Significance
Most cell-cell fusogens have tall ectodomains that drive fusion by undergoing conformational changes that pull two membranes together, akin to the mechanism used by viral fusogens during enveloped virus entry. In contrast, fusion-associated small transmembrane (FAST) proteins from reovirus have short, structurally minimal, membrane-disruptive ectodomains. In this work, we show that evolutionarily distant FAST proteins from aquareovirus and orthoreovirus use different adaptor proteins to hijack host actin assembly and push two membranes together during cell-cell fusion. Despite minimal sequence similarity, the cytoplasmic tails from divergent FAST proteins can be functionally exchanged and even replaced with different actin nucleators while maintaining fusogenicity. This feature suggests a conserved biophysical strategy shared by FAST proteins that could be used by other cell-cell fusogens.
Keywords: actin cytoskeleton, reovirus, cell-cell fusion, FAST proteins
Abstract
Fusion-associated small transmembrane (FAST) proteins are a diverse family of nonstructural viral proteins. Once expressed on the plasma membrane of infected cells, they drive fusion with neighboring cells, increasing viral spread and pathogenicity. Unlike viral fusogens with tall ectodomains that pull two membranes together through conformational changes, FAST proteins have short fusogenic ectodomains that cannot bridge the intermembrane gap between neighboring cells. One orthoreovirus FAST protein, p14, has been shown to hijack the actin cytoskeleton to drive cell-cell fusion, but the actin adaptor-binding motif identified in p14 is not found in any other FAST protein. Here, we report that an evolutionarily divergent FAST protein, p22 from aquareovirus, also hijacks the actin cytoskeleton but does so through different adaptor proteins, Intersectin-1 and Cdc42, that trigger N-WASP–mediated branched actin assembly. We show that despite using different pathways, the cytoplasmic tail of p22 can replace that of p14 to create a potent chimeric fusogen, suggesting they are modular and play similar functional roles. When we directly couple p22 with the parallel filament nucleator formin instead of the branched actin nucleation promoting factor N-WASP, its ability to drive fusion is maintained, suggesting that localized mechanical pressure on the plasma membrane coupled to a membrane-disruptive ectodomain is sufficient to drive cell-cell fusion. This work points to a common biophysical strategy used by FAST proteins to push rather than pull membranes together to drive fusion, one that may be harnessed by other short fusogens responsible for physiological cell-cell fusion.
Aquareovirus and orthoreovirus are two genera of the Reoviridae family of segmented double-stranded RNA viruses that form multinucleated syncytia after infection, which can increase viral spread and pathogenicity (1–4). To drive cell-cell fusion, both aquareovirus and orthoreovirus express a nonstructural, fusion-associated small transmembrane (FAST) protein on the plasma membrane of infected cells. The FAST protein is not required for viral entry, and expression of FAST protein alone is sufficient to cause cells to fuse with naïve neighboring cells, forming large multinucleated syncytium (1, 2, 5–12), confirming they are bona fide cell-cell fusogens. Although they have similar function and topology in the membrane, FAST proteins from aquareovirus and orthoreovirus share minimal sequence identity (13). Based on phylogenetic analysis, they are hypothesized to have evolved from a common, likely nonfusogenic, ancestor 510 million years ago (4, 13, 14). Separate gain-of-function events are believed to have produced fusogenic proteins in both aquareovirus and orthoreovirus, with further divergence or acquisition events resulting in the diversity of FAST proteins found in reoviruses today (13).
Aquareovirus and orthoreovirus FAST proteins are single-pass membrane proteins of fewer than 200 residues comprised of a mostly disordered cytoplasmic tail, a transmembrane domain, and a small ectodomain of fewer than 40 residues (1, 2). The membrane-disruptive ectodomains of FAST proteins typically have solvent-exposed hydrophobic residues and/or myristoylation motifs that are necessary for cell-cell fusion (5, 15–17). In contrast to other cell-cell fusogens that fuse membranes by pulling them together using conformational changes in their ∼10 nm-tall ectodomains, the ectodomains of FAST proteins have minimal predicted secondary structure, are unlikely to undergo conformational changes to drive membrane fusion (1, 2), and extend only ∼1 nm above the bilayer (5, 18). How such short fusogens can overcome the ∼2 nm repulsive hydration barrier and larger barrier presented by cell surface proteins to reach and fuse with an opposing membrane (5, 18) has been a long-standing question for FAST proteins and other short cell-cell fusogens, such as myomixer and myomaker that are involved in myoblast fusion (19–22).
Recently, we found that the FAST protein from reptilian orthoreovirus, p14, hijacks the host cell actin cytoskeleton to drive cell-cell fusion by forming localized branched actin networks (23). This is accomplished through a c-src phosphorylated tyrosine motif, YVNI, in p14’s disordered cytoplasmic tail that binds to a host adaptor protein, Grb2, which then binds to N-WASP and nucleates branched actin assembly. We hypothesize that this directly couples local actin-generated forces to push p14’s short, fusogenic ectodomain into the opposing cell’s plasma membrane (23). While all FAST family proteins have similarly short ectodomains, it is unclear if this is a general strategy used by other FAST proteins to drive cell-cell fusion.
Here, we report that a FAST protein from the divergent aquareovirus, p22, also hijacks the host actin cytoskeleton but does so using a molecular strategy distinct from that of the orthoreovirus FAST protein p14. Instead of binding to Grb2, we find that p22 binds to Intersectin-1 through an SH3 binding motif in its cytoplasmic tail, which binds Cdc42 to activate N-WASP–mediated branched actin assembly. We show that despite minimal sequence identity, the p22 cytoplasmic tail can be functionally swapped with that of p14, suggesting that while the cytoplasmic tails of the two FAST proteins evolved independently, they serve a similar function. By directly coupling the ectodomain to a different actin nucleator, we suggest that actin’s functional role is applying mechanical pressure to a fusogenic ectodomain at the plasma membrane. This biophysical role may be shared across other members of the FAST protein family and could be more generally employed by other cell-cell fusogens.
Results
p22 Localizes to Intracellular Vesicles and the Plasma Membrane.
To explore whether other FAST proteins might hijack the actin cytoskeleton in the same way as reptilian orthoreovirus FAST protein p14, we first examined the primary sequence of the cytoplasmic tails of FAST proteins from aquareovirus and orthoreovirus, two divergent genera of Reoviridae. Surprisingly, the Grb2-binding motif in p14, YVNI, is not found in any other FAST protein (SI Appendix, Fig. S1). While other FAST proteins have one to three tyrosines in their cytoplasmic tails that could be part of motifs used to bind similar cellular adaptor proteins, p22 has no tyrosines (SI Appendix, Fig. S1). In fact, p22 from Atlantic Salmon reovirus-Canada 2009, an aquareovirus-A strain, is the most divergent from p14 from reptilian orthoreovirus (13). To investigate if FAST proteins from evolutionarily distant aquareovirus also hijack the actin cytoskeleton to drive cell-cell fusion—and if so, how—we transiently expressed p22 in Vero cells.
Vero cells expressing mCherry-tagged p22 form multinucleated syncytia, consistent with previous reports (Fig.1 A and B and SI Appendix, Fig. S2A) (10). At 36-h posttransfection, cells with more than 20 nuclei can be readily observed, while cells transfected with mCherry alone have at most only two nuclei (Fig. 1C). We used time-lapse confocal microscopy to directly observe p22-mCherry–expressing Vero cells fusing with neighboring naïve cells (Movies S1, S2, and S3). By labeling the plasma membrane with GFPcaax, we were able to visualize fusion pore expansion and cytoplasmic mixing, as indicated by the exchange of p22-mCherry–labeled intracellular vesicles into the previously nonexpressing cell. To further investigate whether multinucleated cells in our assay are the result of cell-cell fusion, we used a splitYFP cell-cell fusion assay, where two populations of cells expressing either one or the other half of splitYFP are mixed and transfected with p22. We found that when p22-expressing cells formed multinucleated syncytia, their cytoplasm mixed and the two halves of splitYFP associated and fluoresced, as expected for fused cells (SI Appendix, Fig. S2B).
