Abstract
The biological reason(s) behind persistent mother-to-child transmission (MTCT) of HIV (albeit at reduced rate compared to the preantiretroviral therapy era) in spite of the successful implementation of advanced control measures in many African countries remains a priority concern to many HIV/AIDS control programs. This may be partly due to differences in host immunogenetic factors in highly polymorphic regions of the human genome such as those encoding the killer-cell immunoglobulin-like receptor (KIR) molecules which modulate the activities of natural killer cells. The primary aim of this study was to determine the variants of KIR genes that may have a role to play in MTCT in a cohort of infants born to HIV-infected mothers in Yaoundé, Cameroon. We designed a cross-sectional study to molecularly determine the frequencies of 15 KIR genes in 14 HIV-exposed infected (HEI), 39 HIV-exposed/uninfected (HEU), and 27 HIV-unexposed/uninfected (HUU) infants using the sequence specific primer polymerase chain reaction (PCR-SSP) method. We found that all 15 KIR genes were present in our cohort. The frequency of KIR2DL1 was significantly higher in the unexposed (control) group than in the HIV-exposed group (OR = 0.22, P = 0.006). Stratifying analysis by infection status but focusing only on exposed infants revealed that KIR2DL5, KIR2DS1, and KIR2DS5 were significantly overrepresented among the HIV-exposed/uninfected compared to infected infants (OR = 0.20, P = 0.006). Similarly, the frequencies of KIR2DS1, KIR2DS5, and KIR2DL5 were significantly different between infants perinatally infected with HIV (HIV+ by 6 months of age) and HIV-negative infants. Our study demonstrates that KIR genes may have differential effects with regard to MTCT of HIV-1.
1. Introduction
Infection with HIV can be considered a breach to both the innate and adaptive immune systems [1]. NK cells, a key component of the cellular arm of the innate immune response to invasive pathogens, constitute the front line of defence against viral infections including HIV. They operate through two main mechanisms: cytotoxic destruction (using perforin, granzymes, and tumor necrosis factor (TNF)) and release of cytokines that regulate the adaptive arm of the immune system [2]. NK cells can specifically target stress ligands expressed on HIV-infected host cells leading to the autolysis of the infected cell. Furthermore, secretion of chemokines by NK cells in response to HIV infection can block new cycles of infection [3]. The critical role of NK cells in HIV infection control is also evidenced by the expansion of the NK cell population and the change in distribution of NK cell subsets in acute HIV infection [4]. NK cells represent 2-10% of all the leukocytes in the blood [1]. However, they express a variable repertoire of activating and inhibitory receptors on their cell surface including NKG2D, NKp46, CD94/NKG2A, and killer-cell immunoglobulin-like receptors (KIRs) which are the most characterized and representative of them [5].
KIRs are highly polymorphic and are capable of triggering both activating and inhibitory signals in the carrier NK cell to effect its natural killing function. Such regulation affects both innate and adaptive immunities, particularly with regard to antiviral responses to HIV and many other viruses. To date, 16 KIR genes, 1110 alleles, and more than 50 haplotypes have been identified in different populations (KIR release 2.9.0 of December 2019) [6]. Among them, are seven genes encoding receptors transmitting activating signals through their short cytoplasmic tails (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5A, KIR2DS5B, and KIR3DS1) and eight genes encoding inhibitory receptors with long cytoplasmic tails (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5A, KIR2DL5B, KIR3DL1, KIR3DL2, and KIR3DL3). The exception is with KIR2DL4 which has both activating and inhibitory functions [7, 8]. Framework genes KIR3DL3, KIR2DL4, KIR3DL2, and KIR3DP1 are present in almost all human genomes, with rare exceptions [9]. KIR genes are located in 15 different loci on chromosome 19q13.4, and some of them are allelic variants, KIR3DL1/S1, KIR2DL2/L3, and KIR2DS3/S5 [10, 11]. Various evolutionary forces namely the response to pathogens has primarily contributed to the diversity in the repertoire of KIR genes ([12], p. 2). The importance of polymorphism in KIR genes in HIV infection has only been reported in a few populations worldwide [13, 14].
