Significance
The NLRP3 inflammasome is a critical component of the innate immune response to infectious agents, but its overactivation is implicated in numerous pathologies, including atherosclerosis, Alzheimer’s Disease, gout, inflammatory bowel disease, and type 2 diabetes. Our study reveals a mechanism by which cells modulate NLRP3 inflammasome activation and identifies potential therapeutic targets to treat pathologies associated with NLRP3 inflammasome activation.
Keywords: IL-1α, inflammasome, mitochondria, cardiolipin, autophagy
Abstract
The balance between NLRP3 inflammasome activation and mitophagy is essential for homeostasis and cellular health, but this relationship remains poorly understood. Here we found that interleukin-1α (IL-1α)–deficient macrophages have reduced caspase-1 activity and diminished IL-1β release, concurrent with reduced mitochondrial damage, suggesting a role for IL-1α in regulating this balance. LPS priming of macrophages induced pro-IL-1α translocation to mitochondria, where it directly interacted with mitochondrial cardiolipin (CL). Computational modeling revealed a likely CL binding motif in pro-IL-1α, similar to that found in LC3b. Thus, binding of pro-IL-1α to CL in activated macrophages may interrupt CL-LC3b–dependent mitophagy, leading to enhanced Nlrp3 inflammasome activation and more robust IL-1β production. Mutation of pro-IL-1α residues predicted to be involved in CL binding resulted in reduced pro-IL-1α–CL interaction, a reduction in NLRP3 inflammasome activity, and increased mitophagy. These data identify a function for pro-IL-1α in regulating mitophagy and the potency of NLRP3 inflammasome activation.
The two most prominent members of the interleukin-1 (IL-1) superfamily, IL-1α and IL-1β, are equally potent master inflammatory cytokines that bind to the same receptor, and their deregulated signaling leads to severe acute or chronic inflammation (1). Most innate immune cells express either of these cytokines, their receptor, or both, and therefore almost all innate immune cells are affected by IL-1 signaling (2). The activity of these key inflammatory cytokines is tightly regulated both transcriptionally and posttranslationally (3, 4). In addition, various molecules interfere with IL-1 cytokine signaling at different levels to provide tight control of IL-1–mediated inflammatory responses (5). Presently, IL-1β is better studied because of its central involvement in inflammatory diseases, while the biogenesis of IL-1α and its distinctive role in inflammatory processes remains poorly defined.
Unlike pro-IL-1β, caspase-1 and the inflammasome have no direct role in cleaving pro-IL-1α (6, 7). IL-1α functions both as a secreted protein and as a membrane-bound cytokine, and may also have intracellular functions (6). Intracellular localization of pro-IL-1α can change in response to specific stimuli, and both N-terminal pro-IL-1α protein (IL-1α NTP) and pro-IL-1α possess a functional nuclear localization signal and bind the intracellular shuttle protein HAX1 (3), which regulates their translocation into the nucleus (3). The amino acid sequence of the pro-IL-1α N-terminal moiety is highly conserved between mouse and human (68% identity), suggesting that this region may be required for intracellular effects of pro-IL-1α. Indeed, it has been reported that pro-IL-1α and IL-1α NTP exert intracellular effects independent of receptor binding, including cytokine production (8–11). In addition, several studies have described posttranslational modifications of the pro-IL-1α precursor form. Specifically, pro-IL-1α was shown to be phosphorylated at serine 90 (human S90, mouse S87) (12, 13), myristolated on lysine 82 (14), and acetylated on lysine 82 (14). However, the functional significance of these modifications is not fully understood.
Il1a−/− macrophages produce and release significantly less IL-1β in response to activation of the inflammasome by LPS and ATP (15–17), and IL-1β levels are reduced in IL-1α–deficient mice (7). Data from IL-1R–deficient models also indicate that IL-1α may regulate IL-1β activity independent of receptor binding (7, 18), possibly through an intracellular pathway.
Here, we report that loss of IL-1α leads to reduced caspase-1 activity with subsequent reduction in IL-1β and IL-18 secretion. We observed that pro-IL-1α colocalized with mitochondria during LPS stimulation, and loss of IL-1α led to increased mitochondrial damage. We found that pro-IL-1α bound to cardiolipin (CL) on the mitochondrial surface, which required the HAX1 shuttle protein and Ca2+. IL-1α–deficient cells were more susceptible to damage-induced mitophagy. Furthermore, we used computational modeling to reveal that IL-1α interacts with CL via a motif similar to that of light chain 3b (LC3b). Mutation of this binding site in pro-IL-1α led to reduced CL binding, with a subsequent reduction in NLRP3 inflammasome activation and IL-1β secretion, as well as increased mitophagy. We propose that pro-IL-1α binding to mitochondrial CL (mtCL) inhibits LC3b binding and mitophagy, and the subsequent accumulation of damaged mitochondria enhances inflammasome activity, leading to increased IL-1β production.
Results
IL-1α Deficiency Reduces Mitochondrial Damage and Caspase Activation to Diminish IL-1β Release from Activated Macrophages.
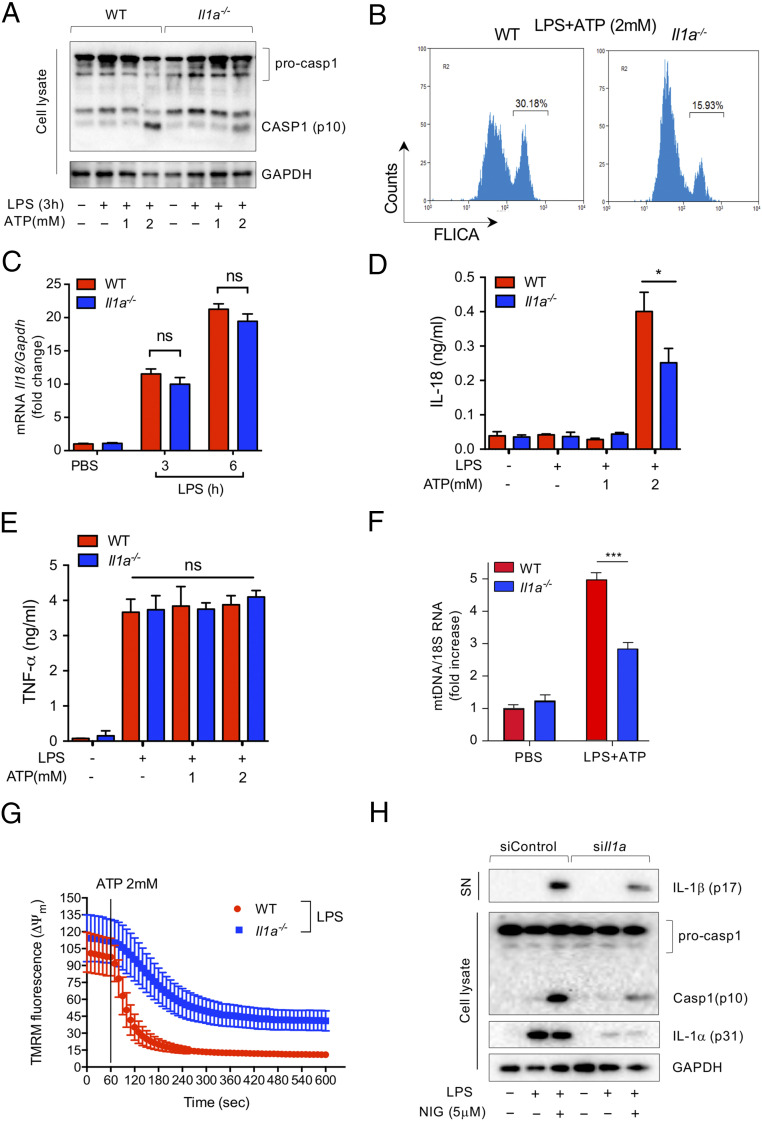
Recent studies to elucidate intracellular pathways by which IL-1α may regulate IL-1β release could not conclusively support the involvement of pro-IL-1α in transcriptional regulation of IL-1β in macrophages (15, 16). Similar to other studies, we observed significantly diminished IL-1β production from Il1a−/− macrophages following LPS and ATP stimulation (SI Appendix, Fig. S1A). Furthermore, caspase-1 activation was reduced in Il1a−/− macrophages compared with WT controls in response to LPS and ATP (Fig. 1 A and B and SI Appendix, Fig. S1B). Importantly, Nlrp3 mRNA expression was not different between WT and Il1a−/− macrophages after LPS simulation (SI Appendix, Fig. S1C), indicating that IL-1α may specifically modulate Nlrp3 inflammasome activation, rather than expression. In support of this, IL-1α deficiency did not affect LPS-induced IL-18 mRNA expression (Fig. 1C), but did impair mature IL-18 release (Fig. 1D). In contrast, TNF-α, which is not processed by the inflammasome, was released in equal measure by WT and Il1a−/− macrophages (Fig. 1E).
Fig. 1.
