Significance
Neuropeptides of the central nervous system regulate neurodevelopment, metabolism, emotion, and stress responses. However, despite previously established recognition of antimicrobial activity in vitro, their precise role(s) in host defense of the brain and other contexts in vivo have been poorly understood. Here, we discovered that the neuropeptide PACAP is an evolutionarily conserved antimicrobial peptide induced in the brain in response to bacterial and fungal infection. Tissue-specific PACAP induction coincided with minimal immune cell infiltration or neuroinflammation. These findings support an emerging paradigm wherein coevolution of the immune and nervous systems is linked through ancient, multifunctional peptides having neurological and immunomodulatory functions that protect the brain. We predict that PACAP is a prototypical example of a general class of endogenous, infection-inducible host-defense neuropeptides and serves as a model template to design next-generation antiinfectives with neurohomeostatic and immunomodulatory capabilities.
Keywords: neuropeptides, innate immunity, antimicrobial peptides, neuroimmunology, host defense
Abstract
Defense of the central nervous system (CNS) against infection must be accomplished without generation of potentially injurious immune cell-mediated or off-target inflammation which could impair key functions. As the CNS is an immune-privileged compartment, inducible innate defense mechanisms endogenous to the CNS likely play an essential role in this regard. Pituitary adenylate cyclase-activating polypeptide (PACAP) is a neuropeptide known to regulate neurodevelopment, emotion, and certain stress responses. While PACAP is known to interact with the immune system, its significance in direct defense of brain or other tissues is not established. Here, we show that our machine-learning classifier can screen for immune activity in neuropeptides, and correctly identified PACAP as an antimicrobial neuropeptide in agreement with previous experimental work. Furthermore, synchrotron X-ray scattering, antimicrobial assays, and mechanistic fingerprinting provided precise insights into how PACAP exerts antimicrobial activities vs. pathogens via multiple and synergistic mechanisms, including dysregulation of membrane integrity and energetics and activation of cell death pathways. Importantly, resident PACAP is selectively induced up to 50-fold in the brain in mouse models of Staphylococcus aureus or Candida albicans infection in vivo, without inducing immune cell infiltration. We show differential PACAP induction even in various tissues outside the CNS, and how these observed patterns of induction are consistent with the antimicrobial efficacy of PACAP measured in conditions simulating specific physiologic contexts of those tissues. Phylogenetic analysis of PACAP revealed close conservation of predicted antimicrobial properties spanning primitive invertebrates to modern mammals. Together, these findings substantiate our hypothesis that PACAP is an ancient neuro-endocrine-immune effector that defends the CNS against infection while minimizing potentially injurious neuroinflammation.
Neuropeptides enable interneuronal communication and signaling (1), mediating diverse functions ranging from endocrine stimulation and homeostatic regulation to immune signaling, pain modulation, and circadian rhythm maintenance. At present, over 100 neuropeptides are known in mammals (2). These peptides originate from neurons in the central, enteric, or peripheral nervous systems and within immune organs (3). Canonically, neuropeptides exert their biological function by binding to a cognate receptor (usually a G-coupled protein receptor [GPCR]), triggering a signal transduction pathway that leads to a functional change in the target cell (1). Neuropeptides are typically considered neurotransmitters or neurohormones, but recent work has illuminated their potential roles in modulating immune responses and neuroinflammation (4–8).
Human innate and adaptive immunity have evolved via two parallel and complementary paradigms in host defense against microbial invasion: molecular and cellular. Molecular defense mediators are secreted or activated rapidly and locally to directly inhibit pathogens. Prototypic examples include host-defense peptides (HDPs), the acute-phase reactants, and the complement cascade. Cellular defense involves infiltration of professional immune phagocytes (neutrophils and macrophages) and lymphocytes into infected tissues. Cellular infiltration into the central nervous system (CNS) is a double-edged sword, given its anatomically confined space and physiologically delicate context. On one hand cellular defense may be necessary to control or clear certain pathogens. On the other hand, neutrophils and other phagocytes can cause counterproductive damage to tissue parenchyma due to production and release of reactive oxygen species and other cytotoxic constituents from phagolysosomes. Thus, molecular defenses that are rapidly deployable in immediate settings of infection to obviate the need for infiltration of potentially harmful immune cells would be of special relevance in context of the CNS.
To explore putative molecular host-defense mediators within the CNS that may have both neuro- and immunomodulatory properties, we used a support vector machine (SVM) trained on HDPs (9, 10) to identify neuropeptides with potential host defense capabilities. Among the human neuropeptides identified as potential HDPs for molecular host defense of the CNS is pituitary adenylate cyclase-activating polypeptide (PACAP). PACAP is a member of the vasoactive intestinal peptide (VIP)/PACAP/secretin family (11) that regulates neurodevelopment (12), metabolism, emotion, mood, and stress responses via GPCRs (13). PACAP is known to interact with the immune system (14, 15) and modulate T helper type 1 (TH1)/TH2 cytokine production (3). Important previous work on structure activity relationships (SAR) of PACAP have also shown that it possess antimicrobial activity in vitro against a range of organisms (16–18), as well as anti-cancer activity against tumor cell lines. (Interestingly, our use of an SVM classifier that can scan different fragments of the same peptide allows us to identify antimicrobial activity in previously identified metabolites of PACAP as well* (19)). However, host defense functions, contextual bioactivity, or pathogen-specific inducibility of PACAP or other neuropeptides regarding antimicrobial activity in vivo are not known. More specifically, the role of PACAP in the larger context of innate immunity and its in vivo relevance to antimicrobial defense of the CNS and in other tissues remains unclear, given that antimicrobial activity is strongly dependent on biochemical and physiological context (20, 21, 22). Here, we examine PACAP inducibility in response to infection in the CNS and other tissues, and whether PACAP exerts antimicrobial activity against relevant organisms in the specific biochemical context relevant to those tissues. Bioinformatic and structural analyses showed PACAP to possess almost identical structural similarity to human cathelicidin LL-37, despite having overall low sequence similarity to other known HDPs. Synchrotron X-ray scattering revealed that PACAP can induce negative Gaussian curvature (NGC) in microbial membranes, a general requirement for membrane-permeating antimicrobial processes such as pore formation, blebbing, and other membrane-perturbing events (23–25). Moreover, extending from prior work (18), antimicrobial assays and mechanistic fingerprinting analyses showed that PACAP exerts potent antimicrobial mechanisms against drug-resistant bacteria and fungi via multiple synergistic pathways, including permeabilization, disruption of cellular energetics, and activation of regulated cell death pathways. In mouse models of bacterial or fungal infection, we demonstrated that PACAP is strongly induced up to 50-fold in brain, spleen, or kidney. Further, in media simulating these tissue contexts, PACAP exerted robust microbiostatic and microbicidal efficacy. Taken together, these findings imply that PACAP is an infection-inducible, tissue-specific host-defense effector that affords rapid and contextual antimicrobial host defense in the CNS and periphery. Beyond immediate contributions to better understanding of antimicrobial defense, the present discoveries reveal specific intersections of neurological and immunological systems and establish insights into antiinfective strategies that preserve critical functions of the CNS.
Results
Machine Learning Predicts PACAP to Be Strongly Antimicrobial.
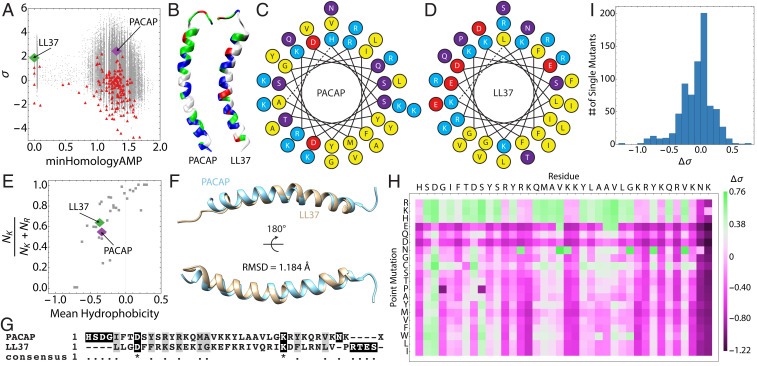
We collated human neuropeptide sequences from the database NeuroPep (2) and screened them for predicted membrane-permeating activity using the MAPT (26). The distance-to-hyperplane (σ) of each sequence was plotted vs. the homology of each sequence to known ɑ-helical antimicrobial peptides (minHomologyAMP) and superimposed on a sequence atlas generated from a Monte Carlo search of the undiscovered sequence space spanning various mutational distances (Fig. 1A). Large, positive values of σ indicate a high probability of antimicrobial activity, while negative values of σ indicate a low probability of antimicrobial activity. To visualize the distribution of sequences based on their antimicrobial potential, histograms of σ and minHomologyAMP were plotted (SI Appendix, Fig. S1). The majority of sequences returned low σ scores, consistent with their low sequence homology to known AMPs. However, a small sequence set (∼10 sequences) received high scores with σ > 0.8. Interestingly, among these the human PACAP isoform 1-38 was the top-scoring neuropeptide sequence (σ = 2.56, minHomologyAMP = 1.31) (Fig. 1A, purple). In comparison, the prototypic ɑ-helical human AMP LL37 received a score of σ = 2.00 (minHomologyAMP = 0) (Fig. 1A, green). Physicochemical and helical wheel analysis of the PACAP sequence revealed a striking similarity to the human cathelicidin LL37, an innate immune peptide with a broad range of host-defense functions (27, 28). Like LL37, PACAP is cationic and facially amphipathic, with a segregation of charged and hydrophobic residues (Fig. 1 B–D). These peptides are also similar in size (PACAP, 38; LL-37, 37) and the overall charge of PACAP is higher than LL37 at pH 7.4 (QPACAP = +9.03 vs. QLL37 = +6.00). The size of the hydrophobic face of LL37 is larger than PACAP (θLL37 ∼ 120° vs. θPACAP ∼ 90°) and have hydrophobic moments of 0.36 and 0.09, respectively (Fig. 1 C and D). Interestingly, PACAP and LL37 contain almost identical proportions of K/K+R and mean hydrophobicity, and both fall close to the saddle-splay selection rule for all known ɑ-helical AMPs (9, 10) (Fig. 1E). As a result, PACAP is predicted to perturb microbial membranes similar to known AMPs, suggesting that early selective forces may have favored multifunctional neuropeptides with host-defense properties.
