Significance
The complex morphology of neurons relies on a tight control of protein localization and activation. How spatiotemporal activity patterns emerge from the interplay between diverse intracellular components is often not known. Here, we addressed this question for the neuronal protein ADAP1, which catalyzes GTP hydrolysis in the small G protein ARF6 and binds to the motor protein KIF13B. Using in vitro reconstitutions from purified proteins, we were able to propose a model where the motor and the signaling lipid PIP3 act in concert to distribute, deposit, and activate ADAP1, which in turn regulates ARF6. This regulatory pathway offers a mechanism for how small GTPases are controlled in space and time to control dendrite branching, which is important for neuronal development.
Keywords: microtubule transport, PIP3 signaling, small GTPases, in vitro reconstitution, neuronal development
Abstract
The differentiation of cells depends on a precise control of their internal organization, which is the result of a complex dynamic interplay between the cytoskeleton, molecular motors, signaling molecules, and membranes. For example, in the developing neuron, the protein ADAP1 (ADP-ribosylation factor GTPase-activating protein [ArfGAP] with dual pleckstrin homology [PH] domains 1) has been suggested to control dendrite branching by regulating the small GTPase ARF6. Together with the motor protein KIF13B, ADAP1 is also thought to mediate delivery of the second messenger phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to the axon tip, thus contributing to PIP3 polarity. However, what defines the function of ADAP1 and how its different roles are coordinated are still not clear. Here, we studied ADAP1’s functions using in vitro reconstitutions. We found that KIF13B transports ADAP1 along microtubules, but that PIP3 as well as PI(3,4)P2 act as stop signals for this transport instead of being transported. We also demonstrate that these phosphoinositides activate ADAP1’s enzymatic activity to catalyze GTP hydrolysis by ARF6. Together, our results support a model for the cellular function of ADAP1, where KIF13B transports ADAP1 until it encounters high PIP3/PI(3,4)P2 concentrations in the plasma membrane. Here, ADAP1 disassociates from the motor to inactivate ARF6, promoting dendrite branching.
Neurons are highly polarized cells with one axon and several highly branched dendrites (1). For their development, the protein ADAP1 (ADP-ribosylation factor GTPase-activating protein [ArfGAP] with dual pleckstrin homology [PH] domains 1; synonym, centaurin-α1) has been found to play an important role as it is thought to connect the major regulatory players involved (2): First, it contains two PH domains that can bind the phosphoinositide phosphatidylinositol (3,4,5)-trisphosphate (PIP3) present in the plasma membrane and in the case of the C-terminal PH domain also to PI(3,4)P2 (Fig. 1A) (3). Second, it has a putative ARF GAP domain at its N terminus that is thought to regulate the activity of the small GTPase ARF6 (2, 4). Third, it was found to interact with the microtubule cytoskeleton and the kinesin-3 member KIF13B (3, 5, 6). How these various interactions are coordinated and contribute to the cellular architecture during neuronal development is not well-understood.
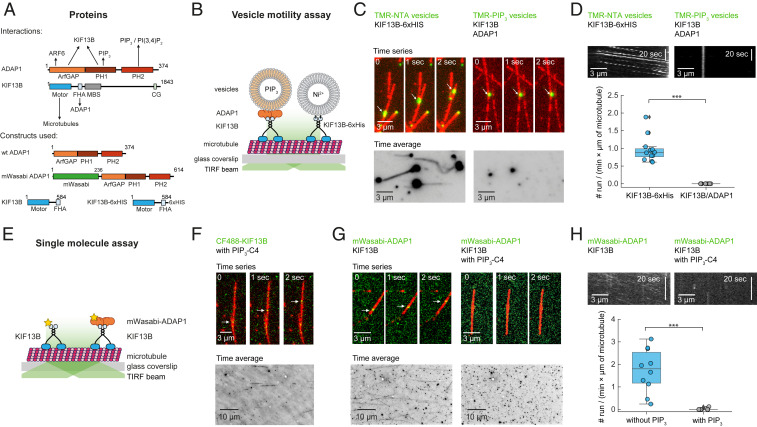
Fig. 1.
KIF13B transport of ADAP1 is inhibited by PIP3. (A) Domain architecture of ADAP1 and KIF13B and known interactions (Top). Architecture of protein constructs used in this study (Bottom). CC, coiled coil; CG, cytoskeleton-associated protein glycine-rich domain; FHA, forkhead-associated domain; MBS, membrane-associated guanylate kinase-binding stalk. (B) Assay scheme for single-vesicle motility assay. Fluorescently labeled vesicles are observed by TIRF microscopy as they are transported along microtubules by molecular motors. (C, Left) Time series of rhodamine-labeled NTA vesicles (green; 3 mol % Ni-NTA) being transported along HiLyte 647-labeled microtubules (red) in the presence of 500 nM KIF13B-6×His (Top) and corresponding time average (100 frames) of the rhodamine channel (Bottom). (C, Right) Time series of rhodamine-labeled PIP3 vesicles (green; 5 mol % PIP3) in the presence of ADAP1 (here 500 nM) and KIF13B (here 500 nM) and corresponding time average (100 frames) of the rhodamine channel (Bottom). (D) Quantification of run frequencies corresponding to C counting run events per minute and per micrometer of microtubule with one representative kymograph per condition. ***P < 0.001. (E) Assay scheme for single-molecule motility assay. Fluorescently labeled proteins are observed by TIRF microscopy along surface-immobilized microtubules. (F, Top) Time series of CF488-labeled KIF13B (green; 10 nM) walking along HiLyte 647-labeled microtubules (red) in the presence of 23 μM water-soluble PIP3-C4. (F, Bottom) Corresponding time average of the 488-nm channel (total duration 100 frames). (G, Top) Time series of 20 nM mWasabi-ADAP1 (green) with 100 nM unlabeled KIF13B in the absence (Left) or presence (Right) of 23 μM water-soluble PIP3-C4. (G, Bottom) Corresponding time average of the 488-nm channel (total duration 100 frames). (H) Quantification of run frequencies corresponding to G with one representative kymograph of the 488-nm channel per condition. ***P < 0.001. Displayed box plots encompass the 25–75th percentiles, the midline indicates the median, the whiskers extend to show the rest of the distribution and to indicate outliers.
A possible GAP function of ADAP1 was suggested to be important in dendrites, where it was found to promote branching and to control the density of spines in the developing neuron (7). Mutations in the putative GAP domain that render other GAPs inactive (8) were found to impede dendrite branching or spine formation when ADAP1 was overexpressed (7). In line with these observations, it has been found that excess ARF6-GTP prevents branching (9) and, at least in some developmental stages, reduces spine density (10, 11). In contrast, several studies in vitro were unable to detect any GAP activity of ADAP1 (4, 12–14). It is therefore not clear how ADAP1 and ARF6 are functionally connected and whether ADAP1 acts as a GAP or as an effector protein (13, 15).
Furthermore, ADAP1 has been implicated in the polarization of neurons by bridging PIP3 vesicles and the motor protein KIF13B, thereby facilitating the polarized transport of PIP3 vesicles to the axon tip (16). This idea was supported by an in vitro reconstitution of a motile complex of GST-ADAP1, KIF13B, and PIP3 (17) and by the observation that inhibition of KIF13B leads to defects in PIP3 polarity and axon specification in cultured hippocampal neurons (17, 18). However, as knockdown experiments with ADAP1 had no axon specification phenotype (7), its role for PIP3 transport in the axon remained elusive.
