Significance
Adenoviruses (AdVs) are DNA viruses that can cause severe respiratory illness in humans. A better understanding of the complex ways AdVs utilize cellular processes in service of their replication is critical to the development of new therapies to counter viral infection and disease. Using a genome-wide screen, we identified a cellular protein Mindbomb 1 (MIB1) required for AdV infection and show that it functions in the AdV entry process to mediate release of the viral genome into the nucleus. We further show that MIB1, as an E3 ubiquitin ligase, mediates AdV capsid disassembly and genome release through ubiquitination of a target protein(s), demonstrating the importance of this rapid posttranslational modification to virus infection.
Keywords: adenovirus, viral entry, host factor, Mindbomb 1, E3 ubiquitin ligase
Abstract
The journey from plasma membrane to nuclear pore is a critical step in the lifecycle of DNA viruses, many of which must successfully deposit their genomes into the nucleus for replication. Viral capsids navigate this vast distance through the coordinated hijacking of a number of cellular host factors, many of which remain unknown. We performed a gene-trap screen in haploid cells to identify host factors for adenovirus (AdV), a DNA virus that can cause severe respiratory illness in immune-compromised individuals. This work identified Mindbomb 1 (MIB1), an E3 ubiquitin ligase involved in neurodevelopment, as critical for AdV infectivity. In the absence of MIB1, we observed that viral capsids successfully traffic to the proximity of the nucleus but ultimately fail to deposit their genomes within. The capacity of MIB1 to promote AdV infection was dependent on its ubiquitination activity, suggesting that MIB1 may mediate proteasomal degradation of one or more negative regulators of AdV infection. Employing complementary proteomic approaches to characterize proteins proximal to MIB1 upon AdV infection and differentially ubiquitinated in the presence or absence of MIB1, we observed an intersection between MIB1 and ribonucleoproteins (RNPs) largely unexplored in mammalian cells. This work uncovers yet another way that viruses utilize host cell machinery for their own replication, highlighting a potential target for therapeutic interventions that counter AdV infection.
Human adenoviruses (hAdVs) are DNA viruses that can cause severe respiratory illness predominantly affecting children and immune-compromised individuals. Aside from respiratory illness, AdVs can also cause meningitis, conjunctivitis, and gastrointestinal disease with recent outbreaks in hospitals and colleges resulting in fatalities (1, 2). The contagiousness of certain AdV serotypes poses a serious public health concern. However, there are currently neither vaccines nor specific antiviral treatments. Adenoviruses can also be reengineered as vectors to treat a variety of other diseases (3, 4). A better understanding of the factors mediating AdV genome delivery to the nucleus has implications for the development of antiviral therapeutics as well as in application of AdV vectors in vaccine development and oncolytic cancer therapies.
AdV entry encompasses all steps from attachment of the virus via its receptor at the plasma membrane to the successful deposition of the viral genome into the nucleus of the host cell. For adenovirus serotype 5 (AdV5), binding of the fiber protein to the coxsackievirus and adenovirus receptor (CAR) triggers endocytosis of the virus particle (5). As this endosome matures and acidifies, the capsid becomes partially degraded, shedding its fibers and exposing the membrane-lytic pVI protein. Puncture of the endosome by pVI releases the compromised yet intact capsid into the cytoplasm where it can couple to microtubule motors (6–8). Microtubules (MTs) are dynamic cytoskeletal components that nucleate predominantly from the microtubule organizing center (MTOC), often synonymous with the nuclear-proximal centrosome. By hijacking MT tracks, the virus scales most of the distance to its nuclear destination (9).
Even upon arrival in the vicinity of the nucleus, a series of coordinated events are still required to successfully deliver the viral genome into the nucleus for early transcription and replication using host cell machinery. The viral capsid must transfer from the MTOC to the nuclear pore where it docks directly onto the nuclear pore complex (NPC) via Nup214 (10, 11). Here, unable to traverse the pore, the capsid must be physically disrupted. The liberated genomic viral DNA (vDNA) and associated proteins are imported into the nucleus while the now-empty capsid shell is relocated toward the periphery. The involvement of numerous cellular proteins including CRM1, histone H1, select nucleoporins, import factors, and MT motors highlight the complexity of these processes (12–16). However, a comprehensive view of the factors governing capsid disassembly and genome release at this interface is still lacking.
In this work, we used a gene-trap screening approach in haploid cells with AdV5 infection and identify the E3 ubiquitin ligase Mindbomb 1 (MIB1) as a host factor required by the virus. We demonstrate that MIB1 functions early in the AdV lifecycle to promote delivery of the viral genome to the nucleus. Specifically, in the absence of MIB1 ubiquitination, capsids successfully arrive at the nucleus but the release of vDNA into the nucleus is blocked. Using complementary proteomic approaches, we identified cellular proteins typically associated with ribonucleoprotein (RNP) granules enriched for ubiquitination and in close proximity to MIB1 during AdV DNA release while observing a depletion of cytoskeletal components. These findings pave the way for future mechanistic work to identify the specific MIB1 ubiquitination target(s) mediating AdV genome delivery.
Results
MIB1 Is a Host Factor for AdV Infection.
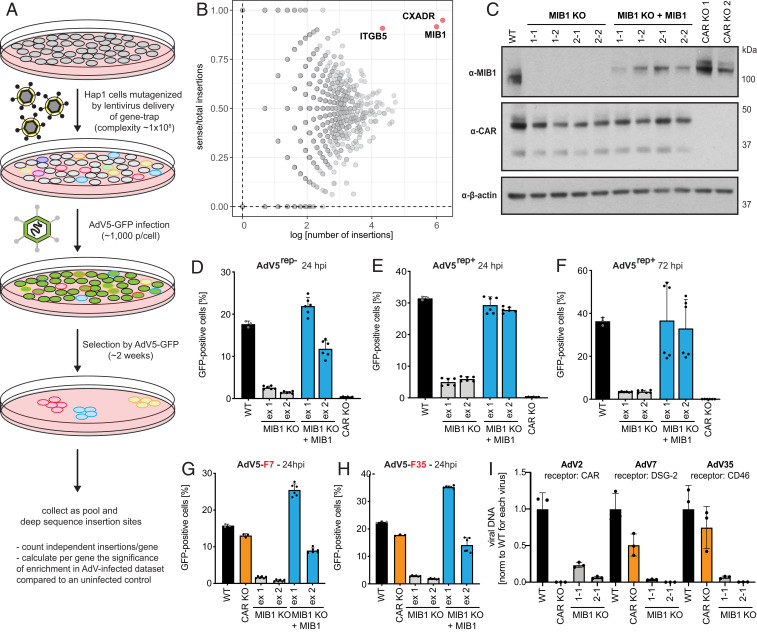
To identify human host factors required for AdV5 infection, we performed a genetic loss-of-function screen in near-haploid (Hap1) cells, which can be efficiently mutagenized via insertional mutagenesis. This approach has been successfully employed in the identification of receptors and host factors for several viruses (17, 18). Retroviral delivery of a gene-trap cassette introducing a strong splice acceptor site and polyadenylation signal was used to disrupt genes and generate a saturated library of loss-of-function mutants (19). Although the orientation in which the gene-trap cassette integrates into genes is stochastic, the cassette was designed to disrupt a gene upon integration in the same transcriptional orientation. Therefore, genes that confer resistance to AdV5 infection upon inactivation should be enriched for disruptive sense orientation gene-trap insertions. We infected this library with replication-competent GFP-expressing adenovirus 5 (AdV5) and isolated surviving cells (Fig. 1A).
Fig. 1.
Haploid genetic screen identifies E3 ubiquitin ligase MIB1 as a host factor necessary for AdV infection independent of AdV receptor. (A) Outline of haploid genetic screen for AdV5 infection. (B) Fishtail plot showing enrichment of genes in haploid screen upon AdV5 selection identified by deep sequencing and plotted as number of distinct gene-trap insertions (x axis) and the ratio of sense over total insertions (y axis) per gene. Genes significantly enriched for sense orientation integrations (FDR < 0.05) labeled by gene name (red). (C) Western blot of MIB1 CRISPR KO clones in Hap1 cells generated by guide RNAs targeted to either exon 1 (1-1, 1-2) or exon 2 (2-1, 2-2), KO clones reconstituted with MIB1, and two CAR KO clones probed for MIB1, CAR (two isoforms), or β-actin expression. (D–F) Infection of Hap1 WT cells, MIB1 KO clones, WT MIB1-reconstituted KO clones, and CAR KO clones with (D) AdV5rep−-GFP (MOI = 2,500 p/cell; 24 hpi). (E and F) AdV5rep+-GFP [(E) MOI = 75 p/cell; 24 hpi and (F) MOI = 50 p/cell; 72 hpi] and (G and H) Infection of Hap1 WT cells, a CAR KO clone, MIB1 KO clones, and WT MIB1-reconstituted KO clones with recombinant AdV5rep−-GFP expressing the fiber protein of (G) AdV7 or (H) AdV35 (MOI = 1 × 104 p/cell; 24 hpi). (I) Hap1 WT cells, a CAR KO clone, and individual MIB1 KO clones (1-1, 2-1), infected with WT AdV serotypes using distinct entry receptors: AdV2 (CAR), AdV7 (DSG-2), and AdV35 (CD46) at MOI = 1 × 103 p/cell (vDNA was harvested at 72 hpi and measured by qPCR). All experiments were performed in triplicate and, if not indicated otherwise, harvested cells were analyzed by flow cytometry and plotted as the percentage of GFP-positive cells averaged across the two selected MIB1 KO clones for guide RNAs targeting either exon 1 (ex 1) or exon 2 (ex 2).
Three genes emerged as significantly enriched for disruptive gene-trap insertions in the infected population, indicating that their disruption protected cells from AdV infection and/or cytotoxicity and marking them as potential host factors necessary for AdV5 infection (Fig. 1B). Two of these genes were the well-known receptors of AdV5: CXADR encoding CAR and integrin ITGB5, validating the effectiveness of the technique in identifying specific determinants of AdV infection (5, 20, 21).
