Significance
The bacterial DNA translocase Mfd binds to stalled RNA polymerases (RNAPs) and is generally thought to facilitate transcription-coupled DNA repair. Most of our knowledge about Mfd is based on data from biochemical studies. However, little is known about Mfd’s function in living cells, especially in the absence of exogenous DNA damage. Here, we show that Mfd modulates RNAP association and alters transcription at a variety of chromosomal loci, especially those containing highly structured, regulatory RNAs. As such, this work improves our understanding of Mfd’s function in living cells and assigns it a function as a transcription regulator.
Keywords: Mfd, small RNAs, TCR, mutagenesis
Abstract
RNA polymerase (RNAP) encounters various roadblocks during transcription. These obstacles can impede RNAP movement and influence transcription, ultimately necessitating the activity of RNAP-associated factors. One such factor is the bacterial protein Mfd, a highly conserved DNA translocase and evolvability factor that interacts with RNAP. Although Mfd is thought to function primarily in the repair of DNA lesions that stall RNAP, increasing evidence suggests that it may also be important for transcription regulation. However, this is yet to be fully characterized. To shed light on Mfd’s in vivo functions, we identified the chromosomal regions where it associates. We analyzed Mfd’s impact on RNAP association and transcription regulation genome-wide. We found that Mfd represses RNAP association at many chromosomal regions. We found that these regions show increased RNAP pausing, suggesting that they are hard to transcribe. Interestingly, we noticed that the majority of the regions where Mfd regulates transcription contain highly structured regulatory RNAs. The RNAs identified regulate a myriad of biological processes, ranging from metabolism to transfer RNA regulation to toxin–antitoxin (TA) functions. We found that cells lacking Mfd are highly sensitive to toxin overexpression. Finally, we found that Mfd promotes mutagenesis in at least one toxin gene, suggesting that its function in regulating transcription may promote evolution of certain TA systems and other regions containing strong RNA secondary structures. We conclude that Mfd is an RNAP cofactor that is important, and at times critical, for transcription regulation at hard-to-transcribe regions, especially those that express structured regulatory RNAs.
Timely and efficient transcription is a fundamental requirement for maintaining cellular homeostasis. Previous work has shown that RNA polymerase (RNAP) encounters a wide range of obstacles making transcription elongation discontinuous (1, 2). These impediments vary in severity, from pause sites that slow the rate of RNAP movement (3–5) to more severe obstacles, such as protein roadblocks and the replication fork. These impediments can induce reverse translocation of RNAP with respect to both DNA and the nascent RNA (RNAP backtracking) (6–9). The impact of these roadblocks on RNAP elongation is prevented and resolved through various mechanisms including the coupling of transcription and translation, as well as various cellular factors that help reestablish transcription elongation (10).
In vitro work shows that the DNA translocase Mfd utilizes its RNAP binding properties and forward translocase activity to rescue arrested RNAPs, restoring transcription elongation (11) as well as promoting transcription termination (12). However, despite decades of research on the biochemical characteristics of Mfd, the endogenous contexts in which its translocase and antibacktracking functions are critical for transcription remain elusive.
Mfd was initially described as a critical DNA repair factor in vivo that promotes transcription-coupled repair (TCR) (13–15). In TCR, Mfd removes stalled RNAPs at bulky DNA lesions and promotes nucleotide excision repair (NER) via UvrA binding (see review in ref. 16). However, cells lacking Mfd show little sensitivity to DNA-damaging agents that promote RNAP stalling (17, 18), especially relative to NER proteins (19, 20). Together with its high degree of conservation, such data suggest that Mfd may have a broader cellular function outside of DNA repair.
Mfd was shown to have other roles in the cell, such as regulating catabolite repression in Bacillus subtilis (21, 22). Recent in vitro experiments show Mfd autonomously translocates on DNA in the absence of a lesion, but whether this occurs in vivo is unclear (23). Interestingly, Mfd functions as an evolvability factor, promoting mutagenesis and rapid evolution of antibiotic resistance in diverse bacterial species (17). However, a comprehensive in vivo study examining what genomic “hotspots” may be prone to Mfd’s mutagenic activity has not been performed.
Despite our limited understanding of Mfd’s cellular functions, its high level of conservation in bacteria implies a fundamental role for Mfd that may be separate from TCR. In addition, Mfd is constitutively expressed, suggesting that it may have a homeostatic role in transcription regulation. Further work has recently suggested that Mfd plays a housekeeping function in cells by associating with RNAP in the absence of exogenous stressors (24). However, the field still lacks a clear understanding of the conditions and chromosomal features that prompt Mfd binding and modulation of RNAP association and function.
In this work, we identify the genomic regions of Mfd association in both B. subtilis and Escherichia coli. We also identify the regions where Mfd modulates RNAP association and transcription. We find that Mfd alters RNAP density predominantly at regions containing highly structured RNAs. Our analysis of RNAP pausing experiments strongly suggests that these sites are hard to transcribe. We find Mfd regulates the transcription of genes involved in various cellular functions including toxin–antitoxin (TA) systems. Importantly, we observe that when we overexpress toxin genes cell viability is significantly compromised without Mfd. Finally, we find that Mfd promotes mutagenesis of at least the txpA toxin gene. We conclude that RNA secondary structure is a major impediment to transcription in vivo and that Mfd is an important RNAP cofactor that regulates transcription and RNAP association with such loci.
Results
Mapping the Genomic Loci Where Mfd Associates.
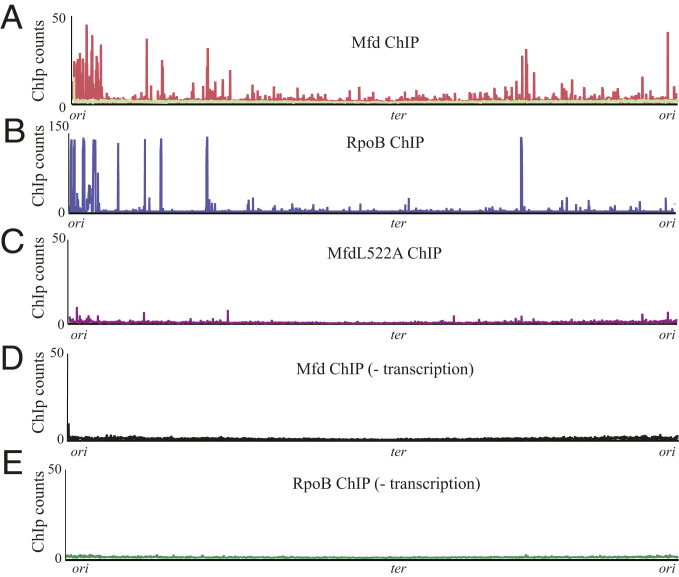
We began by identifying the chromosomal regions where Mfd associates using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). We constructed a B. subtilis strain where Mfd is C-terminally Myc-tagged (SI Appendix, Fig. S1). Prior work has shown that C-terminally tagged Mfd retains its functionality in vivo (24) (below we present data that confirm the functionality of this Mfd-Myc in our system). To identify the chromosomal regions where Mfd associates, we harvested exponentially growing cells and performed ChIP-seq analysis. We controlled for ChIP artifacts by comparing this signal to ChIP-seq performed using Myc antibody against B. subtilis cells lacking Myc-tagged Mfd (Fig. 1A). Under our growth conditions, 489 out of 5,755 genes analyzed exhibited preferential Mfd association (Dataset S1).
Fig. 1.
Mfd functions as an RNAP cofactor and requires transcription elongation for association with DNA. (A) ChIP-seq plot of B. subtilis Mfd tagged with 1× myc (red) and of WT B. subtilis (light green) using myc antibody. (B) ChIP-seq plot of WT B. subtilis RpoB. (C) ChIP-seq plot of B. subtilis MfdL522A-myc point mutant. (D) B. subtilis Mfd-myc and (E) B. subtilis RpoB ChIP-seq after treatment with 50 µg/mL of rifampicin for 5 min. Plots are normalized to total DNA input controls and are the average of at least two independent experiments.