At the same time, previous work also established that ADAP1 localization depends on KIF13B (6). However, this study could not yet explain how exactly ADAP1 gets deposited at its destination and how this localization is compatible with its proposed role for vesicle transport.
To solve this conundrum and to understand the different roles of ADAP1 in the developing neuron, we set out to reconstitute its proposed functions and their interplay in vitro. We used the purified components ADAP1, KIF13B, ARF6, microtubules, and membranes of defined lipid composition and found that in contrast to previous reports, ADAP1 does not allow for directional transport of PIP3 by KIF13B. Instead, we show that PIP3 releases ADAP1 from microtubule-bound KIF13B and therefore acts as a terminator of this transport instead of being a cargo. Furthermore, we show that once engaged to either PIP3 or PI(3,4)P2, ADAP1 stimulates GTP hydrolysis of ARF6 in vitro.
Together, our findings propose a model where KIF13B controls the spatial activity patterns of ARF6 by transporting its corresponding GAP ADAP1 to membranes containing PIP3 or PI(3,4)P2. As ARF6 plays an important role for the regulation of the actin cytoskeleton (8), this mechanism can contribute to the control of neuronal morphology and function.
Results
PIP3 Inhibits ADAP1 Transport by KIF13B Instead of Being Its Cargo.
As ADAP1 was found to bind to both KIF13B (3, 6, 19) and PIP3 (3, 12, 20) (Fig. 1A), we first set out to confirm these interactions. We purified nontagged ADAP1 from SF9 insect cells, which eluted as a monomer with a molecular mass of ∼43 kDa (SI Appendix, Fig. S1A). We then demonstrated its interaction with a motile fragment of KIF13B [amino acids 1 to 584 (17)] in solution (SI Appendix, Fig. S1B) and its binding to PIP3 (SI Appendix, Fig. S1C). We also confirmed the activity of the motor protein in microtubule gliding assays (SI Appendix, Fig. S1D and Movie S1). Thus, these experiments verified the previously identified pairwise molecular interactions relevant for PIP3 vesicle transport.
Next, we set out to study the KIF13B- and ADAP1-mediated transport of PIP3-containing vesicles (Fig. 1B). We incubated all required components on surface-immobilized microtubules and, using total internal reflection fluorescence (TIRF) microscopy, expected to see vesicles being transported along the microtubules. Surprisingly, and in contrast to a previous report (17), we did not observe efficient transport of PIP3-containing vesicles (Fig. 1 C and D and Movie S2). However, we found robust vesicle transport when we bypassed ADAP1 by using KIF13B-6×His and vesicles containing Ni-NTA (Fig. 1 C and D and Movie S3), confirming that vesicle transport is in principle possible in our assay.
To better understand why there was no transport of PIP3 vesicles, we added the components sequentially to our assay. Using CF488-labeled KIF13B, we first performed single-molecule motility assays (Fig. 1E) and observed directed motion of CF488-KIF13B along microtubules in the presence of water-soluble PIP3-C4 (Fig. 1F). This rules out that KIF13B is inhibited by PIP3 and shows that the motor protein is processive (21). Next, we incubated unlabeled KIF13B with fluorescently labeled mWasabi-ADAP1 and found that the motor protein can transport ADAP1 along microtubules (Fig. 1G and Movie S4). Finally, we repeated these experiments in the presence of water-soluble PIP3-C4. Again, there was no KIF13B-mediated transport of ADAP1 under these conditions (Fig. 1 G, Right and Movie S4), as quantified by a dramatic decrease in run frequency (Fig. 1H).
To summarize, our data show that ADAP1 is not an adaptor for the transport of PIP3 vesicles by KIF13B. Instead, and in contrast to a previous report (17), we find that PIP3 inhibits the transport of ADAP1 by KIF13B.
Transport Is Inhibited by a Phosphoinositide-Induced Release of ADAP1 from KIF13B.
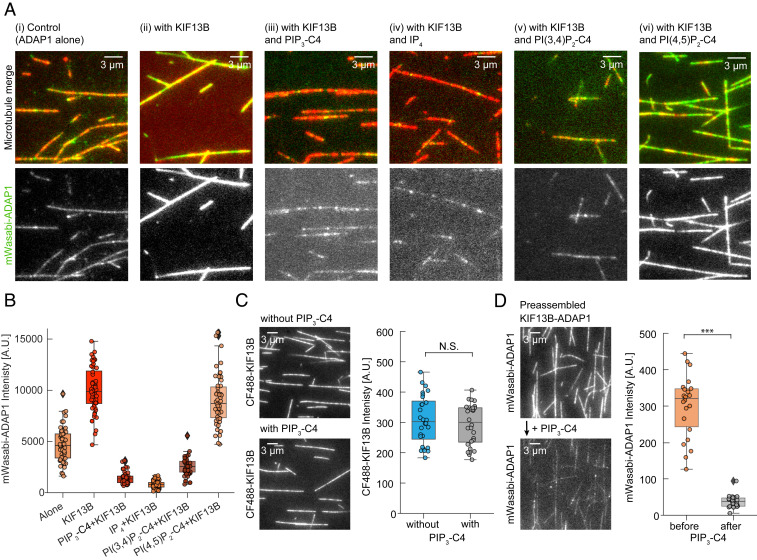
PIP3 could inhibit motility of the ADAP1–KIF13B transport complex on microtubules or prevent its formation. To distinguish between these two scenarios and to understand the mechanism of transport inhibition, we incubated fluorescent mWasabi-ADAP1 with surface-immobilized microtubules. We found that ADAP1 had a weak but detectable affinity toward microtubules (Fig. 2 A, i), consistent with an earlier report (5). The presence of AMP-PNP–bound, unlabeled KIF13B strongly increased the fluorescence signal of mWasabi-ADAP1 (Fig. 2 A, ii), showing that KIF13B recruits ADAP1 to microtubules. However, when we repeated this experiment in the presence of either water-soluble PIP3-C4 (Fig. 2 A, iii) or inositol 1,3,4,5-tetrakisphosphate (IP4) (Fig. 2 A, iv), we found that the intensity signal of mWasabi-ADAP1 along microtubules was strongly reduced to less than 15% of its original value in the presence of the motor (Fig. 2B). In contrast, the signal for KIF13B remained unchanged and was not affected by the presence of PIP3-C4 (Fig. 2C).
Fig. 2.