Africa is the most affected region by HIV/AIDS in the world, with young women disproportionally affected. In 2018, close to 37.9 million people were living with HIV, of whom 1.7 million were children and adolescents under the age of 15 years, the majority of whom (90%) were in sub-Saharan Africa [15]. In 2018, Cameroon had 23,000 (22,000-41,000) new HIV infections and 18,000 (25,000-33,000) AIDS-related deaths [16]. The estimated number of new paediatric HIV infections was 4500 in 2017, and the vertical transmission rate was 13% in breastfed infants [17]. Before the wide availability of antiretroviral therapy (ART), the majority of HIV-infected infants would die under the age of 2 years with 33% dying before their first birthday [18].
In Cameroon, preventive measures to curb the burden and transmission of HIV/AIDS have been undertaken such as the use of antiretroviral prophylaxis and the Prevention of Mother-to-Child Transmission (PMTCT) program. Despite these, few neonates still acquire the infection. However, a majority of infants do not acquire the infection or remain uninfected despite breastfeeding from mothers who are not virally suppressed and who are not on any ART regimen [19]. A growing number of reports from other parts of the world have demonstrated that susceptibility to HIV infection may be associated with a variety of different host immunogenetic factors such as the KIRs ([20–22], p. 2; [14]). Heterogeneity in frequencies and distribution of KIR genes, genotypes, and haplotypes among populations may explain their role in HIV acquisition. The current study describes the frequencies and distribution of KIR genes, genotypes, and haplotype profiles in a cohort of HIV-exposed infants with the aim to determine their role in mother-to-child transmission of HIV-1 among Cameroonians.
2. Materials and Methods
This study was nested within the PREVENT-IT study cohort (CIPHER Pediatric HIV Matters project) which was designed to investigate the impact of in utero exposure to Tenofovir on neonatal tubulopathies. We conducted a cross-sectional study in four hospitals in Yaoundé, Cameroon (Green City Hospital, Efoulan, Mbiyem-Assi and CASS Nkoldongo) over a period of one year from April 2018 to May 2019. Recruitment of participants was done through convenience sampling. Sensitization of participants was done in HIV care and maternity units of the various hospitals. Our study population was made up of three groups: HIV-1-positive infants born of HIV-1-positive mothers (exposed-infected: HEI), HIV-1-negative infants born of HIV-1-positive mothers (exposed-uninfected: HEU), and HIV-1-negative infants born of HIV-1-negative mothers (unexposed-uninfected: HUU). Only infants whose parents had given proxy consent were included in the present study. These infants were all of the same age (6 weeks). Mothers of both groups of infants were screened for Hepatitis B Surface Antigen (HBsAg) and Hepatitis C Core Antigen (HCVcAg) using the ELISA technique (QuickTiter, Cell Biolab, INC, San Diego). This study was approved by the Institutional Ethics Committee for Research on Human Health, of the University of Douala, ethical clearance No. 1639IEC-UD/06/2018/T. Administrative authorization was equally obtained from the various collection sites in accordance with the ethical guidelines of the 1975 Declaration of Helsinki.
2.1. Clinical Characteristics of Mothers and Children
Clinical information from the mother-child dyads was obtained using a structured questionnaire. These included feeding practices of the infants which can be artificial or breast, the type of delivery documented as full-term vaginal delivery (FTVD) or caesarean section, and mother's ART regimen, weight, and height.
2.2. Specimen Collection, Storage, DNA Extraction, and HIV Screening
Six weeks after delivery (mother's visit 1), up to 5 ml of whole blood was collected from both mother and child into ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes. Each sample was processed to isolate the peripheral blood mononuclear cells (PBMCs), buffy coat, and plasma isolated within six hours of collection. PBMCs were stored immediately at -20°C for 2 hours and transferred in -80°C until required for further analysis. Each participant was tested for HIV-1, and their viral load was quantified using the Amplicor HIV-1 DNA PCR assay (Roche Diagnostics, Branchburg, NJ) as previously described [23].