The third gene identified by the screen was Mindbomb E3 ubiquitin ligase 1 (MIB1) (Fig. 1B). MIB1 is a large E3 ubiquitin ligase with well-described roles in the Notch and Wnt signaling pathways during neural development (22, 23). However, there were no reported roles for MIB1 in the context of DNA virus infection. To validate MIB1 as a necessary host factor for AdV infection, we generated Hap1 cells deficient for MIB1 using CRISPR-Cas9 editing and two independent single-guide RNAs (sgRNAs) targeting exons 1 and 2 of MIB1, respectively (Fig. 1C and SI Appendix, Fig. S1A). To rule out the possibility of CRISPR off-target effects, we then reconstituted selected sequence- and Western blot-confirmed MIB1 knockout (KO) clones with wild-type (WT) MIB1. In addition, we generated Hap1 cells deficient for CAR using CRISPR-Cas9 editing to serve as positive controls for AdV5 infection inhibition. This panel of cells was then infected with two different recombinant AdV5-GFP reporter viruses. Replication-competent AdV5 (AdV5rep+), lacking only E4orf3, is capable of viral replication and spread following the initial round of infection at 24 to 30 hours postinfection (hpi). In contrast, the replication-incompetent AdV5 (AdV5rep−) cannot progress to later stages of infection (late mRNA transcription, genome replication, and virion progeny production) due to the absence of viral proteins E1A and E1B. While Hap1 WT cells supported virus infection (GFP expression), all MIB1 KO cells were refractory to GFP expression upon infection with both AdV5rep−- and AdV5rep+-GFP viruses (almost to the degree of inhibition observed for CAR KO cells) (Fig. 1 D–F). Reconstitution with WT MIB1 was sufficient to restore infection (Fig. 1 D–F). Inhibition of both AdV5rep− as well as AdV5rep+ in MIB1 KO cells served as our first indication that MIB1 must act at a step in the viral lifecycle preceding late stages of infection. Additional CRISPR KOs of MIB1 generated in A549 cells and primary STAT1−/− human fibroblasts, as well as partial KO in HeLa cells, also resulted in dramatic inhibition of AdV5 infection, further validating the importance of MIB1 to infection in diverse cellular contexts (SI Appendix, Fig. S1B). Together these results corroborated our findings from the screen supporting MIB1 as a host factor for AdV5.
MIB1 emerged from the screen as the sole additional host factor for AdV5 infection besides known viral receptors. Given MIB1’s well-characterized roles in regulating surface level expression of Notch and Wnt receptors by endocytosis (in the latter case coupled to degradation), we were curious whether the effects of MIB1 were mediated by indirect modulation of CAR expression. Detection of CAR in both whole cell lysates and at the cell surface, assessed by Western blot and flow cytometry, respectively, however, revealed comparable levels between WT, MIB1 KO, and WT MIB1-reconstituted Hap1 cell lines (Fig. 1C and SI Appendix, Fig. S2 A and B). CAR independence of the observed AdV5 inhibition was further confirmed by infection with alternate AdV serotypes that utilize distinct entry receptors. While AdV2 similarly utilizes CAR, AdV7 and AdV35 utilize desmoglein-2 (DSG-2) and CD46, respectively (24). Infections with AdV5rep−-GFP modified to express the fiber proteins of either AdV7 (AdV5-F7) or AdV35 (AdV5-F35), while readily able to infect CAR KO cells, were highly impaired in GFP expression in MIB1 KO cells (25, 26) (Fig. 1 G and H). MIB1 KO cells infected with WT AdV2, AdV7, and AdV35 displayed potent inhibition of vDNA replication across all viral serotypes (Fig. 1I). Based on these data, we concluded that MIB1 plays a role in AdV infection downstream of receptor engagement.
Lastly, infection of MIB1 KO Hap1 cells with a broad panel of both RNA and DNA viruses showed no clear dependence on MIB1 for infection. Therefore, among all viruses tested in Hap1 cells, MIB1 appears to be an AdV-specific host factor (SI Appendix, Fig. S3).
Adenovirus Infection Requires the Ubiquitination Activity of MIB1.
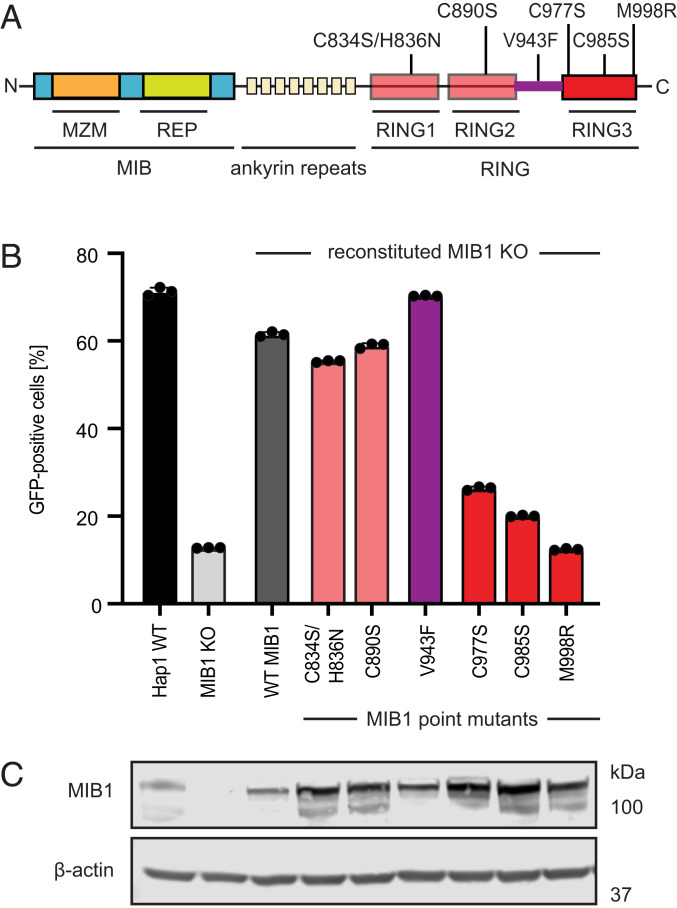
E3 ubiquitin ligases are the effectors of ubiquitination, a posttranslational modification (PTM) characterized by the attachment of ubiquitin to a target protein. Modification can result in rapid degradation of the protein by the proteasome or alter its localization or binding partners. MIB1 is a member of the RING family of E3 ligases. At the N terminus of MIB1 are two substrate-binding domains termed MZM (Mib-Herc2/ZZ Zinc Finger/Mib-Herc2) and REP (consisting of two Mib-repeat domains) that are cumulatively designated the “MIB” domain and which serve as the primary determinants of target specificity. These domains then connect via a series of ankyrin repeats to a C terminus comprised of three RING finger domains (Fig. 2A). These RING domains, the defining feature of these ligases, act by binding E2-conjugating enzymes and facilitating the direct transfer of ubiquitin from E2 onto specific target proteins. Reconstitution of Hap1 MIB1 KO clone 2-1 with domain mutants of MIB1 (lacking combinations of the MIB1 domain, the RING domain, etc.) demonstrated that only full-length MIB1 was capable of rescuing AdV infection (SI Appendix, Fig. S4 A and B). To more specifically determine whether MIB1’s effect on AdV was dependent upon its ubiquitination activity, we reconstituted MIB1 KO clone 2-1 with a set of previously characterized point mutants. These included mutations to RING domains 1 (C834S/H836N) and 2 (C890S), three distinct mutations to RING domain 3 that abrogate ubiquitination activity (C977S, C985S, and M998R), and a patient variant, V943F, which maps to the coiled-coil domain between RING domains 2 and 3. This variant has been shown to eliminate ubiquitination of Notch substrates but is also posited to disrupt MIB1 homodimerization (22, 27, 28). Consistent with previous literature reporting lack of function for RING domains 1 and 2, only those MIB1 constructs with mutations in RING domain 3 failed to restore AdV infection when added back to MIB1 KO cells (22, 29) (Fig. 2 B and C). Therefore, the ubiquitination activity of MIB1 is required for AdV infection in Hap1 cells.
Fig. 2.
The ubiquitination activity of MIB1 is required for AdV infection. (A) Schematic of the MIB1 protein with domains and point mutations annotated. (B) AdV5rep+-GFP infection (MOI = 200 p/cell; 24 hpi) of Hap1 WT cells, MIB1 KO clone 2-1 and KO 2-1 reconstituted with panel of point mutants (annotated in A). Experiments were performed in triplicate and harvested cells were analyzed by flow cytometry and plotted as the percentage of GFP-positive cells. (C) Western blot analysis of MIB1 expression levels in Hap1 WT, MIB1 KO clone 2-1, and KO 2-1 reconstituted with MIB1 constructs in B.
Of note, levels of MIB1 appear to be tightly regulated as reconstitution of WT and MIB1 KO cells with WT MIB1 decreased cell proliferation in the majority of clones as well as in the WT cells (SI Appendix, Fig. S4C). MIB1 KO clone 2-1 and its WT MIB1-reconstituted counterpart were chosen for functional and mechanistic follow-up due to comparable cell growth kinetics and efficiency of AdV rescue upon reconstitution.
MIB1 and the Proteasome Are Required for the Delivery of AdV5 DNA into the Nucleus upon Capsid Arrival at the Nuclear Envelope.
