Abstract
Background
Commercial availability of serological tests to evaluate immunoglobulins (Ig) targeting severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has grown exponentially since the start of the coronavirus disease 2019 (COVID-19) outbreak. Thorough validation of these tests is important before use as epidemiological tools to infer seroprevalence in specific populations and as diagnostic tools to complement molecular approaches (e.g., quantitative reverse transcription-polymerase chain reaction).
Methods
Commercial serological tests from 11 suppliers were assayed side-by-side using 126 samples from SARS-CoV-2-infected inpatients and 36 from healthy and HIV-infected individuals.
Results
The majority of the tests assayed have >95% specificity. For the sensitivity calculation, samples were stratified by days since symptoms onset; sensitivity peaks at 16–21 days for IgM and IgA (maximum 91.2%, Euroimmun) and, dependant on the test, at 16–21 or >21 days for IgG (maximum 94.1%, Snibe). Data from semiquantitative tests show that patients with a severe clinical presentation have lower levels of Ig targeting SARS-CoV-2 at <10 days since symptoms onset and higher levels at >21 days, compared to patients with a non-severe presentation.
Conclusions
This study highlights the heterogeneity of sensitivity and generally high specificity of the serological tests and establishes a basis for their usefulness to complement diagnostic techniques and population seroprevalence studies.
Keywords: COVID-19, Serological tests, Qualitative and semiquantitative tests, Sensitivity, Specificity, Clinical presentation
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a large RNA virus from the Coronaviridae virus family that is currently causing global spread of coronavirus disease 2019 (COVID-19) (Lu et al., 2020). Considering the absence of an effective treatment for the SARS-CoV-2 infection, early diagnosis of infection and isolation of infected individuals is critical to control the ongoing pandemic (Gudbjartsson et al., 2020b). Case detection usually involves collection of swab samples from the upper respiratory tract and the amplification of viral nucleic acids sequences by quantitative reverse transcription-polymerase chain reaction (RT-qPCR) (Corman et al., 2020). Recently tests detecting SARS-CoV-2 proteins from swab samples were developed. While viral detection is essential to identify active infection, detection of antibodies against SARS-CoV-2 is a useful complementary surveillance tool. Following SARS-CoV-2 infection, most patients produce detectable immunoglobulins (Ig) targeting a set of viral antigens, particularly the immunodominant nucleocapsid (N) and spike (S) proteins (Guo et al., 2020, Tang et al., 2020, Walls et al., 2020, Zhao et al., 2020). Current evidence suggests that Ig produced against these antigens may confer protection to SARS-CoV-2 infection (Cao et al., 2020, Ju et al., 2020, Poh et al., 2020). However, there is insufficient data on the timing of Ig production following infection. The literature suggests that IgA and IgM can be detected 6–8 days from symptoms onset (Guo et al., 2020, Montesinos et al., 2020, Okba et al., 2020, Padoan et al., 2020, Yongchen et al., 2020), while IgG seroconversion seems to occur slightly later at 9–10 days. However, many patients appear to seroconvert for both IgM and IgG simultaneously, peaking at around 21 days (Liu et al., 2020b, Sun et al., 2020, Yongchen et al., 2020, Zhang et al., 2020). It is not yet confirmed whether Ig levels vary with disease severity.
The performance (sensitivity and specificity) of serological assays can be affected by many variables including: timing of assessment relative to symptoms onset/infection, course of COVID-19 (from asymptomatic to lethal) and, potentially, population and virus genetics (Osório and Correia-Neves, 2020, Peeling et al., 2020). It is vital to evaluate the performance of commercially available serological tests, comparing their performance side-by-side using samples from well-characterised patients with well-defined clinical presentation and distinct time-points from symptoms onset, to be able to select the most appropriate tests for defined populations.