IL-1α deficiency reduces Nlrp3 inflammasome activation. (A) LPS-primed WT and Il1a−/− peritoneal macrophages were treated with the indicated doses of ATP for 30 min. Immunoblot analysis of caspase-1 and GAPDH in cell lysate. (B) LPS-primed WT and Il1a−/− peritoneal macrophages were treated with ATP for 30 min. Casapase-1 activity was measured by FLICA using flow cytometry. (C) IL-18 mRNA expression was measured by quantitative RT-PCR in peritoneal macrophages following LPS stimulation. mRNA expression was normalized to the reference gene GAPDH. (D and E) ELISA of cytokines in supernatants of LPS-primed WT and Il1a−/− peritoneal macrophages were treated with the indicated doses of ATP for 40 min. (D) IL-18; (E) TNF-α. (F) LPS-primed peritoneal macrophages were treated with ATP for 20 min and cytosolic mtDNA copy number was measured by quantitative RT-PCR. mRNA expression was normalized to the reference gene 18S RNA. (G) LPS-primed peritoneal macrophages were stained with TMRM (200 nM) for 20 min before mitochondrial membrane potential (ΔΨm) was monitored by confocal microscopy. (H) WT peritoneal macrophages were transfected with siControl or siIL-1α for 48h and stimulated with LPS for 3 h before nigericin treatment (5 μM) for 30 min. Immunoblot analysis of IL-1β in supernatant (SN), caspase-1, and GAPDH in cell lysate. Results are representative of three independent experiments. All data represent mean ± SD; (Student’s t test), *P < 0.05, ***P < 0.001; NS, not significant.
Mitochondrial damage is a hallmark of Nlrp3 inflammasome activation (19). To investigate the requirement for IL-1α in mitochondrial damage, we measured cytosolic mitochondrial DNA (mtDNA) content and mitochondrial membrane potential in peritoneal macrophages isolated from WT and Il1a−/− mice. Il1a−/− macrophages exhibited significantly less cytosolic mtDNA and higher mitochondrial membrane potential than WT controls in response to LPS and ATP (Fig. 1 F and G and Movie S1), suggesting that Il1a−/− cells were protected from ATP-induced mitochondrial damage.
To ensure that the effect of IL-1a deficiency on inflammasome activity was not due to an artifact of genomic disruption in the knockout mice, or a developmental defect, we next transfected Il1a small-interfering RNA (siRNA) (siIl1a) into peritoneal macrophages isolated from WT mice (SI Appendix, Fig. S1 D and E). Il1a knockdown led to a slight reduction in Il1b mRNA (SI Appendix, Fig. S1D), which corresponded with less pro-IL-1β protein after LPS stimulation (SI Appendix, Fig. S1F). However, in response to nigericin treatment, caspase-1 cleavage and IL-1β release were dramatically reduced in Il1a knockdown macrophages (Fig. 1H and SI Appendix, Fig. S1G). Overall, these data suggest that intracellular IL-1α (pro-IL-1α) may participate in mitochondrial injury and damage, which enhances NLRP3 inflammasome activation to regulate IL-1β release.
Pro-IL-1α Is Recruited to mtCL upon LPS Activation.
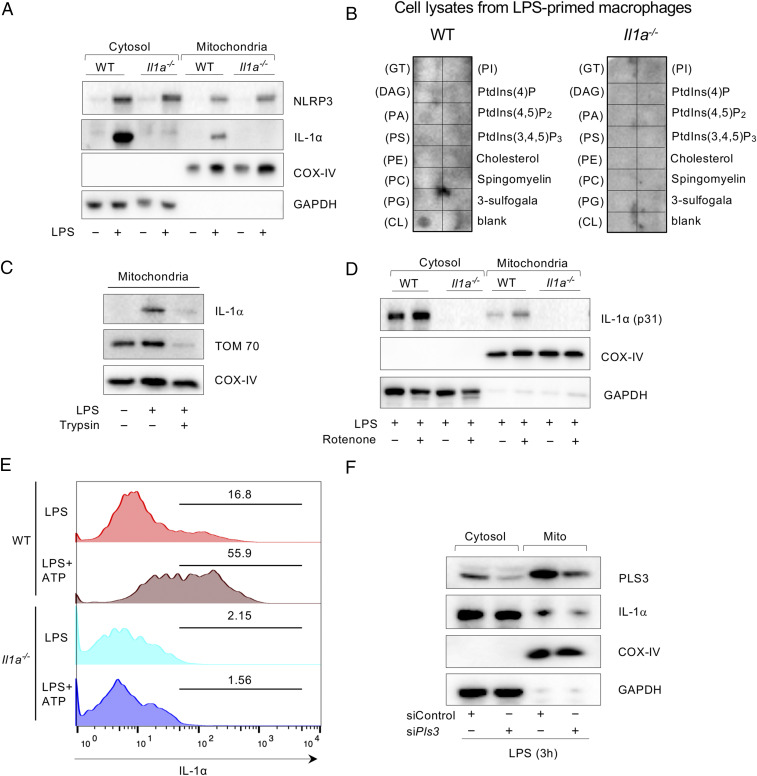
Nlrp3 is localized to the mitochondria and binds to mtCL during LPS stimulation (20, 21). In the setting of Nlrp3 deficiency, cells exhibit reduced mitochondrial damage and lower cytosolic mtDNA in response to LPS and ATP (22). To investigate the role of pro-IL-1α in this process, we first investigated whether IL-1α affects Nlrp3 mitochondrial localization. There was no difference in the amount of Nlrp3 protein in the mitochondrial fraction of LPS-stimulated macrophages from WT and Il1a−/− mice (Fig. 2A). However, to our surprise IL-1α was also present in the mitochondrial fraction of LPS-stimulated WT macrophages (Fig. 2A).
Fig. 2.
Pro-IL-1α localizes to the mitochondria in LPS-stimulated peritoneal macrophages. (A) WT and Il1a−/− peritoneal macrophages were simulated with PBS or LPS for 3 h. Immunoblot analysis for NLRP3, IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fractions. (B) Membrane lipid strips were incubated with cell lysates from LPS-primed WT and Il1a−/− macrophages, then washed and developed with a human anti–IL-1α antibody. GT, triglyceride; DAG, diacylglycerol; PA, phosphatidic acid; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; CL, cardiolipin. (C) Isolated mitochondria from PBS- or LPS-stimulated peritoneal macrophages were treated with or without trypsin (0.05%) for 10 min and then washed. Immunoblots showing IL-1α, TOM 70, and COX-IV in mitochondrial fraction. (D) WT and Il1a−/− peritoneal macrophages were simulated with PBS or LPS for 3 h and treated with Rotenone or DMSO (1 mM) for 1 h. Immunoblot analysis for IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fractions (E) Isolated mitochondria from LPS-stimulated peritoneal macrophages were stained with anti–IL-1α conjugated with APC for 1 h and measured by flow cytometry. (F) WT peritoneal macrophages were transfected with control siControl or siPls3 for 48 h and stimulated with LPS for 3 h before nigericin treatment (5 μM) for 30 min. Immunoblotting for PLS3, IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fraction. Results are representative of three independent experiments.
This observation raises the question of whether binding of pro-IL-1α to mitochondria plays a role in ATP-induced mitochondrial injury and NLRP3 inflammasome activation. Prior studies suggested that pro-IL-1α can selectively bind to acidic phospholipids, such as phosphatidylserine, phosphatidic acid, and phosphatidylinositol in the presence of Ca2+ (23). Thus, we hypothesized that pro-IL-1α also interacts with the phospholipid mtCL. To test this hypothesis, we performed a membrane lipid array experiment using membrane lipid strips. We observed strong binding between pro-IL-1α and CL in WT macrophage lysates following LPS stimulation (Fig. 2B), but not from unstimulated macrophages (SI Appendix, Fig. S2 A and B) or from Il1a−/− macrophages (Fig. 2B). Thus, pro-IL-1α may localize to the mitochondria by binding mtCL.
mtCL is normally an inner mitochondrial membrane phospholipid, but it translocates to the outer mitochondrial membrane (OMM) upon mitochondrial damage (24) or LPS priming of macrophages (21, 25–27) To determine where the binding of pro-IL-1α to mtCL occurs, we treated isolated mitochondria from LPS-stimulated macrophages with trypsin. Pro-IL-1α dissociated from mitochondria (Fig. 2C), suggesting that the binding occurs at the OMM. We next observed that LPS-induced pro-IL-1α mitochondrial localization was increased both when cells were stimulated with rotenone, an inhibitor of electron transport (Fig. 2D), or following stimulation with LPS plus ATP (Fig. 2E). Several studies have shown that the Ca2+-dependent phospholipid scramblase 3 (PLS3) enzyme is responsible for the translocation of mtCL to the OMM (24, 28, 29). Knockdown of PLS3 markedly diminished LPS-induced recruitment of pro-IL-1α to the mitochondria (Fig. 2F and SI Appendix, Fig. S2C), further indicating that pro-IL-1α binds mtCL that has translocated to the OMM.
Recruitment of pro-IL-1α to the Mitochondrial Surface Is HAX-1– and Ca2+-Dependent.