2.3. Genotyping of KIR Genes
Genomic DNA was extracted from buffy coat using the QIA amp DNA Blood Mini kit (Qiagen Ltd, Germany) according to the manufacturer's instructions and quantified using the NanoDrop spectrophotometer. KIR genotyping was carried out using sequence specific primer polymerase chain reaction (SSP-PCR) as previously described [24]. The mix was composed of 2 μl of DNA polymerase (PrimeStar GXL, Takara Bio Europe, France), 270 μl of αQH2O, 60 μl of 5X buffer, and 9 μl of dNTPs (10 mM) per sample. Briefly, two pairs of sequence specific primers were used to amplify each of 14 functional KIR genes: 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 3DS1, 3DL1, 3DL2, 3DL3, and the pseudogene 2DP1. Amplicons were electrophoresed in 2% agarose gel and visualized under ultraviolet light for the presence or absence of each gene.
2.4. KIR Genotypes and Haplotypes
KIR genotypes were assigned according to the allele frequency net database (http://www.allelefrequencies.net). In the assessment of the KIR genotypes, group B genotypes were defined by the presence of one or more of the following genes: KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1. Conversely, the stable group A genotype was defined by the absence of all the above-mentioned genes and the presence of KIR3DL1, KIR2DL1, KIR2DL3, and KIR2DS4 genes. KIR haplotypes AA (A) and Bx were assigned as previously described [8]. Briefly, individuals carrying KIR2DL1, KIR2DL3, KIR2DL4, KIR2DS4, and KIR3DL1 genes in addition to the framework genes were assigned the AA haplotype. Individuals carrying all AA haplotype genes and any one of the following genes, KIR2DL2, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1, were denoted as AB haplotype while individuals lacking any of the following, KIR2DL1, KIR2DL3, KIR3DL1, and KIR2DS4, were BB haplotype. Given the difficulties in distinguishing between AB and BB haplotypes, we coded all AB and BB carriers as Bx [25].
2.5. Statistical Analysis
Statistical analyses were done using STATA v16.1. Frequencies of genes, alleles, genotypes, and haplotypes were determined by direct counting. Differences between groups (HUU, HEU, and HEI) were computed using the Kruskal–Wallis test, Chi-squared, or Fisher exact test as may be appropriate. In multivariate models, the Mantel-Haenszel odd ratios were calculated controlling for sex, type of delivery and feeding practice, and adjusted P values reported. P values < 0.05 were considered significant. Correction for multiplicity testing was performed using the Bonferroni method.
3. Results
3.1. Clinical and Demographic Details
Genotyping of KIR genes was performed on 80 infants (14 HIV-exposed infected, 39 HIV-exposed/uninfected born to HIV-infected mothers, and 27 HIV-unexposed/uninfected born to HIV-uninfected mothers). All infants were recruited at the same age (six weeks). The population and clinical characteristics of HEI, HEU, and HUU infants are presented in Table 1. The median viral load of mothers of HEI infants was 7511 copies/ml while that of mothers of HEU infants was 466 copies/ml (P = 0.012). Breastfeeding was the most represented type of feeding. Female infants were overrepresented in the study population (56.4%).
Table 1.
Demographic and clinical characteristics of the study participants.
| Characteristics | Status | ||
|---|---|---|---|
| HUU, n (%) | HEU, n (%) | HEI, n (%) | |
| Number | 27 (33.6) | 39 (48.8) | 14 (17.5) |
| Female | 15 (55.6) | 22 (56.4) | 6 (42.9) |
| Mother parameters | |||
| On ART | — | 35 (89.7) | 14 (100.0) |
| Mean log HIV | — | 7 (8.5) | 6 (9.6) |
| Delivery | |||
| Normal | 25 (92.6) | 35 (89.7) | 12 (85.7) |
| Caesarian | 2 (7.7) | 4 (10.3) | 2 (14.3) |
| Feeding | |||
| Breast | 22 (81.5) | 30 (76.9) | 10 (71.4) |
| Bottle | 4 (14.8) | 8 (20.5) | 4 (28.6) |
| Mixed | 1 (3.7) | 1 (2.6) | 0 (0.0) |
| Other | |||
| Weight in kg, median (IQR) | 5.2 (4.8-5.5) | 5.0 (4.6-5.7) | 5.5 (4.5-6.0) |
| Height in cm, median (IQR) | 55.0 (53.0-56.0) | 53.0 (52.0-56.0) | 54.0 (53.0-58.0) |
| BMI | 1.8 (1.6-1.9) | 1.8 (1.6-1.9) | 1.7 (1.5-2.1) |
ART, antiretroviral therapy; %, percentage; BMI, body mass index.