Fig. 1.
PACAP is a GPCR-binding neuropeptide with strong physicochemical homology to human cathelicidin from innate immunity. (A) Screening a neuropeptide database (red) using the support vector machine-based MAPT for sequences with hidden membrane-restructuring activity revealed PACAP (purple) to be a hypothesized endogenous HDP. LL37 (green) is shown for comparison alongside sequences generated by a Monte Carlo–based directed search of sequence space (gray). (B) Comparison of the distribution of residue character along the lengths of PACAP and LL37 (blue = cationic, green = hydrophilic, white = hydrophobic, red = anionic). Helical wheel diagrams of (C) PACAP and (D) LL37 show similar distributions of hydrophobic (yellow) and cationic residues (teal). Anionic amino acids (red) and polar amino acids (purple) are also shown. (E) PACAP and LL37 exhibit similar lysine-to-arginine ratios and mean hydrophobicities, which suggests that PACAP (purple) can generate negative Gaussian membrane curvature similar to LL37 (green) and other ɑ-helical AMPs (gray). (F) Structural alignment of PACAP and LL37 yields an rmsd of 1.184 Å, demonstrating close structural similarity, despite (G) very low sequence homology. (H and I) An in silico mutational analysis of the entire PACAP sequence demonstrates that PACAP is locally optimized for membrane-remodeling activity, and that the majority of mutations reduce the microbicidal potential of the peptide.
Despite low primary sequence identity, PACAP and LL37 exhibited high physicochemical similarities. A three-dimensional structural alignment of PACAP and LL37 revealed a strikingly strong structural homology (rmsd = 1.184 Å; Fig. 1F). In contrast, a sequence alignment of PACAP and LL37 showed essentially zero sequence similarity or homology (Fig. 1G), implying these peptides likely evolved independently. The cathelicidin and defensin families of HDPs are known to be heterogeneous and undergo rapid evolution, with substantial variation in both primary sequence and physicochemical characteristics between species (29–31). To study whether PACAP also exhibited such phylogenetic behavior, we conducted multiple sequence alignments of PACAP across 11 organisms spanning 700 My of evolution (SI Appendix, Fig. S2). As compared with the oldest organism, Hydra, the human PACAP sequence shares 92% identity and 97% conservation; these correlates were identical to those of murine PACAP. Furthermore, there is marked conservation of the entire sequence across evolution, with the highest degree of convergence in the N-terminal receptor-binding region and relatively greater diversity in the C-terminal eight amino acid residues (SI Appendix, Fig. S2). Phylogenetic analyses of PACAP sequences indicated they cluster into two clades, with one representing the PACAP sequences in higher vertebrates that exhibit arginine-to-lysine substitutions in positions 32 and 36 (SI Appendix, Fig. S3).
Machine-learning screens of the 11 PACAP homologs predicted that all of the sequences would be antimicrobial, and that differences in primary amino acid sequences across species do not appreciably change this underlying property (SI Appendix, Fig. S2). In light of this observation, we set out to systematically map the membrane activity fitness landscape of PACAP. Using the MAPT, we constructed all single mutants of PACAP and screened them. Deviations of the σ scores of the mutants from the wild-type (WT) sequence (Δσ) were computed and plotted on a heat map (Fig. 1H). The large majority of single mutations reduced or minimized PACAP membrane-permeating properties. Furthermore, the distribution of Δσ is left-skewed, indicating that the PACAP structure is locally optimized for membrane-permeating activity (Fig. 1I). Mutations in the ∼10 C-terminal residue domain are especially prone to reductions in fitness, indicating that membrane activity of PACAP localizes to this region, whereas GPCR-binding capacity is distinctly localized to the N terminus (32), highly reminiscent of mammalian kinocidins (31). The structural compactness of multipotent PACAP peptides suggests some interpenetration between these functional domains.
PACAP Induces NGC in Microbial Membranes to Facilitate Potent Bactericidal and Fungicidal Activity.
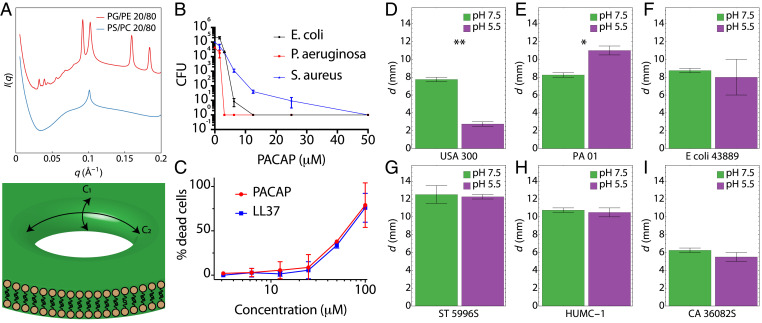
To dissect the biophysical mechanisms of PACAP-mediated membrane permeation, we conducted small-angle X-ray scattering (SAXS) experiments with model membranes mimicking bacterial and mammalian cells (Fig. 2 and SI Appendix, Fig. S4). Consistent with machine learning predictions, PACAP induced NGC in the bacteria-like PG/PE 20/80 membranes (Fig. 2A) but not in the mammalian-like PS/PC 20/80 membranes. PACAP induced the formation of a Pn3m cubic phase with a lattice parameter of a = 28.1 nm and a corresponding magnitude of NGC = 0.8 × 10−2 nm−2, which is comparable to the NGC induced by other HDPs. The fact that PACAP induced NGC comparable to other membrane-remodeling proteins, including AMPs and cell-penetrating peptides (33), implies one mode of antimicrobial activity is pore formation (Fig. 2A). To determine whether PACAP is bactericidal, minimum bactericidal concentration (MBC) plate-killing assays targeting Staphylococcus aureus, Escherichia coli, or Pseudomonas aeruginosa were performed (Fig. 2B). Various concentrations of PACAP were incubated with 2 × 105 colony-forming units (CFU) of bacteria in liquid broth at midlog phase in 96-well plates and incubated at 37 °C for 1 h. Tenfold serial dilutions were made with sterile buffer and spotted onto Luria–Bertani (LB) agar plates and incubated overnight at 37 °C to yield visible colonies. The MBC (≥3-log reduction of CFUs) of PACAP under physiological salt and buffer compositions ranged between ∼2 and 10 μM, which is comparable to other known HDPs. Since PACAP is an endogenous peptide and exhibits membrane-permeating activity, we studied whether it is cytotoxic to mammalian cells. Lactate dehydrogenase-release assays were conducted with murine WT bone marrow-derived macrophages (Fig. 2C). PACAP exhibited a similar cytotoxicity profile to LL37 and was not appreciably toxic relative to control at MBC concentrations. In parallel, radial diffusion assays were also performed to measure the spectrum of antimicrobial activity of PACAP against clinical isolates exhibiting multidrug-resistant (MDR) phenotypes. Fifteen different strains of bacteria and fungi were tested, under neutral and acidic pH conditions simulating CNS and bloodstream (pH 7.5) context vs. those encountered in abcesses or phagolysosomal microenvironments (e.g., pH 5.5; Fig. 2 D–I and SI Appendix, Fig. S5). Notably, PACAP exhibited potent antimicrobial activity against MDR S. aureus (SAUSA300), Acinetobacter baumannii (HUMC-1), Candida albicans (CA 36082S) and Salmonella typhimurium (ST 5996S) (Fig. 2 D–I). For some strains (e.g., SAUSA300), PACAP exhibited pH-dependent antimicrobial activity, while for others no statistical differences were observed in distinct pH conditions (e.g., HUMC-1).
Fig. 2.
PACAP preferentially induces NGC in microbial membranes to facilitate potent bactericidal and fungicidal activity. (A) PACAP recapitulates semiselective membrane remodeling activity of HDPs, inducing NGC in the bacterial model membrane PG/PE 20/80, but not in the eukaryotic-like PS/PC 20/80. A schematic of a transmembrane pore rich in NGC is shown. NGC is present everywhere along the surface highlighted in the dark shaded area spanned by the principal curvature C1 and C2. (B) PACAP was incubated with bacteria grown to midlog phase (1 × 106 CFU/mL). MBC assays were performed on S. aureus (SA113), E. coli (DH5ɑ), and P. aeruginosa (PA01), demonstrating strong antibacterial activity. (C) LDH-release assays with mouse macrophages demonstrate that PACAP is nontoxic to eukaryotic cells at bactericidal concentrations. Assays in B and C are representative of three independent experiments. (D–I) Radial diffusion assays show that PACAP exerts potent antibacterial and antifungal activity against MDR organisms, including MRSA (USA300), A. baumannii (HUMC-1), and C. albicans (CA 36082S). The diameter of induced clearing d is measured in millimeters. *P < 0.05, **P < 0.01. P values were calculated using a two-tailed Student’s t test.