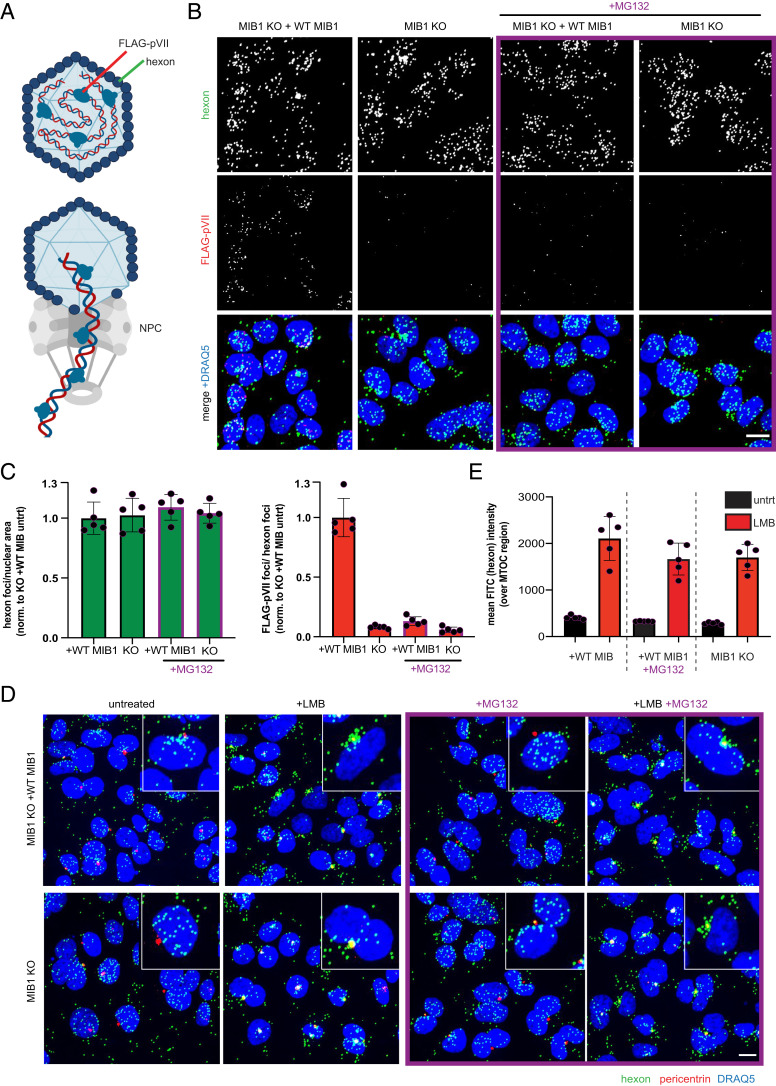
The MIB1 dependence of AdV5rep− which does not progress to late-stage infection allowed us to narrow MIB1’s role to the early window in the viral lifecycle. This early phase encompasses a series of entry steps where the virus traverses the cytosol from the plasma membrane to the nuclear pore, releases its genome, and begins early transcription. To examine the specific step at which MIB1 acts, we utilized a recombinant AdV5 (JR34) modified to express a FLAG-tagged pVII protein (30). The pVII protein is a component of the incoming capsid and arranged in tight complex to the vDNA. Within an intact capsid, the FLAG-tagged pVII is obscured from antibody binding. However, upon release of the vDNA from the capsid into the nucleus, FLAG-tagged pVII (still associated with this vDNA) can be detected by anti-FLAG antibodies and visualized by immunofluorescence (IF) microscopy (Fig. 3A). Detection of pVII has been widely employed as a proxy for genome release (31, 32). MIB1 KO clone 2-1 and clone 2-1 reconstituted with WT MIB1 were infected with JR34 and stained for both capsid (anti-hexon) and released pVII-vDNA complex (anti-FLAG). Strikingly, while MIB1 KO and WT MIB1-reconstituted cells displayed a similar quantity of viral particles in the cytoplasm, the subsequent release of vDNA (quantified as FLAG-pVII foci) into the nucleus by 1.25 hpi was severely compromised in KO relative to reconstituted cells (Fig. 3 B and C).
Fig. 3.
MIB1 and the proteasome are required for AdV DNA delivery upon capsid arrival at the nuclear envelope. (A) Schematic of recombinant AdV5 (JR34) expressing FLAG-tagged pVII protein which is inaccessible to antibody binding within capsid but becomes accessible upon DNA release at the NPC. (B) Representative images of MIB1 KO clone 2-1 and its WT MIB1-reconstituted counterpart infected with JR34 (MOI = 2,000 p/cell; 1.25 hpi) in the presence or absence of MG132 (30 µM) stained for capsid (anti-hexon) and pVII (anti-FLAG) and merged with DRAQ5 nuclear stain. (Scale bar: 10 µm, maximum projected.) (C) Quantification of hexon foci and FLAG foci seen in B normalized to nuclear area and hexon foci, respectively, per field of view (FOV) and averaged across five FOV. (D) Representative images showing distribution of AdV capsids (green: anti-hexon) relative to centrosome (red: anti-pericentrin) merged with DRAQ5 nuclear stain (blue) in MIB1 KO clone 2-1 and its WT MIB1-reconstituted counterpart infected with JR34 (as above) in the presence or absence of LMB (20 nM) and/or MG132 (30 µM). (Scale bar: 10 µm, maximum projected.) (E) Quantification of distribution of AdV capsids relative to MTOC depicted in D as the mean FITC intensity (anti-hexon) over the MTOC region (anti-pericentrin) averaged across five FOV.
Given that MIB1’s ubiquitination activity is required for infection and that one well-characterized consequence of ubiquitination is to target the modified protein to the proteasome for degradation, we were curious whether the proteasome might also play a role in AdV5 genome delivery. Brief pretreatment with the proteasome inhibitor MG132 before and throughout JR34 infection significantly reduced (five- to sixfold) the appearance of FLAG-pVII foci in WT MIB1-reconstituted cells by 1.25 hpi (Fig. 3 B and C). Thus, blocking proteasomal degradation in WT MIB1-reconstituted cells phenocopied MIB1 KO at this stage of infection, suggesting in both cases that aberrant stabilization of some target protein may impede virus infection. Inhibition of vDNA release by MG132, however, reflects a delay rather than the sustained block imposed by lack of MIB1 as FLAG-pVII signal increased over time in MG132-treated cells, although still below the level observed in untreated WT MIB1-reconstituted cells (SI Appendix, Fig. S5 B and C).
Loss of vDNA deposition in MIB1 KO and MG132-treated WT MIB1-reconstituted cells suggested there might be a defect along the trajectory of viral capsids from plasma membrane to nucleus. To resolve first whether MIB1 and the proteasome act upstream or downstream of centrosome arrival, we examined the spatial distribution of JR34 virus particles in MIB1 KO and MG132-treated WT MIB1-reconstituted cells following treatment with leptomycin B (LMB). LMB, an inhibitor of CRM1-mediated export, has been shown to boost virion association with MT motors leading to dramatic aggregation and arrest of virions directly at the MTOC (11). Indeed, in WT MIB1-reconstituted KO cells treated with LMB prior to and throughout JR34 infection we observed robust capsid accumulation at the MTOC upon staining for AdV5 hexon and pericentrin (a defining structural component of the MTOC) (Fig. 3D). Treatment with LMB also blocked vDNA release into the nucleus as quantified by FLAG-pVII staining (SI Appendix, Fig. S5A). Aggregation of capsids at the MTOC was similarly observed for both MIB1 KO cells and MG132-treated WT MIB1-reconstituted cells only upon treatment with LMB, indicating that MIB1 and the proteasome act downstream of arrival at and departure from the centrosome (Fig. 3 D and E). Consistent with this, in untreated MIB1 KO and MG132-treated WT MIB1-reconstituted cells, AdV capsids could be readily observed in the vicinity and along the surface of the nucleus (Fig. 3D). Notably, in MIB1 KO and MG132-treated WT MIB1-reconstituted cells this close association with the nucleus persisted even at 4 hpi, a timepoint when WT MIB1-reconstituted cells show cytoplasmic aggregates of hexon stain indicative of DNA deposition and retrafficking of the emptied capsids away from the nuclear pore (12) (SI Appendix, Fig. S5B). We therefore concluded that both MIB1 and the proteasome (either independently or cooperatively) mediate some critical step upon arrival at the nuclear envelope required for capsid disassembly and release of vDNA into the nucleus.
MIB1 Localizes to Centriolar Satellites in Hap1 Cells.
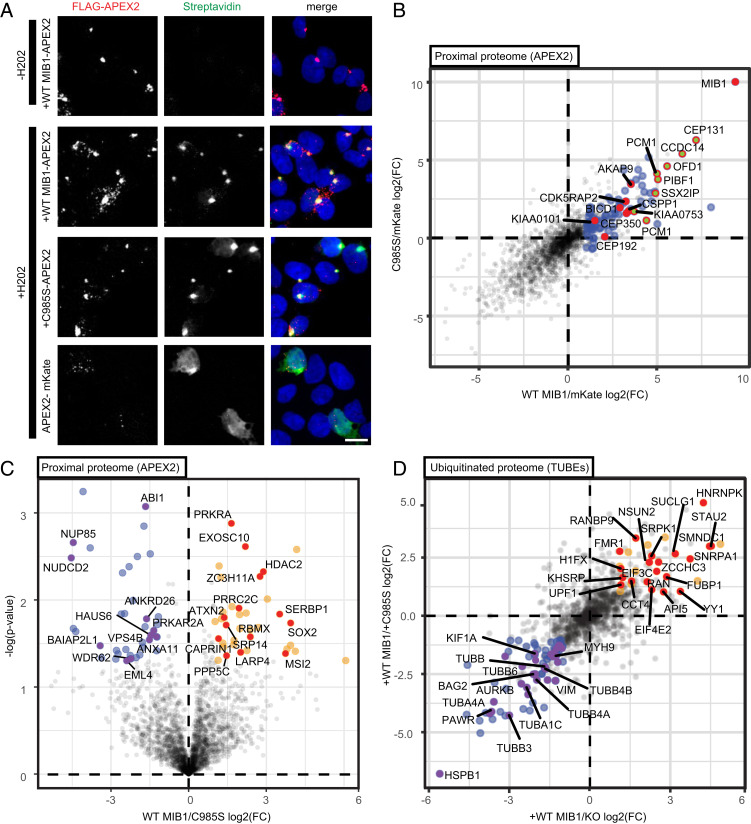
Our microscopy studies with the JR34 virus had placed the block on infection in MIB1 KO cells after arrival at the nuclear envelope but prior to successful vDNA delivery to the nucleus. We were interested in defining the corresponding spatial/cellular context of MIB1 during early AdV infection. To do so we employed APEX2 proximity labeling to biochemically tag proteins proximal (∼10-nm radius) to MIB1 (33). We reasoned that proteins captured in the immediate vicinity of MIB1 would not only provide insight into MIB1’s localization but might also encompass MIB1 ubiquitination targets. MIB1 KO clone 2-1 cells were stably reconstituted with WT MIB1 or the RING3 ubiquitination-defective mutant C985S MIB1 tagged with FLAG-APEX2 and assayed for their capacity to restore AdV infection. Infection was reduced in KO cells reconstituted with APEX2-tagged MIB1 relative to untagged MIB1, possibly due to low levels of functional APEX2-tagged protein (SI Appendix, Fig. S6A). To boost WT MIB1 expression/activity, we introduced the V943F mutation into these constructs which we had found in our MIB1 mutagenesis studies to enhance AdV infection over WT MIB1 (Fig. 2B). Indeed, V943F mutants restored the capacity of APEX2-tagged WT MIB1 to rescue virus infection (SI Appendix, Fig. S6A). All subsequent APEX2 experiments were conducted in the background of the V943F mutation with APEX2 fused to the C terminus of MIB1. In parallel, we generated MIB1 KO cells expressing APEX2 fused to the fluorescent protein mKate. As a ubiquitously localized cytoplasmic protein, the mKate construct served as a generic baseline control over which enrichment for specific proximal partners of MIB1 could be identified.