3.2. Comparative Frequencies of KIR Genes in HUU, HEU, and HEI Infants
We report here the frequencies of 15 KIR genes in an infant Cameroonian cohort (Figure 1). The frequency of individual KIR genes varied from 28.8% to 100.0%. As expected, the framework genes investigated in this study (KIR3DL3, KIR3DL2, and KIR2DL4) were present in all 80 infants. Three inhibitory genes (KIR2DL2, KIR2DL3, and KIR3DL1), and two activating genes (KIR2DS2 and KIR2DS4) were present in >70.0% of the study population. The frequency of KIR2DL1 was significantly higher in the HIV-unexposed/uninfected group compared to their HIV-exposed counterparts, i.e., infants born to HIV+ mothers (85.2% vs. 50.9%, aOR: 0.22, P = 0.006). Interestingly, none of the HEI infants had the activating KIR2DS1 gene which was present in 14 out of 39 (35.9%) HEU and 10 out of 27 (37.0%) HUU infants (Figure 1).
Figure 1.
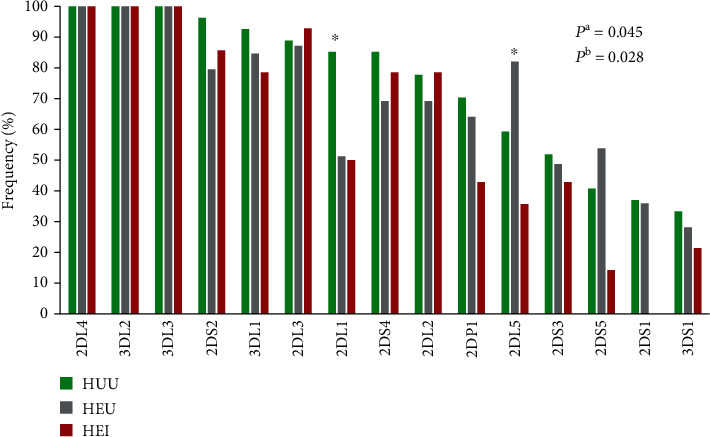
KIR gene frequencies in the study population. P is the P value from the Kruskal–Wallis test comparing all 3 groups. Pa is the P value of KIR2DL1 and Pb is the P value of KIR2DL5.
Comparing infants who acquired HIV-1 through MTCT by 6 weeks of age (n = 14) to their uninfected counterparts of similar age (n = 66), we found that the inhibitory KIR2DL5 and two activating KIR2DS5 and KIR2DS1 were significantly overrepresented in the uninfected group (Table 2), suggesting that these genes may have a role to play in in utero acquisition of HIV-1.
Table 2.
KIR gene frequencies stratified by disease status.
| KIR gene | HIV status | aOR (95% CI) | P | |
|---|---|---|---|---|
| Negative, n (%) | Positive n (%) | |||
| Inhibitory | ||||
| 2DL1 | 43 (65.2) | 7 (50.0) | 0.74 (0.22-2.51) | 0.632 |
| 2DL2 | 48 (72.7) | 11 (78.6) | 1.71 (0.38-7.64) | 0.478 |
| 2DL3 | 58 (87.9) | 13 (92.9) | 1.62 (0.13-20.68) | 0.705 |
| 2DL4 | 66 (100.0) | 14 (100.0) | — | — |
| 2DL5 | 48 (72.7) | 5 (35.7) | 0.31 (0.10 - 0.93) | 0.028 |
| 3DL1 | 58 (87.9) | 11 (78.6) | 0.68 (0.20-2.24) | 0.521 |
| 3DL2 | 66 (100.0) | 14 (100.0) | — | — |
| 3DL3 | 66 (100.0) | 14 (100.0) | — | — |
| Activating | ||||
| 2DS1 | 24 (36.4) | 0 (0.0) | — | 0.023 |
| 2DS2 | 57 (86.4) | 12 (85.7) | 1.16 (0.27-5.09) | 0.843 |
| 2DS3 | 33 (50.0) | 6 (42.9) | 0.82 (0.23-2.92) | 0.762 |
| 2DS4 | 50 (75.8) | 11 (78.6) | 1.75 (0.40-7.59) | 0.451 |
| 2DS5 | 32 (48.5) | 2 (14.3) | 0.14 (0.02-1.11) | 0.029 |
| 3DS1 | 20 (30.3) | 3 (21.4) | 0.70 (0.15-3.31) | 0.647 |
| Pseudogene | ||||
| 2DP1 | 44 (66.7) | 14 (100.0) | 0.45 (0.12 - 1.71) | 0.230 |
The HIV-negative group comprises 66 infants (27 born to HIV-negative mothers and 39 to HIV+ parents). HIV-positive infants n = 14. aOR: the Mantel-Haenszel odds ratio adjusted for sex, type of delivery, and feeding practice. P: adjusted P values.