PACAP Is a Multimodal HDP That Kills Bacteria and Fungi via Synergistic Mechanisms.
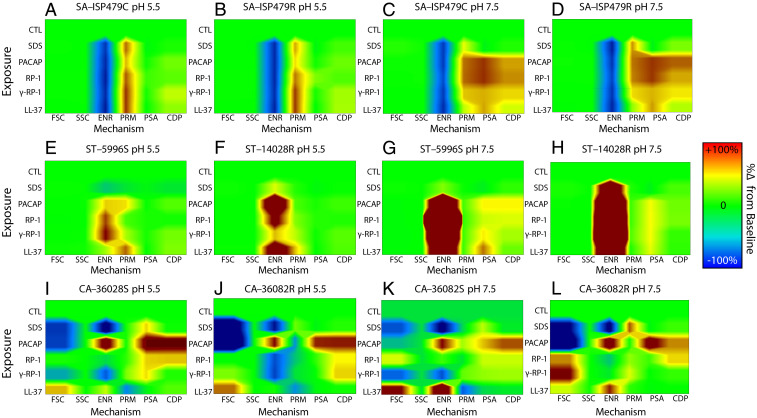
The preceding experiments demonstrated that PACAP has properties of an antibacterial and antifungal peptide that can induce NGC to facilitate target cell membrane disruption. To investigate its mechanisms of action, a multiparametric flow cytometry system was used to compare PACAP and other host-defense peptides by single-cell mechanistic fingerprinting (Fig. 3). Results showed that PACAP exerts antimicrobial activity via multiple mechanisms in an organism- and context-dependent manner, similar to classical HDPs. For example, PACAP kills S. aureus primarily via membrane permeabilization (PRM) at pH 5.5 but induces greater phosphatidylserine accessibility (PSA) and cell death protease (CDP) levels at pH 7.5; its impact on cellular energetics (ENR) is common to both conditions (Fig. 3 A–D). Conversely, PACAP antimicrobial activity against S. typhimurium is mediated less by ENR or PRM perturbation and more by induction of PSA and CDP (Fig. 3 E–H). Its antifungal activity against C. albicans is mediated by both membrane PRM and activation of CDP (Fig. 3 H–K). Collectively, these data confirm that PACAP possesses mechanistic signatures consistent with traditional HDPs that are known to depolarize, permeabilize, and/or induce regulated cell death pathways in human pathogens.
Fig. 3.
PACAP is a multimodal HDP that kills bacteria and fungi via synergistic mechanisms. Mechanistic fingerprinting experiments were conducted using single-cell flow cytometry. SDS (a membrane detergent), PACAP or comparator HDP LL-37, or kinocidin congeners RP-1 and ɣ-RP-1 were incubated with HDP-sensitive or HDP-resistant strains of S. aureus (A–D), S. typhimurium (E–H), or C. albicans (I–L) in the presence of specific fluorescent reporters (Materials and Methods). Six mechanistic parameters were measured in distinct pH conditions mirroring bloodstream (pH 7.5) or abscess/phagolysosomal (pH 5.5) contexts: forward scatter (FSC; cell size), side scatter (SSC; intracellular refractivity indicative of nucleic acid condensation), cellular membrane energetics (ENR; transmembrane potential), cell membrane permeability (PRM), PS accessibility (PSA; membrane turnover indicating early-stage regulated cell death), or activation of CDP (indicative of late-stage regulated cell death) pathways.
PACAP Is Strongly and Selectively Induced in the Brain and in Other Tissues in Response to Infection.
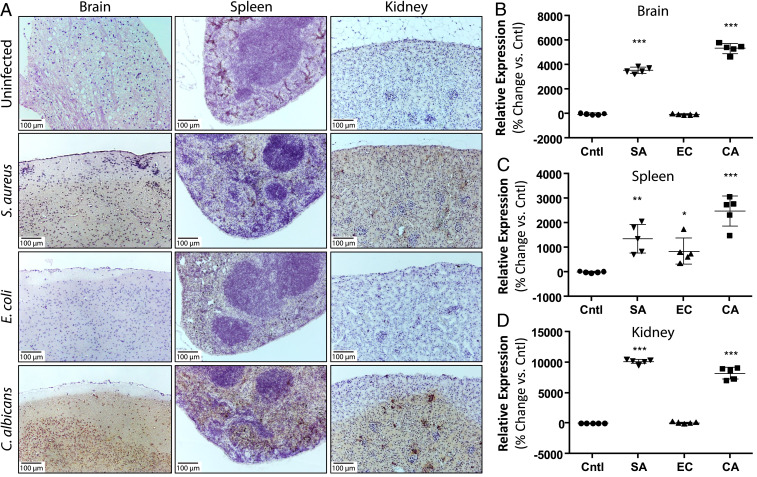
As PACAP exhibits potent antibacterial and antifungal activities, we investigated whether its expression levels in the brain and other tissues are regulated by bacterial and fungal infection. Murine experimental models of infection were selected based on clinically relevant parallels to those in humans, including manifestations in organs such as the brain, spleen, kidney, skin, and lung. We carried out immunohistochemistry of all five tissues in three mouse models of systemic infection (S. aureus, E. coli, or C. albicans). Mice were hematogenously inoculated with bacteria or fungi and relative expression levels of PACAP were quantified. Remarkably, PACAP was potently induced in anatomically relevant tissues within and beyond the CNS in response to infection (Fig. 4). Strikingly, in the brain, PACAP was induced up to 40- to 50-fold in response to S. aureus or C. albicans infection (Fig. 4 A and B and SI Appendix, Fig. S6). By comparison, PACAP induction as a result of E. coli infection was substantially more modest. Importantly, gram-staining of brain abscesses demonstrated strong colocalization of PACAP with S. aureus without infiltration of immune cells like neutrophils (SI Appendix, Fig. S7). Detailed pathology revealed vascular access of brain by S. aureus was associated with a marked up-regulation of perivascular and parenchymal PACAP expression, consistent with a localized PACAP-mediated response to defend against invasion (SI Appendix, Fig. S8). We also observed gross preservation of the parenchymal structure of the tissue despite presence of the microbial infection 48 h after initial seeding, well within the timescale of cellular infiltration. In spleen, PACAP was also surprisingly strongly induced, where it was markedly up-regulated ∼10- to 30-fold in response to S. aureus or C. albicans infection (Fig. 4 A and C). As in brain, E. coli induction of PACAP in spleen was considerably lower. Given that the spleen is a primary lymphoid organ important for lymphocyte-mediated and adaptive immunity, this striking up-regulation in this tissue suggests that PACAP plays a role in direct innate and adaptive host defense. This is consistent with the known hardwiring between the CNS and lymphoid organs via the autonomic nervous system (3). Finally, we found that PACAP expression levels were induced ∼80- to 100-fold in the kidney in response to S. aureus and C. albicans. Interestingly, PACAP expression was intensified in the medulla in response to the fungus C. albicans but more uniformly distributed vs. S. aureus (Fig. 4 A and D). These findings suggest that PACAP activity may be influenced by local conditions such as urea or osmotic tone in these tissue contexts. Expression levels of PACAP in the skin and the lung were less extensive, although modest induction in the lung parenchyma was observed in response to E. coli (SI Appendix, Fig. S9).
Fig. 4.
PACAP is strongly and selectively induced in the brain and other tissues in response to infection. (A) Immunohistochemistry for PACAP in brain, spleen, and kidney was compared in control (uninfected) and hematogenously-infected mice inoculated with S. aureus (SA), E. coli (EC), or C. albicans (CA). Images represent at least five independent experiments. PACAP induction was quantified using image processing in ImageJ. Relative change in expression was calculated vs. control uninfected tissue. All images are shown at 100× magnification. (B) In the brain, PACAP exhibits ∼40- to 50-fold induction in response to S. aureus and C. albicans infection but not E. coli. (C) In the spleen, PACAP is strongly induced in the presence of all three organisms (∼10- to 25-fold). (D) In the kidney, PACAP is induced ∼80- to 100-fold in response to infection with S. aureus and C. albicans, similar to brain. In B–D, n = 5 for all conditions, *P < 0.05, **P < 0.01, ***P < 0.001 relative to control. All other comparisons not significant. P values were calculated using a two-tailed Student’s t test.
PACAP Exerts Context-Dependent Antimicrobial Efficacy in the CNS and Other Tissues.
