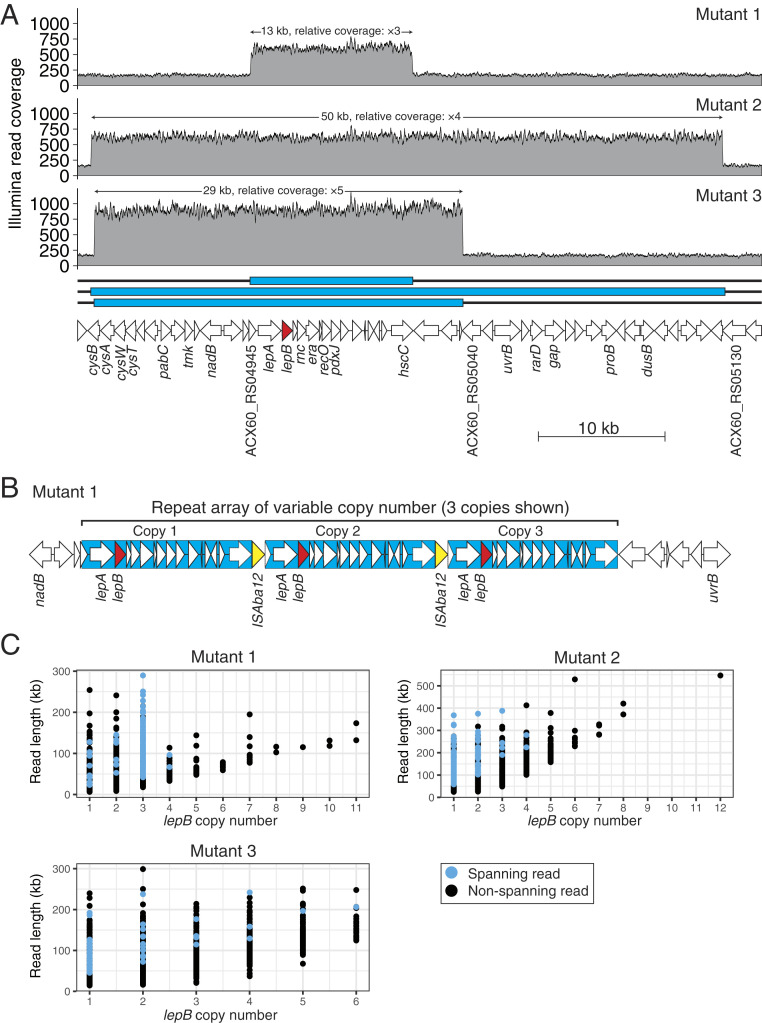
To further investigate CN heterogeneity, we expanded our study to include Acinetobacter baumannii, another clinically important Gram-negative pathogen (15). Resistant mutants were generated by plating A. baumannii ATCC strain 17978 cells on inhibitory concentrations of G0775. Of 35 mutants selected, 23 (66%) contained SNPs in lepB. Ilumina sequencing of the remaining 12 revealed 3 mutants with amplifications encompassing lepB (Fig. 2A and SI Appendix, Table S3). A threefold 13-kb amplification resulted in a fourfold (MIC = 8; WT MIC =2) shift in G0775 susceptibility, while both a fourfold 50-kb amplification and a fivefold 29-kb amplification resulted in an eightfold shift (MIC = 16). Like the amplification found in E. coli, the 50- and 29-kb amplifications exist in direct tandem configuration. In contrast, ONT sequencing showed that the 13-kb amplified region is interleaved with insertion sequence (IS), ISAba12, highlighting the utility of long-read sequencing to identify compound combinations of structural variants—in this case, IS transposition and duplication (Fig. 2B). It is well established that many duplications arise by nonequal homologous recombination between long, direct repeats (8, 9). It is notable that, among the 272 E. coli and 35 A. baumannii resistant mutants, we did not find any amplifications containing lepB flanked by direct repeats. This raises the possibility that there is a genomic feature located in the vicinity of lepB on both the E. coli and A. baumannii chromosomes for which amplification is not tolerated. This hypothesis requires further investigation, but would explain why lepB amplifications are observed infrequently and involve relatively short regions of the genome.
Fig. 2.
Variable CN of amplified regions containing lepB in three arylomycin-resistant A. baumannii mutants. (A) A. baumannii ATCC strain 17978 gene map, with depth of mapped Illumina reads shown. The solid blue rectangles indicate the sizes of the amplified chromosomal regions detected. The lepB gene is shown in red. (B) Schematic representation of the 13-kb amplified region in mutant 1 with interleaving IS element, ISAba12, shown in yellow. Genes interrupted by the ISAba12 insertion are shown with dotted outlines. (C) Number of lepB copies per MinION read, for reads that traverse or partially overlap with repeat arrays.