We first established the expression and labeling specificity of our three different constructs by IF in uninfected cells. MIB1 KO cells reconstituted with WT MIB1-APEX2, C985S MIB1-APEX2, and APEX2-mKate underwent APEX2 labeling. Staining for the introduced FLAG-APEX2-tagged protein revealed that both WT MIB1-APEX2 and C985S MIB1-APEX2 localized predominantly to a single point within each cell. This was corroborated by strong (H2O2 dependent) streptavidin stain for biotinylated proteins in the region surrounding the APEX2-tagged protein (Fig. 4A). The observation of this singular locus per cell aligned with reported roles for MIB1 as a key component of centriolar satellites, substructures in the vicinity of the centrosome involved in regulation of centriole homeostasis and ciliogenesis (34, 35). We confirmed the dominant localization of FLAG-APEX2-tagged WT and C985S MIB1 to the centrosome in AdV-infected cells by costaining for FLAG-tagged MIB1 constructs and pericentrin (SI Appendix, Fig. S6B). By contrast, APEX2-tagged mKate and biotin label was found throughout the cell with no discrete localization (Fig. 4A).
Fig. 4.
Proteomic approaches confirm MIB1 localization to centriolar satellites in AdV-infected Hap1 cells and reveal MIB1 ubiquitination-dependent association with RNPs. (A) Uninfected MIB1 KO clone 2-1 reconstituted with FLAG-APEX2–tagged WT MIB1, C985S MIB1, or mKate were incubated with biotin-phenol for 1 h and pulse labeled with H2O2 followed by fixation and staining with AF488-coupled streptavidin for biotinylated proteins (green), the indicated FLAG-APEX2–tagged construct (red: anti-FLAG), and DRAQ5 nuclear stain (blue). (Scale bar: 10 µm, maximum projected.) (B and C) For LC-MS/MS, MIB1 KO clone 2-1 reconstituted with FLAG-APEX2 fused to either mKate, WT MIB1, or C985S MIB1 were incubated with biotin-phenol and infected with AdV5rep+-GFP (MOI = 200p/cell), pulse-labeled with H2O2 at 70 min pi, and immediately collected for lysate preparation and streptavidin pulldown to isolate biotinylated proteins (n = 3). (B) Proximal protein enrichment (FC) over APEX2-mKate is plotted as log2 of the ratio of protein abundance for WT MIB1/mKate (x axis) and C985S MIB1/mKate (y axis). GO terms analysis of proteins twofold or greater (P value <0.05) in the WT MIB1/mKate comparison (blue) shows enrichment of centriolar satellite proteins (green, red rim), as a subset of centrosome proteins (red). (C) Ubiquitination-dependent proximal proteome determined by direct comparison of APEX2-tagged WT MIB1 and C985S MIB1 (unique peptides ≥1) plotted as log2 of the ratio of protein abundance for WT MIB1/C985S MIB1 (x axis) and −log(P value) (y axis). GO terms analysis of those proteins enriched by twofold or greater (orange) or depleted by twofold or greater (blue) in proximity to WT MIB1 relative to C985S MIB1 (P value <0.05) reveals enrichment of RNA-binding proteins (red with gene names labeled) and depletion of cytoskeletal proteins (purple with gene names labeled). (D) Identification of differentially ubiquitinated proteins between MIB1 KO clone 2-1 and KO reconstituted with either WT MIB1 or C985S MIB1 infected with AdV5rep+-GFP (MOI = 200 p/cell) (n = 3). At 1 hpi, cells were harvested and endogenous ubiquitinated proteins were isolated using TUBE-Dynabeads followed by LC-MS/MS and plotted as log2 of the ratio of protein abundance for WT MIB1/MIB1 KO (x axis) and WT MIB1/C985S MIB1 (y axis). GO terms analysis of those proteins enriched twofold or greater in both WT MIB1/KO and WT MIB1/C985S (P value <0.05 in either) (orange) or depleted by twofold or greater in WT MIB1/KO and WT MIB1/C985S (P value <0.05 in either) (blue) reflect RNA binding (red with all gene names labeled) or cytoskeleton (purple with select proteins significant to both WT MIB1/KO and WT MIB1/C985S condition labeled by gene name), respectively.
To obtain proximal proteomes for each of these constructs in AdV-infected cells, MIB1 KO cells reconstituted with WT MIB1-APEX2, C985S MIB1-APEX2, and APEX2-mKate were infected with 200 virus particles/cell (p/cell) of AdV5rep+-GFP followed by biotin labeling at 70 min postinfection (mpi), streptavidin isolation of biotinylated proteins, and liquid chromatography-tandem mass spectrometry (LC-MS/MS). Corroborating the localization observed by IF, proteins in proximity to both WT and C985S MIB1 in infected cells were significantly enriched (twofold or greater) in centrosome (adjusted [adj.] P value = 6.62 × 10−8 and 1.1 × 10−6 for WT and C985S MIB1, respectively) and, furthermore, centriolar satellite components (e.g., CCDC14, CEP131, KIAA0753, OFD1, PCM1, PIBF1, and SSX2IP) (adj. P value = 9.57 × 10−9 and 2.29 × 10−6 for WT and C985S MIB1, respectively) (Fig. 4B). Interestingly, the Golgi subcompartment and coated vesicles were additional functional categories enriched proximal to both WT and C985S MIB1 relative to the mKate control (SI Appendix, Table S1). No AdV5 virion capsid proteins were detected in any of the samples.
The MIB1 Proximal Proteome Is Ubiquitination-Dependent and Enriched in RNP-Related Factors.
We next compared the proximal proteomes of WT MIB1 against the mutant C985S MIB1 in AdV-infected cells directly (Fig. 4C). We had hypothesized that in WT MIB1-expressing cells, rapid release kinetics and/or subsequent degradation of targets by the proteasome might result in a depletion of direct ubiquitination targets in the vicinity of WT MIB1. Direct targets might then be enriched in proximity to the C985S mutant that, while unable to carry out ubiquitination, should retain the capacity to bind substrates. However, we also considered the possibility that MIB1’s protein partners and localization may in fact depend upon its ability to carry out ubiquitination of a specific target protein(s).
Despite an overarching shared association with centriolar satellites and centrosomes when compared against the mKate control, comparison of WT MIB1 versus the mutant C985S MIB1 revealed key differences in their proximal proteomes (Fig. 4C). Among those proteins enriched in proximity to WT relative to C985S MIB1 were a number of well-characterized cytoplasmic RNA granule components. By gene ontology (GO) terms analysis, we identified significant enrichment of RNA-binding (adj. P value = 0.0003) and RNA transport genes (adj. P value = 0.0375). CAPRIN1, ATXN2, and MSI2 are components of stress granules, cytoplasmic aggregates of stalled translational machinery that form in response to cellular stresses (36). UBQLN2, enriched in proximity to WT MIB1, although not an RNA-binding protein, has been shown to associate with stress granules and act as a bridge between stress granules and the ubiquitin–proteasome pathway (37). EDC4 (classified among the significantly enriched category “cytoplasmic ribonucleoprotein granule”) is a defining factor within constitutively present RNA processing bodies, or P-bodies, sites of RNA storage and degradation (36).
At the same time, proteins enriched in proximity to the mutant C985S MIB1 reflected cytoskeleton components (adj. P value = 0.0201) by GO term analysis (Fig. 4C). Of note, these proteins included NUP85, more commonly referred to as pericentrin, which we had used as a marker for the centrosome in our IF experiments. We concluded that WT and C985S MIB1 possess distinct differences in their proximal proteomes—with WT MIB1 associating preferentially with RNPs while C985S MIB1 remains more tightly associated with certain cytoskeletal and centrosome components.
The MIB1 Ubiquitinated Proteome Is Enriched in RNP Factors and Depleted in Cytoskeletal Factors.
While proximity-labeling proteomics had characterized MIB1’s cellular context during AdV infection, we were also interested in proteins that intersected more directly with MIB1 in its role as an E3 ubiquitin ligase. To characterize possible ubiquitination targets of MIB1 during virus infection, we turned to a complementary proteomics approach. Tandem ubiquitin binding entities (TUBEs) can bind to diverse ubiquitin linkage types (K48, K63, mono, poly, etc.) and have been widely employed to isolate endogenous ubiquitinated proteins from total cell lysates (38). Using this approach, we hoped to identify those proteins enriched for ubiquitination in WT MIB1-expressing cells relative to both cells lacking MIB1 or cells reconstituted with the mutant C985S MIB1 in which all MIB1-mediated ubiquitination events should be absent. MIB1 KO cells, and KO cells reconstituted with either WT MIB1 or C985S MIB1 were infected with AdV5rep+-GFP (200 p/cell) and collected at 1 hpi. Lysates were incubated with TUBE Dynabeads, and the isolated proteins were submitted for LC-MS/MS.
We identified 9 proteins that were significantly (P < 0.05) enriched by greater than twofold in both the comparison between WT MIB1 versus KO cells and between WT MIB1 versus C985S MIB1 following TUBE incubation. These were considered strong potential candidates of direct ubiquitination by MIB1. Several of these MIB1 target candidates, including STAU2 and DRG1, have been reported to localize to the spindle or centrosome and possess MT-binding domains (39, 40). This increased confidence in the capacity of this ubiquitination-based but proximity-blind MS approach to detect plausible targets of centrosome- and satellite-localized MIB1. Reducing our stringency to proteins significantly enriched by more than twofold in a single group comparison (WT MIB1 over MIB1 KO cells or WT MIB1 over C985S MIB1) but enriched by greater than twofold in both yielded an additional list of 22 proteins more strongly observed upon TUBE isolation in the presence of WT MIB1 (SI Appendix, Table S2). GO analysis of this expanded protein list (31 proteins: orange) revealed enrichment of “RNA binding” (8/31 proteins, adj. P value = 0.0028), “RNA transport” (6/31 proteins, adj. P value = 0.001), and “ribonucleoprotein complex” (10/31 proteins, adj. P value = 4.12 × 10−5) categories (Fig. 4D). The emergence of these functional categories corroborated the observation of RNA-related proteins in proximity to WT MIB1 from our proximity-labeling dataset.
We also noted, in parallel, 73 proteins that were depleted upon TUBE incubation in cells expressing WT MIB1 over both MIB1 KO and C985S-reconstituted cells (fold change [FC] < −2 in both, P < 0.05 in either) (SI Appendix, Table S2). GO terms analysis of this set of proteins revealed enrichment for functional categories corresponding to either “cytoskeleton” (29/73 proteins, adj. P value = 2.1 × 10−7) or “proteasome-mediated ubiquitin-dependent protein catabolism” (16/73 proteins, adj. P value = 1.1 × 10−8) (Fig. 4D). This also corroborated our proximity-labeling data in which the C985S mutant displayed enhanced proximity to a subset of cytoskeletal proteins. Due to the mild, nondenaturing conditions of TUBE incubation, we hypothesize that these proteins might reflect broader ubiquitin-dependent changes in the binding partners of TUBE-isolated MIB1-ubiquitinated proteins, specifically, binding partners whose association might be abrogated following ubiquitination of a given target by WT MIB1. Again no viral capsid proteins were detected.