PIP3 displaces ADAP1 from microtubule-bound KIF13B. (A) Representative TIRF images of mWasabi-ADAP1 binding to stabilized and immobilized HiLyte 647-labeled microtubules in the presence or absence of AMP-PNP–bound KIF13B and different phosphoinositides. Merged images are shown (Top) (mWasabi-ADAP1 is in green and microtubules are in red) and the 488-nm mWasabi channel is shown (Bottom) in grayscale. Concentrations were 160 nM mWasabi-ADAP1, 200 nM KIF13B, 28 µM IP4, and 23 µM for all other water-soluble phosphoinositides (SI Appendix, Methods). AMP-PNP was present in all conditions at 0.5 mM. (B) Quantifications for all conditions in A. Intensities of mWasabi-ADAP1 come from at least 20 microtubules from three independent experiments per condition. A.U., arbitrary units. (C, Left) Representative TIRF image of the 488-nm channel showing AMP-PNP–bound CF488-KIF13B binding to stabilized microtubules in the absence (Top) or presence (Bottom) of 23 μM water-soluble PIP3-C4. CF488-KIF13B was present at 100 nM. (C, Right) Corresponding quantification of CF488-KIF13B signals from three independent experiments. N.S., not significant. (D, Left) Representative TIRF images of PIP3-C4 wash-in experiments. mWasabi-ADAP1 signals were recorded in the presence of AMP-PNP–KIF13B without PIP3-C4 (Top) and after (Bottom) solutions were exchanged with a solution containing mWasabi-ADAP1, AMP-PNP–KIF13B, and additionally water-soluble PIP3-C4. Protein and PIP3 concentrations were the same as in A and the time between solution exchange and image recording was ∼30 s. (D, Right) Corresponding quantification of mWasabi-ADAP1 signals before (Left) and after (Right) solution exchange from the same set of microtubules. ***P < 0.001. Displayed box plots encompass the 25–75th percentiles, the midline indicates the median, the whiskers extend to show the rest of the distribution and to indicate outliers.
ADAP1 was also found to bind to PI(3,4)P2, but not to PI(4,5)P2 (20). Interestingly, PI(3,4)P2 is known to promote outgrowth and branching of dendrites from the plasma membrane (22, 23). To test whether these phosphoinositides have an effect on the formation of the ADAP1–KIF13B complex, we repeated our experiment with PI(3,4)P2 or PI(4,5)P2. When we incubated ADAP1 and KIF13B on surface-bound microtubules with soluble PI(3,4)P2-C4, the fluorescence of mWasabi-ADAP1 was reduced to a similar level as found in the presence of PIP3-C4 (Fig. 2 A, v), indicating that also PI(3,4)P2 prevents the formation of a transport complex. This finding also excludes the possibility that ADAP1 can act as a transporter for PI(3,4)P2 vesicles. In contrast, PI(4,5)P2-C4 had only a minor and statistically not significant (at the P < 0.01 level; P = 0.0281) effect on the complex formation of ADAP1 and KIF13B on the microtubule (Fig. 2 A, vi), which also rules out nonspecific effects of phosphoinositides on these interactions.
Next, we wanted to know if this inhibitory effect is limited to water-soluble PIP3. To find out how PIP3 embedded in a lipid membrane acts on ADAP1, we performed pelleting assays. First, we incubated microtubules with AMP-PNP–bound KIF13B and ADAP1 and found that both proteins are recruited to the microtubule pellet (SI Appendix, Fig. S2A), consistent with the presence of a microtubule-bound complex of KIF13B–ADAP1 (Fig. 2). In the presence of PIP3-containing vesicles, however, ADAP1 was selectively recruited to the vesicle pellet, whereas KIF13B remained in the supernatant (SI Appendix, Fig. S2B). Together, these findings show that PIP3 inhibits the interaction between ADAP1 and KIF13B, even in the absence of microtubules, while it does not affect binding of the motor protein to microtubules.
We next wondered whether PIP3 binds to ADAP1 in solution to prevent its recruitment to KIF13B on microtubules and therefore inhibits its transport, or if PIP3 can disassemble the ADAP1–KIF13B complex preformed on microtubules. We therefore first incubated mWasabi-ADAP1 and AMP-PNP–bound KIF13B in the absence of PIP3 together with surface-immobilized microtubules to allow for complex formation. We then replaced the solution in the flow chamber with a buffer containing water-soluble PIP3-C4 in addition to all other components and recorded the fluorescence signals from the same set of microtubules. After the addition of PIP3-C4, we observed a strong decrease of the mWasabi-ADAP1 signal along microtubules (Fig. 2D). In contrast, the intensity of CF488-KIF13B on microtubules was not affected as shown before (Fig. 2C) and the motor also remained motile (SI Appendix, Fig. S3A). Importantly, this effect was reversible, as ADAP1 reattached to the immobilized microtubule when we washed out PIP3-C4 (SI Appendix, Fig. S3B).
Together, these data contradict the idea of a motile PIP3–ADAP1–KIF13B complex on microtubules. Instead, our experiments indicate that binding of IP4, PIP3, or PI(3,4)P2 to at least one of its PH domains releases ADAP1 from its interaction partner KIF13B. Accordingly, our results suggest a role for these second messengers as cargo-release factors that terminate ADAP1 transport by displacing it from its transporter, KIF13B.
ADAP1 Binds Cooperatively to PIP3 in Membranes.
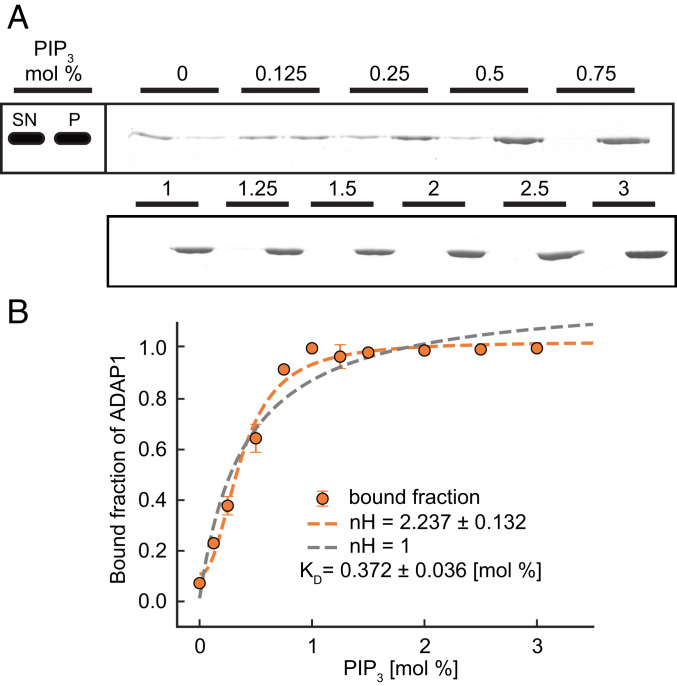
So far, we found that ADAP1 does not allow for the transport of phosphoinositides but that they displace ADAP1 from KIF13B. Next, we wanted to understand how local differences in PIP3 densities, such as its enrichment in cell membranes, could affect the stability of this complex and whether there is a critical density of PIP3 that terminates transport. We therefore performed pelleting assays with vesicles containing different concentrations of PIP3 from 0 to 3% and found an increasing amount of ADAP1 in the pellet fraction with higher PIP3 content (Fig. 3A). Fitting a Hill equation to the data revealed that binding of ADAP1 to PIP3 membranes is cooperative with a Hill coefficient (nH) of 2.2 (± 0.13) (Fig. 3B) and a half-maximal effective concentration of 0.37 (± 0.08) mol % PIP3. This cooperativity can be explained by the presence of two PH domains in ADAP1 and that membrane binding is enhanced when both PH domains are occupied by PIP3 (24, 25). As a consequence, the protein could respond to a local enrichment of PIP3 in the membrane, triggering the release of ADAP1 from the motor complex in a highly sensitive manner.
Fig. 3.