As in E. coli, ONT sequencing (SI Appendix, Table S4) directly showed variable lepB dosage among cells in all three A. baumannii mutant populations (Fig. 2C). Because all mutants showed variable CNs, herein, we focus our discussion predominantly on the mutant containing the 50-kb amplification, the largest of the identified repeat units. Among the 157 spanning reads identified in this mutant, which ranged in size from 58,284 to 388,034 bases, the number of lepB copies is distributed between one and four (Fig. 2C). However, the nonspanning reads support a wider distribution of CNs and indicate the existence of one or more cells in the population with as many as 12 copies of lepB. We identified a read 546,338 bases in length showing 12 adjacent copies of the repeat unit. A further 72 nonspanning supported repeat-unit CNs greater than four, the population average inferred from the Illumina data. There is an interesting preponderance of spanning reads showing three repeat copies in the mutant bearing the 13-kb amplification, indicating that some populations are more heterogenous than others (Fig. 2C). This may be the result of a specific mechanistic limitation related to the involvement of the IS element or reflect a balancing of the cost–benefit equation. Overall, these data show that it is possible to directly observe cell-to-cell CN differences using single-molecule sequencing in populations containing amplifications of at least 50 kb in size and extend our findings to an additional Gram-negative species.
Instability of lepB Amplifications Fuels CN Heterogeneity.
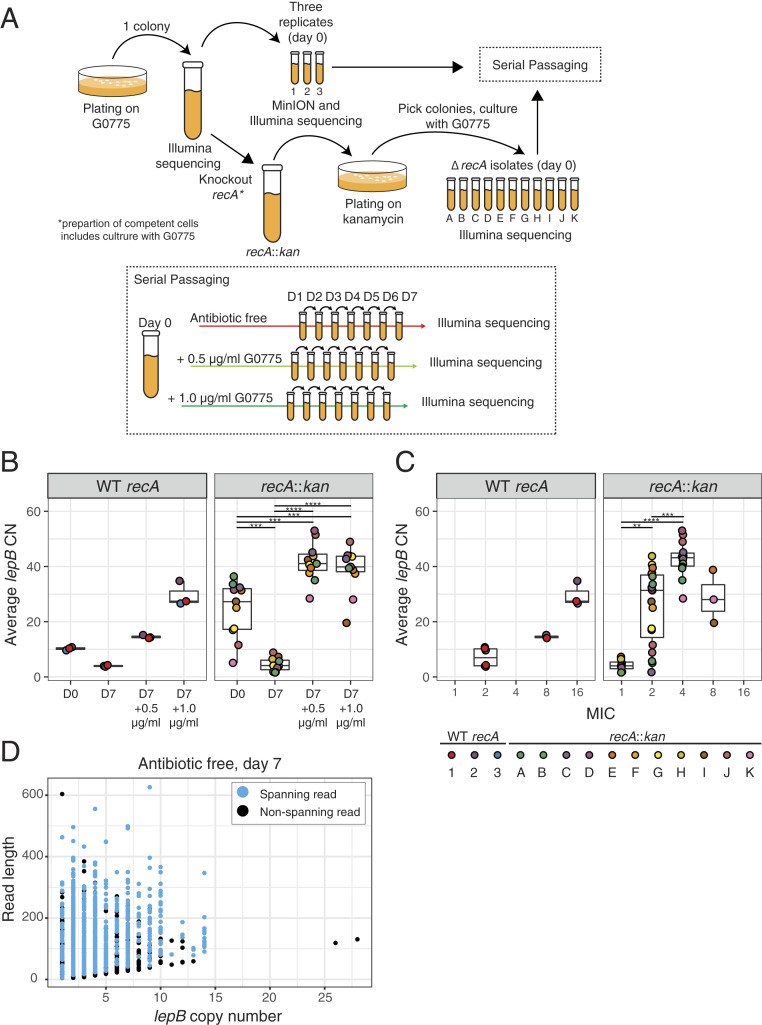
Because heterogeneity is a likely consequence of genomic instability, we sought to better understand the population-level dynamics of lepB CN expansion and contraction over time with different selection regimes in the E. coli amplification mutant. The three samples of the initial population were serially passaged in the presence and absence of subinhibitory concentrations of G0775 for 7 d and subjected to Illumina WGS (Fig. 3A). The amplification segregated to lower average CNs in the absence of selection, which is suggestive of fitness costs associated with carrying extra copies of lepB. By day 7, CNs had decreased from 10 to 11 to 4 (Table 1, Fig. 3B, and SI Appendix, Fig. S5). In contrast, after 7-d serial passage in the presence of 0.5 µg/mL and 1.0 µg/mL G0775, average CNs increased to 14 to 15 and 27 to 35, respectively. This suggests that increased lepB CN leads to increased resistance (Table 1, Fig. 3B, and SI Appendix, Fig. S5). Indeed, we found that higher CNs were associated with higher MICs (Fig. 3C). However, it should be noted that we do not expect a perfect correlation between MIC and CN because the CNs derived from Illumina data reflect the average CN of the population, while MICs as determined here pertain only to the most resistant cells and are, therefore, easily skewed by the presence of high-CN outliers in the population. The passaging results, taken together with the nanopore-based distributional findings, emphasize the importance of selective pressure and the intrinsic instability of amplifications in the ongoing plasticity of genetically heterogenous populations. Cell-to-cell differences in CN can be explained by recurrent gain and loss of amplifications by many cells in the population. Meanwhile, selection with G0775 enriches for cells that maintain or increase resistance, thereby pushing populations toward higher average CNs.