Discussion
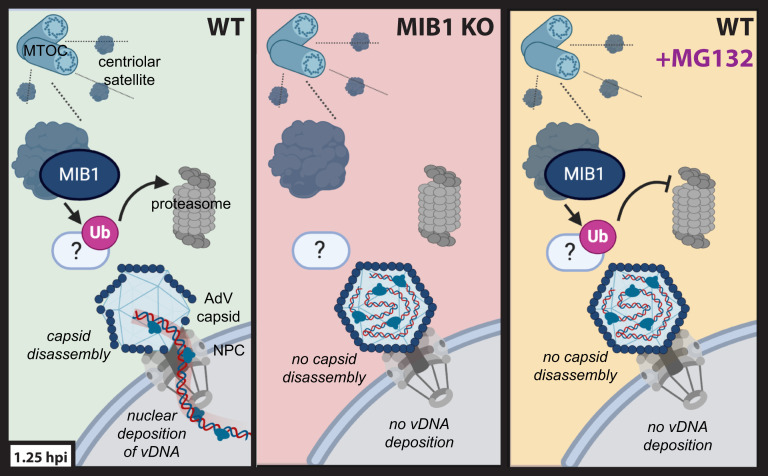
In this study, we carried out a genome-wide gene-trap screen of AdV infection and identified an AdV host factor, the E3 ubiquitin ligase MIB1. Using fluorescence microscopy, we determined that MIB1 is required for the final step in AdV entry: capsid disassembly and vDNA release at the nuclear pore. In the absence of MIB1, capsids successfully reach the nucleus but fail to deposit their vDNA within (Fig. 5). As an E3 ubiquitin ligase, MIB1 acts by ubiquitinating specific substrates resulting in either degradation by the proteasome or alteration of signaling/localization (34, 41). Using a series of point mutants we determined that virus infection depends upon MIB1’s ubiquitination activity. Our subsequent observation that the proteasome is similarly required for vDNA release upon capsid arrival at the nucleus, suggests that ubiquitination by MIB1 mediates AdV genome delivery through proteasomal degradation of a negative regulator of AdV infection (Fig. 5). In this respect, AdVs now emerge as nucleus-targeting viruses, similar to influenza virus and herpes simplex virus (HSV), that require ubiquitin and/or the proteasome for successful vDNA delivery to the nucleus (42, 43).
Fig. 5.
Schematic model of MIB1’s role in AdV genome deposition following capsid arrival at the nuclear pore. In WT cells (Left), MIB1, an E3 ubiquitin ligase, and centriolar satellite component, facilitates successful capsid disassembly and vDNA deposition into the nucleus through the ubiquitination and proteasomal degradation of an as-yet-unidentified target protein (white oval). In MIB1 KO cells (Middle), the MIB1 target is no longer ubiquitinated and degraded resulting in a block on capsid disassembly and genome release. Similarly, upon treatment of WT cells with MG132 (Right), the ubiquitinated MIB1 target, no longer able to be degraded by the proteasome, persists and inhibits capsid disassembly and genome release (graphic created with BioRender.com).
During preparation of this manuscript, work now published by Bauer et al. similarly identified and characterized MIB1 as a host factor for AdV infection required for AdV genome delivery (44). Using electron microscopy, they show that in MIB1 KO cells, AdV capsids arrive at the NPC but remain paralyzed at the pore, intact and unable to release the vDNA into the nucleus. Their observation of transient interactions between MIB1 and NPC-docked virions by live-cell imaging suggests that the critical MIB1 ubiquitination event(s) occurs at this site. Having validated MIB1 as a component of centriolar satellites by proximity-labeling proteomics, we posit that MIB1 interactions near the nuclear pore could occur as part of these centrosomal substructures dynamically sampling the region between centrosome and nuclear membrane (Fig. 5).
The specific target(s) of MIB1 ubiquitination relevant to AdV infection has remained elusive. We had determined that different AdV serotypes were dependent on MIB1 for virus delivery and replication. Sequence diversity of capsid and capsid-associated proteins between these serotypes as well as the lack of APEX2 proximity labeling or TUBE isolation of capsid proteins lead us to favor a cellular protein as the target of MIB1 ubiquitination. However, the possibility that MIB1 acts through a conserved motif across serotypes or through combinatorial action on multiple targets cannot be excluded. By identifying those proteins enriched upon TUBE incubation in WT MIB1-expressing cells, we have generated a list of MIB1 target candidates that may next be probed individually for their effect on AdV infectivity and DNA release.
In the meantime, our unbiased proteomic approaches have uncovered an intriguing balance between functional classes of proteins—RNPs and cytoskeleton—that appear regulated by MIB1 and may therefore be altered in MIB1 KO cells with consequences for AdV DNA delivery. Comparing the proximal proteomes of WT MIB1 relative to the C985S mutant revealed that while a broad association with centriolar satellites is shared, WT MIB1 is enriched in proximity to a number of well-characterized RNA-binding proteins while depleted for certain cytoskeletal components. Isolation of ubiquitinated proteins from MIB1 KO cells and KO cells reconstituted with WT MIB1 or C985S MIB1 recapitulated even more strikingly this MIB1 ubiquitination-dependent enrichment of RNP-related proteins and commensurate depletion of cytoskeletal factors.
An existing role for MIB1 in the regulation of RNPs has emerged very recently in the context of spermatogenesis in Caenorhabditis elegans (45). C. elegans spermatids lack a nuclear envelope and instead delineate nuclear and cytoplasmic compartments via a dense halo of RNA and RNA-binding proteins that encompass chromatin and centrosome. In C. elegans spermatids harboring mutations in MIB1, this perinuclear RNA halo is absent and large tubules are observed in its place (45, 46). An overabundance of tubulin in these mutants is also observed (47). Our findings imply that this RNP-regulatory role for MIB1 may extend to mammalian somatic cells. Taken together, these observations not only suggest a conserved role for MIB1 in regulating a balance between RNPs and filamentous structures but furthermore, specifically at the boundary between cytoplasm and nucleus. This is precisely the interface at which MIB1 appears to act during AdV infection.
We hypothesize that MIB1’s role in AdV genome delivery may intersect with its emerging role in perinuclear RNP regulation. RNPs have, in fact, already been implicated in the relevant step of AdV capsid docking and DNA delivery at the NPC. Trotman et al. observed that rabbit reticulocyte lysates inhibited capsid binding to nuclear envelopes in vitro due to the presence of an inhibitory RNP whose components remain unknown (13). Might any of the RNP granule-associated proteins identified through our proteomic approaches overlap with components of this mystery RNP? We posit that MIB1 might mediate a reorganization or local enrichment of this RNP at the nuclear envelope to assist in the forcible displacement of capsids from the nuclear pore (perhaps by competing for direct binding with or by simultaneously displacing Nup214). This would be compatible with Trotman et al.’s observations in which direct addition of this RNP from a spatially disrupted lysate would preclude capsid binding in the first place.
In C. elegans, the mib-1 mutant phenotype is phenocopied by mutations in a number of endogenous RNA interference (RNAi) factors, including homologs of defining components of mammalian RNPs known as P-bodies (48). Mammalian P-bodies are sites of mRNA sequestration and degradation and have even been shown to migrate along MTs and associate with the centrosome (49). As such, P-bodies could be prime candidates for interaction with MIB1 as a component of centriolar satellites. Indeed, recent work by others to define the proximal proteome of centriolar satellites using a number of different baits (including MIB1), as well as our own proximity-labeling data, have repeatedly uncovered P-body components in proximity to WT MIB1 (50, 51).
Recent studies monitoring the dynamics of P-bodies have observed a phenomenon in which P-bodies that traffic to the surface of the nuclear membrane are rapidly redirected away again (49). These dynamics imply a MT switch triggered upon arrival at the surface of the nucleus reminiscent of that proposed by Strunze et al. for AdV capsids (12). In their model, the simultaneous coupling of AdV capsids to kinesin-1 on periphery-directed MTs and to import factors transiting through the pore generates a shearing force required for capsid disruption and DNA release. We propose that AdV capsids may achieve this MT-mediated disruption by intersecting with a retrafficking pathway of P-bodies regulated by MIB1. Simultaneous live-cell particle tracking of fluorescently labeled P-bodies and AdV capsids will help clarify if and how the dynamics of these species are intertwined and altered in the absence of MIB1.
The work described here encompassing the identification and characterization of MIB1 as a host factor of AdV infection has paved the way for these types of mechanistic studies which will continue to yield new insights into the fascinating complexity of viral genome delivery.
Materials and Methods
Antibodies and Chemicals.
Primary antibodies used for Western blot include rabbit anti-MIB1 (Abcam: 124929 1:2,000), rabbit anti-CAR (Santa Cruz: sc-15405; 1:1,000), and mouse anti-FLAG M2 (Sigma: F3165; 1:2,000). Primary antibodies for IF experiments include mouse anti-hexon (AdV5) (Development Studies Hybridoma: TC21-9C12.C9; 1:200), rabbit anti-FLAG (Cell Signaling Technologies: 14793S; 1:1,000), and rabbit anti-pericentrin (Abcam: ab4448; 1:1,000). Infections with influenza A virus and Zika virus (ZIKV), and cell surface expression of CAR, were quantified by flow cytometry using mouse anti-nucleoprotein (NP) (Millipore: MAB8251; 1:1,000), mouse anti-flavivirus group antigen 4G2 (Millipore: MAB10216; 1:500), and PE-conjugated rabbit anti-CAR (Sino Biological: 10799-R271-P), respectively. Secondary antibodies were Alexa Fluor 488-, Alexa Fluor 594-conjugated anti-mouse and anti-rabbit IgG (Thermo Fisher Scientific), Alexa Fluor 680-conjugated to streptavidin (Thermo Fisher Scientific), goat anti-rabbit and anti-mouse IgG conjugated to horseradish peroxidase (HRP), and mouse anti–β-actin conjugated to HRP (Sigma: A3854; 1:10,000). DRAQ5 (Thermo Fisher Scientific: 62251; 1:1,000) was used for nuclear stain. Drugs were used at concentrations indicated in figure legends and include MG132 (Sigma: M7449), leptomycin B (LMB) (Millipore: L2913), and the deubiquitinating enzyme (DUB)-inhibitor PR-619 (LifeSensors: SI9619).
Cell Culture.
Hap1 cells (obtained from T.R.B.) were cultured in Iscove’s modified Dulbecco’s medium (IMDM, Gibco) supplemented to contain 10% fetal bovine serum (FBS) and 1% nonessential amino acids (NEAAs) and utilized for the haploid gene-trap screen as well as for followup validation and mechanistic studies into MIB1. A549, HeLa, and human STAT1−/− fibroblasts (immortalized by SV40 large T antigen) (for evaluation of MIB1 phenotype in different cellular backgrounds), HEK293 (production of replication-incompetent AdV), and derivative Lenti-X 293T cells (production of lentiviruses) were maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented to contain 10% FBS and 1% NEAAs.