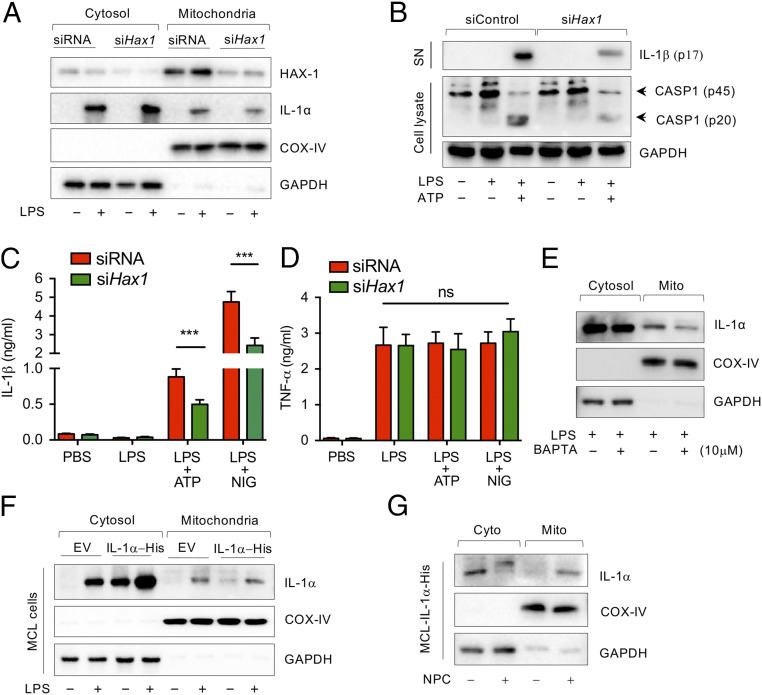
Unlike pro-IL-1β, the N-terminal region of pro-IL-1α has a nuclear localization sequence and can localize into the nucleus, cytosol, or lysosomal compartments, or be displayed on the outer leaflet of the plasma membrane (23, 30). Nuclear localization of pro-IL-1α is regulated by HAX1, an intracellular shuttle protein that binds both pro-IL-1α and IL-1 NTP (31–33). To determine if HAX-1 also controls the translocation of pro-IL-1α to the mitochondrial membrane, we generated HAX-1–deficient macrophages (Fig. 3A). HAX-1 deficiency indeed reduced LPS-induced pro-IL-1α recruitment to the mitochondrial surface (Fig. 3A and SI Appendix, Fig. S3A). Remarkably, caspase-1 activation (Fig. 3B and SI Appendix, Fig. S3B) and IL-1β production and release (Fig. 3C) were also significantly diminished in HAX-1–deficient macrophages following ATP or nigericin treatment, although TNF-α production was comparable between groups (Fig. 3D). This indicates that HAX-1 plays an important role in the recruitment of pro-IL-1α to the mitochondrial surface following macrophage activation for optimal Nlrp3 inflammasome activation and IL-1β release.
Fig. 3.
LPS-induced pro-IL-1α mitochondrial localization is Hax1- and Ca2+-dependent. (A) WT peritoneal macrophages were transfected with siControl or siHax1 for 48 h and stimulated with PBS or LPS for 3 h. Immunoblotting analysis for HAX-1, IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fractions. (B) WT peritoneal macrophages were transfected with siControl or siHax1 for 48 h and stimulated with PBS or LPS for 3 h before ATP (2 mM) treatment for 30 min. Immunoblotting analysis for IL-1b in supernatant (SN), caspase-1, and GAPDH in cell lysate. (C and D) WT peritoneal macrophages were transfected with control siRNA or siHax1 for 48 h and stimulated with PBS or LPS for 3 h before ATP (2 mM) treatment for 30 min. IL-1β (C) and TNF-α (D) production were measured by ELISA. (E) WT macrophages were stimulated with LPS for 1 h and then treated with or without BAPTA-AM for 3 h. Immunoblot analysis for IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fraction. (F) IL-1α-His stable expressing MCL cells were treated with or without LPS for 3 h. Immunoblot analysis for IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fractions. (G) IL-1α-His stable expressing MCL cells were treated with or without NPC for 3 h. Immunoblot analysis for IL-1α, COX-IV, and GAPDH in cytosolic and mitochondrial fractions. Immunoblots are representative of three independent experiments shown in A, B, and E–G. All data represent mean ± SD. Student’s t test was used for (C and D). ***P < 0.001.
A number of studies suggest that HAX-1 also regulates intracellular Ca2+ homeostasis (34–36), and its overexpression depletes endoplasmic reticulum (ER) Ca2+ stores by downregulating the ER Ca2+ pump SERCA2 (37). We also observed that LPS stimulation increased HAX-1 protein levels in the mitochondrial fraction of control-transfected macrophages (Fig. 3A). Therefore, the role of HAX-1 in the recruitment of pro-IL-1α to the mitochondrial surface may be related to intracellular Ca2+ mobilization. Indeed, intracellular Ca2+ mobilization is linked with Nlrp3 inflammasome activation (38, 39). To more directly investigate these pathways, we manipulated intracellular Ca2+ levels. In WT macrophages, LPS-induced pro-IL-1α recruitment to the mitochondrial membrane was diminished during intracellular Ca2+ depletion by BAPTA-AM, an intracellular Ca2+ chelator (Fig. 3E and SI Appendix, Fig. S3C). As expected, pro-IL-1α did not localize to the mitochondrial fraction in the absence of LPS in an IL-1α-His stably expressing macrophage cell line (MCL) (Fig. 3F). However, when we increased the intracellular Ca2+ level in these cells by NPC (an inducer of Ca2+ release from the ER), pro-IL-1α localized to the mitochondrial fraction in the absence of LPS stimulation (Fig. 3G). These data suggest that intracellular Ca2+ mobilization is required for the recruitment of intracellular pro-IL-1α to the mitochondrial membrane and for the interaction between pro-IL-1α and mtCL.
IL-1α Deficiency Leads to Enhanced Mitophagic Flux and Increased Mitochondria-LC3b Colocalization in LPS-Primed Macrophages.
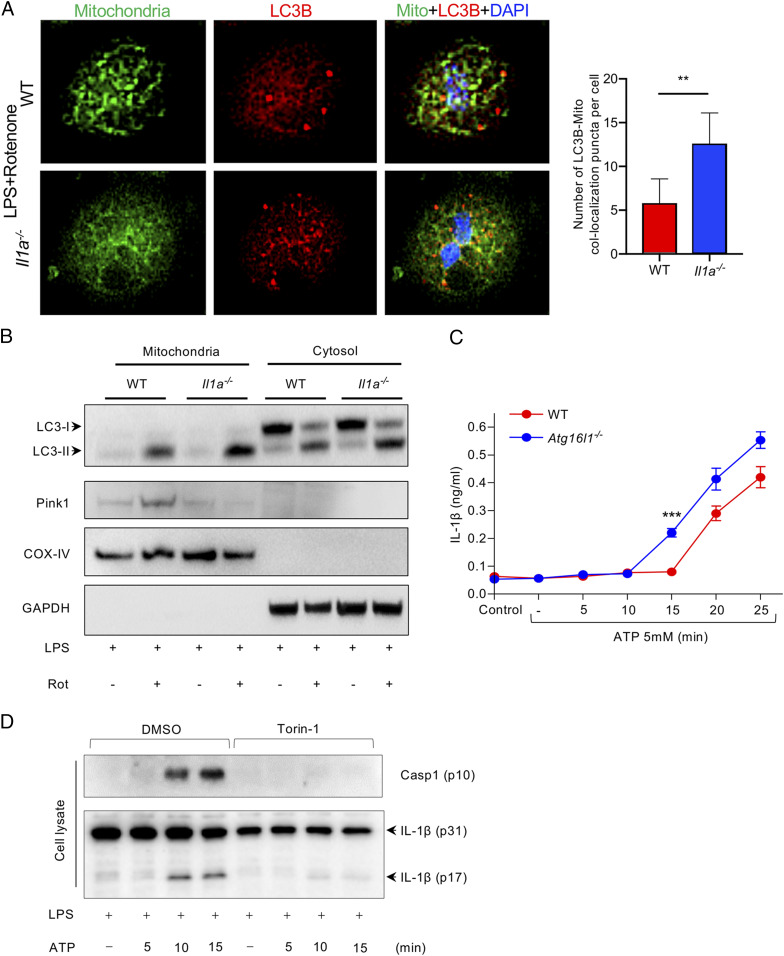
Prior studies suggest that mtCL plays a role in both inflammasome activity and mitophagy. mtCL directly binds to NLRP3 and activates caspase-1 (21). Furthermore, mtCL translocated to the OMM initiates a mitochondrial “eat me” signal, which is recognized by microtubule-associated protein 1 LC3b, promoting mitophagy (24). We found that IL-1α deficiency in macrophages was associated with significantly diminished mitochondrial damage in response to LPS and ATP (Fig. 1 F and G). Interestingly, mitophagic efficiency (flux, as assessed by mitochondria-LC3b colocalization) was higher in LPS-primed IL-1α–deficient macrophages than WT macrophages following treatment with rotenone (Fig. 4A). LC3-II protein expression was also dramatically increased in the mitochondrial fraction of IL-1α–deficient macrophages compared with WT in response to rotenone (Fig. 4B and SI Appendix, Fig. S3D). While LPS treatment increased PINK1 protein in the mitochondrial fraction of WT cells, this did not occur in IL-1α–deficient macrophages, suggesting that the increased mitophagic flux was not due to canonical mitophagy but instead through a direct interaction between mtCL and LC3b, as previously observed (24). Thus, LPS-induced translocation of pro-IL-1α to the mitochondrial membrane may interrupt and attenuate the CL-LC3b–mediated “eat me” mitophagy signal and dampen the inverse relationship between autophagy and inflammasome activity. Indeed, autophagy-deficient macrophages (Atg16l1flox/floxLysMCre) not only exhibited a more robust induction of the inflammasome and IL-1β release than control macrophages (Atg16l1flox/flox), but also a more rapid response (Fig. 4C). Conversely, as expected, autophagy induction with Torin-1 strongly inhibited inflammasome and caspase 1 activation, and IL-1β release (Fig. 4D).