Fig. 3.
Serial passaging of E. coli mutants containing lepB amplifications. (A) Experimental design of serial-passaging experiment in which recA::kan and recA WT amplification mutants were passaged for 7 d with or without LepB inhibitor G0775. Passages performed in the presence of G0775 used a concentration of either 0.5 or 1.0 μg/mL. (B) Boxplots of the average number of lepB copies per cell in mutant populations, as determined by Illumina sequencing before and after 7-d passage—the boxes extend from the 25th to the 75th percentile and encompass the median (horizontal line). **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.001 (Wilcoxon rank-sum test with Holm correction for multiple comparisons); no asterisk means not significant. (C) Boxplots of the average number of lepB copies per cell in mutant populations by the MIC of G0775. No asterisk means not significant (Wilcoxon rank-sum test). (D) Number of lepB copies per MinION read, for reads that traverse or partially overlap with the lepB tandem repeat array after 7 d of passage without G0775.
Table 1.
Genomic variation and resistance of E. coli mutant populations serial passaged for 7 d either with or without LepB inhibitor, G0775
| Strain | Day 0 | Day 7 | ||||||||||
| Passaged in 0 μg/mL | Passaged in 0.5 μg/mL | Passaged in 1 μg/mL | ||||||||||
| MIC† | lepB CN‡ | SVNs§ and short indels¶ | MIC† | lepB CN‡ | SVNs§ and short indels¶ | MIC† | lepB CN‡ | SVNs§ and short indels¶ | MIC† | lepB CN‡ | SVNs§ and short indels¶ | |
| WT | 0.25 | 1 | None | — | — | — | — | — | — | — | — | — |
| ΔrecA | 0.25 | 1 | None | — | — | — | — | — | — | — | — | — |
| Replicate 1 | 2 | 10 | — | 2 | 4 | dgcQ Thr35Asn (74%), ybcM Glu72Asp (70%) | 8 | 14 | — | 16 | 27 | — |
| Replicate 2 | 2 | 11 | — | 2 | 4 | dgcQ Thr35Asn (82%), ybcM Glu72Asp (65%) | 8 | 15 | — | 16 | 35 | yhjK Q329* (68%) |
| Replicate 3 | 2 | 10 | — | 2 | 4 | dgcQ Thr35Asn (75%), ybcM Glu72Asp (78%) | 8 | 14 | yeaJ Ser48Thr (83%), rfbX Val392Gly (67%) | 16 | 27 | — |
| A recA::kan | 2 | 34 | 2 | 6 | — | 4 | 35 | rrmA K43* (60%), wecA Q43* (61%), | 4 | 39 | yhjK Q188* (84%) | |
| B recA::kan | 2 | 36 | — | 1 | 2 | — | 4 | 41 | manC Tyr28* (100%) | 4 | 40 | rpsB Thr102Ala (89%), lrhA Glu148* (96%), rpoC Arg271Leu (93%) |
| C recA::kan | 2 | 32 | — | 2 | 2 | yeaJ Arg254Ser (92%) | 4 | 45 | rplI Met15Gln (90%), lrhA Trp213Arg (97%) | 4 | 43 | — |
| D recA::kan | 2 | 32 | 1 | 3 | g.3112313C > T - upstream of DR76_RS15030 (94%) | 4 | 53 | 4 | 44 | wecA Gln44* (89%), lon p.Ala170fs (91%) | ||
| E recA::kan | 2 | 31 | 1 | 2 | lrhA p.Ala34_Val35insAlaAlaAla (54%) | 2 | 40 | lrhA Gln45His (82%), kpsS Tyr58* (58%) | 8 | 39 | ||
| F recA::kan | 2 | 25 | 1 | 4 | 2 | 38 | acrR Ala163Thr (99%), emrR p.Ile27fs (100%) | 2 | 37 | lrhA Glu148* (90%), g.2910845C > A - upstream of emrR (88%) | ||
| G recA::kan | 2 | 17 | folP Trp92Arg (100%) | 1 | 4 | folP Trp92Arg (100%), yliF D299Y (53%), g.4757212A > C, between clpX and clpP (77%) | 2 | 41 | folP Trp92Arg (100%) | 4 | 44 | folP Trp92Arg (100%) |
| H recA::kan | 2 | 17 | 1 | 6 | 2 | 44 | 4 | 44 | ||||
| I recA::kan | 2 | 27 | g.4953334_4953335insG - upstream of DR76_RS24600 (80%) | 1 | 7 | lrhA p.Gly234fs (81%) | 4 | 42 | mutS p.Asn831fs (81%), fadD p.Gln278fs (88%), ksgA Thr94Pro (81%), tauA Ala140Val(99%), rutA Val148Ile (83%), nrdA Asp598Asn (51%), tmcA Arg203Cys (74%), hycE Thr84Ala (78%), fadh Val52Met (50%), DR76_RS04500 p.Tyr151fs (96%), DR76_RS07985 p.Glu27fs (59%), g.4953334_4953335insG - upstream of DR76_RS24600 (85%), c.4998473del - dowstream of rpiB (74%) | 8 | 20 | g.4953334_4953335insG - upstream of DR76_RS24600 (77%) |