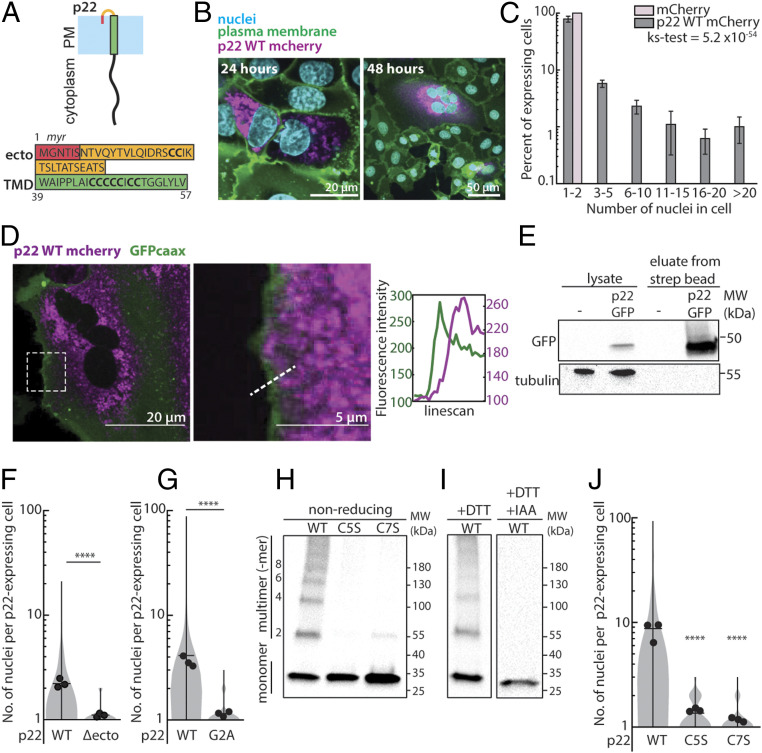
Fig. 1.
p22 is a membrane protein that multimerizes and drives cell-cell fusion. (A) Diagram of p22 topology on the plasma membrane and amino acid sequence of p22 ectodomain, predicted myristoylation, and transmembrane domain. (B) Expression of p22-mCherry (magenta) in Vero cells. Nuclei (Hoechst 33342; cyan) and plasma membrane (CellMaskDeepRed; green) are shown. (C) Nuclei distribution of cells expressing mCherry-tagged p22 and mCherry 36 h posttransfection, error bars indicate SD from three independent replicates, P value of ks-test between the two distributions is shown. (D) Representative confocal images of cells expressing p22-mCherry (magenta) and GFP-caax (plasma membrane marker; green). Contrast was adjusted in the magnified region. Magnified region and fluorescence intensity of line scan of dotted line is shown. (E) Western blot from surface biotinylation of p22-GFP–expressing cells and nontransfected cells. (F) Distribution of number of nuclei in p22-WT– and p22-Δecto–expressing cells from three independent transfections, with mean number of each replicate, average from three independent transfections shown. ****P < 0.0001 using two-tailed Student’s t test. (G) Distribution of number of nuclei in p22-WT– and p22-G2A–expressing cells from three independent transfections, mean number of each replicate, average from three independent transfections shown. ****P < 0.0001 using two-tailed Student’s t test. (H) Western blot of nonreducing SDS/PAGE of myc-tagged p22-WT, p22-C5S, p22-C7S probed with α-myc. (I) Western blot of SDS/PAGE of myc-tagged p22-WT, reduced with DTT and capped with iodoacetamide (IAA), and probed with α-myc. (J) Distribution of number of nuclei in p22-WT–, p22-C5S–, and p22-C7S–expressing cells from three independent transfections, mean number of each replicate, average from three independent transfections shown. ****P < 0.0001 using one-way ANOVA with Dunnett’s test.
Similar to other FAST family proteins, p22 is predicted to be a single-pass transmembrane protein and to localize to the plasma membrane in order to drive membrane fusion with neighboring cells. Consistent with that prediction, prior work has shown that p22 localizes to the membrane fraction of lysates (10). Surprisingly, however, when we imaged mCherry-tagged p22 with confocal microscopy, we found that p22 localized minimally to the plasma membrane (Fig. 1D) and appeared to reside primarily in intracellular structures (SI Appendix, Fig. S2C). By coexpressing p22 with BFP-tagged Rab11 and GFP-tagged EEA1 to label recycling and early endosomes, we found that p22 localizes strongly to endosomes. Similarly, by staining lipid droplets with BODIPY, we found that p22 also localizes to the periphery of lipid droplets (SI Appendix, Fig. S2D). To determine whether any p22 is trafficked to the plasma membrane, we surface biotinylated p22-GFP–expressing cells with cell-impermeable NHS-biotin and enriched for biotinylated proteins with streptavidin beads. We found that p22 eluted from streptavidin beads and, hence, had been biotinylated, confirming its presence on the plasma membrane (Fig. 1E).
p22 Has a Myristoylated Ectodomain and Multimerizes with Its Cysteine-Rich Transmembrane Domain.
A common feature of FAST family proteins is a small, hydrophobic ectodomain. These ectodomains are comprised of less than 40 amino acids, and either are lipidated and/or contain hydrophobic residues, which are necessary for cell-cell fusion and sufficient to disrupt membranes in vitro (1, 2, 16). Similar to other FAST proteins, the putative p22 ectodomain identified using TMHMM (24) is predicted to be myristoylated according to the Eukaryotic Linear Motif prediction tool (Fig. 1A) (25). To determine if the myristoylated ectodomain is needed for p22 to drive cell-cell fusion, as it is for p14 and other FAST proteins, we separately truncated the ectodomain (ΔM1-S38, Δecto) and disrupted the myristoylation motif (p22 G2A). Both perturbations were still trafficked to the plasma membrane, but both p22 Δecto and p22 G2A were nonfusogenic (Fig. 1 F and G and SI Appendix, Fig. S3 A and B). This suggests that despite being short and unstructured, the p22 ectodomain is essential to drive cell-cell fusion and has functional motifs similar to those characterized in FAST proteins from orthoreovirus, including p14 (5, 15, 26).
Surprisingly, the p22 transmembrane domain is predicted, using the same tools as above, to span a cysteine-rich region encoding seven cysteines (Fig. 1A) (24). Other FAST family proteins have been shown to multimerize through membrane-proximal and pH-dependent motifs (27, 28), so we hypothesized that these transmembrane cysteines could be used by p22 to multimerize. Using nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), we found that p22 WT migrated as a monomeric band at a molecular weight of ∼27 kDa, as a dimer at ∼54 kDa, and as multimers of dimers at larger molecular masses (Fig. 1H). When either the first five cysteines (p22 C5S) or all transmembrane cysteines (p22 C7S) were mutated to serines, p22 C5S and C7S were still embedded in the lipid bilayer. However, higher-order multimerization was abrogated, and p22 C5S and C7S migrated primarily as a monomeric band (Fig. 1H and SI Appendix, Fig. S3C). Similarly, when the cysteines were reduced with dithiothreitol (DTT) and capped with iodoacetamide, p22 migrated as a single monomeric band (Fig. 1I). When multimerization was disrupted in p22 C5S and p22 C7S, trafficking to the plasma membrane was unperturbed (SI Appendix, Fig. S3D). However, cell-cell fusion was abrogated (Fig. 1J). Taken together, this data suggests that higher-order multimerization of p22, which could cluster and increase the local concentration of p22 on the plasma membrane, is required for cell-cell fusion.
p22 Hijacks the Actin Cytoskeleton through the ITSN-Cdc42 Pathway.