Fig. 4.
IL-1α interrupts CL–LC3b-mediated mitophagy. (A) WT and Il1a−/− peritoneal macrophages were simulated with LPS for 3 h and treated with Rotenone (1 μM) for 1 h. Cells were stained with Mitotracker green and LC3b-594 (red). Representative confocal images showing colocalization of mitochondria and LC3b (magnification: 40×). (B) WT and Il1a−/− peritoneal macrophages were treated 50 nM bafilomycin A1 for 1 h, then simulated with LPS for 3 h and treated with Rotenone (1 μM) for 15 min. Immunoblots for LC3, Pink1, COX-IV, and GAPDH in cytosolic and mitochondrial fractions are shown. (C) Atg16l1fl/fl Lysmcre and Atg16l1fl/fl BMDM were primed with LPS for 3 h and then treated with ATP for the indicated times. IL-1β production was measured by ELISA. (D) WT peritoneal macrophages were simulated with LPS for 3 h and ATP for the indicated times, followed by Torin-1. Immunoblots for caspase-1 and IL-1β are shown. All data represent mean ± SD. Student’s t test was used for (A). Two-way ANOVA with Bonferonni’s posttest was used for (C). *P < 0.05. ***P < 0.001.
Molecular Simulations Show Pro-IL1α Interacts with CL Embedded in a Membrane.
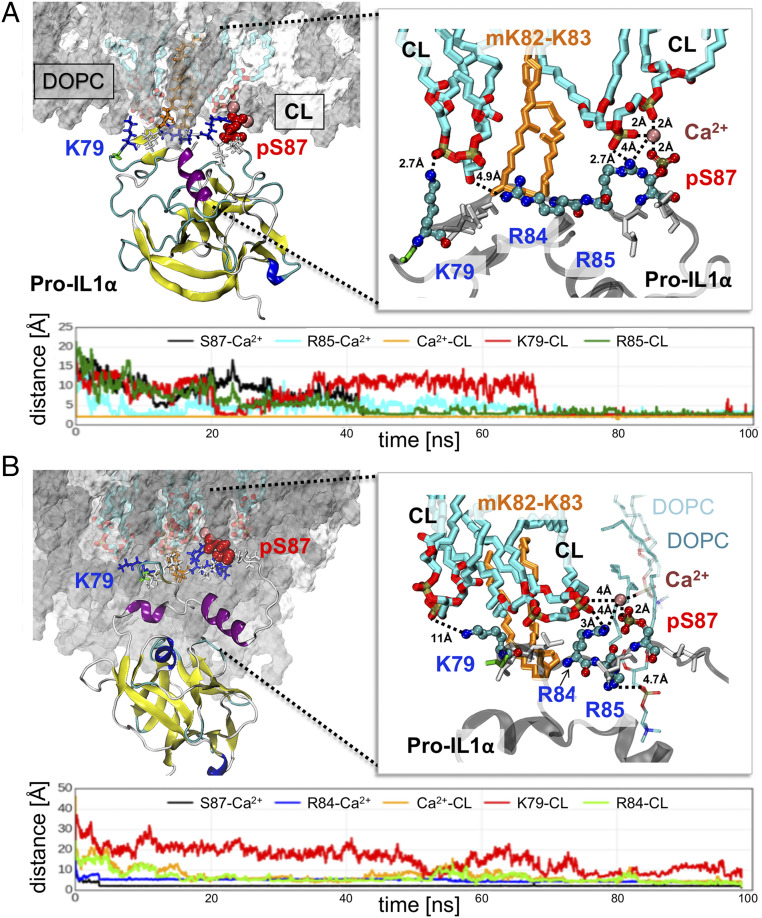
To examine the interactions between pro-IL-1α and mtCL, we generated a highly stable computational model of pro-IL-1α (S70-A271) (Materials and Methods and SI Appendix, Figs. S4 and S5), and performed 12 independent molecular dynamics (MD) simulations of 100 ns each. In each case, the protein was originally located 10 Å away from the membrane. The membrane was composed of two types of lipid molecules: CL and Δ9-Cis phoshatidyl choline (DOPC), in the ratio of 20:100 (comprising 14 CLs and 70 DOPCs). The protein–lipid interactions were observed in the presence of three posttranslational modifications of pro-IL-1α: Phosphorylation of serine 87 (pS87) and myristoylation of lysines at positions 82 and 83 (mK82-K83). The simulation system included free Cl− and Ca2+ ions; Ca2+ ions are known to initiate the insertion of myristoylated proteins into bilayer membranes (40). In 11 of 12 runs, pro-IL-1α readily diffused and positioned itself in close proximity to the membrane. Among the 11 cases, 9 displayed well-defined interactions that could be classified into two types, type I and type II (Fig. 5); two runs did not show any characteristic features that enabled classification.
Fig. 5.
Interaction of pro-IL1α with CL molecules in the lipid bilayer. (A and B) Two types of interactions, I and II, were observed, displayed in A and B, respectively. In both panels, the Upper Left diagram displays pro-IL1α (as ribbon diagram) and the membrane surface, with the DOPC molecules in gray, and CL molecules in cyan stick representation. Residues that play key roles in driving the interactions (K79, R84, R85, and pS87) are displayed in stick representation and enlarged on the Right. Their closest atom–atom distances from CL (cyan stick) are indicated. In both types of interactions, one arginine, R85 in A and R84 in B, is required for mediating the interaction with pS87 (red spheres, Left). Ca2+ ion (dark pink spheres) serves as a bridge that mediates the interaction with CL. Other amino acids from the signal peptide (G78, V80, L81, K82, K83, L86, and L88) are displayed in stick representation; the myristoylated lysines (mK82-K83) are shown in orange stick. The latter fragments insert into the membrane within 100 ns. The Lower panels in each case display the time evolution of selected residues from closest CL atoms or from Ca2+ ion, as well as the distance between Ca2+ and CL.
Type I was the most common type of interaction, observed in six MD runs. At least two CLs took part in the formation of close contacts between pro-IL-1α and the membrane in this type of interaction. In typical type I interactions, the signaling peptide residues (G78-L88) interact with CL (Fig. 5A and SI Appendix, Fig. S6). Residues that play a significant role in the interactions with the lipid molecules are labeled, and the time evolutions of their distance from the closest CL atom are displayed below the molecular model. SI Appendix, Fig. S5 displays similar plots for five other MD runs (MD2 to -6) that exhibited the same type of interactions. In type II interactions, observed in three trajectories, the main interactions came from DOPC and one CL (Fig. 5B).
Myristoylated Lysines Assist in the Initial Protein–Lipid Association, while Phosphorylated Serine Ensures Stable Binding.
The most significant contributions to the association of pro-IL-1α signal peptide with CL were made by K79, as well as R84 and R85. One of these two arginine residues, R85 (Fig. 5A) or R84 (Fig. 5B), consistently interacted with the pS87 and a Ca2+ ion that served as a bridge to mediate the interaction with CL (see expanded view on Right in both Fig. 5 A and B). Moreover, one of the hydrophobic residues, V80 or L81, consolidated the interaction between pro-IL-1α and CL. Hydrophobic interactions—in particular those of leucine, isoleucine, and valine—have been reported to be frequently involved in protein–CL interactions (via the CL’s acyl chains), based on over 20 crystal structures deposited in the Protein Data Bank (PDB) (41). In type II interactions, we observed the ternary interaction pS87-Ca2+-R84 similar to type I; however, the Ca2+ ion did not bridge between the protein and CL but instead, it mediated the interaction with DOPC.