Mfd’s Genomic Association Pattern Correlates with That of RNAP.
Given the physical interaction between Mfd and RNAP (25), we hypothesized that Mfd binding sites may correspond to RNAP binding locations. To test this, we performed ChIP-seq of RpoB, the β subunit of RNAP, using a native antibody. Indeed, we found Mfd association largely overlaps with RpoB occupancy (Pearson coefficient = 0.68) (Fig. 1B and SI Appendix, Fig. S2), consistent with Mfd’s proposed function as an RNAP cofactor in B. subtilis.
Mfd Requires Interaction with RNAP for Its Association with All Genomic Loci.
Mfd is a multimodular protein, consisting of eight domains connected by flexible linkers (25). Of these domains, the RNAP interacting domain (RID) and the translocase module are critical for Mfd’s ability to rescue stalled transcription complexes (26–28). In vitro, Mfd is recruited to the identified genomic regions via interaction with RpoB. We therefore tested whether the interaction of Mfd with RpoB is critical for its recruitment to the genomic loci we identified. Prior in vitro work suggested that RID mutations abrogate the interaction between Mfd and RNAP (25). We therefore constructed a Myc-tagged Mfd strain with a mutation at the L522 residue to disrupt Mfd’s binding to RpoB, without disrupting protein stability, analogous to that described in E. coli (25). Upon confirming that the B. subtilis L522A mutation (analogous to the L499R mutation in E. coli) disrupted Mfd’s interaction with RNAP via a bacterial two-hybrid assay (SI Appendix, Fig. S3), we performed ChIP-seq with this mutant. The ChIP signal was abrogated in the strain expressing the L522A allele of mfd (Fig. 1C). These results suggest that Mfd’s interaction with RNAP is essential for its recruitment to the genomic loci previously identified and that Mfd functions as a genome-wide RNAP cofactor in vivo.
Mfd’s Association with DNA Requires Transcription Elongation.
In vitro, Mfd helps promote the rescue of arrested transcription elongation complexes (TECs), yet how Mfd recognizes stalled RNAPs in vivo remains unclear. We therefore asked whether Mfd association with various genomic loci was facilitated via loading during the transcription initiation or elongation. To distinguish between these two, we utilized rifampicin, which directly blocks transcription initiation (29, 30) and eliminates the formation of TECs. We performed both Mfd and RpoB ChIP-seq in the presence of rifampicin and found that this treatment largely eliminated both Mfd and RpoB ChIP-seq binding signal (Fig. 1 D and E). We find Mfd ChIP signal is decreased across the gene body with rifampicin treatment with no clear accumulation of Mfd at promoters (SI Appendix, Fig. S4). These findings are consistent with biochemical evidence showing Mfd does not release RNAP at initiation sites (11) and suggest that it associates with elongating RNAPs in vivo.
Mfd Decreases RNAP Density at Some Genomic Loci.
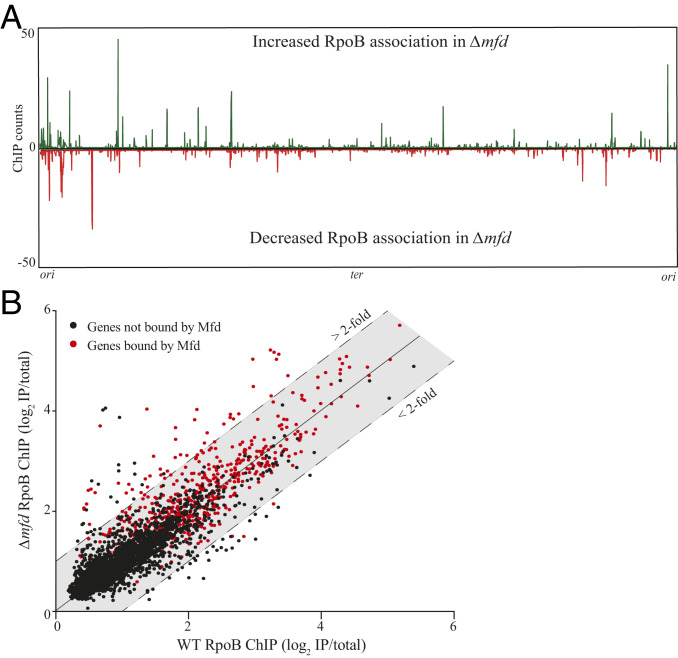
Mfd promotes both transcription elongation and transcription termination, at least in vitro (11, 12). However, the importance of these functions in vivo remains elusive. Because of Mfd’s effects on RNAP in vitro, we wondered if and how Mfd’s close association with RNAP in vivo altered RNAP association. We therefore performed ChIP-seq of RpoB in wild type (WT) and Δmfd and identified where RpoB occupancy is altered in the absence of Mfd. ChIP-seq did not detect alterations in RpoB occupancy at most genes where Mfd associates based on our Mfd ChIP-seq. This may be due to various factors, such as the existence of redundant transcription-associated factors or detection thresholds in an ensemble assay such as ChIP-seq. However, we found a number of genes where RpoB occupancy either increased or decreased in Δmfd compared to WT (Fig. 2A). We noticed more genes exhibited increased, rather than decreased, RpoB occupancy in Δmfd. Quantification revealed 116 genes with at least a twofold increase and 53 genes with at least a twofold decrease in RpoB occupancy without Mfd. Many of these genes are within the same operon and therefore are expressed as single transcripts. Thus, we grouped and analyzed the identified genes as transcription units (TUs) (31). Our analysis revealed that Δmfd contains 71 TUs with at least one gene containing a minimum of twofold increase and 31 TUs with at least one gene containing a minimum of twofold decrease in RpoB occupancy compared to WT (Table 1 and Datasets S2 and S3).
Fig. 2.
Mfd directly promotes release of RNAP in vivo. (A) RpoB ChIP-seq plots showing regions of RpoB enrichment in Δmfd. Top half of graph (counts in green) reflects normalized RpoB ChIP-seq read counts where B. subtilis Δmfd had increased signal relative to WT. Bottom half of graph (counts in red) reflects RpoB ChIP-seq read counts where Δmfd had decreased signal relative to WT B. subtilis. High background signal from ribosomal RNA was removed from plots for better visualization. (B) Scatter plot of WT and Δmfd RpoB ChIP-seq measuring signal at each gene in B. subtilis. For quantification of ChIP signal, read counts for each gene were normalized to total library counts and IP samples were normalized to total DNA input to calculate an IP/total ratio. Ratios were log2-normalized and averaged over at least two independent experiments. Data points above and below colored shading indicate greater than twofold increase and decrease in RpoB signal in Δmfd, respectively. Data points in red indicate genes bound by Mfd, defined as one SD greater than the average ChIP signal across all genes in B. subtilis. Calculation of Mfd binding at each gene was determined as described for RpoB ChIP samples.
Table 1.
Summary list of genes and previously defined TUs bound with changes in RpoB density in Δmfd from ChIP-seq analysis
| Increased RpoB density in Δmfd | Decreased RpoB density in Δmfd | |
| Genes (and TUs) | 116 (71) | 53 (31) |
| Genes (and TUs) bound by Mfd | 52 (35) | 0 (0) |
Changes in RpoB density and Mfd binding at TUs are defined by changes in one or more genes corresponding to its associated TUs.
We next wanted to determine whether the changes in RpoB occupancy observed in Δmfd were directly due to Mfd’s activity at those regions. We therefore looked for a correlation between regions of Mfd association and where there are changes in RpoB association in Δmfd. We only observed Mfd association at the genes that showed increased RpoB occupancy in Δmfd. The genes with decreased RpoB occupancy in Δmfd did not show Mfd association (Fig. 2B). Specifically, we detected Mfd at 52 of the 116 genes with increased RpoB association (35 of the 71 TUs) in Δmfd compared to WT. We did not detect Mfd association at any of the 53 genes (31 TUs) with decreased RpoB occupancy. Because these 31 TUs do not display Mfd association, we concluded that decreased RpoB occupancy at these sites is not directly due to Mfd activity and likely reflects indirect effects on transcription in Δmfd.