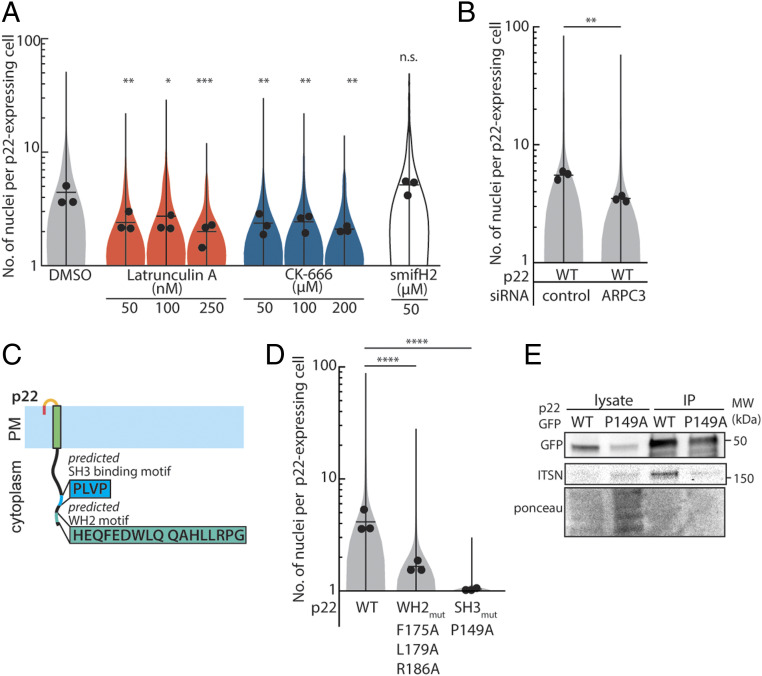
Since the actin cytoskeleton is key to the fusogenicity of p14, we hypothesized that it might similarly contribute to p22-mediated cell-cell fusion. To determine if the host cell’s actin cytoskeleton is essential for p22-mediated cell-cell fusion, we first perturbed the actin cytoskeleton using small-molecule drugs. When actin polymerization is broadly inhibited with latrunculin A, cell-cell fusion was reduced (Fig. 2A). When Arp2/3, a key component of branched actin networks, was inhibited with CK-666, p22-mediated cell-cell fusion was also attenuated (Fig. 2A). Similarly, when ARPC3 expression is reduced with small interfering RNA (siRNA), cell-cell fusion was also attenuated (Fig. 2B and SI Appendix, Fig. S4A). However, when the parallel actin nucleator formin was inhibited with smiFH2, p22-mediated cell-cell fusion was unchanged, even at three times the IC50 (29) (Fig. 2A). These data suggest that branched actin networks are needed for p22-mediated cell-cell fusion.
Fig. 2.
Branched actin cytoskeleton plays a role in p22-mediated cell-cell fusion, and p22 binds to ITSN-1. (A) Distribution of number of nuclei in p22-WT–expressing cells treated with cytoskeletal drugs from three independent transfections, mean number of each replicate, average from three independent transfections shown. P values are one-way ANOVA with Dunnett’s test where not significant (n.s.). P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. (B) Distribution of number of nuclei in p22-WT–expressing cells treated with control siRNA, siRNA targeting ARPC3 from three independent transfections, mean number of each replicate and average from three independent transfections shown. P values are two-way Student’s t test where **P < 0.01. (C) Diagram of predicted SH3 binding motif and predicted WH2 motif in p22 cytoplasmic tail. (D) Distribution of number of nuclei in p22-WT–, p22 WH2mut-, and p22 SH3mut-expressing cells from three independent transfections, mean number of each replicate and average from three independent transfections shown. P values are one-way ANOVA with Tukey’s test where ****P < 0.0001. (E) Western blot of coimmunoprecipitation of GFP-tagged p22-WT and p22-P149A with Intersectin-1.
How does p22 hijack branched actin networks to drive cell-cell fusion? To investigate if p22’s disordered cytoplasmic tail was responsible for hijacking the host cell branched actin cytoskeleton, we truncated the predicted cytoplasmic tail of p22 (ΔT78-T198, Δcyto) and quantified the extent of fusion. Cell-cell fusion was severely attenuated, with only 9 of 235 cells counted having more than 2 nuclei, although Δcyto was not readily trafficked to the plasma membrane, making interpretation difficult (SI Appendix, Fig. S3 E and F). To determine how the p22 cytoplasmic tail might be coupled to branched actin networks, we used the Eukaryotic Linear Motif and Scansite 4.0 prediction tools to identify potential binding motifs (25, 30). A WH2 motif is predicted in the p22 cytoplasmic tail (Fig. 2C). WH2 motifs can bind to actin monomers (31, 32) and when oligomerized can directly nucleate actin comet tails, as in Burkholderia (33). When we disrupted the p22 WH2 motif with point mutations (WH2mut; F175A, L179A, R186A), p22-mediated cell-cell fusion is attenuated but not completely abolished (Fig. 2D). To determine if the predicted WH2 motif can directly bind to actin, we incubated purified GST-tagged WH2 motif with actin monomers in an in vitro binding assay. However, even when incubated with more than 30 times the reported binding affinity of WH2 motifs with actin monomers, there was no detectable binding between WH2 motif and actin (SI Appendix, Fig. S4B). Finally, we observed that p22 WH2mut trafficking to the plasma membrane was significantly reduced, with surface expression of p22 WH2mut only 40% of that of WT p22 (SI Appendix, Fig. S3F). Since the point mutations in WH2mut decreased surface expression, which likely is responsible for the attenuated fusogenicity, we concluded that p22 does not primarily use this predicted WH2 motif to hijack the actin cytoskeleton.