A total of 53 infants were born to HIV-1-infected mothers who were on a combination ART to prevent MTCT of HIV to their babies in utero. HIV testing at 6 weeks of age revealed that the majority 39 out of 53 infants were free of HIV-1, but 26.4% (14 out of 53) had acquired the infection. In this group of HIV-exposed infants, we observed that the same group KIR genes described above (KIR2DL5, KIR2DS1, and KIR2DS5) were significant overrepresented in the exposed-uninfected group and less frequent in the HEI group (Table 3).
Table 3.
KIR distribution among HIV-exposed infants.
| KIR gene | HIV exposed | aOR (95% CI) | P | |
|---|---|---|---|---|
| Uninfected, n (%) | Infected n (%) | |||
| Inhibitory | ||||
| 2DL1 | 20 (51.3) | 7 (50.0) | 1.18 (0.29-4.75) | 0.814 |
| 2DL2 | 27 (69.2) | 11 (78.57) | 1.89 (0.33-10.68) | 0.461 |
| 2DL3 | 34 (87.2) | 13 (92.86) | 1.78 (0.12-25.44) | 0.668 |
| 2DL4 | 39 (100) | 14 (100.0) | — | — |
| 2DL5 | 32 (82.1) | 5 (35.7) | 0.20 (0.05-0.72) | 0.006 |
| 3DL1 | 33 (84.6) | 11 (78.6) | 0.67 (0.18-2.46) | 0.540 |
| 3DL2 | 39 (100.0) | 14 (100.0) | — | — |
| 3DL3 | 39 (100.0) | 14 (100.0) | — | — |
| Activating | ||||
| 2DS1 | 14 (35.9) | 0 (0.0) | — | 0.022 |
| 2DS2 | 31 (79.5) | 12 (85.7) | 1.77 (0.39-8.10) | 0.453 |
| 2DS3 | 19 (48.7) | 6 (42.9) | 0.89 (0.21-3.84) | 0.875 |
| 2DS4 | 27 (69.2) | 11 (78.6) | 2.05 (0.42-9.98) | 0.366 |
| 2DS5 | 21 (53.9) | 2 (14.3) | 0.06 (0.00-1.09) | 0.009 |
| 3DS1 | 11 (28.2) | 3 (21.4) | 1.04 (0.17-6.41) | 0.965 |
| Pseudogene | ||||
| 2DP1 | 25 (64.1) | 14 (100.0) | 0.60 (0.15-2.49) | 0.482 |
Fifty-three (53) infants were born to HIV-infected mothers, 39 were uninfected by 6 weeks of age, and 14 were HIV infected. aOR: the Mantel-Haenszel odds ratio adjusted for sex, type of delivery, and feeding practice. P: adjusted P values.
A total of 61 distinct KIR genotypes were found in the study population, 6 of which have not been previously reported in public databases.