We evaluated the performance of serological tests (3 semiquantitative and 8 qualitative) from 11 suppliers using plasma samples of hospitalised patients with COVID-19 from the Minho region, in the North of Portugal. The particular tests were chosen due to previous reports on their sensitivity, specificity and availability.
Methods
Study population
Patients living in the Minho region of Portugal who were inpatients at Senhora da Oliveira Hospital (Guimarães) or Braga Hospital, admitted with COVID-19 (diagnosed by RT-qPCR at a reference laboratory; at least 2 positive RT-qPCR results were obtained from each patient), were invited to participate in the study. This study was approved by the ethics committees of both participating hospitals (Senhora da Oliveira Hospital: 25/2020; Braga Hospital: 37/2020). An explanation of the project was provided to those individuals and participants signed an informed consent form. The informed consent was prepared according to the Declaration of Helsinki principles, the Oviedo Convention and the General Data Protection Regulation–Regulation (EU) 2016/679. Patients’ blood samples were collected throughout hospitalisation at different time-points following symptoms onset. The number of samples available from each participant varied depending on the duration of their hospitalisation.
For sensitivity calculation, COVID-19 patients were stratified based on the number of days since symptoms onset as follows: <10 days; 10–15 days; 16–21 days; >21 days. Days since symptoms onset were calculated based on each patient's self-report of symptoms manifestation. COVID-19 patients were further categorised according to the severity of their clinical presentation. Patients given oxygen therapy above 10 L/min and/or needing mechanical ventilation (non-invasive or invasive) were considered as having a severe clinical presentation. All other patients (i.e., needing supplementary oxygen therapy below 10 L/min and not requiring mechanical ventilator support) were considered as having a non-severe clinical presentation.
SARS-COV-2 non-infected controls were selected from banked human plasma samples from 2 pre-COVID-19 pandemic studies (the first COVID-19 case in Portugal was reported on 2 March 2020): (i) a study with healthy individuals >55 years old (samples collected between April 2019 and January 2020); (ii) a study with HIV-infected patients on antiretroviral therapy (54–60 months; samples collected between January 2016 and August 2018) (Rb-Silva et al., 2019). Matched samples were selected based on COVID-19 patients’ sex and age. Control samples were collected, processed and preserved at −80 °C using a similar protocol as the one used for samples from COVID-19 inpatients (see below).
Data were handled anonymously. Individuals’ sex, age and comorbidities are summarised in Table 1 .
Table 1.
Clinical and demographic characterisation of the cohort.
| COVID-19 patients |
Pre-COVID-19 controls |
||||
|---|---|---|---|---|---|
| All | Clinical presentationa |
Healthy controls | HIV and other viral infections | ||
| Severe | Non-severe | ||||
| n | 89 | 32 | 57 | 25 | 11 |
| Age (years), Median [min;max] | 71 [30;96] | 75 [45;96] | 67 [30;94] | 71 [59;80] | 57 [33;72] |
| Female, n (%) | 51 (57.3) | 16 (50.0) | 35 (61.4) | 13 (52.0) | 3 (27.3) |
| Hypertension, n (%) | 59 (66.3) | 25 (78.1) | 34 (59.6) | n/a | n/a |
| Diabetes, n (%) | 27 (30.3) | 11 (34.4) | 16 (28.1) | n/a | n/a |
| Obesity, n (%)b | 13 (14.6) | 4 (12.5) | 9 (15.8) | n/a | n/a |
| Neoplasia, n (%) | 8 (9.0) | 4 (12.5) | 4 (7.0) | n/a | n/a |
| Autoimmune disease, n (%) | 5 (5.6) | 2 (6.3) | 3 (5.3) | n/a | n/a |
| Active smokers, n (%) | 2 (2.2) | 0 (0.0) | 2 (3.5) | n/a | n/a |
| Corticosteroids (during hospitalization), n (%) | 6 (6.7) | 3 (9.4) | 3 (5.3) | n/a | n/a |
| Other immunosuppressive drugs, n (%) | |||||
| Prior to hospitalization | 5 (5.6) | 3 (9.4) | 2 (3.5) | n/a | n/a |
| During hospitalization | 1 (1.1) | 1 (3.1) | 0 (0.0) | n/a | n/a |
n/a, not applicable.
