Sir,
Carbapenem-resistant Acinetobacter baumannii (CRAB) are recognized by the Centers for Disease Control and Prevention (CDC) as an urgent threat (1). The most prevalent mechanism of carbapenem resistance in Acinetobacter spp. is due to the presence of carbapenem-hydrolyzing class D β-lactamases (CHDLs) with OXA-23 the most common. The observed dispersion of blaOXA-23 is attributed in part to the spread of successful global clones such as GC1 and GC2 (2). However, in the most recent years an increased emergence of blaNDM-1 harboring Acinetobacter spp. isolates were reported (3).
A surveillance program in Argentina reported the increased occurrence of Acinetobacter NDM-1 isolates (3). A total of 15,621 carbapenem resistant isolates were included in the surveillance 40 of them were blaNDM positive (33 A. baumannii and 7 non-A. baumannii strains). All the A. baumannii isolates belonged to the ST25 and were closely related, suggesting a recent spread of NDM-1. In these strains, blaNDM-1 was located in the chromosome rather than on a plasmid as was found in non-A. baumannii strains. In addition, a resistance island carrying blaPER-7 was found on a plasmid in some A. baumannii strains (3). Reports of NDM-1 positive ST25 isolates have been restricted to some regions, including Latin America, Central and Western Europe, East Africa and Western Asia (3, 8–12).
In the present work, we further characterized the ST25 NDM-1 A. baumannii isolates and compared them to representative strains of the most prevalent clonal complexes (GC1 and GC2) (4, 5). Twenty-two representative strains recovered from different sources, years, and location were selected (Table S1).Among the selected strains, we included eight NDM-1 positive A. baumannii strains, seven OXA-23 positive A. baumannii strains, two strain with upregulation of the endogenous OXA-51-like (OXA-66) carbapenemase, and five NDM-1 positive non-A. baumannii strains (Table S1). Phenotypic assays including motility, macrocolony formation (6), hydrogen peroxide susceptibility, osmotolerance, and desiccation tolerance (7), were performed.
Surface-motility assays were performed on lysogeny broth, LB, agar plates with 0.3% of agarose. None of the tested A. baumannii strains exhibited surface-associated motility (Figure 1). Most of the non-A. baumannii strains also did not exhibit surface-associated motility with the exception of AMA 43. To study biofilm formation, macrocolonies were used as a biofilm model system. The A. baumannii strains were grown on LB agar supplemented with Congo red (6). Macrocolony formation varied among the ST25, GC1, GC2, and non-A. baumannii strains. A few of the ST25 strains produced more extracellular matrix (ECM) than others, i.e. AMA 40 and AMA 46 produced more ECM in comparison to AMA 16 (Figure S1). Among the non-A. baumannii strains, AMA 23 was the one that produced more ECM. In general, we observed that GC1, GC2 and the non-A. baumannii strains produced less ECM compare with the ST25 strains. However, substantial variability of ECM production was observed among the studied strains (Figure S1).
Figure 1.
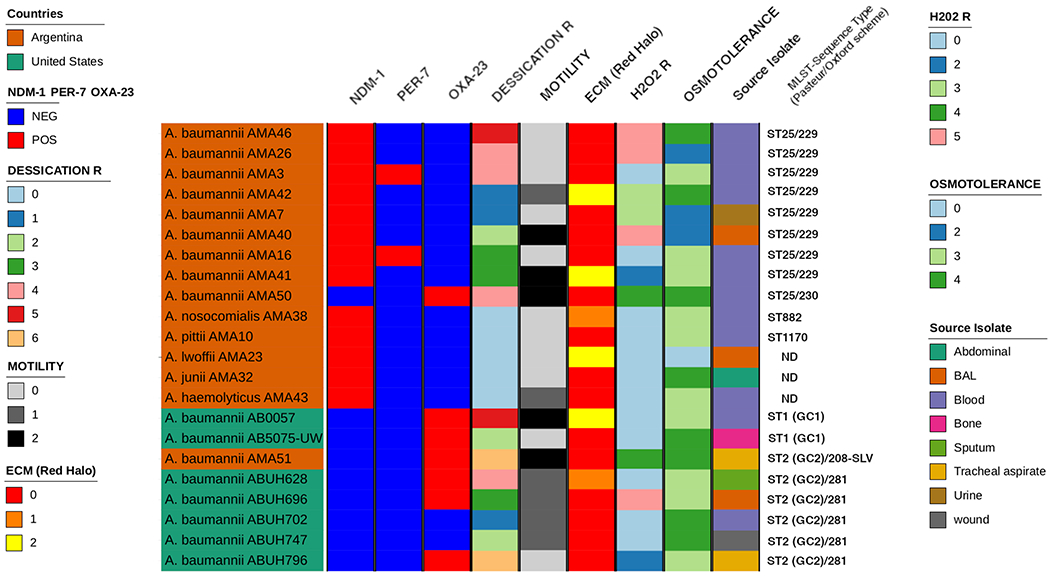
Graphic representation of the different phenotypes of Acinetobacter sp. used in this study. The graphic representation was performed by ComplexHeatmap package (cita_1). The sequence type data was determined by MLST script (https://github.com/tseemann/mlst).