ADAP1 binds to PIP3 vesicles in a cooperative manner. (A) Pelleting assay with ADAP1 (1 µM) and vesicles (375 µM) with the indicated concentration of PIP3. A representative Coomassie brilliant blue-stained sodium dodecyl sulfate gel is shown, where the supernatant (SN) is loaded on the left and the pellet (P) on the right for each PIP3 concentration. Note that different amounts of pellet and supernatant are loaded, which is also accounted for in the quantification (see SI Appendix, Methods for details). (B) Quantification of band intensities. Plotted is the distribution of ADAP1 between pellet and supernatant for each condition together with a Hill fit for the whole dataset (orange dashed line) and compared with a fit where nH was set to 1 (gray dashed line). Error bars are SDs from two independent experiments per condition.
ADAP1 Is an ARF6 GAP.
Due to its N-terminal GAP domain, ADAP1 has been suggested to stimulate GTP hydrolysis in ARF GTPases (2, 14). Indeed, ADAP1 with a mutation in the putative GAP domain was unable to increase spine densities and branching when overexpressed (7). However, several in vitro studies reported a lack of GAP activity of ADAP1 toward ARF6 (4, 12–14). Importantly, these biochemical studies were performed with soluble ARF6 lacking its myristoyl anchor and in the absence of membranes and phosphoinositides, which were found to regulate the activity of GAPs with N-terminal PH domains (25–28). Accordingly, if and under which conditions ADAP1 can activate GTP hydrolysis by ARF6 bound to membranes containing phosphoinositides are not known.
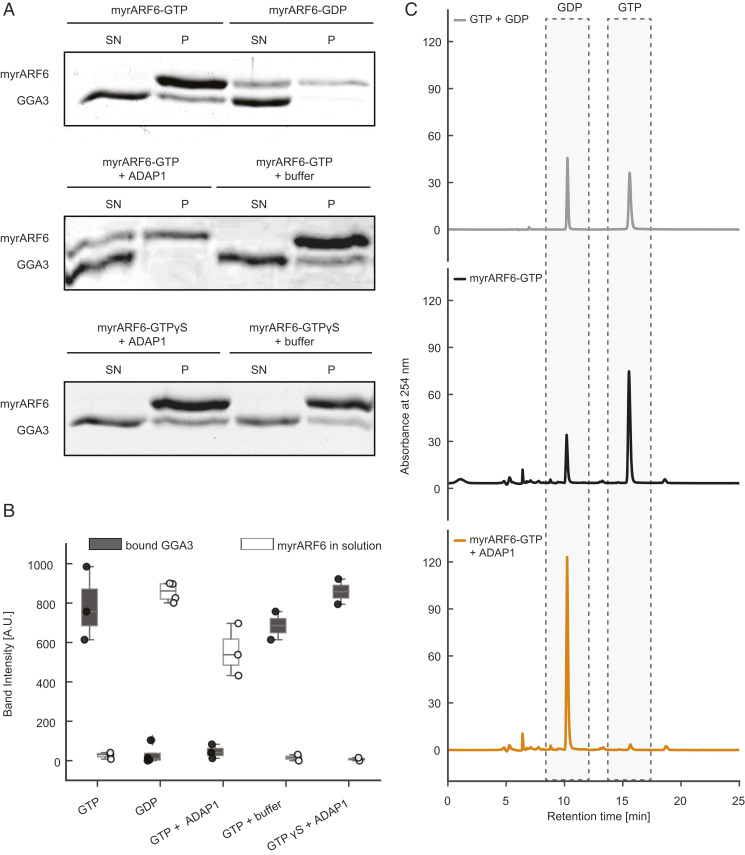
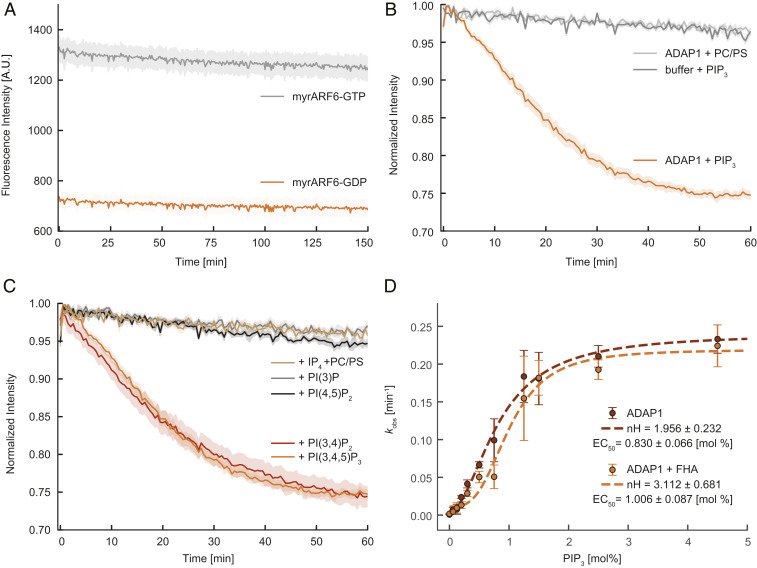
To test the GAP activity of ADAP1 on membrane-bound ARF6, we set up an in vitro assay using myristoylated ARF6 (myrARF6) and vesicles composed of phosphatidylcholine (PC), phosphatidylserine (PS), and PIP3 (Materials and Methods and SI Appendix, Fig. S5A). As a reporter for the nucleotide state of ARF6, we used the GAT domain of GGA3 [amino acids 146–302], an ARF6 effector that specifically binds to membrane-bound ARF6-GTP but not to ARF6-GDP (29). To validate this approach, we prepared myrARF6 in a GTP-bound state or GDP-bound state (30). In vesicle sedimentation experiments, we found that myrARF6-GTP recruited substantial amounts of GGA3 to the vesicle pellet, whereas almost no GGA3 was found in the pellet when myrARF6 was preloaded with GDP (Fig. 4 A, Top, lower band). Additionally, we observed that ARF6 itself bound more tightly to membranes in its GTP-bound form (Fig. 4 A, Top, upper band). Strikingly, addition of ADAP1 to myrARF6-GTP reduced the amount of copelleted GGA3 to the level seen for myrARF6-GDP and increased the amount of soluble myrARF6 (Fig. 4 A, Middle and Fig. 4B). To rule out any competition effect of ADAP1 and GGA3 for membrane-bound ARF6, we performed additional experiments with a nonhydrolyzable GTP analog, GTPγS (29). No significant differences between GGA3 in the pellet fraction or ARF6 in the supernatant were observed in the presence or absence of ADAP1 (Fig. 4 A, Bottom and Fig. 4B). This shows that ADAP1 cannot displace GGA3 from membrane-bound ARF6 in a GTP hydrolysis-independent process and suggests that the decrease of GGA3 in the pellet is due to ADAP1-stimulated GTPase activity. In order to directly confirm GTP hydrolysis, the nucleotide bound to myrARF6 was extracted from the protein and identified in a high-performance liquid chromatography (HPLC)-based assay. GTP-loaded myrARF6 was incubated with PIP3-containing vesicles with or without ADAP1. In the absence of ADAP1, we found that ∼75% of ARF6 was bound to GTP (Fig. 4 C, Middle and SI Appendix, Fig. S4C). Strikingly, in the presence of ADAP1, there was almost no remaining GTP peak observable and only one dominant GDP peak remained (Fig. 4 C, Bottom). Control experiments with either GTPγS or ARF6 lacking its myristoyl anchor did not result in hydrolysis (SI Appendix, Figs. S4 and S5). Together, this firmly establishes ADAP1 as an ARF6 GAP that requires membrane-bound ARF6-GTP as a substrate.