Please see “Study Population” section on Methods. No differences between groups were observed in the percentages of the clinical conditions, as assessed by the Fisher Exact Probability test.
Obesity defined as body mass index over 30 kg/m2.
Sample processing
From each patient, venous blood was collected into K2EDTA collecting tubes and processed on the same day: blood collecting tubes were centrifuged at 2000×g for 15 min, at 20 °C. Plasma was aliquoted into screw-cap tubes and frozen at −80 °C until tested.
Immunoassays
Semiquantitative (enzyme-linked immune-absorbent assays [ELISA] and chemiluminescence immunoassays [CLIA]) and qualitative assays (lateral flow immunoassays [LFIA]) from 11 different suppliers were tested according to the manufacturer’s instructions (Table 2 ). At least 2 different tests were performed for each sample (Supplementary Tables S1 and S2).
Table 2.
Assayed commercial tests to detect immunoglobulins specific for SARS-CoV-2 infection.
| Assay | Supplier | Product | Catalog no. | Technology | Format | Assay Target | Obs |
|---|---|---|---|---|---|---|---|
| Semiquantitative assays | Abbott Diagnostics | SARS-coV-2 IgG assay | 06R86 | CLIA | IgG | N protein | Requires an Abbott Architect i2000 |
| Euroimmun | anti SARS-CoV-2 IgG ELISA kit | EI 2606-9601 G | ELISA | IgG | S1 protein | Requires a regular absorbance microplate reader | |
| anti SARS-CoV-2 IgA ELISA kit | EI 2606-9601 A | ELISA | IgA | ||||
| Snibe Diagnostic | MAGLUMI® 2019-nCoV (SARS-CoV-2) IgM kit | 130219016M | CLIA | IgM | S antigen and N protein | Requires a MAGLUMI chemiluminescence immunoassay system | |
| MAGLUMI® 2019-nCoV (SARS-CoV-2) IgG kit | 130219015M | CLIA | IgG | ||||
| Qualitative assays | Cellex | qSARS-Cov-2 IgG/IgM Cassette Rapid Test | WI5513C | LFIA | IgM/IgG | N and S Proteins | – |
| Getein Biotech, inc | One step test for novel coronavirus (2019-nCoV) IgM/IgG antibody (colloidal gold) | CG2057 | LFIA | Total Ig | N and S Proteins | – | |
| Innovita | 2019-nCoV Ab test (colloidal gold), IgM/IgG whole blood/Serum/Plasma Combo | n/a | LFIA | IgM/IgG | N and S Proteins | – | |
| Liming Bio | StrongStep® SARS-CoV-2 IgM/IgG Test | 502090 | LFIA | IgM/IgG | Not specified | Extremely faint bands. | |
| Leccurate | SARS-CoV-2 antibody test (colloidal gold immunochromatography) |
K-20-RC-CoV-2 | LFIA | IgM/IgG | Not specified | – | |
| Medomics | Rapid IgM-IgG Combined Antibody Test kit for SARS-CoV-2 (ICA) | n/a | LFIA | IgM/IgG | Not specified | – | |
| Render | COVID-19 IgM & IgG Test (immunocromatography) | 3.2.01.0.1701 | LFIA | IgM/IgG | Not specified | – | |
| SD Biosensor | Standard Q COVID-19 IgM/IgG Duo Test | Q-NCOV-01D | LFIA | IgM/IgG | N Protein | – |
CLIA; Chemiluminescence Immunoassay; ELISA: Enzyme Linked Immuno-Sorbent Assay; Ig: Immunoglobulin; LFIA: Lateral Flow Immunoassay; n/a: not available; N: Nucleocapsid; S: Spike.