Haploid Genetic Screen.
Gene-trap retrovirus was produced in 293T cells as previously described (18). To generate the mutagenized library, 1 × 108 Hap1 cells were infected for 3 consecutive days in the presence of protamine sulfate (8 mg/mL) and passaged for 5 d prior to freezing. For the screen itself, a library of 1 × 108 mutagenized cells was infected with AdV5rep+-GFP at a multiplicity of infection (MOI) of 1,000 virus particles/cell. The resistant colonies were expanded for 15 d and ∼1 × 107 cells were used for gDNA isolation.
Sequence Analysis of Gene-Trap Insertion Sites.
Gene-trap insertion sites in AdV5-positive cells were identified by sequencing the gDNA flanking the gene-trap insertion site as previously described (52). Genomic DNA was subjected to a linear amplification PCR using a biotinylated primer followed by single-stranded DNA linker ligation, PCR, and subsequent sequencing using the HiSEq. 2000 platform (Illumina). The acquired reads were mapped to the human genome (hg19) using the Bowtie alignment tool allowing for a single mismatch, and insertion sites located in Refseq genes were identified. For each Refseq gene, the longest transcript was chosen and overlapping gene regions were disregarded, since orientation bias cannot be unambiguously assigned in these areas. The number of disruptive mutations (i.e., in sense with transcriptional orientation) and nondisruptive (i.e., in antisense with transcriptional orientation) mutations per individual gene was counted and genes significantly enriched for disruptive mutations following AdV5 infection were determined using a binomial test and corrected for multiple testing using a Benjamini–Hochberg false discovery rate (FDR). Significantly enriched genes with over 40 integrations in the AdV dataset (National Center for Biotechnology Information [NCBI] Single Read Archive [SRA] accession no: SRP269890 dataset SRX8663593) were compared to a reference dataset of WT Hap1 cells (NCBI SRA accession no: SRP058962 dataset SRX1045464) that was analyzed similarly. Genes already significantly enriched in the WT dataset were subtracted from the identified AdV host factor genes.
Plasmid Generation.
The MIB1 sequence was PCR amplified off of p3HA-hMIB1 plasmid (gift from Vanessa Redecke, St. Jude Children's Research Hospital, Memphis, TN [Addgene plasmid #33317; http://www.addgene.org/33317; RRID: Addgene_33317]) (53) and subcloned into the pDONOR Gateway vector. The MIB1 open reading frame (ORF) was then transferred via Gateway cloning into the lentiviral pLX304 backbone and subsequently used for lentivirus production. MIB1 point mutants were generated using site-directed mutagenesis of the WT MIB1 construct with Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific). The plasmid was then digested with DpnI to remove template plasmid input and PCR purified using the Qiaquick PCR Purification Kit (Qiagen). Domain mutants were generated via amplification of WT MIB1 sequence with primer-mediated incorporation of a 3×FLAG tag sequence at the 5′ end to facilitate detection by Western blot.
Lentivirus Production and Transduction.
Lentivirus stocks were generated in Lenti-X 293T cells (Takara Bio Inc.: 632180) by cotransfection of plasmids expressing 1) the ORF or sgRNA of interest, 2) HIV gag-pol, and 3) the vesicular stomatitis virus glycoprotein (VSV-G) in a ratio of 0.55:0.35:0.1. For blasticidin-selectable lentiviral vectors, we used the pLX304 lentiviral backbone in which the V5 tag was removed, and for puromycin-selectable lentiviral vectors, we used the lentiCRISPRv2 lentiviral backbone (gift from Feng Zheng, Broad Institute of MIT and Harvard, Cambridge, MA). Cells were transfected using Lipofectamine 2000 (Invitrogen) and media were replaced with DMEM containing 3% FBS at 6 h posttransfection. Supernatants were collected at 24 and 48 h, clarified by centrifugation at 2,850 × g, filtered using 0.45-μm syringe filters, and stored at −80 °C. For lentiviral transduction, cells were seeded into six-well plates at a density of 4 × 105 cells/well and transduced with lentiviral pseudoparticles by spinoculation at 930 × g for 60 min at 37 °C in medium containing 3% FBS, 20 mM Hepes, and 4 µg/mL polybrene. Transduced cells were selected at 24 h posttransduction with either 2.5 µg/mL blasticidin or 1 µg/mL puromycin.
Generation and Validation of CRISPR KO Clones.
sgRNA for CRISPR editing targeting MIB1 (sgRNA #1: 5′-CCGGAATAACCGGGTGATGG-3′ and sgRNA #2: 5′-CACTTCCCGGTGTAGTAATT-3′ targeting exon 1 and exon 2, respectively) were designed using http://crispor.tefor.net/ (54) and cloned into pX330 or lentiCRISPRv2 plasmid for subsequent transfection. MIB1 CRISPR KOs in Hap1 cells were generated by transfection with the relevant guides in pX330 and cultured for 4 d prior to single-cell seeding into 96-well plates. MIB1 KOs in all other cell lines (in HeLa, A549, STAT1−/− cells) were generated by transducing with lentiviruses generated from lentiCRISPRv2 containing the relevant guides and selected for 3 d with puromycin (1 µg/mL). Transduced cells were seeded into a 96-well plate at a dilution of 0.7 cells/well to obtain single-cell clonal populations. Clones were expanded and screened for protein knockout by Western blot analysis. Genomic DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit (QIAGEN: 69504) and used as a template for amplification of an ∼800- to 1,000-bp region flanking the PAM site. Discrete bands were gel extracted, cloned into the pCR4-TOPO vector using the TOPO TA Cloning Kit (Invitrogen: 45-0030), and Sanger sequenced. Relative proliferation of WT and verified KO clones was measured using the Cell Titer-Glo Luminescent Cell Viability Assay (Promega: G7570) according to manufacturer’s instructions.
Virus Production and Infections.
Replication-incompetent AdV5rep−-GFP (alternatively Ad5CiG for CMV-driven CAT-IRES-GFP reporter), WT AdV2, AdV7, AdV35, Ad5CiG-F7 (AdV7 fiber), and Ad5CiG-F35 (AdV35 fiber) viruses were provided by E.F.-P. and prepared as previously described (25, 55). Replication-competent AdV5 (AdV5rep+-GFP in which E4-ORF3 ORF has been replaced by EGFP) was generously provided by Patrick Hearing, Stony Brook University, Stony Brook, NY. The protein VII-FLAG-tagged JR34 adenovirus (E1/E3 deleted) was a generous gift from Robin J. Parks, Ottawa Health Research Institute, Ottawa, Ontario, Canada (30).
The generation of viral stocks for additional viruses tested has been previously described: hPIV3-GFP (56) (based on strain JS), YFV-Venus (57) (derived from YF17D-5′C25Venus2AUbi), VEEV-GFP (58) (derived from pTC83-GFP infectious clone), VacV-GFP (59) (based on strain: Western Reserve), HSV-1 US11-GFP (60) (based on strain: Patton), VSV-GFP (61) (based on strain: Indiana), LCMV-GFP (62) (based on strain: Armstrong), and influenza A/WSN/33 virus (generously provided by Peter Palese, Mount Sinai School of Medicine, New York, NY). ZIKV (PRVABC59 obtained from the Centers for Disease Control and Prevention [CDC], Ft. Collins, CO) was amplified in Hap1 cells and titrated by plaque assay on Huh-7.5 cells. Lenti-GFP virus was generated in Lenti-X 293T cells as described. Coxsackievirus CVB-3-GFP (63) (derived from infectious clone pMKS1-GFP) was amplified in HeLa cells and titrated by TCID50 (median tissue culture infectious dose 50).
All GFP-reporter virus infections and quantification were performed using the doses and timepoints reported in the figure legends for quantification by flow cytometry. For the infections, cells were seeded at a density of 2.5 × 104 to 5 × 104 cells/well in 24-well plates. The next day, cells were washed once with Opti-MEM (Gibco) and adsorbed with 200 µL of virus inoculum prepared in Opti-MEM at 37 °C. After 1.5 h, inoculum was removed, cells were washed with Opti-MEM, fresh medium was added, and cells were incubated at 37 °C. At the final timepoint dependent on viral replication kinetics, cells were lifted with Accumax cell dissociation medium (eBioscience: 00-4666-56) and fixed with 4% paraformaldehyde (PFA). Cells were pelleted at 930 × g for 5 min at 4 °C, resuspended in cold phosphate-buffered saline (PBS) containing 3% FBS and stored at 4 °C until analysis using a LSRII flow cytometer (BD Biosciences). FlowJo software (Treestar) was used to obtain the percentage of GFP-expressing cells and their mean fluorescent intensity (MFI). For non-GFP reporter viruses (influenza A virus and ZIKV), cells were washed once in PBS (+2% FBS), permeabilized in 1×BD perm (BD Biosciences: 554723) for 1 h at room temperature (RT) followed by overnight incubation with primary antibody. Cells were then washed an additional three times with BD perm, incubated with secondary antibody for 1 h at RT, washed three times with BD perm, and once with PBS (+2% FBS), and transferred to fluorescence-activated cell sorting tubes for flow cytometry as for reporter viruses.
Alternate WT AdV serotypes of AdV2, AdV7, and AdV35 were quantified by qPCR for vDNA at 72 hpi using the following primers: Ad2 (HAd2prF181: 5′-GCAAACGCTCTGCAACAAGA-3′, HAd2prR181: 5'-CGATCAGCTCGCTCATGACT-3′), Ad7 (HAd7AprF167: 5′-GACTCTGAGCGACGATCTGG-3′, HAd7AprR167: 5′-AGTCCGCTCCCATGTCAAAG-3′) and Ad35 (HAd35prF131: 5′-TTTCATTTTTCG‐CGCACGGT-3′, HAd35prR131: 5′-ACCCCAAAGACGGCCTAATG-3′).
For infection with JR34 AdV5 for imaging of FLAG-pVII DNA release, cells were seeded at 2 × 105 cells per well onto a glass coverslip pretreated with poly-l-lysine in 24-well plates. The next day, cells were washed once with cold Opti-MEM, wash was replaced with 200 µL Opti-MEM, and placed on ice for 10 min to cool before spiking in 50 µL of JR34 virus at the indicated MOI (typically 2,000 p/cell). Infection was synchronized on ice for 45 min and cells were washed once with cold Opti-MEM and incubated with warmed IMDM at 37 °C to initiate infection. For drug treatment experiments unless otherwise stated, cells were pretreated at 37 °C for 30 min prior to virus addition, and drug was included during the 45-min synchronization on ice and over the course of infection.