We next performed RpoB ChIP-qPCR analysis to confirm our ChIP-seq results. We chose two sites from our candidate genes with Mfd binding and increased RpoB occupancy in Δmfd. We found results consistent with our ChIP-seq studies (SI Appendix, Fig. S5). We additionally tested the functionality of the Myc-tagged Mfd using RpoB occupancy as our readout. We did not find changes in RpoB levels in the tagged Mfd strain, confirming its functionality, at least in regulating RNAP association (SI Appendix, Fig. S5).
Based on these findings, we conclude that increased RNAP occupancy at the identified chromosomal sites is a direct result of Mfd’s function as either an RNAP termination (12) or processivity factor (32).
Regions Where Mfd Increased RNAP Density Are Enriched for Regulatory RNAs.
In vitro, Mfd’s translocase activity can release RNAP or assist it with elongation when exposed to different obstacles. However, whether there are endogenous hotspots of RNAP stalling that require Mfd function remains unknown. Furthermore, if such hotspots exist, the nature of the potential obstacles remains to be determined. Intriguingly, 92% of the TUs that showed both an increase in RNAP density in Δmfd and direct Mfd association express a minimum of one regulatory RNA (SI Appendix, Table S1). These regulatory RNAs are a subset of the 1,583 regulatory RNAs in B. subtilis, which encompass various RNAs, including noncoding transcripts, antisense RNAs, and riboswitches (31). In comparison, only 39% of the TUs with decreased RNAP density in Δmfd contain regulatory RNAs (SI Appendix, Table S2), which is consistent with the average percentage of TUs with predicted regulatory RNAs in B. subtilis (31, 33).
Sites of Mfd Function Contain Highly Structured Regulatory RNAs.
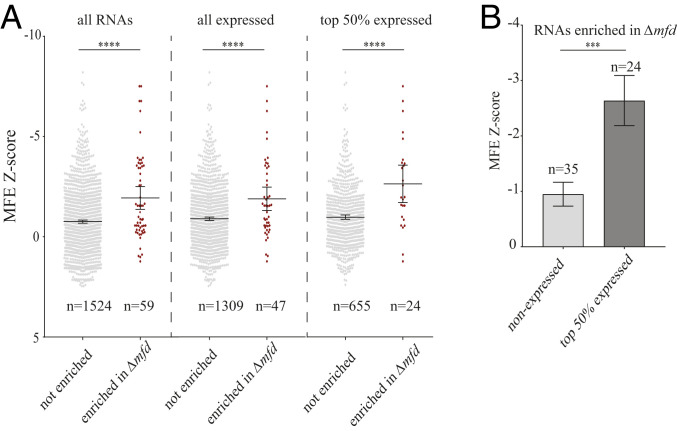
We hypothesized that Mfd function at the identified regions was related to RNA secondary structure impeding RNAP movement. This hypothesis is consistent with changes in RNAP kinetics due to secondary RNA structures, such as hairpins promoting transcription termination (34–36). Previous work characterized the predicted secondary structure for each regulatory RNA in B. subtilis (33). We sought to test whether regions with increased RNAP in Δmfd are more prone to transcribing RNAs with more stable secondary structures. We determined the average minimum free energy (MFE) z-score for each of the RNAs at these regions as a proxy of RNA structure stability (37, 38). Specifically, we examined the regulatory RNAs in TUs that had both increased RpoB density in Δmfd and Mfd association and compared them to TUs that showed no difference in RpoB density between WT and Δmfd. We found that TUs where there is Mfd binding and increased RpoB density in Δmfd have significantly higher predicted RNA secondary structures relative to all other known regulatory RNAs (Fig. 3A).
Fig. 3.
Transcription units with Mfd binding and increased RNAP density in Δmfd are enriched for structured regulatory RNAs. (A) Scatter plot of the MFE z-score for regulatory RNAs in B. subtilis. Data points represent regulatory RNAs within TUs that have no observed change in RpoB density between WT and Δmfd (gray points) and TUs that have increased RpoB density in Δmfd and are also bound by Mfd (red points). The three scatter plots represent all regulatory RNAs (Left), only expressed regulatory RNAs (Middle), and top 50% expressed regulatory RNAs (Right). Expression data determined from RNA-seq analysis. (B) Bar graph of regulatory RNAs within TUs that have increased RpoB density in Δmfd and are bound by Mfd. Nonexpressed RNAs shown in light gray and top 50% of expressed RNAs shown in dark gray. Error bars represent the SEM. Statistical significance was determined using two-tailed z-test for two population means (***P < 0.001, ****P < 0.0001).
Many regulatory RNAs are not transcribed during standard growth conditions. We therefore excluded RNAs not expressed under our growth conditions, as determined from our RNA-seq data (SI Appendix, Fig. S9). Consistent with our global analysis, expressed regulatory RNAs associated with Mfd binding and increased RpoB density in Δmfd have significantly higher MFE z-scores (Fig. 3A and SI Appendix, Fig. S6). We also find that within regulatory RNAs associated with Mfd binding and increased RpoB density in Δmfd, those that were highly expressed (top 50%) have a higher MFE z-score compared to those that were not expressed (Fig. 3B). These findings suggest that Mfd regulates RNAP at regions containing highly structured RNAs.
Sites of Mfd Activity Correlate with Difficult-to-Transcribe Regions.
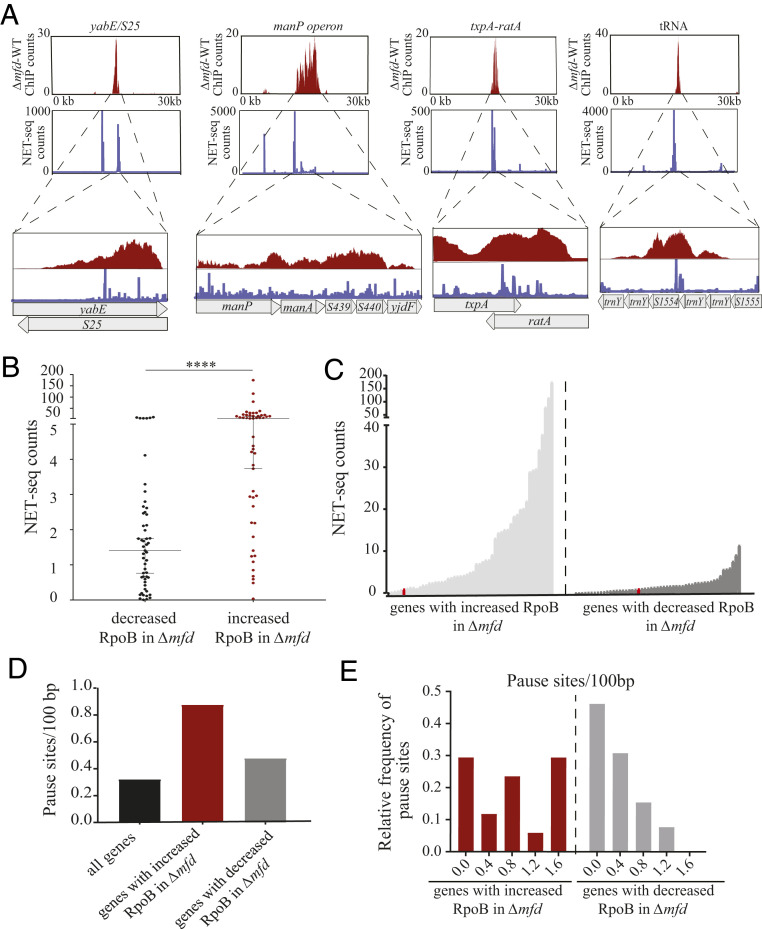
If sites of Mfd association and Mfd alteration of RpoB density were related to RNAP elongation kinetics we would expect these sites to be difficult to transcribe. To identify such regions, we utilized experimental data from native elongating transcript sequencing (NET-seq) in B. subtilis (39). Through the capture of nascent transcripts from tagged immunoprecipitation of RNAP molecules, NET-seq identifies RNAP pausing with high resolution (39). We observed that many genes with increased RpoB levels without Mfd displayed high NET-sig signal and that the corresponding sequencing patterns closely aligned (Fig. 4A). We quantified this pattern by analyzing the NET-seq read count across the previously identified genes where Mfd associates and has increased RpoB signal either with or without Mfd. We found that the median NET-seq signal is significantly higher, by ∼3.7-fold, at genes with both increased RpoB signal in Δmfd and Mfd binding relative to genes with decreased RpoB signal (Fig. 4 B and C).