Data analysis
For each test, specificity was calculated as the percentage of negative tests among the pre-COVID-19 controls, and sensitivity as the percentage of positive tests among the SARS-CoV-2 confirmed cases. Sensitivity was calculated upon stratification in days since symptoms onset (<10, 10–15; 16–21 and >21 days). Whenever the same patient was tested multiple times within the same time range during the course of the disease the results of only 1 plasma sample were considered to calculate sensitivity (Supplementary Table S1). The 95% confidence intervals for sensitivity and specificity were calculated using the Wilson score with continuity correction method. Positive predictive values (PPV) and negative predictive values (NPV) were calculated for 3 prevalence scenarios, as described elsewhere (Tenny and Hoffman, 2020): 3% seroprevalence in a general population, which is the estimated prevalence for the Portuguese population (Rodrigues, 2020); 50% seroprevalence in a high-risk sub-population; and 95% seroprevalence in hospitalised patients suspected for COVID-19. Tests’ agreement was evaluated using Cohen’s Kappa. For the semiquantitative tests, receiver-operator characteristic (ROC) curves were constructed and used to calculate the area under the curve (AUC) of the different serologic tests. All variables analysed had a non-normal distribution, as verified by Shapiro–Wilk tests. Comparisons of the relative amounts of Ig at the various time ranges were performed using the Kruskal–Wallis test, followed by Dunn’s post-hoc tests. Comparisons of the relative amount of Ig in the groups of patients with severe versus non-severe clinical presentation were performed using the Mann–Whitney U test. Comparison of demographic and clinical conditions percentages between groups of patients, with severe and non-severe clinical presentation, were performed using Fisher’s Exact Probability Test.
Differences were considered significant when P < 0.05. Statistical analyses were performed on GraphPad Prism version 8.4.3 (La Jolla, San Diego, CA, USA).
Results
Study population
This study includes 89 inpatients infected with SARS-CoV-2 (diagnosed and re-confirmed during hospitalisation by RT-qPCR). Plasma samples from 40 individuals were tested at different time-points during hospitalisation (Supplementary Table S1). Most participants were women (57%) with a median age of 71 years (Table 1). None of the COVID-19 patients were HIV-positive or had a history of organ transplantation.
Performance of tests
Sensitivity and specificity
Serologic tests from 11 suppliers were assayed for their performance. In the IgM and IgG tests combined, specificity ranges from 95.8% to 100.0%, except for Cellex and Snibe (76.0% and 88.6%, respectively). In 4/10 tests assessing IgG and 5/9 tests assessing IgA or IgM specificity is 100.0% (Table 3 and Supplementary Figure S1). From the 36 samples used as negative controls (collected pre-COVID-19 pandemic), 10 are positive for at least 1 test, 5 of which are positive in more than 1 test (Supplementary Table S2). In the semiquantitative tests (Abbott, Euroimmun and Snibe) 2 of the negative control samples are positive in the Snibe IgG test but have a value close to the negative-to-positive cut-off value (1.124 and 1.652 AU/mL; cut-off 1.000 AU/mL; Supplementary Figure S2 and Supplementary Table S2).
Table 3.
Sensitivity and specificity of the assayed tests to detect immunoglobulins specific for SARS-CoV-2 infection. Sensitivity was evaluated in COVID-19 patients upon stratification by days since symptoms onset. Each colour gradient refers to higher (darker) towards lower (lighter) values of specificity (yellow) or sensitivity (green).