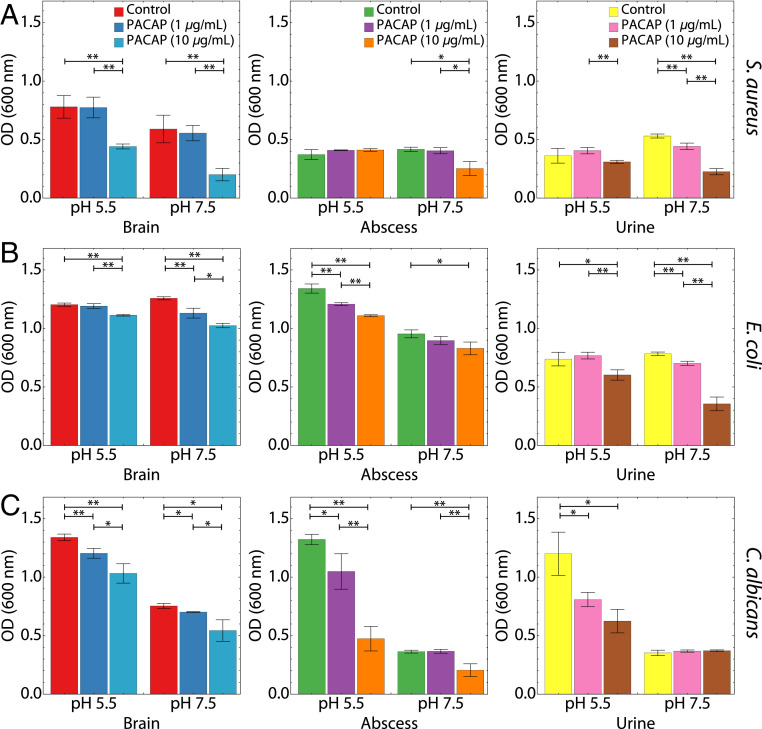
Our experimental in vivo models of infection identified dramatic PACAP up-regulation in multiple tissues in response to both bacteria and fungi. To determine whether PACAP antimicrobial activity varied with biological contexts, we decided to assess the relative efficacy of PACAP against target pathogens in vitro under conditions simulating clinically relevant scenarios, such as abscesses, urinary tract infections, and CNS infections (Fig. 5). Growth inhibition assays were conducted with S. aureus, E. coli, and C. albicans in media mimicking the physiological microenvironment of the brain, urine, and abscess. All three are known to cause septicemia via bloodstream infection. S. aureus is a common cause of brain abscesses and abscesses in general, E. coli is the predominant urinary pathogen and can also cause meningitis, and C. albicans can cause opportunistic infections in all three contexts. We find remarkable evidence for context-dependent antimicrobial activity of PACAP. Overall, PACAP was able to inhibit growth of all three organisms in the brain, urine, and abscess media to varying degrees, but especially against S. aureus in the brain, E. coli in the urine, and C. albicans in the brain, abscess, and urine.
Fig. 5.
PACAP exerts context-dependent antimicrobial efficacy. Growth inhibition assays were conducted in context-dependent media mimicking brain infections, urinary tract infections, and abscesses (Materials and Methods). For each context, 1 or 10 µg/mL of PACAP simulating low or high expression levels were incubated with (A) S. aureus, (B) E. coli, or (C) C. albicans at pH 5.5 and pH 7.5. Growth inhibition was measured after incubation at 37 °C. PACAP inhibited the growth of all organisms in brain-, urine- and abscess-simulating media to varying degrees. PACAP exhibited the strongest bactericidal ability against S. aureus and C. albicans in brain medium and against E. coli in urine medium. n = 3 for all assay conditions. *P < 0.05, **P < 0.01. All other comparisons were not significant. P values were calculated using a two-tailed Student’s t test.
PACAP exhibited the strongest dose-dependent bactericidal activity against S. aureus in brain-simulating conditions at both pH 5.5 and 7.5 (Fig. 5A), whereas it had comparatively weaker activity against S. aureus in media simulating abscess or urine contexts, although there was still significant activity. This pattern of results is consistent with the dramatic increase in PACAP expression observed in the brain upon infection with S. aureus (Fig. 4). S. aureus is a clinically significant pathogen in the brain, suggesting that PACAP is evolved to participate in molecular antimicrobial defense of the CNS while sparing inflammatory cell infiltration. PACAP activity against E. coli was present in brain and abscess media at both pH 7.5 and 5.5, with pronounced anti-E. coli efficacy in urine-simulating medium (Fig. 5B). PACAP also exhibited potent activity against the C. albicans in context of brain and abscesses media, and to a lesser degree in urine medium (Fig. 5C). Taken together, this data profile suggests PACAP is a pathogen-inducible resident antimicrobial neuropeptide specialized for contextual molecular host defense of the CNS and other tissues likely to be challenged by cognate, PACAP-inducible pathogens.
Discussion
Taken together, the present findings elucidate the antimicrobial properties and spectra, context- and organism-specific responses in vivo, empiric antimicrobial mechanisms of action and evolution of PACAP. These findings support the hypothesis that PACAP provides locally induced antimicrobial defense of the CNS from invasive bacteria and fungi without evoking immune cell infiltration and/or parenchymal damage. This strongly conserved neuropeptide possesses striking structural and physicochemical similarities to human cathelicidin LL37, despite low sequence similarity to known HDPs. PACAP exerts broad-spectrum antibacterial and antifungal efficacy, targeting multiple microbial pathways including membrane perturbation, disruption of cellular energetics, and activation of regulated cell-death pathways. Of special importance, current findings establish that PACAP is potently and selectively induced in the CNS and peripheral tissues relevant to molecular defense against metastatic seeding in bacteremia and fungemia. PACAP responses occurred with minimal immune cell infiltration or inflammatory damage.
The fact that PACAP is expressed throughout the central, peripheral, and enteric nervous systems suggests it likely plays a fundamental role in host defense at the interface of nervous and immune homeostasis within and beyond the CNS. It is interesting that PACAP can be expressed at lower levels in immune cells, in lymphoid organs, in blood matrices, and at immune barriers such as skin or mucosa. These insights provide strong evidence to support the hypothesis that PACAP affords rapid and effective host defense optimized to tissue contexts of infection. Consistent with this hypothesis, low plasma PACAP levels are observed in patients with nephrotic syndrome who are at increased risk for bacterial infection (34). PACAP is also known to be neuroprotective in mouse models of Alzheimer’s disease (AD) (35), consistent with hypotheses linking dysfunctions in HDPs or other immune mediators to AD resulting from latent and/or chronic CNS infection (36). Recent work has also shown that bacterial and fungal cryptic infections have been previously linked to AD (37, 38). Collectively, these themes suggest that PACAP is crucial for CNS antimicrobial host defense and that its deficiency or dysregulation renders hosts at risk for spontaneous or microbe-catalyzed neurodegeneration.
The established beneficial antiinflammatory and immunomodulatory functions of PACAP are consistent with its proposed role in CNS defense against invasive microbial infection while preserving CNS and neuronal integrity for cognitive function. PACAP is a potent suppressor of inflammatory cytokines and chemokines (3). PACAP has previously been shown to be antiinflammatory and neuroprotective in mouse models of multiple sclerosis (39). This role aligns to modulation of tissue damage from infection that is not predominantly due to microbe-mediated toxicity but rather from the off-target inflammatory response induced by infection (40). Unchecked, such responses can cause permanent physical and/or cognitive disability due to neuronal demyelination. Infections that can cause irreversible neuronal damage include brain abscesses, encephalitis, meningitis, and others. Such infections often evoke hyperinflammatory responses imposing amplified injurious effects in the confined space of the CNS. Thus, complementing its direct antimicrobial role, PACAP has been shown to down-regulate Th1/Th17 polarization, which usually induces a neutrophil-predominant immune response (41). In parallel, PACAP also shifts the immune axis toward a Th2 phenotype, away from cell-mediated immunology and toward molecular and humoral immunity (42). Interestingly, PACAP also directly inhibits neutrophil chemotaxis (43) and protects against septic shock (44). Thus, our present findings that PACAP is significantly up-regulated in response to S. aureus and Candida infection, which are predominantly cleared via Th17 responses (45) strongly support our hypothesis that PACAP plays crucial and simultaneous roles to defeat pathogens and limit any hyperinflammatory responses. This concept is further supported by the current data demonstrating PACAP to be physicochemically similar to human cathelicidin LL37. Substantial published evidence has shown LL37 to confer both immunomodulatory and antiinflammatory roles, such as modulation of Toll-like receptors by binding to pathogen-associated molecular patterns or damage-associated molecular patterns (28, 46–48).
That PACAP is one of the most conserved peptides throughout evolution (49) supports our hypothesis that PACAP arose at the intersection of evolving nervous, immune, and endocrine systems that emerged in early multicellular organisms and has since evolved into modern humans. Whether its archetypal role first emerged as antimicrobial or neurological, it is logical that the survival advantage afforded by PACAP would have been retained as an essential and multifunctional effector in higher organisms. Moreover, the relatively low rate of mutation observed over evolutionary time in PACAP may reflect selective pressure optimization of multiple integrated constraints to preserve critical functions, including GPCR-mediated signaling and antimicrobial as well as neuromodulatory activities. Notably, while the upstream GPCR-binding domain has been conserved absolutely over the course of evolution, the C-terminal antimicrobial domain has been subject to a greater degree of mutational plasticity (11). This may reflect coevolutionary antimicrobial mechanisms allowing PACAP to keep pace with rapidly evolving microbial targets. For example, from a neuroimmunomodulatory perspective, Hydra species possess rudimentary nervous systems, and a primitive innate immune system with Toll-like receptors (50). Hence, it stands to reason that Hydra species express an evolutionarily early PACAP ancestor exerting immunomodulatory and antiinfective activities. Also, a secreted neuropeptide can regulate the microbiome in Hydra (51), suggesting there are other neuro-AMPs like PACAP that are yet to be discovered or identified (51–53). Furthermore, prior investigations have demonstrated that neuropeptides are central to modulation of neuroinflammation (4) and regulation of the innate (5) and adaptive (7) immune responses, facilitating homeostatic communications between immune, endocrine, and nervous systems (6, 8). In support of these concepts, the AMP NLP-29 regulates neuronal dendrite degeneration in Caenorhabditis elegans (54), of potential relevance to synaptic remodeling during learning. Finally, beyond nervous and lymphoid tissue, PACAP is highly expressed in gut and skin (55), which comprise the two largest surfaces for host interaction with microbes (56). Collectively, the current findings highlight PACAP as a prototype host-defense neuropeptide with reciprocal antimicrobial and antineuroinflammatory activity. We hypothesize that PACAP can serve as a molecular template for design of next-generation antiinfectives with homeostatic capabilities inherently optimal for preventing or treating infections within or beyond the CNS.