Fig. 4.
Genes with Mfd binding and increased RNAP density in Δmfd are enriched for RNAP pause sites. (A) Read counts for RpoB ChIP-seq (red) and NET-seq (blue) across a 30-kb window for four representative genomic regions (yabE/S25, manP operon, txpA/ratA, and a tRNA locus). Zoomed-in plots showing the gene/operon only (y-axis scale is preserved). (B) Scatter plot of the average NET-set read counts for genes with decreased RpoB association in Δmfd (black dots, n = 53) and increased RpoB association in Δmfd (red dots, n = 52). Median and 95% confidence interval shown. Statistical significance was determined using the nonparametric Mann–Whitney U test for two population medians (****P < 0.0001). (C) Line plot showing by genes with increased (light gray) and decreased (dark gray) RpoB association in Δmfd, ranked by increasing signal. Red data point in both plots represents the mean NET-seq read count across the genome. (D) Average number of pause sites (per 100 bp) for all genes in B. subtilis (black bar) and genes with increased (red bar) and decreased RpoB association (gray bar) in Δmfd, respectively. (E) Histogram showing the relative frequency of pause sites (per 100 bp) in genes with increased (red bars) and decreased (gray bars) RpoB association in Δmfd.
Larson et al. identified 9,989 discrete pause sites in B. subtilis (39). We therefore tested whether sites with proposed Mfd activity due to RNA secondary structure contained more pause sites relative to the average number across the genome. We found roughly threefold greater number of discrete pause sites in genes with proposed Mfd activity relative to the average number across the genome, with a notable increase in the frequency of genes with greater than one pause site per 100 base pairs (bp) (Fig. 4 D and E). We did not find this pattern in genes with decreased RpoB association in Δmfd (Fig. 4 D and E). Critically, the discrete pause sites identified by Larson et al. (39) are limited to coding regions and do not include regulatory RNAs, likely underestimating the number of pause sites requiring Mfd activity. From these findings we conclude that Mfd functions mainly at RNAP pause sites in vivo.
Mfd’s Effect on RNAP at Sites of Structured Regulatory RNAs Is Conserved in E. coli.
Mfd is highly conserved across bacterial phyla, and functional homologs exist throughout all domains of life (40–42). To test whether Mfd’s genome-wide coupling with RNAP was conserved in gram-negative species, we performed Mfd and RpoB ChIP-seq in E. coli. As in B. subtilis, Mfd and RpoB occupancy are highly correlated (Pearson coefficient 0.98) in E. coli (SI Appendix, Fig. S7 A–C), showing that Mfd’s function as an RNAP cofactor is likely conserved.
We also determined whether altering RpoB occupancy at sites containing structured RNAs was true in E. coli. We compared the signal of the ChIP-seq data for RpoB in WT and Δmfd in E. coli and looked for genes where RpoB occupancy was altered in Δmfd. As in B. subtilis, more genes had at least a twofold increase in RpoB association in Δmfd, compared to genes with decreased RpoB association (105 versus 24, respectively) (SI Appendix, Fig. S7D and Datasets S4 and S5). Forty-nine of the 105 genes with increased RpoB association also had Mfd association (Dataset S4). Of these 49 sites, roughly 40% contained a regulatory RNAs or a structural element. Three are TA RNAs (symR, sokA, and sokC) (43, 44), one is a transfer RNA (tRNA) (metZ), and two are regulatory RNAs (spf and gadY), all of which contain secondary structures (45–47). Seven sites are repeated extragenic/intragenic palindromes, which contain extensive secondary structure and facilitate transcription termination (48, 49), and six are ribosomal proteins containing structured regulatory elements (50). In contrast, no such RNAs were found in genes with decreased RpoB association in Δmfd (Dataset S5). These findings suggest Mfd’s function at sites of structured regulatory RNAs is conserved across species.
Genome-Wide Mfd Binding Is Correlated with RNA Secondary Structure.
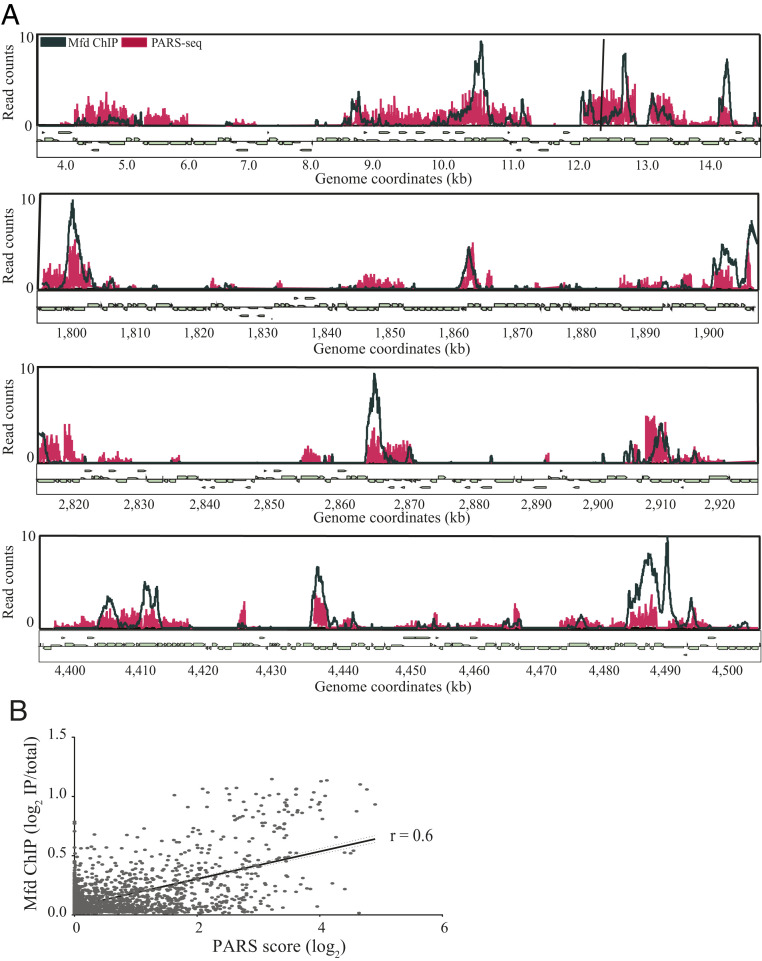
While computational methods exist for predictions of RNA secondary structure, empirical determination of RNA structure is technically challenging, particularly in a high-throughput fashion. The recent development of parallel analysis of the messenger RNA (mRNA) structure (PARS) allows for experimental determination of RNA secondary structure (51, 52). Del Campo et al. utilized PARS to establish the RNA secondary structure for roughly 2,500 genes in E. coli (52). We therefore wanted to determine whether sites with high RNA secondary structure in E. coli correlate with regions of Mfd binding. We observed that across genomic sites containing a high PARS score both the presence of Mfd association and the magnitude of Mfd binding closely aligned with the magnitude of experimentally determined RNA secondary structure (Fig. 5A). To further quantify this, we measured the area under the curve (AUC) for both our Mfd ChIP-seq and the PARS datasets to assess whether these two signals were correlated. We found a strong positive correlation between the AUC for our Mfd ChIP-seq and the PARS score (r = 0.6) across the E. coli genome (Fig. 5B). These data suggest that the presence of RNA secondary structure, at least in E. coli, correlates with Mfd genome-wide association and function.