We observed the insertion of two myristoylated lysines, mK82 and mK83, into the membrane between CL and DOPC molecules in all MD runs. However, none of the 10 MD simulations performed without phosphorylation of S87 but including myristoylated lysines showed stable interactions between pro-IL-1α signal peptide (SI Appendix, Fig. S7A, red arrow) and CL. The distance between S87 and the closest CLs remained 10 Å or higher in all runs (SI Appendix, Fig. S7B). Thus, while the insertion of the myristoylated chain assists in the association between pro-IL-1α and the membrane, it is not sufficient for stable binding of the protein. This is in agreement with previous work showing that the binding energy provided by myristoylated residues is relatively weak (42). Earlier experimental studies also showed that the hydrophobic insertion of the acyl chain into the membrane is necessary but not sufficient for binding of a myristoylated protein to membrane (40, 43). For that reason, an additional interaction is required: Either a polybasic cluster of amino acids or a palmitate moiety. Therefore, both hydrophobic and electrostatic forces are required to synergistically stabilize the bound form (42).
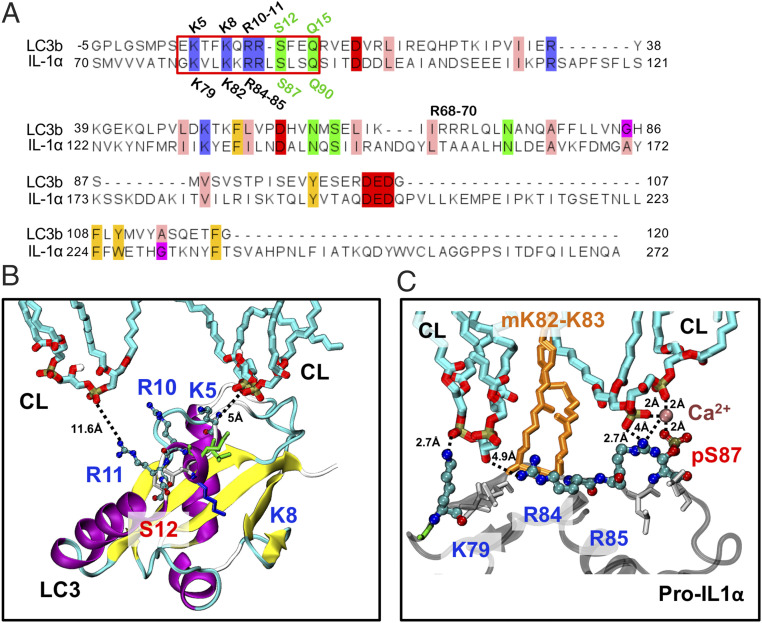
Pro-IL-1α Interaction and LC3 Share a CL-Recognition Motif.
Given our finding that pro-IL-1α may interfere with mtCL-mediated mitophagy, we examined whether this was related to blocking interaction between mtCL and LC3. Our MD results revealed a significant role of K79, myristoylated K82-K83, R84, R85, and pS87 in the pro-IL-1α–CL interaction. Alignment of the LC3 sequence [PDB ID code 1UGM, used in Chu et al. (24)] against that of pro-IL-1α (S70-A271) highlighted residues that are identical between LC3 and pro-IL1α (Fig. 6). Interestingly, among the six pro-IL-1α residues that serve as key interaction sites with CL, five are identical in LC3. Furthermore, a characteristic motif of four positively charged and two polar residues (serine and glutamine) arranged as K–K-RR-S–Q, is shared by the two proteins (Fig. 6A, red box). We also observed similar motifs in the SwissProt database, although their functions are unknown (SI Appendix, Tables S1 and S2).
Fig. 6.
Comparison of pro-IL1α (human) and LC3 (rat) sequences, and interaction motifs with CLs. (A) Sequence alignment of pro-IL1α fragment (res S70-A271) and LC3 (PDB ID code 1UGM). Only residues that are 100% conserved are highlighted in the alignment. Sequence alignment between LC3 and pro-IL1α reveals a common sequence motif of four positively charged residues (K–K-RR) and two polar residues: Serine and glutamine (K–K-RR-S–Q) are shared (red box). (B) Model of LC3 interactions with CL (24). Highly conserved residues are displayed as spheres (K5, R10, R11, and S12) except K8, which is shown in stick representation together with other residues which are not highly conserved in the signal peptide region. (C) Model of pro-IL-1α interactions with CL which shows close interactions with: K79 (counterpart of K5 in LC3) and R85 (counterpart of R11 in LC3).
Previous examination of CL–LC3 interactions showed that two of the charged and conserved residues of LC3, R10-R11, are essential for CL binding (24), and the pairwise sequence alignment between pro-IL-1α and LC3 shows that R10-R11 in LC3 corresponds to R84-R85 in pro-IL1α. Furthermore, CL interacts with the first lysine from the identical motif (Fig. 6A, red box) in both proteins (Fig. 6 B and C). The other two implicated residues, K8 and S12, have not been detected in the simulations of LC3–CL interactions, possibly due to the lack of posttranslational modifications in that study.
Interestingly, among the two myristoylated lysines (K82 and K83) of pro-IL-1α, only one (K82) is present in LC3. Our MD results indicate that this posttranslational modification may serve as an attractor that drives the interactions of pro-IL-1α with lipid bilayer and assists in stabilizing the bound form, thus pro-IL-1α may interact with CL more strongly than LC3.
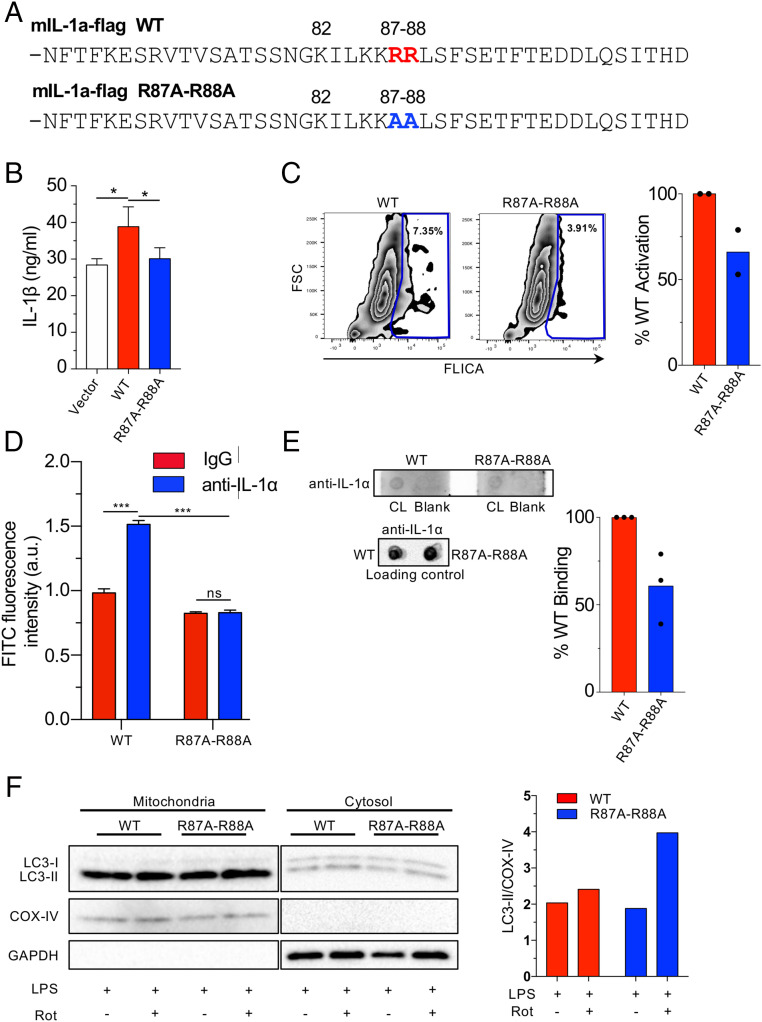
Mutation of the CL Binding Site in pro-IL-1α Reduces NLRP3 Inflammasome Activation and CL Binding.
Finally, in order to confirm that the predicted interactions between pro-IL-1α and CL occur in vivo, we mutated the conserved arginines at positions 87 and 88 in the mouse Il1a gene to alanines (84 and 85 in human) (Fig. 7A), and expressed this mutated version in Il1a−/− bone marrow macrophages. After activation of the NLRP3 inflammasome with LPS and ATP, Il1a−/− macrophages bearing the R87A-R88A mutant secreted significantly less IL-1β than Il1a−/− macrophages bearing WT Il1a (Fig. 7B). We also tested various other combinations of inflammasome activators and found a consistent reduction in IL-1β secretion in cells bearing the mutant IL-1α (SI Appendix, Fig. S8A). We next assessed caspase-1 activity by FLICA (fluorochrome inhibitor of caspases) and similarly found that Il1a−/− macrophages containing the R87A-R88A mutant had less active caspase-1 than WT controls (Fig. 7C). Since mutating R87 and R88 led to a reduction in NLRP3 inflammasome activation and IL-1β secretion, we subsequently investigated the effect of mutating these residues on CL binding to pro-IL-1α. We expressed FLAG-WT and FLAG-R87A-R88A pro-IL-1α in Hek293 cells and immunoprecipitated the WT and mutant proteins using anti-FLAG beads (SI Appendix, Fig. S8B). We observed a significant pro-IL-1α CL interaction with WT protein, but no interaction was observed with the R87A-R88A version (Fig. 7D and SI Appendix, Fig. S8C). Additionally, the mutated pro-IL-1α bound to CL on lipid strips at a reduced rate compared with WT pro-IL-1α (Fig. 7E). Finally, we investigated whether the R87A-R88A mutant would allow more LC3 to bind to CL on damaged mitochondria and found that nearly twofold more LC3-II was observed in the mitochondrial fraction compared with WT pro-IL-1α (Fig. 7F), indicating increased mitophagy. Taken together, these data clearly indicate that R87 and R88 participate in the pro-IL-1α–CL interaction as predicted by our molecular modeling, and that this interaction is key to achieving maximal NLRP3 inflammasome activity.