| g.4953334_4953335insG - upstream of DR76_RS24600 (80%) | ||||||||||||
| J recA::kan | 2 | 12 | — | 2 | 9 | — | 4 | 52 | yfcN Arg76Leu (87%) | 4 | 49 | — |
| K recA::kan | 2 | 5 | 1 | 5 | 4 | 28 | eutT Val217Leu (96%), emrR p.Pro131fs (99%), manC p.Asp259_Ile270del (80%) | 8 | 28 | |||
Mutants were selected as described in Fig. 3A. Only variants present in at least 50% of mapped reads at the site of variation are reported, and the frequency of each variant is indicated in parentheses.
MIC.
Average CN estimated using Illumina sequencing data.
Single nucleotide variants.
Insertions/deletions.
In a clinical setting, subpopulations containing amplifications could slip through standard antibiotic-susceptibility diagnostics, despite a strong predilection toward outgrowth during subsequent antibiotic therapy, leading to the incorrect classification of amplification-containing strains as drug-susceptible (16). For this reason, we used ONT sequencing to investigate the presence of high-CN cells in a sample of the population that was passaged for 7 d without G0775. Consistent with the Illumina sequencing, 958 array-spanning reads had a mean CN of four. However, many reads showed higher CNs. Among the spanning reads, 265 (28%) had CNs > 4, and one read, which only partially traverses the array, showed at least 28 adjacent copies of the repeat (Fig. 3D). Overall, although average CN for the entire population declined in the absence of selection, because of the ongoing dynamism of the amplifications, individual cells continued to harbor higher-level amplifications long after the antibiotic was removed.
Temporal Dynamics of lepB Amplification Are Not Dependent on RecA.
The breakpoints of the E. coli lepB amplification showed no detectable homology that could have been involved in the initial recombination event (SI Appendix, Fig. S6). Although the initial tandem duplication presumably arose in a RecA-independent manner, the extent to which ongoing expansion and contraction of the tandem repeat depends on RecA is not clear. Amplifications are sometimes stabilized by loss-of-function recA mutations, which prevent homologous recombination (5, 17–21). To check for this possibility, we knocked out recA in the E. coli amplification mutant and serially passaged individual colonies picked from the resulting ΔrecA::kan culture in the presence and absence of G0775 (Fig. 3A). Multiple colonies were examined based on the expectation that lepB CN would likely differ between each colony’s founding bacterial cell. Among 11 colonies studied, average lepB CN prior to passaging ranged from 5 to 36 (Table 1 and Fig. 3B), consistent with this expectation. Despite this variable starting point, the overall changes in CN observed in the ΔrecA strains following 7 d of serial passage mirrored that of the clones with an intact copy of recA (Fig. 3B). When bacteria were cultured in the absence of G0775, CNs decreased to 2 to 6, and when the media contained 0.5 µg/mL and 1.0 µg/mL G0775, CN increased to 14 to 53 and 27 to 44, respectively. These results show that the lepB amplification can expand and contract via a RecA-independent mechanism, the molecular details of which remain to be studied.
Secondary Mutations Associated with Drug Efflux.