Desiccation assays were performed for seven days (7). Data collected on days zero, one, four and seven were recorded. Dried cells were incubated at room temperature and at the indicated times dried samples were suspended, and survival was assessed by CFU/mL counts. ST25, GC1, and GC2 A. baumannii strains demonstrated a decrease in viability as the days progressed. After day four, there were no GC2 strains that survived for the day seven collection (Figure 1). Select ST25 strains had an increase in viability from day one to day four. Non-A. baumannii strains were not viable after one day of desiccation (Figure 1).
To assess osmotolerance, bacterial strains were treated and incubated with 0.6 M NaCl (7). Data were collected at hours four, eight and twenty-four. We observed that all of the NDM-1 positive A. baumannii strains and the GC1 and GC2 strains, were able to resist the NaCl treatment and were viable after treatment. Only the AMA23 (non-baumannii) strain was not viable after treatment (Figure 1).
Finally, hydrogen peroxide susceptibility was performed only in A. baumannii strains (7). We observed that hydrogen peroxide treated cells were less viable than non-treated cells. Some strains were not able to resist the peroxide treatment such as AMA3, AMA16, AB0057 (GC2), ABUH796, ABUH628, ABUH747, AB5075 (Figure 1). Collectively, even among the closely related NDM-1 positive ST25 A. baumannii isolates, strain-specific behavior was observed in phenotypic traits. To further explore the characteristics of these isolates, genomic analysis of known Acinetobacter virulence genes, such as iron uptake systems, biofilm-associated proteins, motility, etc., were analyzed using a combination of blastn and sequences of virulence determinants from A. baumannii. The genes that code for the Baumanoferrin siderophore and the feoABC genes related to iron uptake were found in all of A. baumannii and A. nosocomialis genomes studied. Also, feoABC genes were found in A. pittii, A. junii, and A. haemolyticus (Figure S2). The Heme cluster 1 was found only in six genomes of A. baumannii. However, the Heme cluster genes 2 and Acinetobactin genes were identified in A. baumannii ST25 clones, A. baumannii AB0057, and A. baumannii AB5075-UW (Figure S2). However, the genes coding for the Fimsbactin siderophore and the entAB genes were not found in the included Acinetobacter genomes (Figure S2). Genes linked to biofilm formation, motility, and poly-N-acetyl glucosamine (PNAG) production were also analyzed. The presence of pgaABC and bfmSR genes related to biofilm formation were found in A. baumannii, A. nosocomialis, and A. pittii (Figure S2). Strikingly, we identified an orphan bfmR gene in A. junii. The CsuAB/ABCDE genes cluster linked to biofilm formation was also found in fifteen of seventeen genomes of A. baumannii (Figure S2). In A. baumannii and A. nosocomialis the presence of prpABCD genes involved in fimbriae biogenesis and motility were also observed (Figure S2). The K locus (KL) and OC locus (OCL) responsible for the production of capsular polysaccharides and O-antigen, respectively, were identified and characterized using to Kaptive tool (13) and the results were confirmed by BLAST using GenBank database. Seven ST25 genomes contain KL14/OCL6, one has KL14/OCL5 (AMA50), and AMA42 has KL22-like/OCL6 (Figure S2). Among the GC2 genomes included in the study five contain KL22/OCL3-like. However, A. baumannii AMA51 (GC2) recovered from Argentina contained KL2/OCL2. A. nosocomialis and A. junii genomes contained KL48-like/OCL7 and KL38-like/OCL6-like (Figure S2). In the rest of non-baumannii genomes, the KL and OCL genetic structures were not found. Furthermore, we found orphan genes from OCL and KL in A. lwoffii, A. junii and A. haemolyticus with an 80-85 % amino acid identity against to reference sequence of KL and OCL (Kaptive tool, reference sequences) (Figure S2). Overall, no definitive correlations could be made between the observed phenotypes and the presence or absence of specific genes, highlighting the complex nature of most virulence characteristics.