Fig. 7.
Mutated pro-IL-1α reduces IL-1β secretion and CL binding. (A) Mouse pro-IL-1α mutation construct. Changes are indicated. (B and C) Il1a−/− BMDMs were reconstituted with WT or R87A-R88A mouse Il1a and primed with LPS (6 h), then treated with ATP (45 min). IL-1β secretion measured by ELISA (B) and caspase-1 activity was measured by FLICA using flow cytometry (C). (D) HEK293 cells were transfected with mIL-1a-WT-Flag or mIL-1a-R87A-R88A-Flag. Anti-FLAG beads were used to immunoprecipitate the FLAG-proteins. The beads were then incubated with FITC labeled CL and fluorescence was measured using a plate reader. (E) Membrane lipid strips were incubated with cell lysates from immortalized Il1a−/− macrophages expressing flag-tagged WT and R87A-R88A pro-IL-1α, then washed and developed with an anti–IL-1α antibody. Densitometry of pro-IL-1α binding to CL was performed and normalized to total pro-IL-1α in the loading control. (F) Il1a−/− BMDMs were reconstituted with WT or R87A-R88A mouse Il1a and were treated 50 nM bafilomycin A1 for 1 h, then stimulated with LPS for 3 h and treated with Rotenone (1 μM) for 15 min. Immunoblots for LC3, COX-IV, and GAPDH in cytosolic and mitochondrial fractions are shown. Densitometry of LC3-II normalized to COX-IV is shown (mitochondrial fraction). Data shown are representative from two (C, D, and F) to three (B and E) experiments. All data represent mean ± SD. One-way ANOVA with Tuckey’s posttest (B) or two-way ANOVA with Bonferonni’s posttest (D) was used for statistical analyses. *P < 0.05, ***P < 0.001.
Discussion
In this study we found that IL-1α–deficient macrophages have reduced mitochondrial damage and reduced caspase-1 activity, leading to diminished IL-1β release. This supports a role for IL-1α in regulating NLRP3 inflammasome activation. Furthermore, we show that pro-IL-1α directly interacts with mtCL following LPS priming in macrophages, a process that is Ca2+- and HAX1-dependent.
Previous work from several groups indicated a role for IL-1α in modulating IL-1β production or release, but the mechanism had been unclear (7, 15–18). While IL-1β mRNA was known to be reduced in Il1a−/− macrophages following LPS stimulation, no clear role for IL-1α in IL-1β transcription could be determined, and it was suggested that the targeted deletion of Il1a could affect expression from the neighboring Il1b locus (15). Indeed, in Il1a−/− mice, there are no alterations in the levels or activation of IRF3, NF-κB, or p38/JNK; the key factors of Il1b regulation, and in an alternative IL-1α–deficient mouse model, IL-1β mRNA and protein level are not altered (16). However, inflammasome activation and IL-1β release was not investigated in this context (16). In our study, we also observed that although IL-18 mRNA levels were unchanged, IL-18 production was significantly reduced in Il1a−/− macrophages. Additionally, caspase-1 cleavage was reduced despite normal amounts of NLRP3. Taken together, these data indicate that IL-1α plays a transcription-independent role in NLRP3 inflammasome activation. This could also explain why, in a mouse model of LPS toxicity, Il1a−/− mice are more susceptible than Il1b−/− mice (44).
Mitochondria are known to be the critical nexus for NLRP3 inflammasome assembly and activation (45). Not only do the mitochondria provide a scaffold for NLRP3 inflammasome assembly, but mitochondrial damage is also required for NLRP3 inflammasome activation (19, 22, 46). In our model, Il1a−/− macrophages had reduced mitochondrial damage (preserved membrane potential and reduced cytosolic mtDNA), which correlated with decreased inflammasome activation. NLRP3 is known to physically associate with mitochondria (19); similarly, we found that pro-IL-1α also associated with mitochondria after LPS stimulation. Importantly, IL-1α deficiency did not affect NLRP3 association with mitochondria, indicating that these events occur independently.
HAX-1 is a validated IL-1α binding protein (31) required for IL-1α nuclear localization (33). However, HAX-1 is primarily localized at the mitochondria (47), and its overexpression has been shown to be mito-protective (47). We show that HAX-1 deficiency reduced mitochondrial translocation of pro-IL-1α and impaired Nlrp3 inflammasome activation. Notably, HAX-1 overexpression down-regulates SERCA2, resulting in increased intracellular Ca2+ levels (48), which activate the inflammasome (38, 49). We found that intracellular Ca2+ depletion diminished LPS-induced pro-IL-1α mitochondrial translocation in WT macrophages, which may indicate that in addition to shuttling pro-IL-1α to the mitochondria, HAX-1 regulates pro-IL-1α secondary to its effect on intracellular calcium. Additionally, Ca2+ induces a conformational change to the protein interaction C-terminal domain of HAX-1 (50). Therefore, it is also possible that HAX-1 interacts with pro-IL-1α only in the presence of Ca2+ in LPS-stimulated macrophages.
We show that pro-IL-1α interacts with mtCL on the mitochondrial surface. Previous reports indicated that mtCL is required for Nlrp3 inflammasome activation (21). Therefore, it is possible that pro-IL-1α promotes Nlrp3 inflammasome activation by interacting with mtCL. However, we observed that in Il1a−/− macrophages, LPS-induced mitochondrial damage was reduced, indicating that pro-IL-1α mitochondrial translocalization may increase the accumulation of damaged mitochondria. mtCL translocates to the OMM in response to mitochondrial damage and serves as a mitochondrial “eat me” signal recognized by LC3-mediated autophagic machinery (24). Our computational modeling indicates that LC3b and pro-IL-1α share similar CL binding motifs, raising the possibility that pro-IL-1α competes with LC3b for CL binding. Additionally, mutation of the predicted binding motif led to a decreased pro-IL-1α–CL interaction, reduced NLRP3 inflammasome activation, and greater LC3-II binding to mitochondria, all supporting our model. By interrupting the LC3b–CL interaction, pro-IL-1α may allow damaged mitochondria to escape from mitophagy, and the accumulated damage to contribute to Nlrp3 inflammasome activation. Nlrp3 also directly interacts with mtCL (21), and Nlrp3-deficient cells are resistant to mitochondrial damage in response to LPS and ATP (22), further evidence that this process is key to the inverse relationship between inflammasome activity and mitophagy. While both NLRP3 and IL-1α can bind mtCL directly, further studies are required to understand how these interactions take place and cooperate during NLRP3 inflammasome activation.
Prior studies reported that pro-IL-1α is phosphorylated in LPS-stimulated mouse macrophages at S90, most likely by cAMP-dependent kinase (12, 13). This posttranslational modification was suggested to account for its intracellular translocation, and potentially to protect the peptide from proteolytic cleavage (13). In addition, myristylation at lysines 82 and 83 is thought to regulate nuclear localization of pro-IL-1α (8). Our molecular modeling of human pro-IL-1α showed that phosphorylation at S87 (mouse S90) and myristylation at lysines 82 and 83, as well as the presence of Ca2+, were required for optimal interaction between pro-IL-1α and CL.
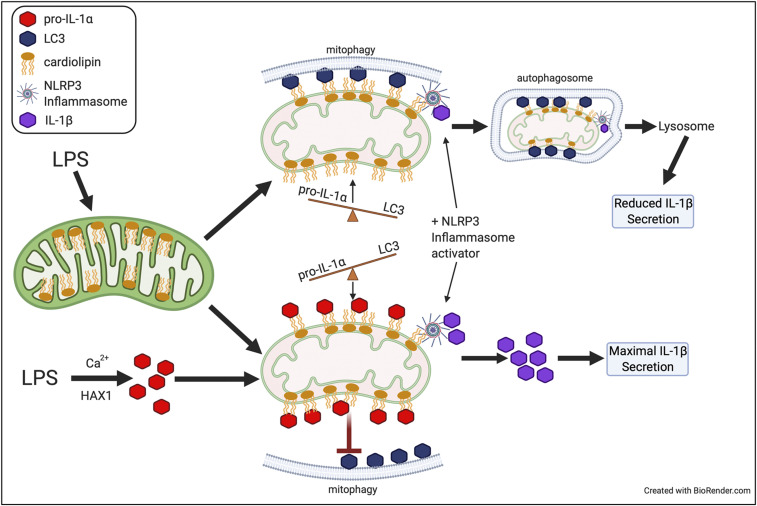
While IL-1α cytokine activity plays important roles in various immune responses and in autoinflammatory diseases, its intracellular regulatory role has not been appreciated. Our data place intracellular pro-IL-1α as a modulator of NLRP3 inflammasome activity and mitophagy (Fig. 8). We show that maximal IL-1β production is achieved by pro-IL-1α binding to mtCL, which prevents LC3b binding and subsequent mitophagy. Thus, in addition to up-regulating expression of various inflammasome components, LPS signaling (signal 1) induces the localization of the necessary NLRP3 inflammasome components, including pro-IL-1α, to the mitochondrial surface, in order to prepare for inflammasome activation after the appropriate danger signal has been detected (signal 2). As CL has already been the target of small-molecule–based therapeutics (51), it is possible that targeting the pro-IL-1α–CL interaction may lead to new inflammasome dampening treatments.