 |
To analyse sensitivity, samples were stratified according to time since symptoms onset (<10, 10–15, 16–21 and >21 days). For each Ig, the lowest sensitivities are observed at <10 days since symptoms onset: the Leccurate IgG test presents the highest sensitivity (71.4%) and Snibe IgM the lowest (33.3%; Table 3 and Supplementary Figure S1). Detection of IgA or IgM reaches maximum sensitivity at 16–21 days in 7/9 assays. The maximum sensitivity for IgG peaks at 16–21 days in 50% of the tests, and >21 days in the other 50% (Abbott, Cellex, Innovita, Leccurate, and Render). The Getein test, which detects total Ig targeting SARS-CoV-2, has a peak detection at 16–21 days. For the combined Ig classes, at >21 days, the Euroimmun test (IgA and IgG) provides the highest sensitivity (91.4%), followed by the Snibe test (IgM and IgG, 90.6%). The combined IgM and IgG Leccurate test provides the lowest sensitivity value (75.0%; Table 3 and Supplementary Figure S1).
Positive and negative predictive values
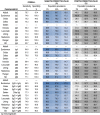
PPV and NPV vary according to the estimated disease seroprevalence. For the assayed tests, 3 distinct population seroprevalence scenarios were tested, from low (3% considered to be the prevalence in the general population and the estimate for the Portuguese population (Rodrigues, 2020)) to high (95%, for a hospitalised setting). To estimate the adjusted PPV and NPV, the overall sensitivity of each test was calculated (i.e., without stratification by days since symptoms onset); Abbot, Euroimmun, Snibe and Cellex present the highest overall sensitivity for the detection of IgG, ranging from 74.3% to 76.7% (Table 4 ). Detection of IgA by the Euroimmun test has an overall sensitivity of 84.9%, and Cellex and SD for IgM of 78.6% and 77.1%, respectively; all the other tests detecting IgM have overall sensitivities <69.4%. For combined detection of IgG and IgA or IgM, semiquantitative tests present the highest overall sensitivity (Euroimmun 86.5%; Snibe 80.3%); all the qualitative tests have an overall sensitivity lower than 78.6% (Table 4). The estimated NPV in a low seroprevalence scenario (3%) varies from 98.6% to 99.6%, dropping drastically in a high seroprevalence scenario (95%) to 10.1%–27.5%. The estimated PPV reached 100% for some tests, irrespective of prevalence. The estimated PPV is >98.4% in high prevalence scenarios, >76.6% in intermediate prevalence scenarios and >9.2% in low prevalence scenarios. Euroimmun, Innovita, Leccurate, Liming, Medomics, Render and SD have PPV of 100%, independent of the Ig type detected; in the combined tests, only Liming and Render have 100% PPV (Table 4).
Table 4.
Negative and positive predictive values (PPV and NPV, respectively) of assayed commercial tests to detect immunoglobulins specific for SARS-CoV-2 infection. PPV and NPV are adjusted for different population prevalence scenarios (3%, 50% and 95%), as described in the “Data analysis” section of material and methods. Each colour gradient refers to higher (darker) towards lower (lighter) values of NPV (blue) or PPV (grey).
 |
Tests result agreement
The 2 tests with the overall highest sensitivity (Euroimmun and Snibe; Table 4) show a moderate agreement index (Cohens’ Kappa = 0.723; Table 5 ). Euroimmun has the best agreement with SD (Cohens’ Kappa = 0.827), and Snibe correlates the best with Getein (Cohens’ Kappa = 0.874). For semiquantitative tests, most of the samples from COVID-19 inpatients with contradicting results present values far from the negative-to-positive cut-off values and therefore do not represent borderline positive results (Supplementary Figure S2).
Table 5.