4. Discussion
Natural killer (NK) cells have the ability to kill virally infected cells without prior sensitisation. They mediate their antiviral activities through a cascade of receptors on their cell surface among which are the killer-cell immunoglobulin-like receptors (KIRs). This study contributes to highlighting further the important role of KIR genes in HIV acquisition. To our knowledge and albeit with a small sample size, this is the first report of KIR diversity in HIV-exposed infants in any sub-Saharan Africa region. The aim of the present study was to determine which of the individual KIR genes or genotypes may be associated with MTCT in a cohort of infants born to HIV+ mothers in Yaoundé, Cameroon. We recruited 80 infants, 53 of whom were born to HIV-infected mothers in combination with ART and 27 born to HIV-negative mothers as an infant population control group. Of the 53 HIV-exposed infants, 14 (26.4%) tested HIV positive at 6 weeks of age (Table 1). DNA was extracted from each consented participant and used for KIR genotyping as described in “Materials and Methods.” Six activating genes (2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1), seven inhibitory genes (2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 3DL2, and 3DL3) in addition to 2DL4 (that has both inhibitory and activating properties), and a pseudogene 2DP1 were investigated in this study population (Figure 1). Many studies have shown that several factors associated with mothers such as viral load and ART uptake can influence the vertical transmission of HIV [26]. However, several other factors related to the child may also be involved in HIV acquisition or resistance. KIR molecules are critical regulators of NK cell effector function and may potentially influence HIV acquisition through NK cell-based innate anti-HIV immune activity [25]. KIR3DS1 and KIR2DS1 were the least frequent in our studied population (Figure 1) and corroborate with findings from other African studies [25, 27]. KIR2DS1 was absent in the HIV-1-infected group but present in 37.0% of the unexposed/uninfected infants (population control group) (Table 2). This finding is difficult to explain but could be suggestive of a role for KIR2DS1 in protecting against in utero acquisition of HIV-1. But given the small sample size, our finding should be interpreted with caution as the significance only became marginal in multivariate models adjusting for sex, delivery mode, feeding practices, and correction for multiple testing using the Bonferroni technique.
We observed consistently that the frequencies of activating KIR2DS1, KIR2DS5, and inhibitory KIR2DL5 genes were higher in the HIV-unexposed/uninfected and the HIV-exposed/uninfected groups compared to infants with perinatally acquired HIV infection (HEI) (Figure 1). Further studies are warranted with larger sample sizes to confirm these findings. KIR2DS1 was the second least frequent gene (30.0%) in this Cameroonian study population. It has previously been reported to be associated with a low viral load in a Zimbabwean study [25]. We observed that KIR2DS1 was only present in infants born to mothers with a low viral load (Table 3). This result corroborate with those of an adult study of phenotypic and functional characterization of natural killer cells in antiretroviral naive HIV+ patients in Cameroon [27]. NK activity is often more pronounced in infected individuals than in healthy population, and it is thought that the innate ability to resist viral infection is linked to activating genes [13]. Contrary to these expectations, we observed that inhibitory KIR2DL1 and KIR2DL5 genes may also be involved in resisting perinatal acquisition of HIV-1 (Figure 1). This is difficult to explain but could be due in part to a low power of inhibition on the activity of NK cells [28]. However, some conflicting reports from Western populations have implicated the 2DL5 gene with both susceptibility to HIV infection [26] and resistance to hepatitis B infection [29]. KIR genes are highly polymorphic and vary across species, race, and ethnicity [30–32], and its activation might also depend on many other factors [26] that are beyond the scope of the present study.
The extensive variation observed in the KIR loci is the origin of the many genotypes and haplotypes reported to date in public databases [33]. We found that the AA genotype was only present in the infected group (Table S1). Some studies have found similar results with higher AA genotype frequency in infected vs. noninfected individuals [25, 26]. In contrast, one study found the AA genotype only in the uninfected population [13]. Further studies with larger sample sizes are warranted to confirm or refute our findings in this and other populations. Investigating the impact of KIR molecules on the function of NK cells in mediating susceptibility to viral infections is important, and we believe our findings will go a long way to help with better design of future studies in our subregion.
5. Conclusion
The present study confirms the diverse nature KIR genes in modulating susceptibility or resistance to viral infections focusing on MTCT of HIV-1 even in this small cohort of Cameroonian population. Our findings suggest that certain KIR genes may have a role to play in in utero and/or perinatal acquisition of HIV-1 irrespective of exposure to combination ART. We found 6 potentially unique KIR genotypes that have not been previously reported.