Fig. 4.
ADAP1 is an ARF6 GAP in vitro. (A, Top) To validate the assay, myrARF6 was brought into a GTP (Left) or GDP (Right) -bound state. Recruitment of GGA3 to the pellet and the amount of ARF6 in the supernatant were monitored. (A, Middle) myrARF6-GTP was incubated with 1 µM ADAP1 (Left) or ADAP1 storage buffer (Right). (A, Bottom) Instead of GTP, myrARF6 was loaded with GTPγS. (B) Quantification of ARF6 band intensity in the supernatant (white) and GGA3 band intensities in the pellet (dark gray). “Buffer” corresponds to ADAP1 storage buffer. Displayed box plots encompass the 25–75th percentiles, the midline indicates the median, the whiskers extend to show the rest of the distribution and to indicate outliers. (C) Representative HPLC chromatograms of pure GTP and GDP nucleotides (Top) compared with nucleotides extracted from PIP3 vesicle-bound myrARF6 incubated without (Middle) or with ADAP1 (Bottom).
ADAP1’s GAP Activity Depends on Binding to PIP3-Containing Membranes.
To shed light on the regulation and kinetics of ADAP1’s GAP activity, we employed a real-time kinetics assay based on the intrinsic tryptophan fluorescence of ARF6 (Materials and Methods). As ARF GTPases have a higher fluorescence signal when loaded with GTP than in their GDP-bound state, this property can be used as a real-time readout of their nucleotide state (30, 31). In the absence of ADAP1, no significant signal changes were observed over a time course of up to 2 h (Fig. 5A), consistent with earlier studies that reported the absence of an intrinsic GTPase activity of ARF6 (32, 33). In the presence of ADAP1 and vesicles containing 2.5% PIP3, we found the fluorescence intensity to continuously decrease following a monoexponential decay, consistent with ADAP1 catalyzing GTPase hydrolysis by ARF6 (Fig. 5B).
Fig. 5.
ADAP1 is a PIP3/PI(3,4)P2 membrane-dependent GAP whose activity is influenced by KIF13B. (A) Intrinsic tryptophan fluorescence of myrARF6 (4.5 µM) preloaded with GTP (gray) or GDP (orange) over time. (B) Time course of normalized tryptophan fluorescence of 4.5 µM myrARF6-GTP in the absence (dark gray) or presence of 1.5 µM ADAP1 with either 375 µM vesicles containing no PIP3 (light gray) or 2.5% PIP3 vesicles (orange). (C) Normalized tryptophan fluorescence of 4.5 µM myrARF6-GTP in the presence of 1.5 µM ADAP1 and 375 µM vesicles with either 2.5% PIP3, PI(3,4)P2, PI(3)P, PI(4,5)P2, or vesicles without phosphoinositides together with 20 µM IP4. Data for the PIP3 condition are the same for B and C. (D) Measurement of the catalytic activity of 1.5 µM ADAP1 (kobs) with 4.5 µM myrARF6-GTP as a function of PIP3 content in vesicles (375 µM) in the absence (red) or presence of 10 µM KIF13B’s FHA domain (orange). kobs values were fitted to a Hill equation (dashed lines) extracting the Hill coefficients of 1.96 (± 0.23) in the absence and 3.11 (± 0.68) in the presence of the FHA domain. Errors are SEMs for A–C and SDs for D and are derived from at least three independent measurements per condition.
Previous studies have found that phosphoinositides activate GAPs with N-terminal PH domains independent of membrane recruitment (27, 28). Instead, binding of phosphoinositides to their PH domains activates the GAP allosterically. To find out whether the localization to a membrane surface or occupation of the PH domains is sufficient to activate ADAP1’s GAP activity, we performed three sets of experiments. First, we asked whether PIP3 is necessary for ADAP1’s GAP activity and recorded fluorescence time courses of myrARF6-GTP with vesicles only containing PC and PS. Under these conditions, fluorescence intensity traces were indistinguishable from the experiments lacking ADAP1 (Fig. 5B), showing that either membrane recruitment or phosphoinositides or both are required to stimulate ADAP1’s GAP activity.
Next, to find out if binding of ADAP1 to phosphoinositides is sufficient to stimulate its GAP activity, we added water-soluble IP4 and vesicles of only PC and PS to our assay. If binding of PIP3 is sufficient for the activation of ADAP1, we expected to see a decrease in fluorescence of myrARF6-GTP. However, we found that the presence of 20 µM IP4 did not activate the GAP activity of ADAP1 (Fig. 5C). In contrast, artificial recruitment of a C-terminally 6×His-tagged ADAP1 to Ni-NTA–containing membranes did result in GAP activity (SI Appendix, Fig. S5B), albeit with reduced efficiency in comparison with vesicles containing 2.5% PIP3.
As ADAP1 was found to also bind to PI(3,4)P2 (3, 20), we then wondered if other phosphoinositides can also stimulate ADAP1’s GAP activity. Consistent with these previous observations, we observed GTP hydrolysis with vesicles containing 2.5% PI(3,4)P2, while PI(4,5)P2 and PI(3)P had no effect on ADAP1’s activity toward ARF6 (Fig. 5C).
To conclude, we found that membrane binding is essential for the GAP activity of ADAP1, and that PIP3 or PI(3,4)P2 has an additional stimulatory effect. Binding of ADAP1 to phosphoinositides via its PH domain could allow for an ideal orientation toward ARF6 on the membrane surface; however, our data cannot rule out an allosteric effect as an explanation for the observed behavior.
We then decided to study the kinetics of this reaction in titration experiments. By titrating either the substrate (myrARF6-GTP) or the enzyme concentration (ADAP1) at a fixed PIP3 concentration, we were able to extract the Michaelis–Menten constant (34), which under our experimental condition was ∼4.5 µM (SI Appendix, Fig. S5C), and the catalytic efficiency kcat/Km, which was 0.27 (± 0.01) µM−1⋅min−1 (SI Appendix, Fig. S5D). We then fixed the ADAP1 and myrARF6-GTP concentrations and varied the PIP3 content of vesicles and plotted the GAP activity against the PIP3 concentration. We observed cooperative activation of the GAP activity in response to increasing PIP3 concentration with a Hill coefficient of 1.96 (± 0.32) and a half-maximal effective concentration of 0.83 (± 0.07) mol % PIP3 (Fig. 5D), in agreement with our membrane-binding data (Fig. 3). This suggests that not only its binding to membranes but also ADAP1’s GAP activity follow a nonlinear response to the concentration of PIP3 in the membrane. A previous study found that overexpression of an FHA domain-containing fragment of KIF13B increased ARF6-GTP levels in cells, possibly via ADAP1 inhibition (6). We therefore tested this idea and, in agreement with this study, found that 10 µM FHA domain of KIF13B resulted in a mild inhibition of GAP activity at intermediate PIP3 concentrations, shifting the half-maximal effective concentration from 0.83 (± 0.07) to 1.01 (± 0.09) mol % PIP3 (Fig. 5D). Interestingly, addition of the FHA domain also increased the Hill coefficient from 1.96 (± 0.32) to 3.11 (± 0.68) (Fig. 5D), enhancing the nonlinearity of ADAP1’s GAP activity in response to increasing PIP3 concentrations.