Fig. 5.
Genome-wide Mfd association is correlated with sites of RNA secondary structure based on PARS seq in E. coli. (A) Overlay of sequencing counts from Mfd ChIP-seq (dark blue) and PARS-seq (red) across four 100-kb regions across the E. coli genome. ChIP-seq plots averaged from at least two independent experiments. (B) Linear regression analysis comparing Mfd ChIP-seq and PARS-seq in E. coli. Read counts from ChIP-seq and PARS-seq were used to calculate the AUC for each gene in the E. coli genome. Pearson’s correlation coefficient for E. coli ChIP-seq and PARS-seq was r = 0.6. Dotted lines represent 95% confidence intervals.
GreA Does Not Preferentially Effect Transcription of Regions Containing Structured RNAs.
Various factors help rescue arrested RNAP through different mechanisms, including GreA, which functions as an RNAP antibacktracking factor (10, 53). GreA also suppresses promoter-proximal pausing during transcription initiation (54). To test whether GreA also contributed to RNAP release at loci transcribing structured RNAs, we performed RpoB ChIP-seq of B. subtilis WT and a ΔgreA strain. We found ΔgreA only had 12 genes (and six TUs) with increased RpoB occupancy (Dataset S6 and SI Appendix, Fig. S8). Two of the six TUs transcribe regulatory RNAs and neither contained significant predicted secondary structure. These results suggest that, unlike Mfd, GreA does not function in decreasing RNAP from sites containing secondary structure. We also observed that in ΔgreA 469 genes exhibited less than twofold RpoB occupancy (Dataset S6 and SI Appendix, Fig. S8). The high number of genes with decreased RpoB occupancy in ΔgreA may be due to decreased efficiency of RNAP promoter escape during initiation and consequently decreased levels of elongating RNAP molecules, consistent with in vitro findings (54, 55).
Mfd Decreases Expression at Structured, Regulatory RNAs.
We next tested the effect of Mfd on transcription at sites containing structured RNAs, first by performing RNA-seq of WT and Δmfd in B. subtilis. Differential expression analysis between WT and Δmfd revealed 378 genes with statistically significant lower RNA levels (Dataset S7). Consistent with RpoB ChIP-seq, more genes were up-regulated in Δmfd (240 genes up-regulated compared to 138 genes down-regulated) (SI Appendix, Fig. S9 and Dataset S7). When comparing our RpoB ChIP-seq findings to RNA-seq, we found that of the 116 genes with greater than twofold RpoB ChIP-seq signal in Δmfd, 30 had increased expression, while none show decreased expression. Genes with increased expression in Δmfd showed an equal increase throughout the gene body, suggesting Mfd suppresses full-length transcripts (SI Appendix, Fig. S10). Because standard RNA sequencing protocols often do not accurately measure small RNAs (56), we wondered if expression of some regulatory RNAs with increased RpoB density in Δmfd were not accurately measured in our RNA-seq. We therefore measured RNA levels using qRT-PCR at three loci containing regulatory RNAs (the trnY locus, txpa-ratA, and bsrH-asBsrH) with Mfd binding and increased RpoB signal in Δmfd. We found all three loci have increased RNA levels in Δmfd compared to WT (SI Appendix, Fig. S11). To confirm that Δmfd does not globally repress transcription, we performed qRT-PCR on two control loci, rpoB and yolA, and found no difference in RNA between WT and Δmfd (SI Appendix, Fig. S11). These findings suggest that Mfd’s in vivo effects on RNAP lead to decreased transcription.
Cells Lacking Mfd Are Highly Sensitized to TxpA and BsrH Overexpression.
Aside from decreased rates of evolution, there are few phenotypic defects that have been detected in the absence of Mfd, even upon exposure to DNA damage (17, 18, 57–59). We wondered whether the transcriptional regulation activity of Mfd at regions we detected were physiologically relevant. To address this, we focused on the highest structured regulatory RNAs from our dataset, which included two pairs of type I TA loci: the txpA/ratA locus and the bsrH/as-bsrH locus. Type I TA loci contain a small toxic peptide and a noncoding RNA that neutralizes toxin expression by inhibiting translation or promoting degradation of the toxin mRNA (60). The cellular functions of type I TA loci remain unclear, but they appear to influence diverse aspects of physiology, including persister formation (61), biofilm formation (62), and prophage maintenance (63). Five type I TA loci have been identified in B. subtilis (60)—we found three have both Mfd binding and a minimum of twofold increase in RpoB density in Δmfd, while a fourth locus, yonT/as-YonT, showed an increase in RpoB density in Δmfd (SI Appendix, Table S3).
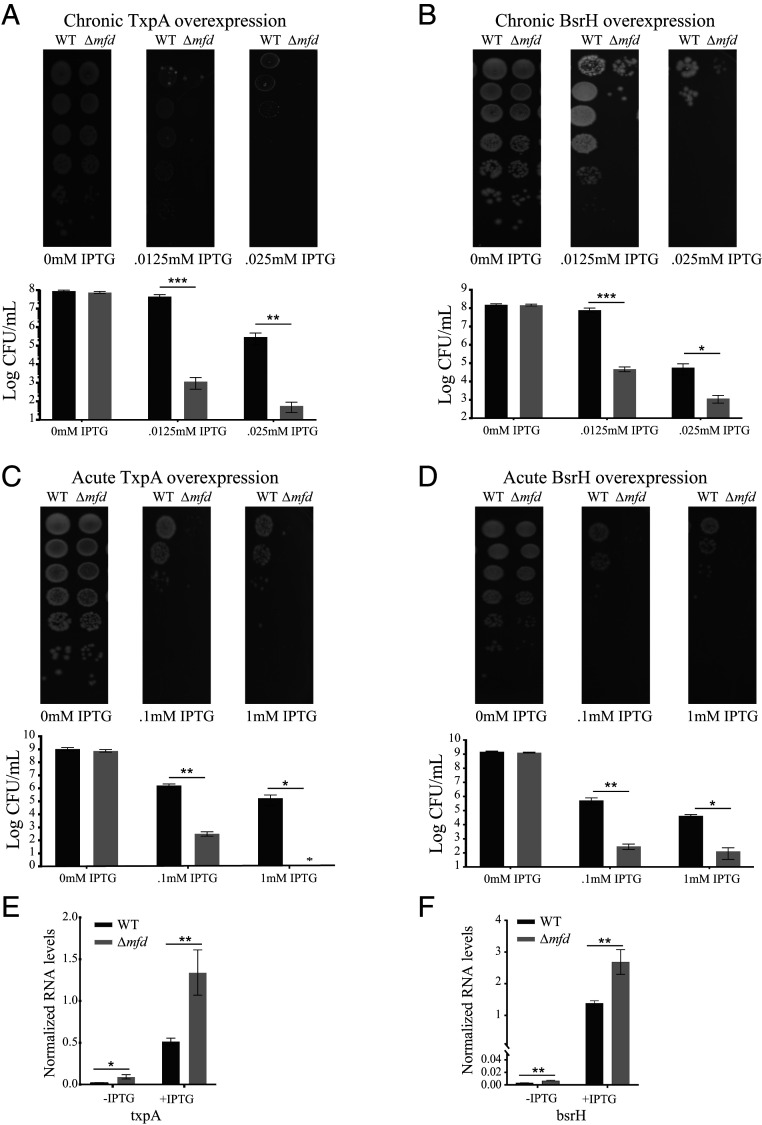
Overexpression of type I toxins can be lethal (64). Based on our ChIP-seq and RNA-seq, we wondered if Mfd’s effect on transcription at TA sites would alter sensitivity to selective toxin overexpression. We therefore constructed integrative plasmids with either the txpA and bsrH toxins under an IPTG (isopropyl β-d-1-thiogalacto-pyranoside)–inducible promoter in both WT and Δmfd and performed cellular viability assays. We found Δmfd was highly sensitized (up to five orders of magnitude) to both chronic (Fig. 6 A and B) and acute (Fig. 6 C and D) overexpression of either TxpA or BsrH. To test whether this effect was due to toxin overexpression, we performed qRT-PCR in WT and Δmfd strains containing the overexpression constructs and found both toxins had increased expression by approximately two- to threefold in Δmfd (Fig. 6 E and F). To confirm the expression effects observed were not specific to our integration site or promoter, we performed qRT-PCR on strains containing an IPTG-inducible lacZ gene and found no expression difference between WT and Δmfd (SI Appendix, Fig. S12). Our data suggest that cells with Mfd are more resistant to toxin expression via transcriptional repression; however, we cannot rule out that Mfd alters toxin sensitivity via posttranslational effects or other indirect means. Interestingly, Clostridium difficile cells lacking Mfd have morphologic differences and viability defects likely related to toxin overexpression (65), underscoring the conserved role Mfd may play on expression of regulatory RNAs.