Acknowledgments
The work was supported by CIPHER Pediatric HIV Matters, Biomedsup Cameroon, and the Biotechnology Center of Yaoundé I. We would like to thank the staff of Green City, Efoulan, Mbiyem-Assi, and CASS Nkoldongo hospitals for their tremendous supports and collaborations.
Data Availability
The clinical, biological, and demographic data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflicting financial interests.
Supplementary Materials
Table S1: represents the entire genotypes found in the study population with their frequencies. Common forms represent genotypes already present in others countries of the world, and novel forms are unique in Cameroon. This table represents also different genes composing each genotype and their frequencies in the study population. We have two types of genotype: AA and Bx. Given the difficulties in distinguishing between AB and BB haplotypes, we coded all AB and BB carriers as Bx.
References
- 1.Abbas A., Lichtman A. H., Pillai S. Cellular and Molecular Immunology. 9th. USA: Elsevier; 2017. [Google Scholar]
- 2.Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nature Immunology. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 3.Scully E., Alter G. NK cells in HIV disease. Current HIV/AIDS Reports. 2016;13(2):85–94. doi: 10.1007/s11904-016-0310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavilio D., Lombardo G., Benjamin J., et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houchins J. P., Yabe T., McSherry C., Bach F. H. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. Journal of Experimental Medicine. 1991;173(4):1017–1020. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson J., Barker D. J., Georgiou X., Cooper M. A., Flicek P., Marsh S. G. E. IPD-IMGT/HLA database. Nucleic Acids Research. 2019;48 doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faure M., Long E. O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. Journal of Immunology. 2002;168(12):6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 8.González-Galarza F. F., Takeshita L. Y. C., Santos E. J. M., et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Research. 2015;43(D1):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niepiekło-Miniewska W., Żuk N., Dubis J., et al. Two new cases of KIR3DP1, KIR2DL4-negative genotypes, one of which is also lacking KIR3DL2. Archivum Immunologiae et Therapiae Experimentalis. 2014;62(5):423–429. doi: 10.1007/s00005-014-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baer J., Jiang K. How does human resource management influence organizational outcomes? A meta-analytic investigation of mediating mechanisms. Consulté. 2019;55 https://www.academia.edu/2370757/How_does_human_resource_management_influence_organizational_outcomes_a_meta-analytic_investigation_of_mediating_mechanisms. [Google Scholar]
- 11.Carrington M., Martin M. P., van Bergen J. _KIR-HLA_ intercourse in HIV disease. Trends in Microbiology. 2008;16(12):620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Borra J., Khakoo S. I. Speed and selection in the evolution of killer-cell immunoglobulin-like receptors. International Journal of Immunogenetics. 2008;35(2):89–96. doi: 10.1111/j.1744-313X.2008.00756.x. [DOI] [PubMed] [Google Scholar]
- 13.Chavan V. R., Ahir S., Ansari Z., et al. Diversity in KIR gene repertoire in HIV-1 exposed infected and uninfected infants: a study from India. Journal of Medical Virology. 2016;88(3):417–425. doi: 10.1002/jmv.24348. [DOI] [PubMed] [Google Scholar]
- 14.Paximadis M., Minevich G., Winchester R., et al. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PloS One. 2011;6(2, article e16541) doi: 10.1371/journal.pone.0016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UNAIDS. Global HIV & AIDS statistics—2019 fact sheet. 2019. https://www.unaids.org/en/resources/fact-sheet.
- 16.UNAIDS. Cameroun. 2018. https://www.unaids.org/fr/regionscountries/countries/cameroon.