Cohen’s kappa coefficient for the strength of agreement between assayed tests to detect immunoglobulins specific for SARS-CoV-2 infection. Colour gradient refers to higher (darker) towards lower (lighter) values Cohen’s kappa coefficient (purple).
 |
Recipient-operating characteristic (ROC) curves
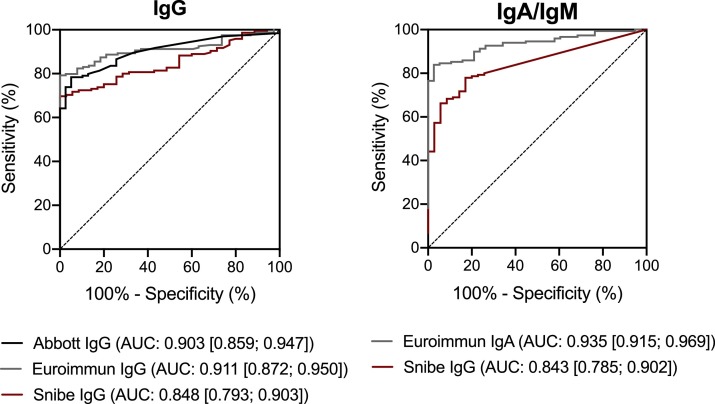
Analysis of the ROC curves of the semiquantitative tests reveals that Euroimmun is the test that best distinguishes SARS-CoV-2 non-infected controls from the SARS-CoV-2 confirmed cases, both for IgA and IgG (AUC [IgG] = 0.911; AUC [IgA] = 0.935; Figure 1 ).
Figure 1.
Receiver operator characteristic (ROC) curves of the assayed semiquantitative tests. AUC: Area Under the Curve. Ig: Immunoglobulin.
Comparison of immunoglobulins levels in COVID-19 inpatients with distinct clinical presentation
To further understand the immune response profile to SARS-CoV-2, Ig levels were associated with clinical presentation. COVID-19 inpatients were classified as having a severe or non-severe clinical presentation based on the need, or not, for oxygen therapy above or below 10 L/min, respectively, as specified in the Methods section. When comparing COVID-19 patients with a severe or non-severe clinical presentation, no differences were observed on their clinical and demographic characteristics (i.e., gender, hypertension, diabetes, obesity, neoplasia, autoimmune disease, smoking habits, and administration of corticosteroids or other immunosuppressive drugs) as assessed by the Fisher Exact Probability test (Table 1).
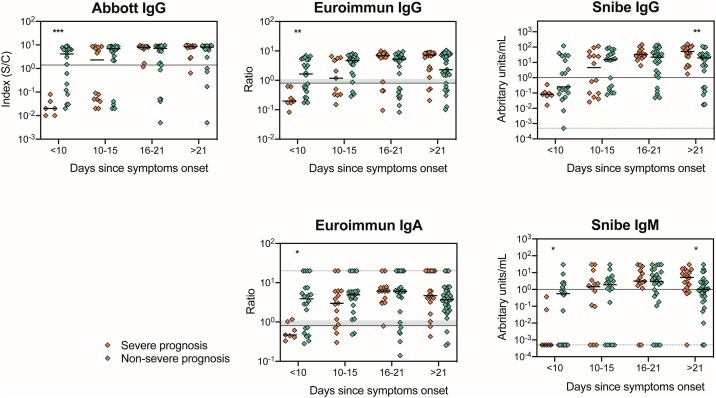
Independent of clinical presentation, all Ig measured in the semiquantitative tests are detected at <10 days since symptoms onset. IgM and IgG plateau from 16–21 days forwards, and a peak in the relative amount of IgA is observed at 16–21 days since symptoms onset (Supplementary Figure S3). When comparing COVID-19 inpatients with a severe versus a non-severe clinical presentation, IgG tends to be lower in the severe group at <10 days since symptoms onset, reaching statistical significance for Abbot and Euroimmun (Abbott: U = 7.5, P = 0.0006; Euroimmun IgG: U = 23.5, P = 0.0045; Figure 2 ). Euroimmun IgA and Snibe IgM also present lower relative amounts in the severe group at <10 days since symptoms onset when compared with the non-severe group (Euroimmun IgA: U = 29.5, P = 0.0133; Snibe IgM: U = 33.5, P = 0.0446). For Snibe, both IgM and IgG levels at >21 days are higher in the severe group (Snibe IgM: U = 145.5, P = 0.0106; Snibe IgG: U = 110, P = 0.0064). No differences are observed in the other tests or time ranges analysed (Figure 2).