In summary, we found that ADAP1’s GAP activity depends on membrane binding and is additionally activated by PIP3. In contrast, there is only a minor inhibitory influence of the binding partner KIF13B.
Discussion
ADAP1 and Regulated Transport.
Intracellular transport and GTPase regulation are important processes that define essential cellular functions (1, 35). For many tasks, such as cell polarization and differentiation, these two processes mutually regulate each other in dynamic biochemical networks (35, 36). But because of the complexity of the living cell, their independent systematic characterization is often impossible in vivo. Our study provides details of such coordination, exemplified by the reconstituted protein interaction network of ADAP1 with PIP3, KIF13B, and ARF6.
We found that KIF13B transports ADAP1 along microtubules, consistent with previous observations that the two proteins physically interact with each other (3, 6, 19) and that KIF13B plays a role for the intracellular location of ADAP1 (6). In some cases, KIF13B has been shown to be dendrite-selective (37), which might explain the observation that the majority of ADAP1 is localized to the somatodendritic compartment (7).
We show that PIP3 triggers release of ADAP1 from its complex with KIF13B on microtubules and therefore provide evidence that phosphoinositides can act as a stop signal for microtubule-based transport. So far, Ca2+, several protein kinases, and kinesin-binding proteins have been described to trigger cargo release but, in most cases, their spatial control is not very well understood (38–41). In the case of ADAP1, PIP3 and PI(3,4)P2 not only release the protein from the motor but also activate its enzymatic function, possibly at a well-defined intracellular location that is defined by a targeted molecular transport and the local lipid composition. Our results establish phosphoinositides as a cargo-displacement factor for the ADAP1–KIF13B complex and it will be interesting to see if similar mechanisms are applied for other PH domain-containing proteins to displace them from their interaction partners.
ADAP1 and PIP3 Polarity.
The prevalent model of PIP3 polarity in neurons assumes that it depends on two complementary processes: first, its local production and degradation (42, 43), and second, targeted delivery to the axon tip (17, 44). Herein, ADAP1 and KIF13B would be part of a positive feedback loop involving MARK2/Par1 and aPKC that delivers PIP3 to the axon tip by microtubule-based transport (16, 18). This idea was supported by the observation that inhibition of KIF13B caused failures in PIP3 polarity (17, 18, 45) and by an in vitro study that reconstituted a ternary complex of PIP3 vesicles, GST-ADAP1, and KIF13B (17). Our data contradict this suggested function of ADAP1 during PIP3 vesicle transport. We believe that the discrepancy from our data can be explained by the previous use of the GST-tagged ADAP1 in in vitro experiments. As GST is known to dimerize with Kd ∼20 pM (46), simultaneous binding of ADAP1 to PIP3 and KIF13B could have been due to a stably linked GST dimer, where one ADAP1 monomer binds PIP3 and the other KIF13B. Consistent with our results, the structure of ADAP1 showed two PIP3 molecules in the absence of the FHA domain of KIF13B, while in its presence only 0.5 PIP3 was found per ADAP1, indicative of competitive binding (3). The binding interfaces for FHA and PIP3 on ADAP1 do not directly overlap and it therefore is not obvious why they cannot bind at the same time. However, binding of the FHA domain to ADAP1 induces a conformational change in the PH2 domain, which indicates an allosteric mode of inhibition. Importantly, genetic inhibition of ADAP1 did not cause an axonal phenotype and, in fact, ADAP1 is mainly present in dendrites (7). It is therefore likely that for the directed transport of PIP3 vesicles, KIF13B uses different adaptors as it was found to transport early endosomes independent of ADAP1 (47). Alternatively, it is also possible that PIP3 polarity solely depends on local production and degradation by phosphoinositide kinases and phosphatases. To conclude, we believe that our results from biochemical reconstitution experiments provide evidence that support observations made in the living cell.
ADAP1 and ARF6 GAP Activity.
We demonstrate here in vitro that ADAP1 has ARF6 GAP activity, that this activity is coupled to the presence of membranes containing PIP3 or PI(3,4)P2, and that it does not require additional proteins. This reconciles observations made in cells that suggested that ADAP1 has GAP activity toward ARF6 (6, 14, 48) and biochemical studies that reported the lack of GAP activity in the absence of membranes (4, 12, 13). In fact, other GAPs and guanine nucleotide-exchange factors (GEFs) are known to be regulated by phosphoinositides (25, 26, 33, 49, 50), which highlights the important role of these lipids for the regulation of ARF GTPases. The kinetic rates obtained for ADAP1 were similar to other ARF6 GAPs such as ACAPs (0.025 to 0.25 hydrolyses per minute) and slower in comparison with ARF1 GAPs such as ASAP1 (50). Also, the obtained Km in the low micromolar range is comparable to other known GAPs (26, 49). It should be noted that these values refer to solution concentrations but that the actual reactions happen on the membrane surface. Accordingly, GAP-stimulated GTP hydrolysis by ARF GTPases is the result of a more complex reaction pathway that includes several membrane-binding events. Furthermore, local concentrations of proteins on the membrane surface can differ considerably compared with their concentration in solution.
We found cooperative binding of ADAP1 to PIP3-containing membranes and a similar nonlinear response of its GAP activity. This behavior is often seen in proteins with tandem PH domains (24) and likely ensures that activation happens only above a certain phosphoinositide threshold in the membrane to fine-tune cellular responses to external cues. Addition of the FHA domain of KIF13B mildly enhanced the degree of cooperativity, which is in line with theoretical and experimental work that showed an increase in ultrasensitivity in the presence of sequestering inhibitors (51). Our data also agree with studies in cells, where overexpression of a nonmotile KIF13B fragment leads to increased ARF6-GTP levels (6), likely via ADAP1 inhibition.
ADAP1 promotes dendrite branching (7) whereas ARF6-GTP inhibits this process via recruitment of its effector phosphatidylinositol 4-phosphate 5 kinase and the activation of the Rac1 pathway (9, 52, 53). Interestingly, high PI(3,4)P2 concentrations coincide with branching points (23). Therefore, we speculate that local PI(3,4)P2 accumulation could release ADAP1 from KIF13B to inactivate ARF6 to promote dendrite branching. In fact, previous studies could demonstrate such local PIP enrichment in response to extracellular cues (54).
Consistent with a previous study in cells that indicated that artificial membrane recruitment of ADAP1 by the RAS membrane-targeting sequence leads to ARF6 GAP activation independent of PIP3 binding (14), we did not observe any GAP activity in solution, regardless of whether ADAP1’s PH domains were bound to IP4 or not. Instead, artificial recruitment of ADAP1 only allowed for moderate activity. These experiments show that ADAP1’s ability to stimulate GTP hydrolysis by ARF6 is restricted to membranes. They also indicate that the orientation of both proteins on the membrane is critical for this GTPase stimulation. KIF13B (18) and ADAP1 (55) are phosphorylated by different kinases. As it has been shown that lipid affinity and specificity of PH domains can be regulated by phosphorylation (25, 56), it will be interesting to study how these phosphorylations can alter the activity of the proteins used in this study.