Fig. 6.
Transcriptional regulation by Mfd at TA loci is critical for cell survival. Survival assays under chronic (A) and acute (C) overexpression of TxpA toxin (MFE z-score = −6.77) in WT and Δmfd and survival assays under chronic (B) and acute (D) overexpression of BsrH toxin (MFE z-score = −7.51) in WT and Δmfd. For all figures, representative images are shown above and quantification of data is shown below. Error bars represent the SEM from at least three independent experiments. (E and F) qRT-PCR analysis of txpA and bsrH overexpression in WT and Δmfd strains. RNA values normalized to ribosomal RNA. Error bars represent the SEM from at least two independent experiments. Statistical significance was determined using a two-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Mfd Promotes Mutagenesis of the txpA Gene.
Mfd is an evolvability factor (17) that promotes mutagenesis and antimicrobial resistance under various conditions (57–59, 66). Most of these studies utilize engineered reporter systems to study Mfd-mediated mutagenesis, and it remains unclear what endogenous sites are prone to this mutagenesis. We wondered if sites with high RNA secondary structure were particularly prone to Mfd-mediated mutagenesis. We tested this hypothesis by performing Luria–Delbrück fluctuation assays (67) to select for toxin-resistant mutants and assess the mutation rate at our ectopic txpA locus. We found the mutation rate of txpA is nearly sevenfold lower in Δmfd compared WT (SI Appendix, Fig. S13). This reduction is greater than previously reported mutation rate reductions in Δmfd (17, 59). We conclude Mfd promotes mutagenesis at TA loci and given our prior findings likely accelerates evolution at many of its endogenous targets.
Discussion
In this work we identified the endogenous targets of Mfd and how Mfd impacts RNAP association and transcription in vivo. We found most sites where Mfd associates and modulates transcription contain highly structured regulatory RNAs. We found RNAP modulation and transcriptional control by Mfd predominantly occur at regions of frequent RNAP pausing. Our experiments demonstrated that cells lacking Mfd are sensitive to expression of at least some type I toxins which contain highly structured RNAs. We also showed that Mfd promotes mutagenesis at a TA locus. Based on these findings, we propose Mfd is a RNAP cofactor which helps regulate transcription at many chromosomal regions, especially those with highly structured RNAs.
Although Mfd is nonessential under many conditions, we found cells lacking Mfd are sensitive to selective overexpression of the TxpA and BsrH toxins. Such findings suggest there may be other situations when transcriptional regulation by Mfd is important. It is not unreasonable to expect Mfd would be critical under conditions related to the functions of the genes where Mfd binds and regulates RNAP association. For example, Mfd targets a mannose utilization operon, suggesting that efficient growth on mannose may require transcriptional regulation by Mfd. By identifying genomic regions where Mfd binds, our study has provided key information regarding the conditions under which Mfd function may become critical.
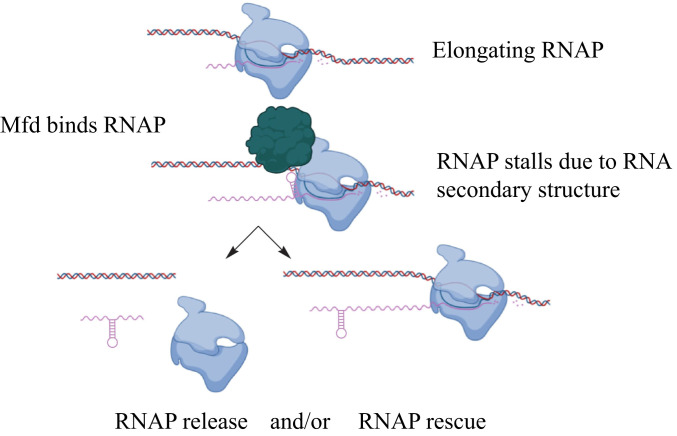
Although Mfd clearly reduces RNAP density at regions containing structured RNAs, it is not clear if this is due to RNAP rescue, early transcription termination, or a combination of both (Fig. 7). Although Mfd can rescue or terminate transcription, the decision about which activity will occur is thought to be related to the magnitude of the roadblock faced by RNAP, with more significant roadblocks favoring termination (10, 21, 22, 32, 68, 69). Additionally, recent work from Le et al. using in vitro translocase assays showed that Mfd rescues RNAP at pause sites but that more severe obstacles to RNAP movement lead to eventual termination (23).
Fig. 7.
Model of Mfd activity at structured regulatory RNAs. During transcription, elongating RNAP (shown in blue) transcribes a highly structured RNA sequence. The formation of a structured RNA within the exit channel of RNAP can pause RNAP on DNA, leading to Mfd binding (shown in red). Two (nonmutually exclusive) models are shown to explain Mfd’s function once RNAP encounters a highly structured RNA. (Bottom Left) The arrested complex is recognized by Mfd, which releases RNAP from the template and in doing so represses transcription. (Bottom Right) Mfd recognizes the arrested or stalled RNAP complex and functions as an elongation factor, accelerating RNAP release off of the DNA template.
Existing studies show that RNA secondary structure is capable of impeding RNAP elongation. For example, RNAP pausing can be promoted by the formation of stable RNA hairpin structures in the exit channel of RNAP and inhibit its movement (5, 36, 70, 71). Pausing via RNA secondary structure is a mechanism that can regulate gene expression at riboswitches and promote coupling of transcription and translation (72, 73) and is critical for transcription termination (35, 74).
The severity of stalling at structured RNAs is likely dependent on multiple factors including the stability of the structure, length, and expression level. At certain sites, it is possible that such pausing may induce RNAP backtracking, but mechanistic studies suggest that pausing more commonly induces a “half-translocated” state of RNAP (70, 75) inhibiting its elongation (76). To our knowledge, RNA secondary structure is not a significant enough roadblock to cause RNAP termination. These findings would favor a model whereby Mfd promotes RNAP rescue and elongation at structured RNAs, leading to the decreased RNAP levels observed in our ChIP studies. However, we cannot rule out that the regulatory RNAs in our study promote a more severe impediment to RNAP and that decreased RNAP levels reflect RNAP termination by Mfd.
Given the range of biological functions regulated by structured RNAs, Mfd’s activity at the sites we identified may have broad phenotypic effects. For example, we identified multiple riboswitches containing TUs where Mfd binds and alters RNAP. These TUs are involved in various processes, ranging from beta-glucoside metabolism (bglP-bglH-yxiE) (77, 78) to glycerol utilization (glpF-glpK and glpT-glpQ) (79) to purine metabolism (purEKBCSQLFMNHD) (80, 81). We also identified Mfd activity at a locus containing a long cis-acting antisense RNA (yabE/S25) thought to play a role in cell wall maintenance (82). Finally, we identified Mfd activity at tRNA loci (the trnY locus containing a highly structured RNA). The relevance of Mfd’s activity at these sites requires further investigation.