- 17.Sofeu C. L., Tejiokem M. C., Penda C. I., et al. Early treated HIV-infected children remain at risk of growth retardation during the first five years of life: results from the ANRS-PEDIACAM cohort in Cameroon. PLoS One. 2019;14(7, article e0219960) doi: 10.1371/journal.pone.0219960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesh K. K., de Bruyn G., Marinda E., et al. Morbidity and mortality among infants born to HIV-infected women in South Africa: implications for child health in resource-limited settings. Journal of Tropical Pediatrics. 2011;57(2):109–119. doi: 10.1093/tropej/fmq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekali G. L., Jesson J., Enok P. B., Leroy V. Effect of in utero exposure to HIV and antiretroviral drugs on growth in HIV-exposed uninfected children: a systematic review and meta-analysis protocol. BMJ Open. 2019;9(6, article e023937) doi: 10.1136/bmjopen-2018-023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballan W. M., Vu B.-A. N., Long B. R., et al. Natural killer cells in perinatally HIV-1-infected children exhibit less degranulation compared to HIV-1-exposed uninfected children and their expression of KIR2DL3, NKG2C, and NKp46 correlates with disease severity. The Journal of Immunology. 2007;179(5):3362–3370. doi: 10.4049/jimmunol.179.5.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chavan V. R., Chaudhari D., Ahir S., Ansari Z., Mehta P., Mania-Pramanik J. Variations in KIR Genes: a study in HIV-1 serodiscordant couples. BioMed Research International. 2014;2014:11. doi: 10.1155/2014/891402.891402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennes W., Verheyden S., Demanet C., et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. Journal of Immunology. 2006;177(10):6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 23.Torimiro J. N., Mafopa N. G., Ndongo A., et al. Evaluation of virologic methods for early detection of HIV-1 in a resource-limited setting: performance and cost analysis. Health Sciences And Disease. 2013;14:p. 6. [Google Scholar]
- 24.Martin M. P., Carrington M. Innate Immunity. 2008. KIR Genotyping and Analysis; pp. 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mhandire K., Zijenah L. S., Yindom L.-M., et al. KIR gene content diversity in a Zimbabwean population: does KIR2DL2 have a role in protection against human immunodeficiency virus infection? Omics: A Journal of Integrative Biology. 2016;20(12):727–735. doi: 10.1089/omi.2016.0154. [DOI] [PubMed] [Google Scholar]
- 26.Zwolińska K., Błachowicz O., Tomczyk T., et al. The effects of killer cell immunoglobulin-like receptor (KIR) genes on susceptibility to HIV-1 infection in the Polish population. Immunogenetics. 2016;68(5):327–337. doi: 10.1007/s00251-016-0906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sake Ngane C. S. Phenotypic and functional characterization of natural killer cells in anti-retroviral naïve HIV-1 infected people in Cameroon. UNIVERSITE DE YAOUNDE I; 2019. [Google Scholar]
- 28.Moesta A. K., Norman P. J., Yawata M., Yawata N., Gleimer M., Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. The Journal of Immunology. 2008;180(6):3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 29.Zhi-ming L., Yu-lian J., Zhao-lei F., et al. Polymorphisms of killer cell immunoglobulin-like receptor gene: possible association with susceptibility to or clearance of hepatitis B virus infection in chinese han population. Croatian medical journal. 2007;48(6):800–807. doi: 10.3325/cmj.2007.6.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook M. A., Moss P. A. H., Briggs D. C. The distribution of 13 killer-cell immunoglobulin-like receptor loci in UK blood donors from three ethnic groups. European Journal of Immunogenetics. 2003;30(3):213–221. doi: 10.1046/j.1365-2370.2003.00394.x. [DOI] [PubMed] [Google Scholar]
- 31.Denis L., Sivula J., Gourraud P.-A., et al. Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens. 2005;66(4):267–276. doi: 10.1111/j.1399-0039.2005.00473.x. [DOI] [PubMed] [Google Scholar]
- 32.Norman P. J., Carrington C. V. F., Byng M., et al. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes & Immunity. 2002;3(2):86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 33.Marsh S. G. E., Parham P., Dupont B., et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55(4):220–226. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- 34.Jennes W., Verheyden S., Demanet C., et al. Low CD4+ T cell counts among African HIV-1 infected subjects with group B KIR haplotypes in the absence of specific inhibitory KIR ligands. PloS One. 2011;6(2, article e17043) doi: 10.1371/journal.pone.0017043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: represents the entire genotypes found in the study population with their frequencies. Common forms represent genotypes already present in others countries of the world, and novel forms are unique in Cameroon. This table represents also different genes composing each genotype and their frequencies in the study population. We have two types of genotype: AA and Bx. Given the difficulties in distinguishing between AB and BB haplotypes, we coded all AB and BB carriers as Bx.
Data Availability Statement
The clinical, biological, and demographic data used to support the findings of this study are included within the article.