Finally, the demonstration of a fundamental link between antimicrobial functions of the immune and nervous systems via ancient neuropeptides has potentially broader implications. In nature, fitness can be enhanced through multiplexed functions within the same molecule (57). This theme is illustrated by kinocidins (10, 58–60), which exert direct microbicidal activity, modulate innate receptors (61), and mediate immune cell chemotaxis. That molecules can integrate reciprocal immune and neurological functions raises intriguing possibilities (62, 63). For example, interestingly, in PACAP knockout models, mice have mood and psychotic disorders (64), and genetic variants of PACAP are associated with schizophrenic-like disorders in humans (65). Moreover, PACAP plasma levels correlate with symptom severity in posttraumatic stress disorder in humans (66). For example, GPCR-mediated outcomes such as mood can be connected to increased host-defense responses or stressors as occur in relation to microbial challenge or infection (3, 67). In this light, it is intriguing that the absence of nervousness, anxiety, and depression-like symptoms in mice (68) is correlated to the availability of PACAP (64). Importantly, our current findings support an emerging paradigm in which evolution of immune and nervous systems is linked (57, 69). The present findings suggest that microbial infection can lead to a spectrum of GPCR-activated consequences mediated via neuropeptides that have multiple functions in host defense and homeostasis. Thus, the striking rapid and extensive up-regulation of an ancient neuropeptide with GPCR-mediated activity in essential tissues (∼50-fold increase in the infected brain) may impact neuropsychiatric health and disease, which are emergent properties of the evolved CNS. Mounting evidence suggests that mental illness, behavioral stimuli, and sleep abnormalities may also be associated with consequences of chronic infection (70) beyond disruption of the microbiome. Conversely, behavioral interventions that modulate neuropeptide expression could be exploited to alleviate autoimmunity by suppressing unwanted immune responses (3). In summary, the specialized need to defend the CNS with resident neuropeptides rather than immune cells may ultimately reveal essential intersections between host neurological and immunological functions, and the microbiome.
Our work demonstrates a clear role for PACAP in context-dependent host defense in the CNS and other tissues, but the limitations of the present study should be understood. In order to more carefully control for experimental variables, our context-dependent antimicrobial assays were conducted in vitro under conditions that closely mimic the in vivo physiologic environment. We did not directly seed microbes into specific tissues in vivo or administer PACAP as an exogenous therapeutic. However, our in vivo bacteremia/fungemia model showed how marked up-regulation of endogenous PACAP production in tissues correlates with its context-dependent antimicrobial activity. Brain infections and abscesses are often the result of hematogenous spread rather than direct invasion from the external environment; thus, our study design more faithfully recapitulated actual clinical scenarios. We did not explore whether the dramatic up-regulation of PACAP resulted from the release of preformed PACAP or increased transcription of the gene. Another aspect that is not addressed experimentally here is the potential link between PACAP induction during infection and resulting changes in neuropsychiatric behavior, which would require behavioral studies in relevant experimental models. While studies addressing these intriguing hypotheses are beyond the scope of the current work, they are logical extensions of the present findings and ongoing in our laboratories.
Materials and Methods
Machine-Learning Screen for Membrane-Active Peptides.
A previously developed machine-learning classifier was used to discover hidden membrane activity in known neuropeptides. Full details of dataset curation, physicochemical descriptor selection, and model training and validation have been reported previously (9, 26, 33). Briefly, the classifier is based on a linear SVM that takes as input n = 12 physicochemical descriptors generated from the peptide sequence and outputs a score σ specifying the distance of the peptide from an (n − 1) = 11-dimensional hyperplane trained to optimally separate 243 known α-helical curvature-generating peptides from 243 decoy peptides. The 12 descriptors provided to the classifier were identified by principled feature selection as those most discriminatory in classification accuracy. A large, positive σ score correlates with increased ability to induce NGC in membranes, whereas a negative σ score indicates a lack of membrane-disruptive activity. This score can be converted through a monotonic function into a probability 0 < P(+1) < 1 that the peptide induces membrane curvature. Large, positive values of σ [P(+1) > 0.95] represent a high likelihood of membrane activity (ability to generate NGC), and negative values of σ [P(+1) < 0.50] indicate a low probability of membrane activity. Testing of the classifier on a blind balanced test set of 86 peptides demonstrated 91.9% prediction accuracy, 93.0% specificity, and 90.7% sensitivity. Experimental validation of computational predictions was carried out using SAXS. A subset of ⍺-helical test peptides was incubated with model membranes, and induced NGC was measured. A strong correlation between the ability to generate NGC in membranes and the distance-to-hyperplane SVM metric σ was observed. Classification of a single peptide requires ∼0.1 s of central processing unit time, permitting high-throughput computational screening for membrane-active peptide discovery and design.
Here, we used the machine-learning-based tool to screen an annotated neuropeptide database (2) for α-helical neuropeptides with potential antimicrobial activity in a similar manner to prior work (71). Neuropeptide sequences with lengths between 7 and 40 amino acids were considered, in concordance with the parameters of the training data. Using Python, physicochemical characteristics were calculated for each neuropeptide sequence and σ and P(+1) values were tabulated for each sequence. The top helical sequences were sorted by descending σ scores. For each peptide, the minimum homology to known AMPs was calculated (minHomologyAMP) using the Jukes–Cantor distance metric. The distance was calculated to 301 known helical AMPs pulled from the APD3 database (72).
Sequence Alignment of PACAP Family Sequences and Phylogenetic Reconstruction.
PACAP sequences from multiple organisms were obtained using a search on the UniProt database. Sequence alignment of PACAP sequences were conducted using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) (73). Sequence similarity and identity were calculated from the aligned sequences using an online web tool (www.bioinformatics.org/sms2/ident_sim.html). Phylogenetic trees were constructed using the PhyML method in SeaView 4.6.3 (74). Sequences were screened using the machine-learning classifier described above and σ and P(+1) scores were reported.
Mutational Analysis of PACAP.
An in silico exploration of the membrane activity fitness landscape of the PACAP sequence was conducted by generating all 722 unique single mutants (38 × 19) and screening them using the SVM-based membrane activity classifier (9). The σ values of all single mutants were quantitatively compared to the WT sequence by the metric Δσ = σmutant – σWT. An increase in σ upon mutation (Δσ > 0) corresponds to conservation of NGC-generating activity, and a decrease in σ upon mutation (Δσ < 0) predicts a loss of NGC-generating activity. The sign and magnitude of Δσ for each single mutant is plotted on a heat map and the distribution of Δσ is quantified.
Comparison of Amino Acid Content in PACAP to That of AMPs.
The amino acid composition of PACAP is compared to the compositions of known AMPs, including LL37. Previous work demonstrates that a relationship exists between the lysine-to-arginine ratio and mean hydrophobicity in membrane-active AMPs and that the relative amounts of these two quantities are major determinants of membrane remodeling required for topological changes (75). A set of 301 cationic helical AMP sequences were obtained from the AMP database as comparators (72). The mean hydrophobicity of a given peptide is calculated as
where n is the number of amino acids in the peptide and hi is the hydrophobicity of the ith amino acid in the peptide using the Eisenberg consensus hydrophobicity scale (76). The minimum and maximum mean hydrophobicity values within the set of AMP sequences were used to define a range. This range was then divided into 31 equal bins, into which the peptides were partitioned. For m peptides in a given bin, we sum over the jth peptide:
where NK/(NK + NR) is the ratio of the number of lysines to the total number of lysines plus arginines. For each bin, NK/(NK + NR) vs. mean hydrophobicity was plotted using Mathematica. PACAP and LL37 were analyzed and plotted using the same procedure (77).
PACAP/LL37 Sequence and Three-Dimensional Structural Alignment and Protein Data Bank–Wide Search for Structural Homologs.
Initial sequence alignment of PACAP and LL37 were carried out using the MatchMaker tool in Chimera (University of California, San Francisco). The Needleman–Wunsch alignment algorithm was used and the BLOSUM-62 matrix was used for residue similarity. Subsequently, the aligned residue pairs were fitted to generate a structural alignment. To search for other peptide and protein motifs with similar secondary structures, a Protein Data Bank–wide protein structural alignment analysis using DaliLite (78) was conducted on PACAP and LL37. The results of the analysis were first filtered by their Z-score and rmsd, which assesses the difference in the spatial arrangement of the amino acids between the peptide of interest and other proteins. Those with a Z-score greater than 3.0 and an rmsd value less than 4.0 were kept. The results were then sorted by percent physicochemical similarity. The functions of other diverse families of proteins and peptides related in structure were tabulated.
Host-Defense Peptides.
Comparative peptides used in this study are LL37, RP-1, and ɣ-RP-1. LL37 is a prototypical ⍺-helical human cathelicidin HDP with previously reported antimicrobial activity (79). The ⍺-helical mimetic peptide RP-1 is a synthetic 18-amino-acid congener engineered in part from the microbicidal ⍺-helix domains of the platelet factor 4 family of kinocidins (80). By comparison, ɣ-RP-1 is an ⍺-helical β−sheet peptide also based upon platelet factor 4 family of kinocidins. Both RP-1 and ɣ-RP-1 were synthesized, purified, authenticated by mass spectroscopy, and demonstrated to have strong antimicrobial activity in vitro and in vivo (81–83).