Resistance mediated by gene amplification can allow cell populations to expand sufficiently to accumulate rarer secondary mutations either in the amplified gene or elsewhere in the genome (8). If these secondary mutations yield high-level resistance, the amplification may be lost, even with ongoing antibiotic selection. We do not observe this to be the case in our E. coli mutant. Despite the acquisition of secondary mutations, with only one exception, 7-d serial passage of both recA WT and ΔrecA::kan lineages with subinhibitory concentrations of G0775 led to populations with increased average lepB CN and maintained or increased levels of G0775 resistance (Table 1 and Fig. 3 B and C). It is particularly notable that no on-target LepB point mutations were identified. To investigate this further, we evaluated the relative fitness costs of lepB CN changes compared to lepB coding changes by growth-rate determination. We found no substantial difference between the growth rate of the lepB amplification mutant and two previously selected mutants harboring LepB mutations P88L and K146_V149dup (SI Appendix, Fig. S7) (11). We attribute the lack of secondary lepB mutations to low selective pressure and small population size. Strains were serial-passaged in sub-MIC concentrations of G0775 with ∼106 colony-forming units (CFU) transferred per day. The selection of lepB mutations was statistically unlikely given the small population size, and, under low selective pressure, any lepB mutants that did arise were unlikely to have a competitive advantage.
The phenotypic consequences of many of the secondary mutations we identified are unknown, but in several lineages, serial passaging resulted in the selection of subclones harboring point mutations either within or directly upstream of genes associated with efflux, suggesting efflux-based contribution to drug resistance (Table 1). These include acrB, which is a multidrug efflux pump subunit; emrR, a negative regulator of the multidrug resistance pump EmrAB (22); and lon, an adenosine triphosphate-dependent serine protease that has been shown to be involved in the induction of the AcrAB–TolC pump (23). In all cases, selection of efflux mutations was associated with either maintained or at most twofold increases in the level of resistance, and this is consistent with the previous finding that a mutation within the AcrB efflux pump confers a modest fourfold increase in the G0775 MIC (11).
Conclusion and Outlook
The use of MinION sequencing has provided unprecedented insight into CN variation within an antibiotic-resistant bacterial isolate. Previous studies have hypothesized that CN heterogeneity can explain variable resistance phenotypes (3), but ours uses single-molecule sequencing to directly examine the cell-to-cell distribution of CN within an antibiotic-resistant population. Although the initial selection of the lepB amplification with G0775 occurred at a low frequency, we have shown that, once selected, tandem amplifications containing lepB expand and contract at a high rate, even in the absence of RecA. These processes lead to the formation of distinct CN alleles segregating within the population. Antibiotic resistance is a growing problem for effective antimicrobial therapy, but because intraisolate genetic heterogeneity is difficult to detect, the clinical significance of this phenomenon is not well understood (24). It is, therefore, timely that we have explored this heterogeneity in the context of the optimized arylomycins, a promising new class of Gram-negative antibiotic (11).
The utility of the method presented here for variable CN detection hinges on the recovery of ultralong reads. Because ONT sequencing directly detects the input DNA molecule without amplification, there is no apparent technical limit to the length of DNA that can be sequenced, but read lengths do depend on DNA fragment size. As the field continues to optimize DNA-extraction and library-preparation protocols, as well as bioinformatics methods for analyzing ONT data, we anticipate that the ability to span longer and higher CN repeats will become increasingly tractable. Indeed, the recovery of a 2.3-Mb read shows that, although challenging, it is possible to generate bacterial chromosome-sized reads with ONT (25, 26). Recent developments in real-time adaptive sequencing represent another promising avenue for increasing yields of repeat-spanning reads. This approach allows nanopore devices to selectively eject individual reads from the pore in real time and could, therefore, be used to enrich for strands containing known repeat arrays (27). In sum, our results demonstrate the important role of CN variation in driving the emergence of genetically heterogeneous bacterial populations and, in certain cases, could be applicable to other heterogeneous cell populations.
Materials and Methods
Isolation of a Mutant Containing Extensive Amplification of lepB.
During the chemical optimization and development of a bacterial type I signal peptidase inhibitor (11), candidate molecules were evaluated for potency and resistance frequency against E. coli ATCC strain 25922 and A. baumannii ATCC strain 17978. Colonies that formed due to mutations that confer resistance were restreaked on plates containing the same drug concentration as the plates from which the mutants first appeared. The streaks were used to perform MICs, and resistant mutants displaying a fourfold or higher MIC shift against molecule G0775 were cultured in the absence of drug and frozen at −80 °C for further characterization.
MinION Sequencing.