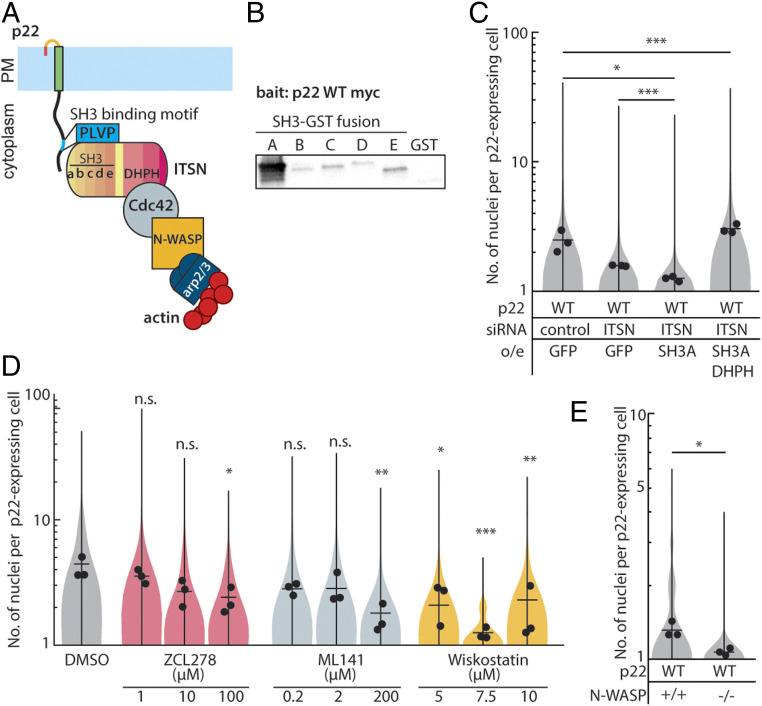
We returned to the Eukaryotic Linear Motif and Scansite4.0 prediction tools and found that they also predicted a SH3-domain binding motif (PXXP) in p22’s cytoplasmic tail (Fig. 2C). This motif was predicted to bind to Intersectin-1. Using coimmunoprecipitation, we found that Intersectin-1 does indeed directly bind to p22 WT (Fig. 2E). When the SH3 binding motif is disrupted with a point mutation (P149A), binding to Intersectin-1 is abrogated (Fig. 2E). To determine if p22 WT colocalizes with Intersectin-1 in live cells, we transfected genome-edited cells expressing endogenously mEGFP-tagged Intersectin-1 with HaloTag-tagged p22 WT (Dataset S1). Because fluorescently labeled p22 WT is undetectable on the plasma membrane via confocal microscopy, we imaged endosomes and the periphery of lipid droplets, where p22 is primarily localized, and found that Intersectin-1 colocalizes there (SI Appendix, Fig. S4C). When the SH3 domain binding motif is disrupted (P149A), p22 P149A is still present in these Intersectin-1 endosomes and lipid droplets (SI Appendix, Fig. S4C). Trafficking of p22 P149A to the plasma membrane is unperturbed (SI Appendix, Fig. S3G), but p22-mediated cell-cell fusion is abrogated (Fig. 2D). Of 952 p22-expressing cells imaged, only 1 cell had more than 2 nuclei (Fig. 2D).
Intersectin-1 has five SH3 domains (Fig. 3A). The p22 SH3 domain binding motif can bind to any or all of the five SH3 domains in Intersectin-1, coupling p22 to branched actin assembly (Fig. 3A). To determine which of the five SH3 domains p22 binds to, we purified each SH3 domain as a GST-fusion protein and incubated it with immunoprecipitated, myc-tagged p22 (SI Appendix, Fig. S4D). We found that p22 strongly binds to SH3 domain A and weakly to the other four SH3 domains of Intersectin-1 (Fig. 3B). To confirm that direct coupling of p22 to Intersectin-1 is needed for p22-mediated cell-cell fusion, we first reduced endogenous expression of Intersectin-1 with siRNA and overexpressed the SH3 domain A to compete with residual Intersectin-1 (Fig. 3C and SI Appendix, Fig. S4E). This decreased the extent of p22-mediated cell-cell fusion (Fig. 3C).
Fig. 3.
Cdc42 and N-WASP signaling downstream of ITSN-1 is needed for p22-mediated cell-cell fusion. (A) Diagram of p22 binding to Intersectin-1 through a SH3 binding motif in its cytoplasmic tail. (B) In vitro binding assay with myc-tagged p22-WT as bait for purified GST-tagged SH3 domain A–E. (C) Distribution of number of nuclei in p22-WT–expressing cells treated with control siRNA or siRNA targeting ITSN-1 and/or overexpressing SH3A and SH3A-DHPH from three independent transfections, mean number of each replicate and average from three independent transfections shown. P values are two-way Student’s t test where *P < 0.05, ***P < 0.001. (D) Distribution of number of nuclei in p22-WT–expressing cells treated with drugs from three independent transfections, mean number of each replicate, average from three independent transfections shown. P values are one-way ANOVA with Dunnett’s test where not significant (n.s.) P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. (E) Distribution of number of nuclei N-WASP null mouse embryonic fibroblasts and control cells expressing p22-WT from three independent transfections, with mean number of each replicate, average from three independent transfections shown. P values are two-tailed Student’s t test where *P < 0.05.
Intersectin-1 is an endocytic scaffolding protein that could couple p22 to other downstream effectors through its five SH3 domains, C2 domain, two EH domains, or DH and PH domains that act as a guanine-exchange factor (GEF) for the small GTPase, Cdc42. This GEF activity has been implicated in actin comet tail formation in Vaccinia viruses and is sufficient to trigger assembly of actin comet tails from purified proteins in vitro (34–36). To determine if binding of Intersectin-1 with Cdc42 is necessary for p22-mediated cell-cell fusion, we targeted the Intersectin-Cdc42 binding site with the inhibitor ZCL278 (37) and found that cell-cell fusion was impaired (Fig. 3D). To determine if Intersectin-1 GEF activity is sufficient to drive p22-mediated cell-cell fusion, we overexpressed a fusion protein consisting of Intersectin-1 SH3 domain A and the DH and PH domains (SH3A-DHPH). This construct was sufficient to restore the extent of p22-mediated cell-cell fusion (Fig. 3C and SI Appendix, Fig. S4E), indicating that only the GEF activity of Intersectin-1 was required.
We next determined if Cdc42 GTPase activity and downstream N-WASP promotion of branched actin network nucleation is needed for p22-mediated cell-cell fusion. We inhibited Cdc42 GTPase activity allosterically with ML141 and found that it also blocked p22-mediated cell-cell fusion (Fig. 3D). When we inhibited N-WASP with Wiskostatin, p22-mediated cell-cell fusion was also inhibited (Fig. 3D). To confirm that N-WASP was specifically required for nucleation of branched actin networks by p22, we expressed p22 in N-WASP-null mouse embryonic fibroblasts (MEFs). Despite the lower transfection efficiency in MEFs, we found that p22-mediated cell-cell fusion was attenuated in N-WASP-null MEFs compared to WT MEFs (Fig. 3E). Altogether, these data show that p22, like p14, hijacks branched actin network assembly to drive cell-cell fusion but uses a distinct pathway. Instead of using a phosphorylation-dependent motif to bind to adaptor proteins, p22 binds to Intersectin-1 through an SH3 binding motif and then relies on Intersectin-1 GEF activity to activate Cdc42 activity to locally trigger N-WASP–mediated actin assembly.
p14 and p22 Are Modular Cell-Cell Fusogens.
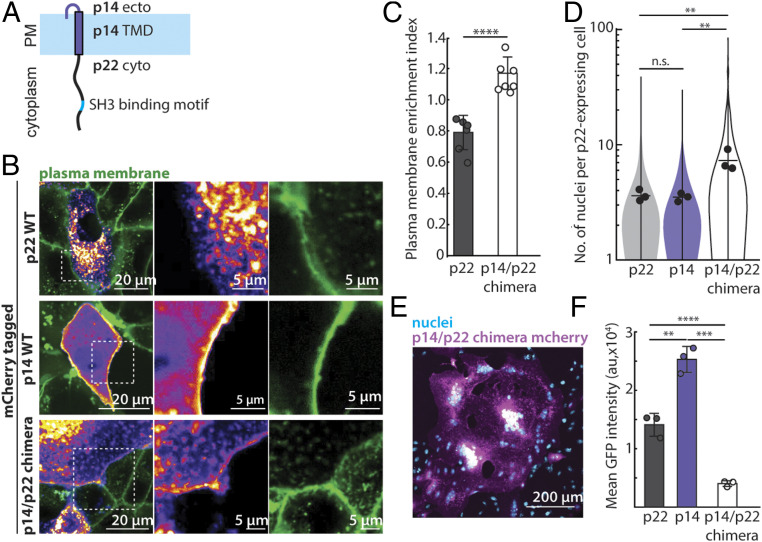
The functional similarities of the molecular pathways used by p22 and p14 raised the possibility that the cytoplasmic tails of FAST proteins from aquareovirus and orthoreovirus might be modular components of a minimal cell-cell fusogen. To directly test if p22 and p14 are modular, we swapped p22’s cytoplasmic tail with that of p14 to create a chimeric fusogen that is comprised of the p14 ectodomain, p14 transmembrane domain, and p22 cytoplasmic tail (p14/p22 chimera, Fig. 4A). Since trafficking to the plasma membrane for type III integral membrane proteins is primarily determined by their transmembrane domain, we hypothesized that the p14/p22 chimera, with p14’s transmembrane domain that localizes readily to the plasma membrane (Fig. 4B), will be expressed at higher levels on the plasma membrane. Consistent with this, the p14/p22 chimera showed higher plasma membrane localization than WT p22 (Fig. 4 B and C). Remarkably, we found that the p14/p22 chimera expressing cells have significantly more nuclei and fuse more readily than those expressing WT p14 or p22, forming large syncytia with more than 80 nuclei at 24 h posttransfection (Fig. 4 D and E). Due to extensive fusion, merging of transfected cells with nontransfected cells diluted the p14/p22 chimera and decreased its expression (Fig. 4F and SI Appendix, Fig. S5A). Despite lower expression, the p14/p22 chimera is so potent that we had to measure cell-cell fusion 12 h earlier than in our standard assay in order to be able to quantify the number of nuclei in p14/p22 chimera-expressing cells.