SAXS Experiments with Model Membranes.
Peptides were purchased and synthesized in high purity (>95% high-performance liquid chromatography) using solid-phase synthesis (Anaspec). Liposomes were prepared as previously described elsewhere (9). Briefly, lyophilized DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), DOPG (1,2-dioleoyl-sn- glycero-3-[phospho-rac-(1-glycerol)]), and DOPS [1,2-dioleoyl-sn-glycero-3-phospho-l-serine (sodium salt)] were purchased from Avanti Polar Lipids. Stock lipid solutions were dissolved in chloroform to 20 mg/mL. Mixtures of these lipids were used as models for bacterial membranes. DOPS and DOPE were mixed at a 1:4 molar ratio (DOPS/DOPE = 20/80), and DOPG and DOPE were mixed at a 1:4 molar ratio (DOPG/DOPE = 20/80). Immediately after mixing, chloroform was evaporated under nitrogen gas, and the lipids were further dried for 24 h under vacuum. Dried lipids were resuspended and vortexed in 140 mM NaCl + 10 mM Hepes at pH 7.4 to a final concentration of 20 mg/mL. Aqueous lipid solutions were incubated overnight at 37 °C. Liposomes were prepared by sonication of resuspended lipids until clear. Monodispersity in size was obtained via extrusion through a 0.2-μm filter.
Before use, peptides are solubilized in pure nuclease-free water. Liposomes (DOPS/DOPE = 20/80, DOPG/PE 20/80) and peptides were mixed at a peptide-to-lipid charge ratio (P/L) of 1/2 and equilibrated at room temperature overnight. Precipitated peptide–lipid complexes were loaded into 1.5-mm glass quartz capillaries (Mark-tubes; Hilgenberg GmbH) and hermetically sealed. Samples were incubated at 37 °C and centrifuged down at 6,000 rpm for 20 min before measurement. SAXS experiments were conducted at the Stanford Synchrotron Radiation Laboratory (BL 4-2) with monochromatic X-rays of energy 9 keV. Scattered radiation was collected using a Rayonix-MX225-HE detector (pixel size 73.2 μm). The two-dimensional diffraction patterns were integrated using the Nika 1.76 package (84) for Igor Pro-6.37 (Wavemetrics) and FIT2D (85). For all samples, multiple measurements of different samples were taken to ensure mutual consistency. No changes were observed over multiple experiments and exposures.
SAXS data were analyzed by plotting the integrated scattering intensity I(q) vs. q using Mathematica. To determine the lipid phases induced in each sample, the Bragg peak positions were measured, and their ratios were compared to the permitted reflections for different liquid-crystalline phases (e.g., lamellar, hexagonal, and cubic). Curvature-generating ability of the peptides were determined by calculating the lattice parameter a for the measured cubic phases. Cubic phases observed belonged to the Im3m and Pn3m space groups. Measured q positions for the Bragg peak reflections were fitted to the equation qmeasured = 2π√(h2 + k2 + l2)/a, where (hkl) are the Miller indices (86). The quantity of average induced NGC in the unit cell was calculated using the equation <k> = 2πχ/A0d2, where χ is the Euler characteristic and A0 is the surface area per cubic unit cell for each phase. Parameter values are χ = −4 and A0 = 2.345 for Im3m and χ = −2 and A0 = 1.919 for Pn3m.
Bacterial Killing Assays.
Plate killing assays were carried out as previously described (87). Briefly, S. aureus (SA113), P. aeruginosa (PA01), and E. coli (DH5⍺) cells from freshly streaked LB broth agar plates were inoculated into LB and grown overnight at 37 °C in a shaker incubator into the stationary phase. The next day, a 1/100 dilution was made of the overnight culture into fresh LB and was incubated at 37 °C for 2 to 3 h to midlog growth phase (optical density at 600 nm = 0.4 to 0.6). The midlog phase culture was diluted to 107 CFU/mL in a sterile filtered buffer of 100 mM NaCl + 10 mM Hepes (pH 7.4). We inoculated a 20-μL suspension of each bacteria into 180 μL of buffer containing various concentrations of PACAP in 96-well plates for a total of 2 × 105 CFU per well (1/2 serial dilutions from 50 μM to 3.125 μM with a buffer-only control). The 96-well killing plates were sealed with Parafilm and incubated on microplate shakers at 37 °C for 1 h. Postincubation, 10-fold serial dilutions were made with sterile buffer. Half of each dilution (100 μL) was spotted onto LB agar plates and incubated overnight at 37 °C to yield visible colonies. Colonies were quantified and plotted as a function of antimicrobial concentration. All killing assays were done in three replicates under identical conditions.
Antimicrobial Radial Diffusion Assays.
Peptides were assayed for antimicrobial activity using a well-established radial diffusion method adjusted to pH 7.5 or 5.5 (88). The efficacy of these peptides was evaluated against a panel of human pathogens, including AMP-susceptible and -resistant isogenic pairs: S. aureus [ISP479C/ISP479R (89)]; S. typhimurium [MS5996S/MS14028 (83, 90)]; P. aeruginosa (PA01), A. baumanni (19606); and C. albicans [36082S/36082R (91)]. Bacteria were grown to midlog phase and inoculated at 106 CFU/mL into warm, liquid buffered molecular-grade agarose plates. Peptides (10 µg) were introduced into wells in the seeded matrix and incubated for 3 h at 37 °C. Nutrient overlay medium was applied, and the agarose plates were incubated at 37 °C or 30 °C for bacteria or fungi, respectively. After 24 h, zones of inhibition were measured in millimeters. Independent experiments were repeated a minimum of two times.
Cytotoxicity Assays.
Cytotoxicity of peptides against WT bone-marrow-derived macrophages was measured using a lactate dehydrogenase (LDH) release assay kit (Promega). Macrophages were grown to confluency in RPMI 1640 + bovine growth serum + penicillin/streptomycin + l-glutamine (Gibco), washed, and seeded into 96-well microplates with a final concentration of 105 cells per well. Peptides were diluted into 140 mM NaCl + 10 mM Hepes (pH 7.4) buffer and 1:2 serial dilutions were made starting at 100 μM, including a buffer-only control. Fifty microliters of each peptide at each dilution was transferred into 96-well microplates containing the macrophages and incubated at 3 h in a 37 °C shaker. After incubation, supernatant from the cells were removed and measured for LDH release. Fifty microliters of supernatant was placed into a fresh 96-well plate, 50 μL of LDH substrate mix was added, and the plate was incubated at room temperature for 2 min under foil. Fifty microliters of stop solution was added, and the plate was centrifuged briefly to remove air bubbles. The plate absorbance was read at 490 nm and LDH release vs. peptide concentration was quantified.
Flow Cytometry and Mechanistic Fingerprinting.
Mechanisms of PACAP and comparative host-defense peptide action were assessed as previously described (88). PACAP was incubated with a gram-positive (S. aureus), gram-negative (S. typhimurium), and a fungus (C. albicans). Six-parameter multicolor flow cytometry was used to analyze four specific mechanisms of HDP action and two generalized effects associated with the study peptides: 1) perturbation of cell membrane (CM) energetics (ENR) (e.g., transmembrane potential), 2) CM permeabilization (PRM), 3) annexin V binding/PSA (negatively charged phospholipids and CM turnover), 4) induction of caspase-like/metacaspase-like regulated CDP, 5) osmodisruption (forward scatter [FSC]), and 6) CM invagination/nucleic acid condensation (side scatter [SSC] granularity). The following fluorophores were used with a FACSCalibur instrument (Becton Dickinson): 3,3-dipentyloxacarbocyanine (DiOC5) (excitation, 484 nm; emission, 660 nm) (Invitrogen) for ENR, propidium iodide (PI) (excitation, 535 nm; emission, 620 nm) (Sigma) for PRM, annexin V-allophycocyanin conjugate (ANX-V) (excitation, 650 nm; emission, 660 nm) (Invitrogen) for ANX, and CellEvent caspase-3/7 green (C-3/7) (excitation, 502 nm; emission, 530 nm) (Invitrogen) for CDP. FSC and SSC were measured in parallel. Logarithmic-phase organisms were adjusted to 106 CFU/mL in Pipes (pH 7.5) or MES (pH 5.5) and exposed to 20 μg/mL of each HDP of interest for 1 h at 37 °C. Based on extensive pilot data, this peptide concentration was used to achieve ∼50% survival given the high inoculum of bacteria exposed. The HDPs tested included PACAP, LL37, and kinocidin congeners RP-1 and ɣ-RP-1. After addition of the HDP to each organism of interest, a triple-stain mixture containing DiOC5 (0.5 μM), PI (5.0 μg/mL), and ANX-V (2.5 μL/mL) in 50 mM potassium-containing MEM (K+ MEM) (without phenol red; Sigma) was added to each sample following incubation. Samples were stained at room temperature for 15 min before flow cytometry. Parallel samples were incubated with 30 μL of C-3/7 reagent for 30 min at 37 °C following peptide exposure. After incubation, 400 mL of phosphate-buffered saline (PBS) was added to remove any background signal. Sodium dodecyl sulfate (SDS) (10%, wt/vol; Ambion) (a nonspecific perturbant of ENR and PRM) or buffers alone (K+ MEM or PBS) were included as controls in each experiment. Fluorescence of a minimum of 10,000 cells was acquired from each sample, and results from a minimum of two independent studies conducted on different days were used for statistical analyses. Resulting mechanistic fingerprints (mechanigrams) were visualized by KyPlot software.