DNA was extracted by using a modified phenol–chloroform extraction method (28). One milliliter of an overnight culture was pelleted and resuspended in 400 μL of lysis buffer containing 100 mM NaCl, 10 mM Tris-Cl (pH 8.0), 25 mM ethylenediaminetetraacetic acid (EDTA) with pH 8.0, 0.5% (weight/volume) sodium dodecyl sulfate, and 20 μg/mL RNase A (Qiagen), and incubated for 1 h at 37 °C. Proteinase K was then added to a final concentration of 200 μg/mL, and the lysate was incubated for 2 h at 50 °C. The lysate was transferred to an Eppendorf tube containing phase-lock gel along with 200 μL of BioUltra phenol saturated with tris(hydroxymethyl) aminomethane and EDTA buffer (Sigma-Aldrich catalog no. 77607). After gently mixing, the solution was centrifuged at 10,000 × g for 5 min, and the aqueous layer was transferred to an Eppendorf tube containing phase-lock gel. After the addition of 100 μL of phenol saturated with tris(hydroxymethyl) aminomethane and EDTA buffer, as well as 100 μL of chloroform-isoamyl alcohol (Sigma-Aldrich catalog no. 25666), the solution was again mixed gently and centrifuged at 10,000 × g for 5 min. The aqueous layer was transferred to a new tube, and the genomic DNA was ethanol-precipitated and then resuspended in 50 μL of 10 mM Tris-Cl (pH 8.0).
The quality of the genomic DNA was determined by Nanodrop (Thermo Fisher Scientific) to ensure an optical density (OD) of 260/280 of 1.8 to 2.0 and an OD 260/230 of 2.0 to 2.2 for all library input. Genomic DNA was quantified by using the Qubit double-stranded DNA (dsDNA) Broad-Range (BR) assay kit (Thermo Fisher Scientific). The library for E. coli mutant replicate 1 was generated by using 15 μg of DNA and the ONT Rapid Sequencing kit, SQK-RAD004, following a previously reported protocol (29). The library was sequenced by using a R9.4 flowcell on a MinION device running MinKnow (version [v] 2.1) to generate fast5 files that were subsequently base-called by using ONT’s Albacore Sequencing Pipeline Software (v 3.2.1) with default parameter values. Libraries for the remaining E. coli mutant replicates and WT sample were generated by using 400 ng of DNA and the Rapid Sequencing kit, according to the manufacturer’s protocol. Each of these libraries was sequenced on an R9.4 flowcell on a MinION device running MinKnow (v 3.1.3) to generate fast5 files that were subsequently base-called by using ONT’s Guppy (v 2.3.1) with default parameter values.
Libraries for the A. baumannii mutants and the passaged E. coli mutant were generated by using 1.2 to 5 μg of DNA and the ONT Ligation Sequencing kit, SQK-LSK109, following the protocol reported by John Tyson (https://dx.doi.org/10.17504/protocols.io.7eshjee). Each of the libraries was sequenced with a R9.4 flowcell on a MinION device running MinKnow (v 20.6.04) to generate fast5 files. Base-calling was performed by using ONT’s Guppy with default parameter settings: v 4.0.15 for the A. baumannii mutants and v 4.2.2 for the E. coli mutant.
Illumina Sequencing.
Genomic DNA was isolated by using a DNeasy Blood and Tissue Kit (Qiagen), according to the instructions of the manufacturer. The quality of the genomic DNA was determined by using the Genomic DNA Screen Tape and Tapestation 4200 (Agilent Technologies). Genomic DNA was quantified by using the Qubit dsDNA BR assay kit (Thermo Fisher Scientific). For library preparation, the Nextera DNA Flex kit (Illumina) was used with an input of 100 ng of genomic DNA. The resulting libraries were multiplexed and sequenced on NovaSeq (Illumina) to generate 5 million 75-bp paired-end reads for each sample.
Variant Detection Inference of Repeat CN Using Illumina Reads.
Illumina reads were mapped onto the E. coli ATCC strain 25922 and A. baumannii ATCC strain 17978 reference genomes (GenBank accession nos. NZ_CP00907 and NZ_CP012004, respectively) by using GSNAP (v 2013-10-10) (30). Single-nucleotide variant detection was carried out by using in-house R scripts, which utilized the Bioconductor packages, GenomicRanges (31), Genomic Alignments (31), VariantTools, and gmapR. Only base calls with phred quality score ≥30 were used for variant calling. The lepB amplification was inferred by comparing relative read coverages across the reference genome. Regions with higher relative coverage than the surrounding region corresponded to amplified DNA. Average CN was estimated by dividing the sequence coverage of the lepB gene by the mean coverage of single-copy multilocus sequence type genes (adk, fumC, gyrB, icd, mdh, and purA [E. coli]; and gyrB, gltA, gdhB, recA, cpn60, gpi, and rpoD [A. baumannii]).