Fig. 4.
p14 and p22 are modular cell-cell fusogens, and their cytoplasmic tails can be swapped. (A) Diagram of chimeric fusogen with p14 ectodomain, p14 transmembrane domain, and p22 cytoplasmic tail. (B) Confocal images of mCherry-tagged p22-WT, p14-WT, and p14/p22 chimera with plasma membrane labeled with CellMaskDeepRed (green). Boxed regions are magnified. (C) Average plasma membrane enrichment index of mCherry-tagged p22-WT and p14/p22 chimera from three independent transfections, and error bars represent SD 24 h posttransfection. P values are two-tailed Student’s t test where ****P < 0.0001. (D) Distribution of number of nuclei in p22-WT, p14-WT, and p14/p22 chimera expressing cells 24 h posttransfection from three independent transfections, with mean number of each replicate, average from three independent transfections shown. P values are one-way ANOVA with Tukey’s test where not significant (n.s.) P > 0.05, **P < 0.01. (E) Representative confocal image of p14/p22 chimera mCherry (magenta) cell with nuclei labeled with Hoechst 33342 (cyan) at 24 h posttransfection. (F) Mean GFP intensity in each cell expressing GFP-tagged p22-WT, p14-WT, and p14/p22 chimera 24 h posttransfection from three independent transfection and error bars represent SDs. P values are two-tailed Student’s t test where **P < 0.01, ***P < 0.001, ****P < 0.0001.
Why is the p14/p22 chimera more fusogenic than either WT p14 or p22? As we previously showed, p14 drives cell-cell fusion by binding to Grb2 through a motif in its cytoplasmic tail that needs to be phosphorylated by c-src at the plasma membrane (23), and its fusogenicity is attenuated when this binding motif is dephosphorylated. Furthermore, the cytoplasmic tails of a fraction of p14 molecules are cleaved, rendering it nonfusogenic (23). In contrast, p22 is not significantly cleaved (SI Appendix, Fig. S5B) and binds to its adaptor protein, Intersectin-1, in a constitutively active manner. However, its fusogenicity appears to be reduced by limited trafficking to the plasma membrane, likely due to the nonconventional, cysteine-rich transmembrane domain (Fig. 1A). Therefore, when the p22 cytoplasmic tail is ligated with p14’s fusogenic ectodomain and transmembrane domain, the p14/p22 chimera is readily trafficked to the plasma membrane (thanks to p14) with constitutively active coupling to actin assembly (thanks to p22), resulting in a more potent fusogen than either individually. Other permutations of p14, p22 ectodomain, transmembrane domain, and cytoplasmic tails chimeras were nonfusogenic and had large intracellular puncta, suggesting trafficking defects (SI Appendix, Fig. S5C).
Replacing the p14/p22 Chimera’s Branched Actin Nucleator with Formin Preserves Cell-Cell Fusion.
The fusogenicity of the p14/p22 chimera suggests that FAST proteins of both aquareovirus and orthoreovirus are modular cell-cell fusogens, with only the requirement to hijack actin-generated forces to drive cell-cell fusion. Although the cytoplasmic tails of p22 and p14 use different molecular strategies to accomplish the same task of coupling actin assembly with a membrane-disruptive ectodomain, they can be swapped and remain fusogenic. If the primary role of actin assembly is to apply localized pressure at the plasma membrane and “push” the membrane-disruptive ectodomain into the opposing plasma membrane, could other force-generating actin nucleators also drive cell-cell fusion? Branched actin networks can exert up to 5 nN or about 1,250 nN/μm2 as they grow, a filopodium can exert up to only 10 pN (38–41). However, the diameter of a filopodium is 100–300 nm and, hence, can exert localized pressures ranging from ∼140–1,270 nN/μm2 (41–44), comparable to branched networks.
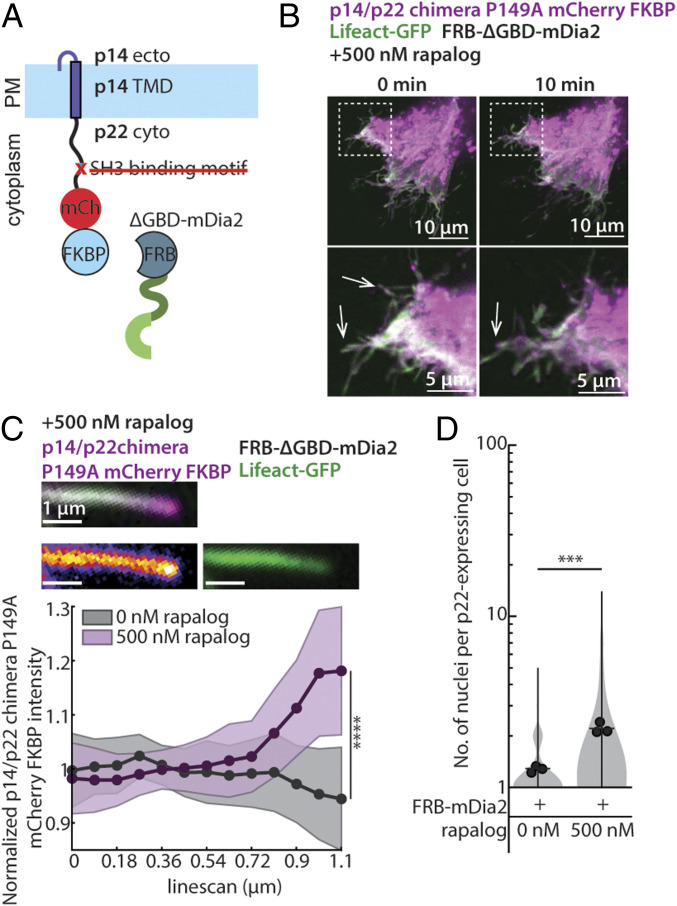
To determine if the pressure generated by formin-based actin nucleation is sufficient to drive cell-cell fusion, we first rendered p14/p22 chimera nonfusogenic by disrupting the SH3 binding motif. We then coupled the nonfusogenic mutant of p14/p22 chimera to a constitutively active formin (ΔGBD-mDia2), which is involved in filopodia formation, using FKBP-FRB tags (Fig. 5A). In the absence of the rapamycin analog (rapalog), coexpression of p14/p22 chimera-P149A-FKBP with an FRB-ΔGBD-mDia2 led to filopodia formation (Fig. 5B), consistent with previous reports expressing ΔGBD-mDia2 (45). Upon addition of the rapamycin analog, the p14/p22 chimera-P149A-FKBP clustered and enriched at the tip of the filopodia (Fig. 5 B and C and Movies S4 and S5) and the filopodia elongated (Movies S4 and S5), consistent with the localization of formins to the ends of actin filaments where they drive filament polymerization and subsequent filament bundling (46–49). Remarkably, this coupling of ΔGBD-mDia2 with the p14/p22 chimera-P149A-FKBP is sufficient to drive cell-cell fusion (Fig. 5D). Overall, this suggests that the local assembly actin, independent of the specific architecture, is sufficient to drive cell-cell fusion, supporting the hypothesis that FAST proteins are minimal fusogens that couple membrane-disruptive ectodomains with localized pressure exerted on the plasma membrane.