Fig. 8.
Role of pro-IL-1α in regulating NLRP3 inflammasome activity.
Materials and Methods
Reagents.
For a detailed list of reagents and primers, see SI Appendix, Table S3.
Mice.
Il1a−/− mice were kindly provided by Yoichiro Iwakura, University of Tokyo, Japan. Atg16l1fl/fl Lysmcre mice were obtained from David Shih, Cedars-Sinai Medical Center, Los Angeles, CA. All experiments were conducted according to Cedars-Sinai Medical Center Institutional Animal Care and Use Committee guidelines.
Culture and Stimulation of Peritoneal Macrophages.
As reported previously (52), elicited macrophages were harvested from WT or Il1a−/− mice, which had received a 1.5 mL intraperitoneal administration of 3% Brewer thioglycollate broth (Sigma) 72 h prior to collection. After extensive washing, cells were cultured for 4 h and nonadherent cells removed, and 24 h later stimulated with 0.5 μg/mL LPS (3 h) and treated with ATP (1 to 2 mM) or nigericin (5 μM) for 0.5 to 1 h.
Mitochondrial Isolation.
To extract cytosolic and mitochondrial fractions, peritoneal macrophages were homogenized and mitochondria were isolated with a mitochondria isolation kit (Abcam) according to the manufacturer’s protocol. The remaining supernatant was centrifuged at 10,500 × g for 5 min to obtain the cytosolic fraction. The proteins of cytosolic and mitochondrial fractions were analyzed by Western blotting.
Western Blot Analysis.
As described previously (53), cytosolic and mitochondrial fractions were suspended in SDS sample buffer before separation by SDS/PAGE and transferred onto PVDF membrane. Membranes were blocked in 5% nonfat milk in TBST for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C. Membranes were then washed and incubated with a secondary antibody conjugated with horseradish peroxidase (HRP) for 1 h at room temperature. After washing the membranes, signal was detected by using chemiluminescent substrate (Thermofisher Scientific) and ChemiDoc XRS Imaging system (Bio-Rad).
Mitochondrial Membrane Potential.
LPS-primed WT and Il1a−/− peritoneal macrophages were cultured with tetramethylrhodamine methyl ester (TMRM) (200 nM) containing PBS-based solutions for 20 min at 37 °C. Solutions were removed, serum-free growth media was reapplied under sterile conditions, and TMRM intensity was measured and analyzed by confocal microscopy and Leica Microsystems LAS AF Lite (Leica Microsystems).
Immortalization of Il1a−/− Bone Marrow-Derived Macrophages.
Il1a−/− bone marrow-derived macrophages (BMDMs) were immortalized as described previously (54) using the AMJ2C-11 (ATCC: CRL-2456) virus-containing supernatant. Once a homogenous population of immortal cells has been established (4 to 6 wk), cells were transduced as described in the next section. Immortal BMDMs (iBMDMs) were maintained in identical culture media as primary BMDMs, containing 20% L929 cells supernatant.
Transduction of iBMDMs and Cell Sorting Upstream of Lipid-Binding Experiments.
Il1a−/− iBMDMs were transduced with either IL-1a WT or IL-1aRR-encoding lentiviruses, which coexpress IRES-dTomato. Seventy-two to 96 h after transduction, cells were sorted by flow cytometry on a BD Influx system to select for high-expressing fractions (dTomato), and further grown.
Protein–Lipid Assay.
Binding of proteins to lipid strips (Echelon Biosciences) was performed according to the manufacturer’s protocol and Iyer et al. (21). Strips were blocked in 3% fatty acid-free BSA in TBS, and incubated with cell lysates of LPS-stimulated (3 h) WT or Il1a−/− peritoneal macrophages in 1% fatty acid-free BSA in TBS for 8 to 16 h at 4 °C. In some experiments, immortalized Il1a−/− BMDMs transduced with lentivirus expressing either WT or R84A R85A mutant Il1a were used. Additionally, human IL-1α–Flag stably expressing HEK293 cell lysates were used. Strips were washed extensively in TBST (0.05% Tween-20 in TBS) buffer to remove unbound proteins and incubated with anti–IL-1a or anti-Flag primary antibody for 16 h at 4 °C. After washing with TBST, strips were incubated with secondary antibody conjugated with HRP for 1 h at room temperature. After washing the strips, signal was detected using chemiluminescent substrate (Thermofisher Scientific) and ChemiDoc XRS Imaging system (Bio-Rad). For WT and R84A R85A mutant Il1a analyses, lipid strips were developed together and cardiolipin binding was normalized to total IL-1α in the cell extract as measured by dot blot.
Immunoprecipitation and Protein–Lipid Overlay Assay.
For immunoprecipitation and protein–lipid overlay, assay see SI Appendix, SI Methods.
LC3 Western Blot.
For LC3 Western blot, see SI Appendix, SI Methods.
Immunofluorescence Staining.
Peritoneal macrophages were cultured on coverslips. After simulation with LPS (3 h) and ATP (15 min), medium was removed and cells were directly fixed with fixation/permeabilization solution kit (BD Biosciences) for 20 min on ice and blocked by Sea block serum-free PBS (Abcam) for 1 h at room temperature. Cells were incubated with LC3b-DyLight650 and mouse LAMP1 primary antibodies for overnight at 4 °C, and after washing cells were incubated with anti-mouse Alexa-488 secondary antibody for 1 h at room temperature, and localization measured by fluorescence microscopy with BZ-X analyzer (Keyence).
Total RNA Isolation and Real-Time Quantitative PCR.
Total RNA was extracted from peritoneal macrophages by RNeasy plus mini kit (Qiagen) according to the manufacturer’s protocol. Total RNA was used as a template for reverse-transcription (RT) with a High-Capacity cDNA reverse-transcription kit (Takara). Real-time RT-PCR was performed in CFX96 (Bio-Rad) using SYBER green dye (Clontech). Genes were normalized by GADPH a housekeeping gene.
Cytokine Assay.
Flow Cytometry.
Mitochondria were isolated from LPS-stimulated WT and Il1a−/− peritoneal macrophages following ATP treatment, and stained with anti–IL-1α antibody conjugated with PE for 1 h on ice. Samples were analyzed on a Cyan flow cytometry system using Submit v4.3 software (Dako).
Constructs and siRNA Transfections.
IL-1α, HAX-1, and PLS3 genes in the peritoneal macrophages were silenced by IL-1α siRNA, HAX-1 siRNA, and PLSCR3 siRNA (Mybioscourse) according to the manufacturer’s protocol. Briefly, peritoneal macrophages were cultured in 12-well plates 24 h at 37 °C, 5% CO2 and transfected with 60 pmol of the indicated siRNA using 4 μL Lipofectamine RNAiMAX (Life technologies). Cells were treated 48 h after siRNA transfection.
Mutagenesis.
Mouse IL-1a expression construct (mIL-1a-Flag from SinoBiological) was used as a template to mutate R84 and R85 to alanine (mIL-1a-AA-Flag). The QuikChange Site Directed Mutagenesis Kit (Agilent) was used to create the mutant Il1a.
Lentivirus Constructs and IL-1a Expression in iBMDM.
mIL-1a-Flag and mIL1a-AA-Flag were subcloned into the lentivirus vector pHIV-dTomato. PCR amplification primers introduced an N-terminal EcoRI site and a C-terminal BamHI site, which were used for subcloning of mIL-1a-Flag and mIL1a-AA-Flag into the EcoRI and BamHI sites of pHIV-dTomato. Lentivirus was generated by cotransfecting Lenti-X 293 cells with pHIV-dTomato-mIL-1a-Flag or pHIV-dTomato-mIL-1a-AA-Flag, and pMD2.G and psPAX2 packaging vectors. Lentivirus containing cell supernatants were harvested after 48 h and used for infection of BMDM.
Differentiation, Transduction, and Functional Assays on BMDM.