Inference of Repeat CN Using MinION Reads.
To determine the lepB CN distribution in the population using the MinION data, we needed to count the number of lepB copies on each read found to span the entire tandem array. First, spanning reads were identified by comparing 450 bp of flanking sequence on either side of the tandem array to all MinION reads using BLASTN 2.4.0+ (32). A read was classified as “spanning” if both left and right flanking sequences yielded one or more high-scoring segment pairs (HSPs) satisfying <0.01. In general, using local alignment-based methods, such as BLAST, to identify specific sequences within MinION reads is potentially problematic because single-molecule sequencing reads are known to have significantly higher error rates compared with Illumina sequencing (10), and the errors often cluster into low-quality segments within the reads (33). The absence of an HSP could reflect a true lack of homology, but it could also be the result of low sequencing quality obscuring underlying similarity. Interestingly, regions of dramatically reduced sequencing quality have recently been reported to coincide with inverted duplicated DNA sequences (34).
For the E. coli mutant, we deemed it prudent to visualize the base-to-base phred quality scores of all spanning reads and compare these side by side with DNA dot plots comparing each read to the ∼4.8-kb amplified region (SI Appendix, Figs. S1 and S2). Dot plots were generated by using the R package dotplot (https://github.com/evolvedmicrobe/dotplot) with a sliding window of size 14. As expected, quality plots showed complex and variable error profiles that included regions of contiguous low-quality phred scores ≤7 (SI Appendix, Figs. S1 and S2). Although reads are composed of both high- and low-quality segments, it is notable that local variability in quality is not position-specific relative to the duplicated sequence, indicating that low quality is not an artifact associated with sequencing tandemly repeated DNA. Comparison of the error profiles of the spanning reads with that of 60 random reads (SI Appendix, Fig. S8) provided further evidence that low-quality base calls are not characteristic specific to tandem repeats.
The side-by-side visualization of quality plots and dot plots showed that, despite the presence of contiguous low-quality segments, there was, in all cases, sufficient sequence similarly to unambiguously determine the CN of an ∼4.8-kb region for each spanning read. We took advantage of the dot matrices already generated by the dotplot package to infer CN for all E. coli and A. baumannii MinION reads containing lepB using a custom R script. The x axis of each matrix represents base position along a read sequence, and the y axis represents the same along the amplified reference region. Matrices were constructed by comparing nucleotides pairwise using a sliding window of size 14 and recording matches. Discrete copies of the repeated unit were identified based on regions of homology of at least 1,000 bp yielding 100 or more window matches. These criteria were validated by visual verification of all 60 spanning reads. These criteria were then used to infer CN among reads that overlap with, but do not entirely span, the repeat array (i.e., nonspanning reads). As dot-plot analyses are computationally intensive, nonspanning reads were initially identified by comparing the lepB gene sequence to all reads of size >5,000 bp using BLASTN (32). Dot-plot analysis was carried out on reads yielding one or more HSPs satisfying e-value <0.01.
Deletion of recA.
The recA gene was deleted from the E. coli 25922 lepB amplification mutant by using lambda Red recombination (35). Subinhibitory concentrations of G0775 were added to the media during the preparation of electrocompetent cells to delete recA. The ΔrecA::kan gene, along with ∼500 bp of upstream and downstream region, was amplified by PCR from E. coli BW25113 in the Keio collection (36) using primer pair ACGCGG‐ATTTGTCACCTACA (forward) and AGCGGTGCTCTTGCTCATAA (reverse). The amplificon was electroporated into the E. coli 25922 amplification mutant expressing lambda Red recombinase from pKD46. Transformants were selected on Luria–Bertani agar containing 50 μg/mL kanamycin. Colonies were screened by PCR for the successful replacement of recA with the kanamycin-resistance cassette and subsequently cultured in the presence of subinhibitory concentrations of G0775 prior to DNA extraction and sequencing.
Serial Passage of Isolates.
Amplification-containing E. coli mutants were serially passaged in Mueller–Hinton II cation-adjusted broth (MHB) in the presence or absence of G0775. Specifically, three replicates of the initial mutant culture, as well as isogenic recA::kan isolates, were grown to stationary phase, and then 3 μL of these cultures was used to inoculate 1.5 mL of MHB (0.2% inoculum) containing either 0, 0.5, or 1.0 μg/mL G0775. These cultures were incubated overnight at 37 °C and serially passaged each day for 7 d using a 0.2% inoculum while maintaining identical drug concentrations as the previous growth cycle.
Determination of MICs.