Fig. 5.
Replacing the p14/p22 branched actin nucleator with a formin is sufficient to drive cell-cell fusion. (A) Diagram of p14/p22 chimera with FKBP at C terminus and P149A mutation with FRB-tagged ΔGBD-mDia2. (B) Confocal images of p14/p22 chimera P149A FKBP-mCherry (magenta) coexpressed with FRB-ΔGBD-mDia2 and Lifeact-GFP (green) cell at 0 min and 10 min after addition of 500 nM rapalog. Filopodia-like protrusions before and after rapalog addition is denoted with arrows. Box regions magnified. (C) Representative confocal merged image of a filopodia-like protrusion from cell expressing p14/p22 chimera-P149A-FKBP-mCherry (magenta), Lifeact-GFP (green), and FRB-ΔGBD-mDia2 10 min after addition of 500 nM rapalog. Each channel is shown with p14/p22 chimera-P149A-FKBP-mCherry (fire) and Lifeact-GFP (green). Average normalized fluorescence intensity of p14/p22 chimera-P149A-FKBP-mCherry along the length before (n = 33 filopodia-like protrusions) and 10 min after (n = 36 filopodia-like protrusions) addition of 500 nM rapalog. SD above and below the average are shown. ****P < 0.0001 by Student’s t test on the last datapoint. (D) Distribution of number of nuclei in p14/p22 chimera-P149A-FKBP– and FRB-ΔGBD-mDia2–expressing cells with and without rapalog from three independent transfections, with mean number of each replicate, average from three independent transfections shown. P values are two-tailed two-sample Student’s t test where ***P < 0.001.
Discussion
FAST proteins are a unique family of fusogens that lack large ectodomains and the ability to undergo conformational changes like those of better-studied fusogens, such as the viral fusogen hemagglutinin and the cell-cell fusogens Eff-1 and Hap2 (50–55). The orthoreovirus FAST protein p14 nucleates branched actin network assembly by binding the adaptor protein Grb2 to a phospho-tyrosine motif in its cytoplasmic tail, which subsequently binds the nucleation-promoting factor N-WASP (23). While the FAST family proteins are thought to have evolved from a common ancestor over 500 million years ago and now show little sequence similarity, we hypothesized that other fusogens in the FAST family also harness the actin cytoskeleton to push their similarly short ectodomains into the opposing bilayer to drive cell-cell fusion. However, analysis of the cytoplasmic domains of all known FAST proteins revealed that none had the required Grb2-binding sequence. This led us to investigate how the most evolutionary distant FAST protein from p14, the aquareovirus p22, is able to drive cell-cell fusion without using the molecular strategy employed by p14.
In this study, we found that p22 uses a phosphorylation-independent SH3 binding motif to bind to Intersectin-1 and subsequently Cdc42 to hijack N-WASP–mediated actin assembly for cell-cell fusion. Interestingly, this SH3 motif is conserved in the FAST protein of Turbot reovirus, another aquareovirus-A strain (NS22, SI Appendix, Fig. S6). However, such an Intersectin-1 binding motif is not found in other orthoreovirus and aquareovirus species, which highlights the divergence of FAST proteins and raises the possibility that the other FAST proteins (p16, p10, p13, p15) may hijack different, yet unidentified host factors to drive cell-cell fusion.
Although neither the Grb2 nor Intersectin-1 binding motifs are conserved beyond closely related aquareovirus and orthoreovirus fusogens, FAST proteins appear to be modular cell-cell fusogens with a common functional role for their cytoplasmic tails. Both p22 and p14 cytoplasmic tails couple to N-WASP–nucleated branched actin assembly and can be swapped to create a functional p14/p22 chimeric fusogen. Interestingly, the p14/p22 chimera reported here is substantially more potent than WT p14 or p22. This suggests that selective pressures may limit the potency of FAST protein fusion during viral infection in an organism (3). Although the actin cytoskeleton has yet to be implicated in other FAST proteins, similar chimeric fusogens combining p10 from avian reovirus and p15 from baboon reovirus have also been shown to be functional (1, 27), further supporting the idea that FAST family fusogen domains are modular.
The modularity of FAST family proteins indicates that the specific identity of host molecular players hijacked by the fusogens is secondary to their biophysical roles. Both p14 and p22 hijack N-WASP, which promotes the nucleation of branched actin networks, which can exert pressures up to 1,250 nN/μm2. However, when we replace N-WASP with a formin, which nucleates parallel actin bundles instead of branched actin networks, we find that cell-cell fusion is preserved. While branched actin structures have been implicated during other physiological and pathological cell-cell fusion processes, this is a demonstration (albeit a synthetic one) that filopodia-mediated forces can drive cell-cell fusion.
Overall, our findings support a model of short cell-cell fusogens in which the minimal requirements for fusion are mechanical pressure to bring two plasma membranes together, regardless of how the force is generated, coupled to a multimerized, membrane-disruptive ectodomain. Localized assembly of actin beneath a membrane-disruptive ectodomain could be a fundamental strategy for overcoming the fusion energy barrier, one that is used not only by FAST family proteins but also by other cell-cell fusogens that are not able to use conformational changes to pull membranes together, such as myomaker and myomixer.
Materials and Methods
A full description of methods is available in SI Appendix.
Molecular Cloning.
Aquareovirus fusion associated small transmembrane protein, p22 (Atlantic salmon reovirus Canada-2009, accession no. C0L0N0), inserted into mammalian expression vector pcDNA3.1 with Kozak sequence and C terminus tags.
ΔGBD-mDia2 (amino acids 258–1171) was amplified from pCMV-eGFP-mDia2 (a kind gift from Scott Hansen, University of Oregon, Eugene, OR) and inserted with a N terminus FRB tag into pcDNA3.1. Codon-swapped versions of SH3A domain from human Intersectin-1 (amino acids 740–816) and DH and PH domains from human Intersectin-1 (amino acids 1226–1573) was amplified and inserted downstream of SH3 A domain and eGFP into pcDNA3.1
Cell Culture, Plasmid, and siRNA Transfection.
Vero cells were obtained from the University of California-Berkeley Cell Culture Facility. Vero cells were grown in DMEM (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies), 10% nonessential amino acids (Life Technologies), and 1% Pen-Strep (Life Technologies), at 37 °C, 5% CO2. N-WASP−/− and N-WASP+/+ mouse embryonic fibroblasts cells were a kind gift from Scott Snapper (Harvard University, Cambridge, MA) and were grown in DMEM (Life Technologies) supplemented with 10% heat-inactivated FBS (Life Technologies) and 1% Pen-Strep (Life Technologies), at 37 °C, 5% CO2.