BM was isolated from femurs of Il1a−/− mice and differentiated for 7 d in 20% L929 cell-supplemented media (DMEM high-glucose, 10% FBS, 1% penicillin-streptomycin, 10 mM Hepes). At day 7 and day 9, BMDMs were transduced overnight with lentivirus-containing cell supernatants. After transduction, IL-1aWT- or IL-1aAA mutant-reconstituted (Rec-WT and Rec-AA) BMDMs were washed extensively and left to rest for 48 to 72 h before functional assays.
FLICA assay.
BMDMs were primed for 1 h with LPS (1 ug/mL) followed by activation by Nigericin (8 uM) for 45 min. Cells were extensively washed, fixed, stained for caspase-1 activation and analyzed by flow cytometry.
ELISA.
BMDM were primed with LPS (1 ug/mL) or Pam2CSK4 (1 ug/mL) for 6 h, then treated with ATP (5 mM) or Nigericin (8 to 10 uM) for 45 min before being assayed for IL-1b by ELISA.
Molecular Modeling and Sequence Alignments.
For molecular modeling and sequence alignments, see SI Appendix, SI Methods.
Quantification and Statistical Analysis.
All statistical analysis was performed using GraphPad Prism 6.0. Data are reported as mean ± SD. Statistical differences were assessed by Student’s t test between two groups and one-way ANOVA for three or more groups (Tukey’s posttest), and values of P < 0.05 were considered significant. For data with two independent variables, two-way ANOVA was used with Bonferonni’s posttest. Statistical parameters can be found in the figure legends.
Supplementary Material
Acknowledgments
We thank Wenxuan Zhang, Ganghua Huang, and Debbie Moreira for excellent technical assistance. This work was supported by NIH Grants AI072726, 2R56AI067995, and 1R21AI105845-01A1 (to M.A.), AI112826 (to T.R.C.), and P30 DA035778 and P41 GM103712 (to I.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015632118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Garlanda C., Dinarello C. A., Mantovani A., The interleukin-1 family: Back to the future. Immunity 39, 1003–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber A., Wasiliew P., Kracht M., Interleukin-1 (IL-1) pathway. Sci. Signal. 3, cm1 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Di Paolo N. C., Shayakhmetov D. M., Interleukin 1α and the inflammatory process. Nat. Immunol. 17, 906–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson K. V., Deng M., Ting J. P. Y., The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P., The TLR and IL-1 signalling network at a glance. J. Cell Sci. 127, 2383–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik A., Kanneganti T.-D., Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol. Rev. 281, 124–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross O., et al. , Inflammasome activators induce interleukin-1α secretion via distinct pathways with differential requirement for the protease function of caspase-1. Immunity 36, 388–400 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Hu B., et al. , A nuclear target for interleukin-1alpha: Interaction with the growth suppressor necdin modulates proliferation and collagen expression. Proc. Natl. Acad. Sci. U.S.A. 100, 10008–10013 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werman A., et al. , The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc. Natl. Acad. Sci. U.S.A. 101, 2434–2439 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahon G. A., Garfinkel S., Prudovsky I., Hu X., Maciag T., Intracellular precursor interleukin (IL)-1alpha, but not mature IL-1alpha, is able to regulate human endothelial cell migration in vitro. J. Biol. Chem. 272, 28202–28205 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Pollock A. S., Turck J., Lovett D. H., The prodomain of interleukin 1alpha interacts with elements of the RNA processing apparatus and induces apoptosis in malignant cells. FASEB J. 17, 203–213 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y., et al. , Phosphorylation of intracellular precursors of human IL-1. J. Immunol. 140, 2279–2287 (1988). [PubMed] [Google Scholar]
- 13.Beuscher H. U., Nickells M. W., Colten H. R., The precursor of interleukin-1 alpha is phosphorylated at residue serine 90. J. Biol. Chem. 263, 4023–4028 (1988). [PubMed] [Google Scholar]
- 14.Stevenson F. T., Bursten S. L., Fanton C., Locksley R. M., Lovett D. H., The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc. Natl. Acad. Sci. U.S.A. 90, 7245–7249 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamacchia C., Rodriguez E., Palmer G., Gabay C., Endogenous IL-1α is a chromatin-associated protein in mouse macrophages. Cytokine 63, 135–144 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Almog T., et al. , Knockdown of interleukin-1α does not attenuate LPS-induced production of interleukin-1β in mouse macrophages. Cytokine 73, 138–143 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Kamari Y., et al. , Reduced atherosclerosis and inflammatory cytokines in apolipoprotein-E-deficient mice lacking bone marrow-derived interleukin-1α. Biochem. Biophys. Res. Commun. 405, 197–203 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Shemesh S., et al. , Interleukin-1 receptor type-1 in non-hematopoietic cells is the target for the pro-atherogenic effects of interleukin-1 in apoE-deficient mice. Atherosclerosis 222, 329–336 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Zhou R., Yazdi A. S., Menu P., Tschopp J., A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Subramanian N., Natarajan K., Clatworthy M. R., Wang Z., Germain R. N., The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer S. S., et al. , Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahira K., et al. , Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y., Oppenheim J. J., Matsushima K., Calcium-dependent binding of phosphorylated human pre interleukin 1 alpha to phospholipids. J. Biochem. 107, 666–670 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Chu C. T., et al. , Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y., Kelley R. I., Blanck T. J., Schlame M., Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278, 51380–51385 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Daum G., Vance J. E., Import of lipids into mitochondria. Prog. Lipid Res. 36, 103–130 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Elliott E. I., et al. , Cutting edge: Mitochondrial assembly of the NLRP3 inflammasome complex is initiated at priming. J. Immunol. 200, 3047–3052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., et al. , Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol. Cancer Res. 1, 892–902 (2003). [PubMed] [Google Scholar]
- 29.Van Q., et al. , Phospholipid scramblase-3 regulates cardiolipin de novo biosynthesis and its resynthesis in growing HeLa cells. Biochem. J. 401, 103–109 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wessendorf J. H., Garfinkel S., Zhan X., Brown S., Maciag T., Identification of a nuclear localization sequence within the structure of the human interleukin-1 alpha precursor. J. Biol. Chem. 268, 22100–22104 (1993). [PubMed] [Google Scholar]
- 31.Yin H., et al. , Evidence that HAX-1 is an interleukin-1 alpha N-terminal binding protein. Cytokine 15, 122–137 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Suzuki Y., et al. , HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J. Immunol. 158, 2736–2744 (1997). [PubMed] [Google Scholar]
- 33.Kawaguchi Y., et al. , Intracellular IL-1alpha-binding proteins contribute to biological functions of endogenous IL-1alpha in systemic sclerosis fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 103, 14501–14506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton T. A., Adams D. O., Molecular mechanisms of signal transduction in macrophages. Immunol. Today 8, 151–158 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Matzner N., et al. , Ion channels modulating mouse dendritic cell functions. J. Immunol. 181, 6803–6809 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Zanoni I., Granucci F., Differences in lipopolysaccharide-induced signaling between conventional dendritic cells and macrophages. Immunobiology 215, 709–712 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Vafiadaki E., et al. , The anti-apoptotic protein HAX-1 interacts with SERCA2 and regulates its protein levels to promote cell survival. Mol. Biol. Cell 20, 306–318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami T., et al. , Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 109, 11282–11287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horng T., Calcium signaling and mitochondrial destabilization in the triggering of the NLRP3 inflammasome. Trends Immunol. 35, 253–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka T., Ames J. B., Harvey T. S., Stryer L., Ikura M., Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376, 444–447 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Planas-Iglesias J., et al. , Cardiolipin interactions with proteins. Biophys. J. 109, 1282–1294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Resh M. D., Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 (1999). [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin S., Aderem A., The myristoyl-electrostatic switch: A modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 20, 272–276 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Joosten L. A. B., et al. , Differential susceptibility to lethal endotoxaemia in mice deficient in IL-1α, IL-1β or IL-1 receptor type I. APMIS 118, 1000–1007 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Elliott E. I., Sutterwala F. S., Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 265, 35–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimada K., et al. , Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam C. K., et al. , HAX-1 regulates cyclophilin-D levels and mitochondria permeability transition pore in the heart. Proc. Natl. Acad. Sci. U.S.A. 112, E6466–E6475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao W., et al. , The anti-apoptotic protein HAX-1 is a regulator of cardiac function. Proc. Natl. Acad. Sci. U.S.A. 106, 20776–20781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee G.-S., et al. , The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca(2+) and cAMP. Nature 492, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirasaka K., et al. , UCP3 is associated with Hax-1 in mitochondria in the presence of calcium ion. Biochem. Biophys. Res. Commun. 472, 108–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szeto H. H., First-in-class cardiolipin-protective compound as a therapeutic agent to restore mitochondrial bioenergetics. Br. J. Pharmacol. 171, 2029–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Goncalves R., Mosser D. M., The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. chap 14, Unit 14 11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dagvadorj J., et al. , Lipopolysaccharide induces alveolar macrophage necrosis via CD14 and the P2X7 receptor leading to interleukin-1α release. Immunity 42, 640–653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratsimandresy R. A., Indramohan M., Dorfleutner A., Stehlik C., The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell. Mol. Immunol. 14, 127–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.