MICs were measured by performing twofold serial dilutions of inhibitor in 0.1 mL of MHB in round-bottom 96-well plates (Corning Life Sciences catalog no. 3788). Each well was inoculated with 5 × 105 CFU/mL of log-phase cultures, and plates were incubated without agitation at 37 °C. Plates were scored by eye after 18 h for the presence or absence of growth. The MIC was determined to be the lowest drug concentration that prevents visible growth.
Supplementary Material
Acknowledgments
We thank Thomas Wu for helpful suggestions regarding analysis of error-prone long reads and Man-Wah Tan for helpful discussions and advice.
Footnotes
Competing interest statement: P.A.S. is listed as an inventor on patent WO2017084630. H.S.G., C.D.D., J.R., J.L., S.D., Y.L., J.K., P.A.S. and E.S. are employees of Genentech, a member of the Roche Group, and are shareholders in Roche. J.G. is employed by Oxford Nanopore Technologies Ltd.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021958118/-/DCSupplemental.
Data Availability.
DNA sequencing data have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information (PRJNA672356).
References
- 1.Band V. I., Weiss D. S., Heteroresistance: A cause of unexplained antibiotic treatment failure? PLoS Pathog. 15, e1007726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson D. I., Nicoloff H., Hjort K., Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 17, 479–496 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hjort K., Nicoloff H., Andersson D. I., Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol. Microbiol. 102, 274–289 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Schechter L. M., et al. , Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. mBio 9, e00583-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson S. E., Sherman E. X., Weiss D. S., Rather P. N., Aminoglycoside heteroresistance in Acinetobacter baumannii AB5075. mSphere 3, e00271-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicoloff H., Hjort K., Levin B. R., Andersson D. I., The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Pantua H., et al. , Unstable mechanisms of resistance to inhibitors of Escherichia coli lipoprotein signal peptidase. mBio 11, e02018-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandegren L., Andersson D. I., Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Anderson P., Roth J., Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl. Acad. Sci. U.S.A. 78, 3113–3117 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain M., et al. , Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith P. A., et al. , Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 561, 189–194 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Brochet M., Couvé E., Zouine M., Poyart C., Glaser P., A naturally occurring gene amplification leading to sulfonamide and trimethoprim resistance in Streptococcus agalactiae. J. Bacteriol. 190, 672–680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer A. C., Kishony R., Opposing effects of target overexpression reveal drug mechanisms. Nat. Commun. 5, 4296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen K. H., et al. , Resistance to piperacillin/tazobactam in Escherichia coli resulting from extensive IS26-associated gene amplification of blaTEM-1. J. Antimicrob. Chemother. 74, 3179–3183 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Peleg A. Y., Seifert H., Paterson D. L., Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Band V. I., et al. , Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 4, 1627–1635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galitski T., Roth J. R., Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics 146, 751–767 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haack K. R., Roth J. R., Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141, 1245–1252 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R. J., Capage M., Hill C. W., A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177, 1–18 (1984). [DOI] [PubMed] [Google Scholar]
- 20.Anderson R. P., Roth J. R., Tandem chromosomal duplications in Salmonella typhimurium: Fusion of histidine genes to novel promoters. J. Mol. Biol. 119, 147–166 (1978). [DOI] [PubMed] [Google Scholar]
- 21.Hill C. W., Foulds J., Soll L., Berg P., Instability of a missense suppressor resulting from a duplication of genetic material. J. Mol. Biol. 39, 563–581 (1969). [DOI] [PubMed] [Google Scholar]
- 22.Lomovskaya O., Lewis K., Matin A., EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177, 2328–2334 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicoloff H., Andersson D. I., Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol. Microbiol. 90, 1233–1248 (2013). [DOI] [PubMed] [Google Scholar]
- 24.El-Halfawy O. M., Valvano M. A., Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne A., Holmes N., Rakyan V., Loose M., BulkVis: A graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35, 2193–2198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kono N., Arakawa K., Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 61, 316–326 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Loose M., Malla S., Stout M., Real-time selective sequencing using nanopore technology. Nat. Methods 13, 751–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY, 3rd Ed., 2001). [Google Scholar]
- 29.Quick J., Ultra-long read sequencing protocol for RAD004. Protocols. Io (2018). [Google Scholar]
- 30.Wu T. D., Nacu S., Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence M., et al. , Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin S., et al. , Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 25, 1750–1756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spealman P., Burrell J., Gresham D., Inverted duplicate DNA sequences increase translocation rates through sequencing nanopores resulting in reduced base calling accuracy. Nucleic Acids Res. 48, 4940–4945 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing data have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information (PRJNA672356).