Immunofluorescence.
Unless otherwise described, cells were fixed in 4% PFA prepared in PBS at RT for 10 min followed by blocking with PBTG (PBS containing 10% normal goat serum, 1% BSA, 0.1% Triton X-100) at RT for 1 to 2 h. Cells were then incubated with primary antibodies diluted in PBTG (concentrations above) at 4 °C overnight. Following four washes with PBS with 0.1% Tween-20 (PBST), cells were stained with Alexa Fluor-conjugated secondary antibodies in PBTG (concentrations above) at RT for 1 h. Following one wash with PBST, cells were incubated with nuclear stain DRAQ5 diluted in PBS at RT for 15 min, followed by washes with PBST and a final wash with PBS. Coverslips were mounted on SuperFrost Plus Microscope Slides (Thermo Fisher Scientific: 12-550-15) using ProLong Gold antifade reagent (Invitrogen: P36934). Images were acquired using a 63× oil immersion objective on an inverted Zeiss Axiovert 200 spinning disk confocal microscope using solid-state 491, 561, and 644 lasers (Spectral Applied) for excitation for collection with an Andor iXon 512 × 512 electron multiplying charge-coupled device camera through MetaMorph software. Acquired images were analyzed as described using Fiji software (64).
Western Blot.
Cell lysates were prepared in RIPA buffer (50 mM Tris⋅HCl [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% Nonidet P-40 with addition of cOmplete Mini EDTA (ethylenediaminetetraacetic acid)-free protease inhibitor tablet; Roche), incubated on ice for 30 min and clarified at 14,000 × g for 20 min at 4 °C. Protein concentration was determined by bicinoninic (BCA) protein assay (Thermo Fisher Scientific: 23227) and samples were resolved on 4 to 12% Bis-Tris gels (Invitrogen) followed by transfer onto nitrocellulose 0.45 µm (Amersham Protran: GE10600002) membrane. Membranes were blocked with 5% milk in PBST and incubated with primary antibody at 4 °C overnight in 5% milk PBST. Membranes were washed three times with PBST and incubated with secondary antibody. For chemiluminescent readout, membranes were incubated with HRP-conjugated secondary antibody and exposed using SuperSignal West Pico PLUS Chemiluminescent Substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific: 34577 and 34095) by film.
Ubiquitinated Proteome Isolation Using TUBES.
Hap1 cells were seeded at 6 × 106 cells per 10-cm dish (quadruplicate per condition). For infection, cells were washed once with Opti-MEM, wash was replaced with 3 mL of Opti-MEM, and placed on ice for 10 min before spiking in 200 p/cell of AdV5rep+-GFP prepared in Opti-MEM. Following synchronization for 45 min at 4 °C, inoculum was removed and cells were washed with cold Opti-MEM. Prewarmed Opti-MEM was added and cells were incubated at 37 °C. At 1 hpi cells were placed on ice, washed with PBS, and lysed by adding 500 µL TUBE lysis buffer (50 mM Tris⋅HCl [pH 7.5], 0.15 M NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol) freshly supplemented with protease inhibitor tablet (Roche) and 50 µM DUB inhibitor PR-619 (LifeSensors: SI9619) and stored at −80 °C. In preparation for incubation with TUBE Dynabeads, lysates were thawed and clarified by centrifugation at maximum speed (21,000 × g) in a table-top centrifuge at 4 °C. Supernatants were transferred to fresh tubes, protein concentrations were quantified by BCA protein assay, and lysate input was normalized to 2 mg of protein lysate in a 500-µL volume for each condition for addition to the TUBE beads.
A total of 100 µL of control magnetic beads (LifeSensors: UM400M) and TUBE2 magnetic beads (LifeSensors: UM402M) per sample were prepared in separate low-fluid-retention tubes by washing three times with TBST (20 mM Tris⋅HCl [pH 8.0], 0.15 M NaCl, 0.1% Tween-20). To remove proteins that nonspecifically bind to the TUBE beads, the 500 µL of normalized input lysate was added to the control beads and nutated for 1 h at 4 °C. To isolate ubiquitinated proteins, this supernatant was transferred directly to the prepared TUBE2 beads and nutated for 3 h at 4 °C. After incubation, beads were washed three times with TBST, twice with TBS, and transferred to fresh tubes before two final washes with TBS. Supernatants were removed and the beads were stored dry at −20 °C prior to analysis by mass spectrometry.
Proximity Labeling.
WT and C985S MIB1 constructs were C-terminally tagged with a FLAG-tagged APEX2 protein via Gibson assembly of overlapping PCR-amplified MIB1 fragment and FLAG-APEX2 fragments into the recipient pLX304 lentivirus expression vector followed by site-directed mutagenesis to introduce the V943F mutation. APEX2-mKate was PCR amplified off of plasmid pcDNA3 APEX2-NES (a gift from Alice Ting, Stanford University, Stanford, CA, Addgene plasmid #49386; http://www.addgene.org/49386; RRID: Addgene_49386) (65) and cloned into the pLX304 backbone. Lentiviruses from these three constructs were used to reconstitute Hap1 MIB1 KO clone 2-1 as described.
For proteomics, cells were seeded at 1.5 × 107 cells per 15-cm dish in quadruplicate per condition. On the day of infection, cells were preincubated with 500 µM biotin-phenol (BP: sold as biotin tyramide, Iris Biotech: LS-3500.1000) at 37 °C for 30 min. Media were then replaced with cold Opti-MEM containing BP (500 µM) and 500 µL of AdV5rep+-GFP (MOI = 200 p/cell) or plain Opti-MEM (for uninfected controls) was spiked into the medium. Cells were incubated at 4 °C for synchronization of infection. At 40 min postadsorption, inoculum was replaced with 25 mL of prewarmed IMDM (+BP) and plates were incubated at 37 °C for 75 min. At 70 min pi, MIB1-proximal proteins were labeled by a 1-min incubation by spiking in H2O2 (Thermo Fisher Scientific: H325) to 100 mM, immediately followed by media removal and addition of 10 mL of a freshly prepared “quencher” solution (10 mM sodium ascorbate [Spectrum Chemical: S1349], 5 mM Trolox [Sigma: 238813] in PBS) for 2 min at RT. Solution was aspirated and cells were incubated with another 10 mL of quencher and collected into 15-mL conical tubes on ice by scraping. Cells were then pelleted at 1,200 × g, washed with 1 mL of quencher, and transferred to 1.2-mL Eppendorf tubes. Cells were pelleted at 5,000 × g, supernatants were removed, and pellets were stored at −80 °C until further processing.
To enrich proximity-labeled proteins, pellets were lysed in 1 mL of RIPA-Quench (Quench prepared in RIPA buffer and supplemented with protease-inhibitor tablet), and disrupted with pipetting. After a 15-min incubation on ice, samples were sonicated for 15 s and clarified by centrifugation at 15,000 × g at 4 °C. Meanwhile, 400 µL of High-Capacity Neutravidin Agarose Resin bead slurry (Thermo Fisher Scientific: 29284) was transferred to low-fluid retention 1.5-mL tubes (National Scientific: CN1700L-BP) and washed twice in 1 mL RIPA buffer prior to addition of proximity-labeled samples. Supernatants from clarified samples were added to the neutravidin beads and nutated overnight at 4 °C. Supernatants were removed and beads were washed twice with RIPA buffer, once with 1 M potassium chloride (KCl), once with 0.1 M sodium carbonate (Na2CO3), once with freshly prepared 2 M urea in 10 mM Tris⋅HCl (pH 8.0), and three times in 10 mM Tris⋅HCl (pH 8.0) with transfer to a fresh tube before the final wash. Samples were eluted either directly in lysis buffer with reducing agent for Western blot or beads were stored in 500 µL of Tris⋅HCl at −20 °C prior to mass spectrometry. Ten percent input and 10% post-IP fractions for each condition were analyzed by Western blot to ensure H2O2 labeling and streptavidin-mediated depletion.
For IF readout, cells were seeded onto glass coverslips treated with poly-l-lysine in 24-well plates. The following day, cells were preincubated with BP and infection and H2O2 labeling steps were carried out as described above except cells were fixed by addition of 4% PFA immediately following the first quencher incubation and processed for IF as described.
Proteomics Collection and Analysis.
Sample digestion.
Ubiqutin isolation mass spectrometry.
Beads were incubated with 8 M urea in 50 mM ammonium bicarbonate (AMBIC, Fluka Chemicals: 09830) for 1 h at RT. The supernatant was withdrawn and the process was repeated. The two supernatants were pooled and disulfide bonds were reduced by additon of DTT (dithiothreitol; EMD Chemicals) to 10 mM final concentration and incubation for 1 h at RT with vigorous shaking. Iodoaceamide (IAA, Sigma) was added to 20 mM and alkylation proceeded for 1 h at RT in the dark. Samples were diluted to reduce urea concentration to 3.5 M and digested using Lysyl Endopeptidase (Lys-C, Wako Chemicals) overnight at RT. The following day samples were further diluted to 1.5 M urea and digested with 1 µg sequencing grade trypsin (Promega). Digestion was stopped by acidification using trifluoroacetic acid (TFA, Thermo Fisher Scientific), and peptides were purified by in-house constructed reversed-phase micropurification tips.
APEX2 mass spectrometry.
Decanted agarose beads were subjected to on-bead digestion by the addition of 1 µg sequencing grade trypsin (Promega). The supernatant was withdrawn, evaporated to dryness and dissolved in 50 mM AMBIC (Fluka Chemicals), 10 mM DTT (EMD Chemicals) in water, and disulfide bonds were reduced for 1 h at room temperature with vigorous shaking. IAA (Sigma) was added to 20 mM and alkylation proceeded for 1 h at RT in the dark. Samples were further digested for 6 h at RT using trypsin. Digestion was stopped by acidification using TFA (Thermo Fisher Scientific), and peptides were purified by in-house constructed reversed-phase micropurification tips.
LC-MS/MS.
Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid, 80% acetonitrile (ACN) in water. All LC-MS solvents are of LC-MS grade and purchased from Thermo Fisher Scientific. Liquid chromatography was performed by a Dionex 3000 Ultimate HPLC (high-performance liquid chromatography) equipped with a NCS3500RS nano- and microflow pump (Dionex). Peptides were loaded onto a 100 µm × 20 mm Acclaim PepMap C18 trap column (Thermo Fisher Scientific) at 3 µL/min. Separation was achieved using a 75 µm × 120 mm pulled-emitter nanocolumn (Nikkyo Technos). Solvent B went from 1 to 38% over 90 min, followed by a sharp 1-min increase to 90% where it was kept for 8 min. Peptides were analyzed using a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). Data were recorded in positive mode with top 20 data-dependent acquisition. MS1 resolution was set to 60,000 and an MS2 resolution of 30,000. Automatic gain control targets of 3e6 (MS1) and 2e5 (MS2) were applied.