Animal Models of Infection.
All animal studies were performed in adherence to Institutional Animal Care and Use Committee review and approval at the Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center. For bacteremia models, immunocompetent Balb/C mice (22 g) were acclimated for 2 d prior to infection. S. aureus (strain LAC-USA300), E. coli (strain 43889), or C. albicans (strain 36082) were used as prototypic gram-positive bacterium, gram-negative bacterium, and fungal pathogens, respectively. Each of these microbial strains has been well-characterized previously. Each strain was cultured from virulence-validated master cell banks and grown to log phase in appropriate media at 37 °C (bacteria) or 30 °C (fungus). Resulting log-phase cells were harvested, washed, suspended in PBS, sonicated, quantified by spectrophotometry, and diluted to the desired CFU inoculum in PBS. Infection was achieved via intravenous tail vein injection using a standard, nonlethal inoculum of 106 CFU (100 μL PBS). At 48 h postinfection, brain, kidney, spleen, lung, and skin were excised and processed for quantitative microbial culture, histology, and immunohistochemistry.
Histology and Immunohistochemistry.
Tissue morphology and infection were assessed by hematoxylin/eosin and gram staining, respectively. PACAP expression relative to infection was assessed in tissues at the study endpoint as previously described (92). In brief, tissues were aseptically dissected from animals, fixed in 10% zinc formalin, and embedded in paraffin. Next, 3-μm sections were cut, dewaxed, and rehydrated, followed by heat-induced antigen retrieval (Dako). Samples were blocked with dual endogenous enzyme block (Dako) and 2 to 10% normal serum corresponding to the secondary antibody. Sections were then incubated with a primary antibody targeting PACAP (BS-0200R; Bioss), followed by staining with a secondary antibody conjugated to horseradish peroxidase or biotin (Santa Cruz Biotechnology). Colorimetric development was achieved by reaction with streptavidin–horseradish peroxidase (Dako) and 3,3′-diaminobenzidine (DAB; Vector Laboratories), and the samples were counterstained with hematoxylin. Images were visualized using a Zeiss BX43 microscope with a DP21 digital camera.
Quantification of PACAP Induction from Immunohistochemistry.
Immunohistochemistry Images were quantitatively analyzed using ImageJ software (https://imagej.nih.gov/ij/) (93) as previously described (92). Outcomes were enumerated as an integral of staining in a given tissue sample compared to uninfected control. A minimum of five images with identical total areas were quantified for each study group; areas containing no tissue were subtracted as background to afford internal control. PACAP expression indices were calculated by comparison to uninfected control values. Data are presented as mean values ± SDs. Differences in experimental results were compared by Student’s t test and ANOVA, where appropriate. P values of P < 0.05 were considered statistically significant. Data were visualized using GraphPad Prism software.
PACAP Efficacy in Media Simulating Infection Context.
In parallel with experimental murine models of infection in vivo, studies were performed to assess relative efficacy of PACAP against target pathogens in conditions simulating cognate physiological contexts in vitro. Organisms were grown to midlog phase and inoculated at 106 CFU/mL into prewarmed media simulating respective contexts of infection investigated in vivo: 1) brain medium: RPMI medium (92.5%) containing 5% vol/vol of brain–heart infusion (BHI), 2% vol/vol PBS, and 0.5% mouse serum vol/vol for bacteria (S. aureus or E. coli) or RPMI medium (67%) containing 30% vol/vol of BHI, 2% vol/vol PBS, and 0.001% mouse serum vol/vol for C. albicans; 2) abscess medium: RPMI medium (92.5%) containing 5% vol/vol nutrient broth (NB), 2% vol/vol PBS, and 0.5% vol/vol mouse serum for bacteria or RPMI medium (84.9%) containing 15% vol/vol yeast nitrogen broth (YNB with 1% glucose), 2% PBS, and 0.01% mouse serum for C. albicans; 3) urine medium: HBSS medium (89.5%) containing 10% vol/vol NB, 38 mM urea, and 6.8 mM sodium chloride for S. aureus or HBSS medium (94.5%) containing 5% vol/vol NB, 38 mM urea, and 6.8 mM sodium chloride for E. coli or HBSS medium (84.9%) containing 15% vol/vol YNB (with 1% glucose), 7.5 mM urea, and 1.4 mM sodium chloride for C. albicans. PACAP was introduced into media in concentrations simulating low (1 µg/mL) vs. high (10 µg/mL) expression, corresponding to findings determined in vivo as induced by distinct organisms in respective tissues (Fig. 5). Media were incubated at 37 °C and cultured for survival/nonsurvival at indicated time points. Independent experiments performed identically were repeated a minimum of three times and results were analyzed for statistical significance.
Statistical Analyses.
Unless otherwise indicated, statistical analysis used an unpaired Student’s t test. Differences were considered statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Ling Wang and Hong-Kyu Lee, M.S. for their efforts in antimicrobial assessment of PACAP. E.Y.L. acknowledges support from the Systems and Integrative Biology Training Program (NIH T32GM008185), the Medical Scientist Training Program (NIH T32GM008042), and the Dermatology Scientist Training Program (NIH T32AR071307) at the University of California, Los Angeles. E.Y.L. also acknowledges an Early Career Research Grant from the National Psoriasis Foundation. G.C.L.W. is supported by NIH R01AI143730, NIH R01AI052453, NSF DMR1808459, and the National Psoriasis Foundation (20194384). M.R.Y. acknowledges grant support from the National Institute of Allergy and Infectious Diseases (NIAID) AI-124319 and AI-111661. K.-F.L. is supported by grants from the NIH (OD023076, MH114831, NS115183, AG054714, AG062232, and AG064049), a grant to the Salk core facility (CA014195), the Clayton Foundation and the Freeburg Foundation, and by funding from the Helen McLoraine Chair. X-ray research was conducted at Stanford Synchrotron Radiation Lightsource, SLAC National Laboratory, supported by the US Department of Energy (DOE) Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
Competing interest statement: M.R.Y is founder and shareholder of NovaDigm Therapeutics, Inc., which pursues novel antiinfective agents and strategies; he holds patents in the area of antimicrobial peptides and related molecules.
This article is a PNAS Direct Submission.
*We have run the SVM classifier on the PACAP metabolite identified here and found it to be antimicrobial but with a different score than the parent PACAP peptide, which is consistent with the findings of the authors.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917623117/-/DCSupplemental.
Data Availability.
All study data are included in the paper and SI Appendix.
References
- 1.van den Pol A. N., Neuropeptide transmission in brain circuits. Neuron 76, 98–115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., et al. , NeuroPep: A comprehensive resource of neuropeptides. Database (Oxford) 2015, bav038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinman L., Elaborate interactions between the immune and nervous systems. Nat. Immunol. 5, 575–581 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Mykicki N., et al. , Melanocortin-1 receptor activation is neuroprotective in mouse models of neuroinflammatory disease. Sci. Trans. Med. 8, 362ra146 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Zugasti O., et al. , Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nat. Immunol. 15, 833–838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardoso V., et al. , Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klose C. S. N., et al. , The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallrapp A., et al. , The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee E. Y., Fulan B. M., Wong G. C. L., Ferguson A. L., Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl. Acad. Sci. U.S.A. 113, 13588–13593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yount N. Y., et al. , Unifying structural signature of eukaryotic α-helical host defense peptides. Proc. Natl. Acad. Sci. U.S.A. 116, 6944–6953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherwood N. M., Krueckl S. L., McRory J. E., The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon superfamily. Endocr. Rev. 21, 619–670 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Chao M. V., Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat. Rev. Neurosci. 4, 299–309 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Zhou C. J., et al. , PACAP and its receptors exert pleiotropic effects in the nervous system by activating multiple signaling pathways. Curr. Protein Pept. Sci. 3, 423–439 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Abad C., Waschek J. A., Immunomodulatory roles of VIP and PACAP in models of multiple sclerosis. Curr. Pharm. Des. 17, 1025–1035 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Delgado M., et al. , PACAP in immunity and inflammation. Ann. N. Y. Acad. Sci. 992, 141–157 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Lugo J. M., et al. , Evidence for antimicrobial and anticancer activity of pituitary adenylate cyclase-activating polypeptide (PACAP) from North African catfish (Clarias gariepinus): Its potential use as novel therapeutic agent in fish and humans. Fish Shellfish Immunol. 86, 559–570 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Semple S. L., et al. , PACAP is lethal to Flavobacterium psychrophilum through either direct membrane permeabilization or indirectly, by priming the immune response in rainbow trout macrophages. Front. Immunol. 10, 926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr C. G., Maderdrut J. L., He J., Coy D. H., Wimley W. C., Pituitary adenylate cyclase-activating polypeptide is a potent broad-spectrum antimicrobial peptide: Structure-activity relationships. Peptides 104, 35–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debbabi S., et al. , Antibacterial properties of the pituitary adenylate cyclase-activating polypeptide: A new human antimicrobial peptide. PLoS ONE 13, e0207366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan L. C., et al. , Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc. Natl. Acad. Sci. U.S.A. 115, E11111–E11119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yount N. Y., et al. , Context mediates antimicrobial efficacy of kinocidin congener peptide RP-1. PLoS ONE 6, e26727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yount N. Y., Yeaman M. R., Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 52, 337–360 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Schmidt N. W., Wong G. C. L., Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr. Opin. Solid State Mater. Sci. 17, 151–163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M. W., et al. , Molecular motor Dnm1 synergistically induces membrane curvature to facilitate mitochondrial fission. ACS Cent. Sci. 3, 1156–1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silvestre-Roig C., et al. , Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature 569, 236–240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E. Y., Wong G. C. L., Ferguson A. L., Machine learning-enabled discovery and design of membrane-active peptides. Bioorg. Med. Chem. 26, 2708–2718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E. Y., Lee M. W., Wong G. C. L., Modulation of toll-like receptor signaling by antimicrobial peptides. Semin. Cell Dev. Biol. 88, 173–184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee E. Y., et al. , Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 10, 1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semple C. A., Gautier P., Taylor K., Dorin J. R., The changing of the guard: Molecular diversity and rapid evolution of beta-defensins. Mol. Divers. 