Cells were negative for mycoplasma as verified with Mycoalert mycoplasma detection kit (Lonza).
Cells were transfected with plasmids with FuGENE HD (Promega) according to manufacturer’s instructions.
siRNA Knockdown.
Cells were transfected with endoribonuclease-prepared siRNA (esiRNA) targeting ARPC3 (EHU107121, Sigma-Aldrich), esiRNA targeting ITSN-1 (EHU102031, Sigma-Aldrich), or control esiRNA (EHUEGFP, Sigma-Aldrich) with Lipofectamine RNAiMAX (ThermoFisher) according to manufacturer’s instructions. Twenty-four hours posttransfection, cells were transfected with p22-WT-mCherry alone or with rescue plasmids, codon-swapped SH3A, SH3A-DHPH, or GFP using FuGENE HD (Promega). The extent of cell-cell fusion was assayed 36 h postplasmid transfection/66 h post-siRNA transfection.
Nuclei Count.
Twenty-five to thirty fields of view from each of three independent transfections were collected with a spinning disk confocal microscope using either a 20× or 40× objective. Cytoplasmic p22 fluorescence intensity was used to make a binary mask of cell bodies. Nuclei masks were made from cells stained with 5 μM Syto11 (Thermo Fisher), Hoechst 33342 (Thermo Fisher), or from the inverted mask of cytoplasm fluorescence. Nuclei per cell expressing fluorescently tagged constructs were counted either manually or using the Speckle Inspector FIJI plugin (https://www.biovoxxel.de/). Cells expressing the p14/p22 chimera were masked using pixel classification in Ilastik (https://www.ilastik.org/), and nuclei were counted as above. Cells with more than two nuclei were considered multinucleated.
Plasma Membrane Enrichment Quantification.
The plasma membrane of p22-mCherry– and p14/p22-Chimera-mCherry–expressing cells were labeled with CellMaskDeepRed (ThermoFisher). Cells were imaged with confocal microscopy with a 60× objective. A linescan spanning the plasma membrane and 600–1000 nm proximal to the plasma membrane was analyzed in ImageJ. The plasma membrane enrichment index is defined as the fluorescence intensity of mCherry-tagged proteins at the plasma membrane, normalized to the average fluorescence intensity of the protein in the cytosol.
Filopodia Enrichment Quantification.
Vero cells coexpressing p14/p22 chimera-P149A-FKBP, FRB-ΔGBD-mDia2, and Lifeact-GFP were imaged with confocal microscopy. Five hundred nanomolar rapalog (AP21967, Takara Bio) was added to the media, and cells were imaged for 15 min. Linescans spanning 1.1 μm from the tip of filopodia were analyzed using ImageJ. The last 270 nm of filopodia is defined as the tip of the filopodia, and the remaining 900 nm is defined as the length of the filopodia. Fluorescence intensity of p14/p22 chimera-P149A-mCherry-FKBP along the length of the filopodia is normalized to the average fluorescence intensity of the entire length of the filopodia. Approximately 30 filopodia at 0 min and 30 filopodia at 10 min after addition of rapalog, all selected at random, were analyzed.
Surface Biotinylation.
Vero cells expressing GFP-tagged or myc-tagged p22 was labeled with Sulfo-NHS-Biotin at 4 °C. The reaction was quenched with 100 mM glycine, and the cells were lysed with RIPA buffer. Biotinylated proteins were enriched with streptavidin beads (ThermoFisher). Beads were washed and boiled in sample buffer. Samples were analyzed on Western blot with primary antibodies α-GFP (1:5000, g1544, Sigma Aldrich), α-myc (1:5000, 9e10, Sigma Aldrich), α-tubulin (1:5000, Clone YL1/2, Thermo Fisher), and secondary antibodies, α-rabbit HRP (1:5000, 65–6120, Thermo Fisher), α-mouse HRP (1:5000, Jackson Labs), α-rat AlexaFluor 647 (1:5000, Life Technologies). Densitometry of eluate and lysate lanes of Western blots was used to quantify and normalize surface expression of p22 mutant to that of WT p22.
Nonreducing SDS-PAGE, Reduction, and Alkylation.
For nonreducing SDS-PAGE, Vero cells expressing myc-tagged p22 WT, C5S, and C7S were lysed, enriched with myc-Trap beads (Chromotek), and boiled in sample buffer without reducing agents. To reduce p22, samples were boiled in Laemmli sample buffer with 350 mM DTT. To cap cysteines in p22 WT, cysteines were first reduced with 50 mM DTT and capped with 100 mM iodoacetamide (Sigma).
Coimmunoprecipitation.
Vero cells expressing GFP-tagged p22 and p22-P149A were lysed and immunoprecipated with GFP-Trap beads. The beads were washed, boiled in sample buffer, and separated by SDS/PAGE. Proteins were transferred onto a nitrocellulose membrane and probed with primary antibodies, α-ITSN (1:1000, Clone 29, BD Biosciences), α-GFP (1:5000, gp1544, Life Technologies), and secondary antibodies, α-mouse HRP (1:5000, Jackson Labs), α-rabbit HRP (1:5000, 65–6120, Thermo Fisher), α-rat AlexaFluor 647(1:5000, Life Technologies).
Protein Purification and SH3 and Actin Binding Assay.
GST-tagged WH2 motif was purified from Rosetta using affinity chromatography. Actin was purified from rabbit skeletal muscle (56). GST-tagged WH2 motif and GST bound to glutathione beads were incubated with 20 μM actin. Beads were washed, boiled in sample buffer, and analyzed with SDS-PAGE.
N terminus GST-tagged SH3A, SH3B, SH3C, SH3D, and SH3E domains from human Intersectin-1 were expressed in Rosetta and purified using affinity chromatography. HEK293T cells were transfected with myc-tagged p22 and immunoprecipitated with myc-Trap beads (Chromotek). Beads were washed and incubated with 30 μM GST-tagged SH3 domains. Beads were washed, boiled in sample buffer, and analyzed on a Western blot with α-GST (1:5000, abcam).
Flow Cytometry.
Cells were transfected with GFP-tagged p14, p22, and chimera. Twenty-four hours posttransfection, the cells were lifted and analyzed with Attune (Thermo Fisher). GFP-expressing population was identified by comparing with nontransfected cells, and the average GFP intensity of the GFP-expressing population was calculated using FlowJo.
Statistical Analysis.
Two-way Student’s t test was used to compare between two conditions, one-way-ANOVA with Dunnett’s test when we are comparing across more than two conditions to a control. Kolmogorov–Smirnov test was used compare distributions of the number of nuclei in p22-expressing cells. Kolmogorov–Smirnov test was calculated using SciPy and other statistics were calculated using Prism 8 (GraphPad).