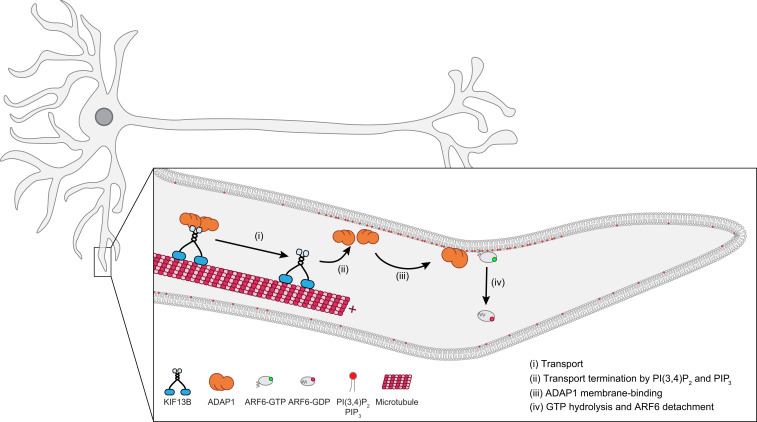
To summarize, our data do not support ADAP1’s proposed role in axonal PIP3 transport. Instead, we favor a model for the function of ADAP1 where this protein is transported to dendrites by the motor KIF13B until it binds to membranes enriched in PIP3/PI(3,4)P2, which terminate cargo transport. The phosphoinositide-enhanced GAP activity of ADAP1 and the deactivation of ARF6 could then initiate dendrite branching and contribute to the maintenance of dendritic spines (Fig. 6). Our findings extend previous models for the regulation of GTPases that typically assume a diffusion and capture process for the localization of GAPs and GEFs (33). Instead, our data support a microtubule-based deposition of a GAP that contributes to the spatial regulation of its cognate ARF GTPase. While a similar mechanism has been described for the localization of the Tea1–Tea4 complex to the cell ends of the fission yeast Schizosaccharomyces pombe (57), it has so far not been identified to play a role for the regulation of small GTPases in vertebrate systems.
Fig. 6.
Proposed model for the action of ADAP1. KIF13B transports ADAP1 along microtubules in dendrites (i) until it reaches regions of high PIP3 or PI(3,4)P2 concentration. Here, ADAP1 is released from KIF13B (ii) and recruited to the membrane (iii), where it stimulates GTP hydrolysis and membrane detachment of ARF6 (iv).
As many neurological diseases are linked to misregulation of neuronal transport and small GTPase signaling (58, 59), shedding new light on these processes is important. Due to the pleiotropy of PIP3’s functions (60) and the difficulty of imaging ADAP1 under native expression levels for longer times, experiments to disentangle these processes in the living cell are currently challenging. Accordingly, we believe that the in vitro reconstitution approach applied here is important to further dissect the molecular events that shape neurons and better understand their regulation to get a more complete picture of neuronal development.
Materials and Methods
Protein Biochemistry.
Proteins were overexpressed in either SF9 insect cells or BL21 Escherichia coli cells and purified by immobilized metal-affinity chromatography and size-exclusion chromatography. Sortase or NMT1 was used to attach dyes or lipid modifications. Enzyme kinetics (30) and nucleotide extractions (61) are based on previously published protocols. Protein–protein and protein–lipid interaction experiments were performed essentially as described before (3, 17, 24, 61). See SI Appendix, Methods for details and modifications.
Microscopy.
Microscopy experiments were performed on an Olympus IX83 or an iMIC TIRF microscope with a 100× 1.49 numerical aperture oil objective on functionalized glass coverslips (62), where biotinylated microtubules were attached via neutravidin. Fluorescent proteins or fluorescent lipids were then imaged in TIRF mode by time-lapse microscopy in the presence of an oxygen scavenger system.
Analysis.
Numerical values from experiments were extracted with the appropriate software (e.g., ImageJ for microscopy movies) and subsequent analysis and data visualization were performed in Python as detailed in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Urban Bezeljak, Natalia Baranova, Mar Lopez-Pelegrin, Catarina Alcarva, and Victoria Faas for sharing reagents and helpful discussions. We thank Veronika Szentirmai for help with protein purifications. We thank Carrie Bernecky, Sascha Martens, and the M.L. lab for comments on the manuscript. We thank the bioimaging facility, the life science facility, and Armel Nicolas from the mass spec facility at the Institute of Science and Technology (IST) Austria for technical support. C.D. acknowledges funding from the IST fellowship program; this work was supported by Human Frontier Science Program Young Investigator Grant RGY0083/2016.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.J.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010054118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Schelski M., Bradke F., Neuronal polarization: From spatiotemporal signaling to cytoskeletal dynamics. Mol. Cell. Neurosci. 84, 11–28 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Stricker R., Reiser G., Functions of the neuron-specific protein ADAP1 (centaurin-α1) in neuronal differentiation and neurodegenerative diseases, with an overview of structural and biochemical properties of ADAP1. Biol. Chem. 395, 1321–1340 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Tong Y., et al. , Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1. Proc. Natl. Acad. Sci. U.S.A. 107, 20346–20351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkateswarlu K., Oatey P. B., Tavaré J. M., Jackson T. R., Cullen P. J., Identification of centaurin-alpha1 as a potential in vivo phosphatidylinositol 3,4,5-trisphosphate-binding protein that is functionally homologous to the yeast ADP-ribosylation factor (ARF) GTPase-activating protein, Gcs1. Biochem. J. 340, 359–363 (1999). [PMC free article] [PubMed] [Google Scholar]
- 5.Borrmann C., Stricker R., Reiser G., Tubulin potentiates the interaction of the metalloendopeptidase nardilysin with the neuronal scaffold protein p42IP4/centaurin-α1 (ADAP1). Cell Tissue Res. 346, 89–98 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Venkateswarlu K., Hanada T., Chishti A. H., Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J. Cell Sci. 118, 2471–2484 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Moore C. D., et al. , The neuronal Arf GAP centaurin alpha1 modulates dendritic differentiation. J. Cell Sci. 120, 2683–2693 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sztul E., et al. , ARF GTPases and their GEFs and GAPs: Concepts and challenges. Mol. Biol. Cell 30, 1249–1271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernández-Deviez D. J., Casanova J. E., Wilson J. M., Regulation of dendritic development by the ARF exchange factor ARNO. Nat. Neurosci. 5, 623–624 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki H., et al. , The small GTPase ADP-ribosylation factor 6 negatively regulates dendritic spine formation. FEBS Lett. 579, 6834–6838 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kim Y., et al. , ADP-ribosylation factor 6 (ARF6) bidirectionally regulates dendritic spine formation depending on neuronal maturation and activity. J. Biol. Chem. 290, 7323–7335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka K., et al. , A target of phosphatidylinositol 3,4,5-trisphosphate with a zinc finger motif similar to that of the ADP-ribosylation-factor GTPase-activating protein and two pleckstrin homology domains. Eur. J. Biochem. 245, 512–519 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Thacker E., et al. , The Arf6 GAP centaurin α-1 is a neuronal actin-binding protein which also functions via GAP-independent activity to regulate the actin cytoskeleton. Eur. J. Cell Biol. 83, 541–554 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Venkateswarlu K., Brandom K. G., Lawrence J. L., Centaurin-α1 is an in vivo phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating protein for ARF6 that is involved in actin cytoskeleton organization. J. Biol. Chem. 279, 6205–6208 (2004). [DOI] [PubMed] [Google Scholar]