In summary, we found that ST25 isolates had similar characteristics to GC1 and GC2 strains. The majority of strains did not exhibit surface-associated motility. Macrocolony biofilms assays showed variation in structural characteristic among the different strains. All selected strains were able to resist desiccation for up to seven days and some strains were able to resist hydrogen peroxide treatment. In addition, ST25 strains were able to resist NaCl treatment. We did not observe a clear difference between NDM-1 ST25 strains and the isolates from the two major GC. However, in addition to carbapenem-resistance, we identified different traits, such as desiccation, osmotolerance, etc, that can allow the strains to persist in the hospital environment and can in part explain the potential of ST25 to spread as GC1 and GC2. Further in vivo studies will shed more light on the success and pathogenicity of ST25 A. baumannii.
Supplementary Material
Figure S1. Macrocolony biofilm of A. baumannii strains grown on LB agar supplemented with Congo red. Differences were observed among CC25, GC1 and GC2.
Figure S2. Graphic representation of the presence and absence of virulence genes on Acinetobacter sp. used in this study. The graphic representation was performed by ComplexHeatmap package (14).
Table S1. Characteristics of strains used in this study.
Acknowledgments
Funding: Authors’ work cited in this review article was funded by Public Health Service Grants SC3GM125556 (MSR), R01AI100560 (RAB and AJV), R01AI063517 (to RAB), and R01AI072219 (to RAB) from the National Institutes of Health and VA 1I01BX001974 (to RAB) form the Cleveland Department of Veterans Affairs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. DR has a MARC U*STAR fellowship by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T34GM008612.
Footnotes
Transparency declarations. None to declare
Reference
- 1.CDC. 2019. Antibiotic Resistance Threats in the United States. Atlanta, GA: US Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 2.Bush K, Bradford PA. 2020. Epidemiology of beta-lactamase-producing pathogens. Clin Microbiol Rev 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MD, Pasteran F, Traglia GM, Martinez J, Huang F, Liu C, Fernandez JS, Lopez C, Gonzalez LJ, Albornoz E, Corso A, Vila AJ, Bonomo RA, Ramirez MS. 2020. Distinct mechanisms of dissemination of NDM-1 metallo- beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob Agents Chemother. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–9. [DOI] [PubMed] [Google Scholar]
- 5.Adams MD, Wright MS, Karichu JK, Venepally P, Fouts DE, Chan AP, Richter SS, Jacobs MR, Bonomo RA. 2019. Rapid Replacement of Acinetobacter baumannii Strains Accompanied by Changes in Lipooligosaccharide Loci and Resistance Gene Repertoire. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wermser C, Lopez D. 2018. Identification of Staphylococcus aureus genes involved in the formation of structured macrocolonies. Microbiology 164:801–815. [DOI] [PubMed] [Google Scholar]
- 7.Farrow JM 3rd, Wells G, Pesci EC. 2018. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One 13:e0205638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TP, Carvalho-Assef AP, Asensi MD. 2014. First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother 58:7592–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rafei R, Pailhories H, Hamze M, Eveillard M, Mallat H, Dabboussi F, Joly-Guillou ML, Kempf M. 2015. Molecular epidemiology of Acinetobacter baumannii in different hospitals in Tripoli, Lebanon using bla(OXA-51-like) sequence based typing. BMC Microbiol 15:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18:E362–5. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez CH, Nastro M, Famiglietti A. 2018. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev Argent Microbiol 50:327–333. [DOI] [PubMed] [Google Scholar]
- 12.Revathi G, Siu LK, Lu PL, Huang LY. 2013. First report of NDM-1-producing Acinetobacter baumannii in East Africa. Int J Infect Dis 17:e1255–8. [DOI] [PubMed] [Google Scholar]
- 13.Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. 2020. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb Genom 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Macrocolony biofilm of A. baumannii strains grown on LB agar supplemented with Congo red. Differences were observed among CC25, GC1 and GC2.
Figure S2. Graphic representation of the presence and absence of virulence genes on Acinetobacter sp. used in this study. The graphic representation was performed by ComplexHeatmap package (14).
Table S1. Characteristics of strains used in this study.