Data Analysis.
Data were analyzed with MaxQuant v. 1.6.6.0 (66). Spectra were queried against the human proteome (downloaded from uniprot.org on February 12, 2019) concatenated with a custom adenovirus database and common contaminants. A false discovery rate of 1% at both peptide and protein levels was applied. Further statistical analysis was performed within the Perseus framework (67). Significant changes were determined using the Student’s t test (P < 0.05). GO terms enrichment analysis was performed using STRING protein–protein interaction networks functional enrichment analysis (https://string-db.org).
Supplementary Material
Acknowledgments
We thank the following investigators for contributing viral molecular clones, viral stocks, and cell lines: P. Hearing (replication-competent AdV5-GFP), R. J. Parks (AdV5 JR34), I. Mohr (HSV-1-GFP), P. L. Collins (hPIV-3-GFP), I. Frolov (VEEV-GFP), J. K. Rose (VSV-GFP), J. C. de la Torre (LCMV-GFP), P. Palese (influenza A virus), J. L. Whitton (CVB-3-GFP), P. Traktman (VacV-GFP), the CDC (ZIKV), and J. L. Casanova (human STAT1−/− fibroblasts). Thanks to the resource centers at The Rockefeller University: the technical advice of S. Mazel, S. Semova, A. Keprova, and S. Han at the Flow Cytometry Resource Center and the expertise of the Proteomics Resource Center and the technical advice of A. North, K. Cialowicz, and C. Pyrgaki at the Bio-Imaging Facility. Thanks to B. Razooky for image analysis advice. We are also grateful for the administrative and/or experimental support of S. M. Pecoraro Di Vittorio, G. Santiago, A. O’Connell, M. E. Castillo, A. Webson, and S. Shirley. Thanks to A. W. Ashbrook and L. C. Aguado for valuable feedback on the manuscript and to all in the C.M.R. laboratory whose advice over the years helped guide this project. This work was supported by a grant from the Robertson Foundation (to H.-H.H.). The project was also supported in part by grant R01AI143295 (to C.M.R.) from the National Institute of Allergy and Infectious Disease, NIH. S.L.S. was supported by a David Rockefeller Graduate Student Fellowship and Rockefeller University Women and Science Fellowship. T.R.B. is supported by the Oncode Institute and the Cancer Genomics Center (GCG.nl) and a Vici grant from the Netherlands Organization for Scientific Research (NWO; 016.170.033).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015794118/-/DCSupplemental.
Data Availability.
Sequencing data from the haploid genetic screen for both AdV5-selected cells (NCBI SRA accession no. SRP269890 dataset SRX8663593) and unselected WT Hap1 reference (NCBI SRA accession no. SRP058962 dataset SRX1045464) are deposited on the NCBI SRA. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD020542. Processed data/analysis can be viewed within supplementary excel file Dataset S1.
References
- 1.Khanal S., Ghimire P., Dhamoon A. S., The repertoire of adenovirus in human disease: The innocuous to the deadly. Biomedicines 6, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crenshaw B. J., Jones L. B., Bell C. R., Kumar S., Matthews Q. L., Perspective on adenoviruses: Epidemiology, pathogenicity, and gene therapy. Biomedicines 7, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ungerechts G., et al. , Moving oncolytic viruses into the clinic: Clinical-grade production, purification, and characterization of diverse oncolytic viruses. Mol. Ther. Methods Clin. Dev. 3, 16018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemann J., Kühnel F., Oncolytic viruses: Adenoviruses. Virus Genes 53, 700–706 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bergelson J. M., et al. , Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275, 1320–1323 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Wiethoff C. M., Wodrich H., Gerace L., Nemerow G. R., Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79, 1992–2000 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wodrich H., et al. , A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog. 6, e1000808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bremner K. H.et al., Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6, 523–535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leopold P. L., et al. , Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 11, 151–165 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Cassany A., et al. , Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J. Virol. 89, 1719–1730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang I.-H., Burckhardt C. J., Yakimovich A., Morf M. K., Greber U. F., The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J. Cell Sci. 130, 2185–2195 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Strunze S., et al. , Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 10, 210–223 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Trotman L. C., Mosberger N., Fornerod M., Stidwill R. P., Greber U. F., Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3, 1092–1100 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Strunze S., Trotman L. C., Boucke K., Greber U. F., Nuclear targeting of adenovirus type 2 requires CRM1-mediated nuclear export. Mol. Biol. Cell 16, 2999–3009 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlon-Andres I., et al. , Nup358 and transportin 1 cooperate in adenoviral genome import. J. Virol. 94, 725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wodrich H., et al. , Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 80, 9608–9618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillay S., Carette J. E., Hunting viral receptors using haploid cells. Annu. Rev. Virol. 2, 219–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann H.-H., et al. , Diverse viruses require the calcium transporter SPCA1 for maturation and spread. Cell Host Microbe 22, 460–470.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carette J. E., et al. , Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Bergelson J. M., Receptors mediating adenovirus attachment and internalization. Biochem. Pharmacol. 57, 975–979 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Wickham T. J., Filardo E. J., Cheresh D. A., Nemerow G. R., Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127, 257–264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh M., et al. , Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Berndt J. D.et al., Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling. J. Cell Biol. 194, 737–750 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., et al. , Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 17, 96–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoggins J. W., Gall J. G. D., Falck-Pedersen E., Subgroup B and F fiber chimeras eliminate normal adenovirus type 5 vector transduction in vitro and in vivo. J. Virol. 77, 1039–1048 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall J., Kass-Eisler A., Leinwand L., Falck-Pedersen E., Adenovirus type 5 and 7 capsid chimera: Fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70, 2116–2123 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo B., McMillan B. J., Blacklow S. C., Structure and function of the Mind bomb E3 ligase in the context of Notch signal transduction. Curr. Opin. Struct. Biol. 41, 38–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luxán G., et al. , Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 19, 193–201 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Choe E.-A., et al. , Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J. Neurosci. 27, 9503–9512 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith A. C., Poulin K. L., Parks R. J., DNA genome size affects the stability of the adenovirus virion. J. Virol. 83, 2025–2028 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu T., Dacheux D., Kreppel F., Nagata K., Wodrich H., A method for visualization of incoming adenovirus chromatin complexes in fixed and living cells. PLoS One 10, e0137102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y., Johnson J. S., Ornelles D. A., Lieberman J., Engel D. A., Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol. 79, 2474–2483 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung V., et al. , Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Čajánek L., Glatter T., Nigg E. A., The E3 ubiquitin ligase Mib1 regulates Plk4 and centriole biogenesis. J. Cell Sci. 128, 1674–1682 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Villumsen B. H., et al. , A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 32, 3029–3040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov P., Kedersha N., Anderson P., Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 11, a032813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dao T. P., et al. , Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopitz-Otsoa F., et al. , Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs). J. Proteomics 75, 2998–3014 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Schellhaus A. K., et al. , Developmentally Regulated GTP binding protein 1 (DRG1) controls microtubule dynamics. Sci. Rep. 7, 9996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y., et al. , RNA- binding protein Stau2 is important for spindle integrity and meiosis progression in mouse oocytes. Cell Cycle 15, 2608–2618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizoguchi T., Ikeda S., Watanabe S., Sugawara M., Itoh M., Mib1 contributes to persistent directional cell migration by regulating the Ctnnd1-Rac1 pathway. Proc. Natl. Acad. Sci. U.S.A. 114, E9280–E9289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee I., et al. , Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346, 473–477 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Delboy M. G., Nicola A. V., A pre-immediate-early role for tegument ICP0 in the proteasome-dependent entry of herpes simplex virus. J. Virol. 85, 5910–5918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauer M., et al. , The E3 Ubiquitin ligase mind bomb 1 controls adenovirus genome release at the nuclear pore complex. Cell Rep. 29, 3785–3795.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Herrera L. A., Starr D. A., The E3 ubiquitin ligase MIB-1 is necessary to form the nuclear halo in Caenorhabditis elegans sperm. G3 (Bethesda) 8, 2465–2470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward S., Argon Y., Nelson G. A., Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91, 26–44 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratliff M., et al. , MIB-1 is required for spermatogenesis and facilitates LIN-12 and GLP-1 activity in Caenorhabditis elegans. Genetics 209, 173–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conine C. C., et al. , Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 3588–3593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aizer A., et al. , The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19, 4154–4166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gheiratmand L., et al. , Spatial and proteomic profiling reveals centrosome-independent features of centriolar satellites. EMBO J. 38, e101109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dho S. E., et al. , Proximity interactions of the ubiquitin ligase Mind bomb 1 reveal a role in regulation of epithelial polarity complex proteins. Sci. Rep. 9, 12471–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blomen V. A., et al. , Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Stempin C. C., et al. , The E3 ubiquitin ligase mind bomb-2 (MIB2) protein controls B-cell CLL/lymphoma 10 (BCL10)-dependent NF-κB activation. J. Biol. Chem. 286, 37147–37157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haeussler M., et al. , Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoggins J. W., Falck-Pedersen E., Fiber and penton base capsid modifications yield diminished adenovirus type 5 transduction and proinflammatory gene expression with retention of antigen-specific humoral immunity. J. Virol. 80, 10634–10644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., et al. , Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79, 1113–1124 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones C. T., et al. , Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28, 167–171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrakova O., et al. , Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J. Virol. 79, 7597–7608 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoggins J. W., et al. , Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benboudjema L., Mulvey M., Gao Y., Pimplikar S. W., Mohr I., Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides. J. Virol. 77, 9192–9203 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalton K. P., Rose J. K., Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279, 414–421 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Ngo N., Cubitt B., Iwasaki M., de la Torre J. C., Identification and mechanism of action of a novel small-molecule inhibitor of arenavirus multiplication. J. Virol. 89, 10924–10933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Feuer R., Mena I., Pagarigan R., Slifka M. K., Whitton J. L., Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J. Virol. 76, 4430–4440 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam S. S., et al. , Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from the haploid genetic screen for both AdV5-selected cells (NCBI SRA accession no. SRP269890 dataset SRX8663593) and unselected WT Hap1 reference (NCBI SRA accession no. SRP058962 dataset SRX1045464) are deposited on the NCBI SRA. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD020542. Processed data/analysis can be viewed within supplementary excel file Dataset S1.