- 15.East M. P., Kahn R. A., Models for the functions of Arf GAPs. Semin. Cell Dev. Biol. 22, 3–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Insolera R., Chen S., Shi S. H., Par proteins and neuronal polarity. Dev. Neurobiol. 71, 483–494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horiguchi K., Hanada T., Fukui Y., Chishti A. H., Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J. Cell Biol. 174, 425–436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimura Y., Terabayashi T., Miki H., Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol. Cell Biol. 30, 2206–2219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada K. H., Hanada T., Chishti A. H., The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry 46, 10039–10045 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanck T., et al. , Recombinant p42IP4, a brain-specific 42-kDa high-affinity Ins(1,3,4,5)P4 receptor protein, specifically interacts with lipid membranes containing Ptd-Ins(3,4,5)P3. Eur. J. Biochem. 261, 577–584 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Huckaba T. M., Gennerich A., Wilhelm J. E., Chishti A. H., Vale R. D., Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J. Biol. Chem. 286, 7457–7467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S. X., Duan L. H., He S. J., Zhuang G. F., Yu X., Phosphatidylinositol 3,4-bisphosphate regulates neurite initiation and dendrite morphogenesis via actin aggregation. Cell Res. 27, 253–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler A. B., Tavosanis G., Glycerophospholipids—Emerging players in neuronal dendrite branching and outgrowth. Dev. Biol. 451, 25–34 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Lu Q., Yu J., Yan J., Wei Z., Zhang M., Structural basis of the myosin X PH1(N)-PH2-PH1(C) tandem as a specific and acute cellular PI(3,4,5)P(3) sensor. Mol. Biol. Cell 22, 4268–4278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy N. S., Yohe M. E., Randazzo P. A., Gruschus J. M., Allosteric properties of PH domains in Arf regulatory proteins. Cell. Logist. 6, e1181700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo R., et al. , Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 402, 439–447 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campa F., et al. , A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J. Biol. Chem. 284, 28069–28083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kam J. L., et al. , Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. Evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 275, 9653–9663 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Dell’Angelica E. C., et al. , GGAs: A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–94 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padovani D., Zeghouf M., Traverso J. A., Giglione C., Cherfils J., High yield production of myristoylated Arf6 small GTPase by recombinant N-myristoyl transferase. Small GTPases 4, 3–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonny B., Huber I., Paris S., Chabre M., Cassel D., Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 272, 30848–30851 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Ismail S. A., Vetter I. R., Sot B., Wittinghofer A., The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism. Cell 141, 812–821 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Cherfils J., Zeghouf M., Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Michaelis L., Menten M. L., Johnson K. A., Goody R. S., The original Michaelis constant: Translation of the 1913 Michaelis–Menten paper. Biochemistry 50, 8264–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kjos I., Vestre K., Guadagno N. A., Borg Distefano M., Progida C., Rab and Arf proteins at the crossroad between membrane transport and cytoskeleton dynamics. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1397–1409 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Hansen A. H., Duellberg C., Mieck C., Loose M., Hippenmeyer S., Cell polarity in cerebral cortex development—Cellular architecture shaped by biochemical networks. Front. Cell. Neurosci. 11, 176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins B., Decker H., Bentley M., Luisi J., Banker G., A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J. Cell Biol. 198, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Schwarz T. L., The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kevenaar J. T., et al. , Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 26, 849–861 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Gibbs K. L., Greensmith L., Schiavo G., Regulation of axonal transport by protein kinases. Trends Biochem. Sci. 40, 597–610 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Tsai M. Y., Morfini G., Szebenyi G., Brady S. T., Release of kinesin from vesicles by hsc70 and regulation of fast axonal transport. Mol. Biol. Cell 11, 2161–2173 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi S. H., Jan L. Y., Jan Y. N., Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112, 63–75 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Jiang H., Guo W., Liang X., Rao Y., Both the establishment and the maintenance of neuronal polarity require active mechanisms: Critical roles of GSK-3β and its upstream regulators. Cell 120, 123–135 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Xiao Q., Hu X., Wei Z., Tam K. Y., Cytoskeleton molecular motors: Structures and their functions in neuron. Int. J. Biol. Sci. 12, 1083–1092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarbashevich K., Dzementsei A., Pieler T., A novel function for KIF13B in germ cell migration. Dev. Biol. 349, 169–178 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Fabrini R., et al. , Monomer-dimer equilibrium in glutathione transferases: A critical re-examination. Biochemistry 48, 10473–10482 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Bentley M., Decker H., Luisi J., Banker G., A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J. Cell Biol. 208, 273–281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson A. C., Humphreys D., Brooks A. B. E., Hume P. J., Koronakis V., The Arf GTPase-activating protein family is exploited by Salmonella enterica serovar Typhimurium to invade nonphagocytic host cells. MBio 6, e02253-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitale N., et al. , GIT proteins, a novel family of phosphatidylinositol 3,4,5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 275, 13901–13906 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Jackson T. R., et al. , ACAPs are Arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151, 627–638 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ricci F., Vallée-Bélisle A., Plaxco K. W., High-precision, in vitro validation of the sequestration mechanism for generating ultrasensitive dose-response curves in regulatory networks. PLoS Comput. Biol. 7, e1002171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernández-Deviez D. J., Roth M. G., Casanova J. E., Wilson J. M., ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol. Biol. Cell 15, 111–120 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda A., et al. , Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99, 521–532 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Ménager C., Arimura N., Fukata Y., Kaibuchi K., PIP3 is involved in neuronal polarization and axon formation. J. Neurochem. 89, 109–118 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Zemlickova E., et al. , Centaurin-alpha(1) associates with and is phosphorylated by isoforms of protein kinase C. Biochem. Biophys. Res. Commun. 307, 459–465 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Li J., Lambright D. G., Hsu V. W., Coordination of Grp1 recruitment mechanisms by its phosphorylation. Mol. Biol. Cell, E20030173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huisman S. M., Brunner D., Cell polarity in fission yeast: A matter of confining, positioning, and switching growth zones. Semin. Cell Dev. Biol. 22, 799–805 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Gabrych D. R., Lau V. Z., Niwa S., Silverman M. A., Going too far is the same as falling short: Kinesin-3 family members in hereditary spastic paraplegia. Front. Cell. Neurosci. 13, 419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevalier-Larsen E., Holzbaur E. L. F., Axonal transport and neurodegenerative disease. Biochim. Biophys. Acta 1762, 1094–1108 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Raghu P., Joseph A., Krishnan H., Singh P., Saha S., Phosphoinositides: Regulators of nervous system function in health and disease. Front. Mol. Neurosci. 12, 208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manser E., Leung T., Eds., GTPase Protocols: The Ras Superfamily (Springer Science & Business Media, 2002). [Google Scholar]
- 62.Bieling P., Telley I. A., Hentrich C., Piehler J., Surrey T., Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 95, 555–580 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.