10, 575–584 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Tennessen J. A., Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J. Evol. Biol. 18, 1387–1394 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Yeaman M. R., Yount N. Y., Unifying themes in host defence effector polypeptides. Nat. Rev. Microbiol. 5, 727–740 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Cao Y. J., et al. , Identification of binding domains of pituitary adenylate cyclase activating polypeptide (PACAP) for its type 1 receptor by photoaffinity labeling. Ann. N. Y. Acad. Sci. 865, 82–91 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Lee E. Y., Lee M. W., Fulan B. M., Ferguson A. L., Wong G. C. L., What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus 7, 20160153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eneman B., et al. , Pituitary adenylate cyclase-activating polypeptide deficiency associated with increased platelet count and aggregability in nephrotic syndrome. J. Thromb. Haemost. 13, 755–767 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Rat D., et al. , Neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP) slows down Alzheimer’s disease-like pathology in amyloid precursor protein-transgenic mice. FASEB J. 25, 3208–3218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sochocka M., Zwolińska K., Leszek J., The infectious etiology of Alzheimer’s disease. Curr. Neuropharmacol. 15, 996–1009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso R., et al. , Fungal infection in patients with Alzheimer’s disease. J. Alzheimers Dis. 41, 301–311 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Dominy S. S., et al. , Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5, eaau3333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Y.-V., et al. , Pituitary adenylyl cyclase-activating polypeptide is an intrinsic regulator of Treg abundance and protects against experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 106, 2012–2017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Neutrophils in tissue injury and repair. Cell Tissue Res. 371, 531–539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waschek J. A., VIP and PACAP: Neuropeptide modulators of CNS inflammation, injury, and repair. Br. J. Pharmacol. 169, 512–523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delgado M., Leceta J., Sun W., Gomariz R. P., Ganea D., VIP and PACAP induce shift to a Th2 response by upregulating B7.2 expression. Ann. N. Y. Acad. Sci. 921, 68–78 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Kinhult J., Uddman R., Laan M., Lindén A., Cardell L. O., Pituitary adenylate cyclase-activating peptide inhibits neutrophil chemotaxis. Peptides 22, 2151–2154 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Martinez C., et al. , Anti-inflammatory role in septic shock of pituitary adenylate cyclase-activating polypeptide receptor. Proc. Natl. Acad. Sci. U.S.A. 99, 1053–1058 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin L., et al. , Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog. 5, e1000703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee E. Y., et al. , A review of immune amplification via ligand clustering by self-assembled liquid-crystalline DNA complexes. Adv. Colloid Interface Sci. 232, 17–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee E. Y., et al. , Crystallinity of double-stranded RNA-antimicrobial peptide complexes modulates toll-like receptor 3-mediated inflammation. ACS Nano 11, 12145–12155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi T., et al. , Cathelicidin promotes inflammation by enabling binding of self-RNA to cell surface scavenger receptors. Sci. Rep. 8, 4032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.On J. S. W., Chow B. K. C., “Molecular evolution of pituitary adenylyl cyclase-activating polypeptide subfamily and cognate receptor subfamily” in Pituitary Adenylate Cyclase Activating Polypeptide—PACAP, Reglodi D., Tamas A., Eds. (Springer International Publishing, ed. 1, 2016), pp. 3–17. [Google Scholar]
- 50.Franzenburg S., et al. , MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. U.S.A. 109, 19374–19379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Augustin R., et al. , A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat. Commun. 8, 698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimovich A. V., Bosch T. C. G., Rethinking the role of the nervous system: Lessons from the Hydra holobiont. BioEssays 40, e1800060 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Klimovich A., et al. , Prototypical pacemaker neurons interact with the resident microbiota. Proc. Natl. Acad. Sci. U.S.A. 117, 17854–17863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.e L., et al. , An antimicrobial peptide and its neuronal receptor regulate dendrite degeneration in aging and infection. Neuron 97, 125–138.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y. E., Fischbach M. A., Belkaid Y., Skin microbiota-host interactions. Nature 553, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brogden K. A., Guthmiller J. M., Salzet M., Zasloff M., The nervous system and innate immunity: The neuropeptide connection. Nat. Immunol. 6, 558–564 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Lee M. W., Lee E. Y., Wong G. C. L., What can pleiotropic proteins in innate immunity teach us about bioconjugation and molecular design? Bioconjug. Chem. 29, 2127–2139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meller S., et al. , T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat. Immunol. 16, 970–979 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan A., et al. , Direct antimicrobial activity of IFN-β. J. Immunol. 198, 4036–4045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dishman A. F., et al. , Switchable membrane remodeling and antifungal defense by metamorphic chemokine XCL1. ACS Infect. Dis. 6, 1204–1213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lande R., et al. , CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis. Nat. Commun. 10, 1731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee E. Y., et al. , Functional reciprocity of amyloids and antimicrobial peptides: Rethinking the role of supramolecular assembly in host defense, immune activation, and inflammation. Front. Immunol. 11, 1629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stolzenberg E., et al. , A role for neuronal alpha-synuclein in gastrointestinal immunity. J. Innate Immun. 9, 456–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto H., et al. , Altered psychomotor behaviors in mice lacking pituitary adenylate cyclase-activating polypeptide (PACAP). Proc. Natl. Acad. Sci. U.S.A. 98, 13355–13360 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hashimoto R., et al. , Pituitary adenylate cyclase-activating polypeptide is associated with schizophrenia. Mol. Psychiatry 12, 1026–1032 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Ressler K. J., et al. , Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470, 492–497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black P. H., Central nervous system-immune system interactions: Psychoneuroendocrinology of stress and its immune consequences. Antimicrob. Agents Chemother. 38, 1–6 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hashimoto H., et al. , Depression-like behavior in the forced swimming test in PACAP-deficient mice: Amelioration by the atypical antipsychotic risperidone. J. Neurochem. 110, 595–602 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Lee M. W., Lee E. Y., Ferguson A. L., Wong G. C. L., Machine learning antimicrobial peptide sequences: Some surprising variations on the theme of amphiphilic assembly. Curr. Opin. Colloid Interface Sci. 38, 204–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsiao E. Y., et al. , Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yount N. Y., et al. , Discovery of novel type II bacteriocins using a new high-dimensional bioinformatic algorithm. Front. Immunol. 11, 1873 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang G., Li X., Wang Z., APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gouy M., Guindon S., Gascuel O., SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Schmidt N. W., et al. , Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133, 6720–6727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eisenberg D., Weiss R. M., Terwilliger T. C., Wilcox W., Hydrophobic moments and protein structure. Faraday Symp Chem Soc 17, 109 (1982). [Google Scholar]
- 77.Realegeno S., et al. , S100A12 is part of the antimicrobial network against Mycobacterium leprae in human macrophages. PLoS Pathog. 12, e1005705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nizet V., et al. , Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Yeaman M. R., et al. , Modular determinants of antimicrobial activity in platelet factor-4 family kinocidins. Biochim. Biophys. Acta 1768, 609–619 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeaman M. R., Platelets: At the nexus of antimicrobial defence. Nat. Rev. Microbiol. 12, 426–437 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Yount N. Y., et al. , Structural correlates of antimicrobial efficacy in IL-8 and related human kinocidins. Biochim. Biophys. Acta 1768, 598–608 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Yeaman M. R., Gank K. D., Bayer A. S., Brass E. P., Synthetic peptides that exert antimicrobial activities in whole blood and blood-derived matrices. Antimicrob. Agents Chemother. 46, 3883–3891 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ilavsky J., Nika: Software for two-dimensional data reduction. J. Appl. Cryst. 45, 324–328 (2012). [Google Scholar]
- 85.Hammersley A. P., “FIT2D: An introduction and overview” (European Synchrotron Radiation Facility internal report ESRF97HA02T, 1997).
- 86.Sankhagowit S., Lee E. Y., Wong G. C. L., Malmstadt N., Oxidation of membrane curvature-regulating phosphatidylethanolamine lipid results in formation of bilayer and cubic structures. Langmuir 32, 2450–2457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt N. W., et al. , Engineering persister-specific antibiotics with synergistic antimicrobial functions. ACS Nano 8, 8786–8793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaili S., et al. , The GraS sensor in Staphylococcus aureus mediates resistance to host defense peptides differing in mechanisms of action. Infect. Immun. 84, 459–466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yount N. Y., Yeaman M. R., Structural congruence among membrane-active host defense polypeptides of diverse phylogeny. Biochim. Biophys. Acta 1758, 1373–1386 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Fields P. I., Groisman E. A., Heffron F., A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243, 1059–1062 (1989). [DOI] [PubMed] [Google Scholar]
- 91.Sheppard D. C., et al. , Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279, 30480–30489 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Chan L. C., et al. , Innate immune memory contributes to host defense against recurrent skin and skin structure infections caused by methicillin-resistant Staphylococcus aureus. Infect. Immun. 85, e00876-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the paper and SI Appendix.