Various mechanisms of transcription-associated mutagenesis exist (83, 84). Based on our findings, we propose that the inherent structure of RNA may be an additional mechanism by which transcription promotes mutagenesis, at least partially through Mfd. Interestingly, RNA secondary structure has been reported to enhance mutation rates in replicating retroviruses (85), suggesting that evolution via RNA secondary structure may be universal. In addition, recent work revealed that tRNAs have higher mutation rates relative to other parts of the genome (86). By promoting mutagenesis at sites of highly structured RNAs, Mfd may inherently alter the secondary structure encoded at the site of its activity, leading to novel or altered functions of the RNA. Additionally, noncoding RNAs are well known to evolve very quickly (87) via unknown mechanisms. Our results suggest that Mfd may contribute to the evolution of these regions via its mutagenic activity. Addressing this possibility would further contribute to our understanding of how noncoding RNAs evolve.
Materials and Methods
Strain Constructions.
Strains and plasmids used in this study are listed in SI Appendix, Table S4 and primers are listed in SI Appendix, Table S5. B. subtilis strains used in this study were derivates of the HM1 (JH642) parent strain (88). E. coli strains used were derivates of K-12 MG1655 (89). Transformations into B. subtilis HM1 were performed as previously described (90). Plasmids used in this study were grown in E. coli DH5α. Plasmids were cloned using chemical transformations of competent E. coli. Plasmid purification was performed by growth of E. coli overnight at 37 °C in Luria–Bertani (LB) medium supplemented with the appropriate antibiotic. Plasmids were purified using the GeneJet Plasmid Miniprep Kit (Thermo). Further details on strain construction can be found in SI Appendix.
Growth Conditions.
For experiments in B. subtilis and E. coli, cultures were grown as described unless otherwise indicated. Overnight cultures from single colonies were grown at 37 °C in LB at 260 rpm and the following day cells were diluted back to optical density at 600 nm (OD600) 0.05 and grown to exponential phase (OD600 0.3 to 0.5) before harvesting. For rifampicin ChIP experiments, cultures were grown in identical fashion until they reach OD600 0.3 to 0.5 and rifampicin was added at a concentration of 100 µg/mL for 5 min before harvesting.
ChIP-Seq and ChIP-qPCR Experiments.
For B. subtilis Mfd ChIPs, c-Myc mouse monoclonal antibody (clone 9E10) was used (Thermo). For E. coli ChIPs, custom polyclonal E. coli Mfd rabbit antisera was produced and purified by Covance. Enzyme-linked immunosorbent assay was used to confirm Mfd titers from antisera. For RpoB experiments, RNAP beta mouse monoclonal antibody (clone 8RB13) was used (Thermo). ChIP experiments were performed as previously described (91, 92). Further details can be found in SI Appendix. Library preparation for ChIP-seq was performed using Nextera XT DNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. For ChIP-qPCR, Sso Advanced Universal SYBR Green Supermix (Bio-Rad) was used according to the manufacturer’s instructions.
NET-Seq Analysis.
Data from Larson et al. (39) were analyzed for determination of average NET-seq read counts in addition to identification of pause sites. Data were collected from Gene Expression Omnibus (accession number GSE56720) and plotted as a wiggle (.wig) file on the B. subtilis 168 genome. Details of pause site determination are defined in the supplementary methods of Larson et al. (39). To calculate the average read count for each gene, gene coordinates were determined and NET-seq signal was averaged across these nucleotide positions. Average NET-seq signal for each gene was calculated from the strand (plus or minus strand) with the highest NET-seq signal. The number of pause sites for each gene of interest was normalized to gene length.
PARS-Seq Analysis.
PARS data were kindly provided by Zoya Ignatova. Original PARS score data were refined to remove duplicate data points. All positive PARS scores were then plotted as a wiggle file on the E. coli K-12 MG1655 genome sequence (GenBank accession number U00096.2). The average PARS score for each annotated feature in the E. coli chromosome was calculated using custom scripts.
Whole-Genome Sequencing Analysis.
ChIP-seq and RNA-seq samples were sequenced using the Illumina NextsEq. 500/550 Sequencing system at the University of Washington Northwest Genomics Center and the VANTAGE Sequencing Core at Vanderbilt University. Further details on sequencing analysis can be found in SI Appendix.
qRT-PCR Assays.
For qRT-PCR, B. subtilis cultures were grown as previously described. For IPTG-inducible constructs, expression was induced with 1 mM IPTG for 5 min and RNA was subsequently extracted and processed as described in SI Appendix, RNA-seq and qRT-PCR Experiments.
Cell Survival Assays.
For chronic survival assays, B. subtilis cultures were grown in 2 mL LB until they reached an OD600 of 0.5 to 1.0. Cultures were normalized to OD600 0.3 and serial dilutions were performed in 1× Spizizen’s salts. Five microliters of cells were plated LB agar only and on LB agar plates containing IPTG (see figure legends for concentrations). Plates were grown at 30 °C overnight and colony-forming units enumerated the following day.
For acute survival assays, cultures were grown until they reached an OD600 of 0.5 to 1.0 and then diluted to OD600 0.05. Either 1 mM or 0.1 mM IPTG was added and cells were grown to OD600 ∼0.3. Cells were then washed twice with 1× Spizizen’s salts and serially diluted. Five microliters of cells were plated on LB agar and grown at 30 °C overnight for enumeration. Images were taken using the Bio-Rad Gel Doc Molecular Imager.
Supplementary Material
Acknowledgments
We thank Patrick Nugent and Ankunda Kariisa for assistance with strain construction and two-hybrid assays, Kevin Lang for helpful discussions, Sirena Tran for assistance with E. coli ChIP-seq, Zoya Ignatova for PARS data, and Charlie Lee at the University of Washington for sequencing assistance. This work was supported by the NIH National Institute of Allergy and Infectious Diseases AI127422 and Training Program in Environmental Toxicology T32 ES007028 to K.B.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008498118/-/DCSupplemental.
Data Availability.
Sequencing data have been deposited in National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) at https://www.ncbi.nlm.nih.gov/sra/SUB8702796.