Supplementary Material
Acknowledgments
We thank D.A.F. Laboratory members, especially Andrew Harris, for useful feedback and technical consultation; Conklin Laboratory at the University of California, San Francisco for providing WTC10 hiPSC line; Lavis Laboratory at Janelia for providing JF635 HaloTag ligand; Dr. Sun Hae Hong for generating the AP2-tagRFP-T hiPSC line; the UC Berkeley QB3 MacroLab for purified S. pyogenes NLS-Cas9; and Cancer Research Laboratory Flow Cytometry Facility for hiPSC sorting. This work is supported by National Institute of General Medical Sciences Grants R01GM114671 and R01GM134137 (to D.A.F.), the Chan Zuckerberg Biohub (D.A.F.), NSF Grant DBI-1548297 (to D.A.F.), and NIH Grant MIRA R35GM118149 (to D.G.D.). K.M.C.C. was funded by an NSF Graduate Research Fellowship Program fellowship. D.A.F. is a Chan Zuckerberg Biohub Investigator. A.L.A. was supported by NIH F31 Grant 1F31GM128325-01. J.M. is supported through NIH Grant F99CA253758, the New York University (NYU) Margaret and Herman Sokol Fellowship, NYU Horizon Fellowship, and German Academic Fellowship Foundation (Studienstiftung) funding. M.J. is supported by American Heart Association Postdoctoral Fellowship 18POST34000029. D.S. was supported by a Boehringer Ingelheim Fonds PhD Fellowship. J.M. and D.S. thank the Boehringer Ingelheim Fonds for generously supporting their participation in Marine Biological Laboratory Physiology Course 2019. S.S. was funded by a Life Sciences Research Foundation fellowship. We thank the Marine Biological Laboratory Physiology Course 2019 for providing an opportunity to explore initial ideas that formed the basis of this study, and we thank the Burroughs Wellcome Fund for supporting postcourse fellowships for A.L.A. and J.M. to continue the project at University of California, Berkeley.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007526118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Ciechonska M., Duncan R., Reovirus FAST proteins: Virus-encoded cellular fusogens. Trends Microbiol. 22, 715–724 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Duncan R., Fusogenic reoviruses and their fusion-associated small transmembrane (FAST) proteins. Annu. Rev. Virol. 6, 341–363 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kanai Y., et al. , Cell-cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses. PLoS Pathog. 15, e1007675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefkowitz E. J.et al., Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46, D708–D717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corcoran J. A., et al. , Myristoylation, a protruding loop, and structural plasticity are essential features of a nonenveloped virus fusion peptide motif. J. Biol. Chem. 279, 51386–51394 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Dawe S., Duncan R., The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 76, 2131–2140 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan R., Corcoran J., Shou J., Stoltz D., Reptilian reovirus: A new fusogenic orthoreovirus species. Virology 319, 131–140 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Duncan R., Murphy F. A., Mirkovic R. R., Characterization of a novel syncytium-inducing baboon reovirus. Virology 212, 752–756 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Guo H., Sun X., Yan L., Shao L., Fang Q., The NS16 protein of aquareovirus-C is a fusion-associated small transmembrane (FAST) protein, and its activity can be enhanced by the nonstructural protein NS26. Virus Res. 171, 129–137 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Racine T., et al. , Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site. J. Virol. 83, 5951–5955 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmulevitz M., Epand R. F., Epand R. M., Duncan R., Structural and functional properties of an unusual internal fusion peptide in a nonenveloped virus membrane fusion protein. J. Virol. 78, 2808–2818 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmulevitz M., Duncan R., A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 19, 902–912 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nibert M. L., Duncan R., Bioinformatics of recent aqua- and orthoreovirus isolates from fish: Evolutionary gain or loss of FAST and fiber proteins and taxonomic implications. PLoS One 8, e68607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attoui H., et al. , Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 83, 1941–1951 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Top D., Read J. A., Dawe S. J., Syvitski R. T., Duncan R., Cell-cell membrane fusion induced by p15 fusion-associated small transmembrane (FAST) protein requires a novel fusion peptide motif containing a myristoylated polyproline type II helix. J. Biol. Chem. 287, 3403–3414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Top D., et al. , Liposome reconstitution of a minimal protein-mediated membrane fusion machine. EMBO J. 24, 2980–2988 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Key T., Sarker M., de Antueno R., Rainey J. K., Duncan R., The p10 FAST protein fusion peptide functions as a cystine noose to induce cholesterol-dependent liposome fusion without liposome tubulation. Biochim. Biophys. Acta 1848, 408–416 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Corcoran J. A., et al. , The p14 fusion-associated small transmembrane (FAST) protein effects membrane fusion from a subset of membrane microdomains. J. Biol. Chem. 281, 31778–31789 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Millay D. P., et al. , Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi P., et al. , Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q., et al. , The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 8, 15664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn M. E., et al. , Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 8, 15665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K. M. C., Son S., Schmid E. M., Fletcher D. A., A viral fusogen hijacks the actin cytoskeleton to drive cell-cell fusion. eLife 9, e51358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Gouw M., et al. , The eukaryotic linear motif resource–2018 update. Nucleic Acids Res. 46, D428–D434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry C., Key T., Haddad R., Duncan R., Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides. J. Biol. Chem. 285, 16424–16433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Key T., Duncan R., Compact A., A compact, multifunctional fusion module directs cholesterol-dependent homomultimerization and syncytiogenic efficiency of reovirus p10 FAST proteins. PLoS Pathog. 10, e1004023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran J. A., Clancy E. K., Duncan R., Homomultimerization of the reovirus p14 fusion-associated small transmembrane protein during transit through the ER-Golgi complex secretory pathway. J. Gen. Virol. 92, 162–166 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Rizvi S. A., et al. , Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16, 1158–1168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obenauer J. C., Cantley L. C., Yaffe M. B., Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Symons M., et al. , Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell 84, 723–734 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Machesky L. M., Insall R. H., Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347–1356 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Benanti E. L., Nguyen C. M., Welch M. D., Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell 161, 348–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Co C., Wong D. T., Gierke S., Chang V., Taunton J., Mechanism of actin network attachment to moving membranes: Barbed end capture by N-WASP WH2 domains. Cell 128, 901–913 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries A. C., Donnelly S. K., Way M., Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J. Cell Sci. 127, 673–685 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Snetkov X., Weisswange I., Pfanzelter J., Humphries A. C., Way M., NPF motifs in the vaccinia virus protein A36 recruit intersectin-1 to promote Cdc42:N-WASP-mediated viral release from infected cells. Nat. Microbiol. 1, 16141 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Friesland A., et al. , Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. U.S.A. 110, 1261–1266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bieling P., et al. , Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parekh S. H., Chaudhuri O., Theriot J. A., Fletcher D. A., Loading history determines the velocity of actin-network growth. Nat. Cell Biol. 7, 1219–1223 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Prass M., Jacobson K., Mogilner A., Radmacher M., Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 174, 767–772 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cojoc D., et al. , Properties of the force exerted by filopodia and lamellipodia and the involvement of cytoskeletal components. PLoS One 2, e1072 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogilner A., Rubinstein B., The physics of filopodial protrusion. Biophys. J. 89, 782–795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atilgan E., Wirtz D., Sun S. X., Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 90, 65–76 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris A. R., Jreij P., Fletcher D. A., Mechanotransduction by the actin cytoskeleton: Converting mechanical stimuli into biochemical signals. Annu. Rev. Biophys. 47, 617–631 (2018). [Google Scholar]
- 45.Yang C., et al. , Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, e317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignjevic D., et al. , Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863–875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S., Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13, 534–545 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Romero S., et al. , Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Liu A. P., et al. , Membrane-induced bundling of actin filaments. Nat. Phys. 4, 789–793 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison S. C., Viral membrane fusion. Virology 479-480, 498–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez-Vargas J., et al. , Structural basis of eukaryotic cell-cell fusion. Cell 157, 407–419 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Zeev-Ben-Mordehai T., Vasishtan D., Siebert C. A., Grünewald K., The full-length cell-cell fusogen EFF-1 is monomeric and upright on the membrane. Nat. Commun. 5, 3912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fédry J., et al. , The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168, 904–915.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valansi C., et al. , Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 216, 571–581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng J., et al. , Fusion surface structure, function, and dynamics of gamete fusogen HAP2. eLife 7, e39772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spudich J. A., Watt S., The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 10, 2131–2140 (2002). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.