References
- 1.Komissarova N., Kashlev M., Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. U.S.A. 94, 1755–1760 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGlynn P., Savery N. J., Dillingham M. S., The conflict between DNA replication and transcription. Mol. Microbiol. 85, 12–20 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch I., Landick R., Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 97, 7090–7095 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer A., Landry H. M., Churchman L. S., Pause & go: From the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 46, 72–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan C. L., Wang D., Landick R., Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J. Mol. Biol. 268, 54–68 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Erie D. A., The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim. Biophys. Acta 1577, 224–239 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Cheung A. C. M., Cramer P., Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Nudler E., Mustaev A., Lukhtanov E., Goldfarb A., The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89, 33–41 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Nudler E., RNA polymerase backtracking in gene regulation and genome instability. Cell 149, 1438–1445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta D., Shatalin K., Epshtein V., Gottesman M. E., Nudler E., Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J. S., Marr M. T., Roberts J. W., E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Park J.-S., Roberts J. W., Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. U.S.A. 103, 4870–4875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellon I., Spivak G., Hanawalt P. C., Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Selby C., Sancar A., Molecular mechanism of transcription-repair coupling. Science 260, 53–58 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Selby C. P., Mfd protein and transcription-repair coupling in Escherichia coli. Photochem. Photobiol. 93, 280–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Ragheb M. N., et al. , Inhibiting the evolution of antibiotic resistance. Mol. Cell 73, 157–165.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamarthapu V., et al. , ppGpp couples transcription to DNA repair in E. coli. Science 352, 993–996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epshtein V., et al. , UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505, 372–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen S. E., et al. , Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 15517–15522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalieckas J. M., Wray L. V. Jr, Ferson A. E., Fisher S. H., Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol. Microbiol. 27, 1031–1038 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Zalieckas J. M., Wray L. V. Jr, Fisher S. H., Expression of the Bacillus subtilis acsA gene: Position and sequence context affect cre-mediated carbon catabolite repression. J. Bacteriol. 180, 6649–6654 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le T. T., et al. , Mfd dynamically regulates transcription via a release and catch-up mechanism. Cell 173, 1823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho H. N., van Oijen A. M., Ghodke H., The transcription-repair coupling factor Mfd associates with RNA polymerase in the absence of exogenous damage. Nat. Commun. 9, 1570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deaconescu A. M., et al. , Structural basis for bacterial transcription-coupled DNA repair. Cell 124, 507–520 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Chambers A. L., Smith A. J., Savery N. J., A DNA translocation motif in the bacterial transcription–repair coupling factor, Mfd. Nucleic Acids Res. 31, 6409–6418 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A. J., Savery N. J., RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 33, 755–764 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brugger C., et al. , Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd. Nat. Commun. 11, 3740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure W. R., Cech C. L., On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253, 8949–8956 (1978). [PubMed] [Google Scholar]
- 30.Campbell E. A., et al. , Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 104, 901–912 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Nicolas P., et al. , Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Washburn R. S., Wang Y., Gottesman M. E., Role of E.coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 329, 655–662 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Mars R. A. T., Nicolas P., Denham E. L., van Dijl J. M., Regulatory RNAs in Bacillus subtilis: A gram-positive perspective on bacterial RNA-mediated regulation of gene expression. Microbiol. Mol. Biol. Rev. 80, 1029–1057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray-Soni A., Bellecourt M. J., Landick R., Mechanisms of bacterial transcription termination: All good things must end. Annu. Rev. Biochem. 85, 319–347 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Gusarov I., Nudler E., The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Kang J. Y., et al. , RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le S. Y., J. V. Maizel, Jr, A method for assessing the statistical significance of RNA folding. J. Theor. Biol. 138, 495–510 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Freyhult E., Gardner P. P., Moulton V., A comparison of RNA folding measures. BMC Bioinformatics 6, 241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larson M. H., et al. , A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selby C. P., Sancar A., Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanawalt P. C., DNA repair. The bases for Cockayne syndrome. Nature 405, 415–416 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Menoni H., Hoeijmakers J. H. J., Vermeulen W., Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J. Cell Biol. 199, 1037–1046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen K., Gerdes K., Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32, 1090–1102 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Kawano M., Aravind L., Storz G., An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64, 738–754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potts A. H., et al. , Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8, 1596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenri T., Imamoto F., Kano Y., Three tandemly repeated structural genes encoding tRNA(f1Met) in the metZ operon of Escherichia coli K-12. Gene 138, 261–262 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Bækkedal C., Haugen P., The spot 42 RNA: A regulatory small RNA with roles in the central metabolism. RNA Biol. 12, 1071–1077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khemici V., Carpousis A. J., The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51, 777–790 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Liang W., Rudd K. E., Deutscher M. P., A role for REP sequences in regulating translation. Mol. Cell 58, 431–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y., Deiorio-Haggar K., Anthony J., Meyer M. M., Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Res. 41, 3491–3503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan Y., Qu K., Ouyang Z., Chang H. Y., Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 8, 849–869 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Del Campo C., Bartholomäus A., Fedyunin I., Ignatova Z., Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 11, e1005613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borukhov S., Polyakov A., Nikiforov V., Goldfarb A., GreA protein: A transcription elongation factor from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 8899–8902 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu L. M., Vo N. V., Chamberlin M. J., Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 92, 11588–11592 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stepanova E., et al. , Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 189, 8772–8785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ozsolak F., Milos P. M., RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin H. A., Pedraza-Reyes M., Yasbin R. E., Robleto E. A., Transcriptional de-repression and Mfd are mutagenic in stressed Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol. 21, 45–58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross C., et al. , Novel role of mfd: Effects on stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 188, 7512–7520 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Million-Weaver S., et al. , An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 112, E1096–E1105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brantl S., Jahn N., sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 39, 413–427 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Dörr T., Vulić M., Lewis K., Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8, e1000317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Domka J., Lee J., Bansal T., Wood T. K., Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9, 332–346 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Durand S., Jahn N., Condon C., Brantl S., Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 9, 1491–1497 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Silvaggi J. M., Perkins J. B., Losick R., Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 187, 6641–6650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willing S. E., et al. , Increased toxin expression in a Clostridium difficile mfd mutant. BMC Microbiol. 15, 280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wimberly H., et al. , R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat. Commun. 4, 2115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luria S. E., Delbrück M., Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selby C. P., Sancar A., Transcription-repair coupling and mutation frequency decline. J. Bacteriol. 175, 7509–7514 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hung S. C., Gottesman M. E., Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J. Mol. Biol. 247, 428–442 (1995). [DOI] [PubMed] [Google Scholar]
- 70.Kireeva M. L., Kashlev M., Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 106, 8900–8905 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tadigotla V. R., et al. , Thermodynamic and kinetic modeling of transcriptional pausing. Proc. Natl. Acad. Sci. U.S.A. 103, 4439–4444 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkler M. E., Yanofsky C., Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry 20, 3738–3744 (1981). [DOI] [PubMed] [Google Scholar]
- 73.Yakhnin A. V., Yakhnin H., Babitzke P., RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol. Cell 24, 547–557 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Epshtein V., Cardinale C. J., Ruckenstein A. E., Borukhov S., Nudler E., An allosteric path to transcription termination. Mol. Cell 28, 991–1001 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Vos S. M., Farnung L., Urlaub H., Cramer P., Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saba J., et al. , The elemental mechanism of transcriptional pausing. eLife 8, e40981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krüger S., Gertz S., Hecker M., Transcriptional analysis of bglPH expression in Bacillus subtilis: Evidence for two distinct pathways mediating carbon catabolite repression. J. Bacteriol. 178, 2637–2644 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irnov I., Sharma C. M., Vogel J., Winkler W. C., Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38, 6637–6651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilsson R.-P., Beijer L., Rutberg B., The glpT and glpQ genes of the glycerol regulon in Bacillus subtilis. Microbiology (Reading) 140, 723–730 (1994). [DOI] [PubMed] [Google Scholar]
- 80.Mandal M., Boese B., Barrick J. E., Winkler W. C., Breaker R. R., Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Ebbole D. J., Zalkin H., Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262, 8274–8287 (1987). [PubMed] [Google Scholar]
- 82.Eiamphungporn W., Helmann J. D., Extracytoplasmic function σ factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J. Bacteriol. 191, 1101–1105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jinks-Robertson S., Bhagwat A. S., Transcription-associated mutagenesis. Annu. Rev. Genet. 48, 341–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim N., Jinks-Robertson S., Transcription as a source of genome instability. Nat. Rev. Genet. 13, 204–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pathak V. K., Temin H. M., 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 66, 3093–3100 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thornlow B. P., et al. , Transfer RNA genes experience exceptionally elevated mutation rates. Proc. Natl. Acad. Sci. U.S.A. 115, 8996–9001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dutcher H. A., Raghavan R., Origin, evolution, and loss of bacterial small RNAs. Microbiol. Spectr. 6, 10.1128/microbiolspec.RWR-0004-2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brehm S. P., Staal S. P., Hoch J. A., Phenotypes of pleiotropic-negative sporulation mutants of Bacillus subtilis. J. Bacteriol. 115, 1063–1070 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blattner F. R., et al. , The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
- 90.Harwood C., Cutting S. M., Molecular Biological Methods for Bacillus (Wiley, 1990). [Google Scholar]
- 91.Lang K. S., et al. , Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merrikh H., Machón C., Grainger W. H., Grossman A. D., Soultanas P., Co-directional replication-transcription conflicts lead to replication restart. Nature 470, 554–557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) at https://www.ncbi.nlm.nih.gov/sra/SUB8702796.