Figure 2.
Comparison of the Ig levels in patients with a severe and non-severe clinical presentation using the semiquantitative tests Abbott, Euroimmun and Snibe. Each dot represents one sample and the solid thick lines correspond to the group’s median. Solid thin lines represent each test’s cut-off value. In the case of Euroimmun test, the shadowed grey banner refers to borderline values according to the manufacturer (between 0.8 and 1.1). Since the y-axis has a log scale that does not allow the representation of zero, in those situations (Snibe IgG and IgM) arbitrary values were attributed, and a dashed grey line was represented in this value. For values above the detection limit (Euroimmun IgA), a random value of 20 was attributed and represented as a dashed grey line. Groups median were compared using a Mann–Whitney U-tests; significant differences were represented as: * for P < 0.05; ** for P < 0.01; ***, P < 0.001.
Discussion
Serological testing for SARS-CoV-2 infection is a rapid, inexpensive and easy-to-perform diagnostic approach complementary to RT-qPCR and an essential tool to access the presence and profile of Ig against the virus, both individually and at the population level. The present study compares the performance of serological tests for SARS-CoV-2 using well-characterised COVID-19 inpatients with a diverse presentation of clinical severity, at various time-points across the disease course. Many studies exploring the production of Ig against SARS-CoV-2 have been reported, however, few compare different tests applied to the same set of plasma samples. Comparison of test performance, as presented here, is relevant because it depends on inherent population genetic variations, on the time-point in the disease course and on disease severity.
Regardless of the test methodology applied (ELISA, CLIA, LFIA), we observe here, as reported previously, that test sensitivity is dependent on time since symptoms onset. In addition, the combined detection of IgG and IgA/IgM against SARS-CoV-2 leads, in the majority of cases, to a better performance than measurements of a single Ig class, irrespective of the time range evaluated. These observations were previously reported by others and are in agreement with the establishment of the immune response, where IgA, IgM and IgG have different dynamics throughout disease progression (Alter and Seder, 2020, Guo et al., 2020, Liu et al., 2020b, Montesinos et al., 2020, Okba et al., 2020, Padoan et al., 2020, Sun et al., 2020, Yongchen et al., 2020, Zhang et al., 2020). This dynamic nature of test sensitivity needs to be considered when interpreting performance. Inconsistent reporting by manufactures or lack of details about the time-points used to establish test performance may create confusion regarding the sensitivity levels achievable. This has contributed to the recent decision by the US Food and Drug Administration to remove some tests from the Emergency Use Authorization.
Previous reports concluded that N-protein/peptide-based tests achieve better sensitivities than those based on the S-protein/peptide. This difference may result from an earlier immune response against the N-protein/peptide in comparison to the S-protein/peptide or it may be related to the higher specificity of Ig towards the N-protein/peptide (Caini et al., 2020, Kontou et al., 2020, Liu et al., 2020a). It is not possible to compare these results with the present study since 4 of the assayed tests displayed no information regarding the target antigen, 4 targeted both proteins, 2 targeted only the N-protein/peptide and 1 targeted only the S-protein.
This study reports that Ig targeting SARS-CoV-2 are detectable soon after symptoms onset for some patients (3–5 days for IgM, IgA or IgG; e.g., Patients 012, 033 and 070; Supplementary Table S1) while for others detection (with the same tests) occurred much later (>21 days; e.g., Patients 004, 037 and 064; Supplementary Table S1). Several factors might account for these differences; antibody production kinetics during SARS-CoV-2 infection is not yet fully elucidated nor are the factors responsible for the differences in patient response (Chen et al., 2020, Gudbjartsson et al., 2020a, Liu et al., 2020b, Wang et al., 2020, Zhao et al., 2020). Identifying the variable(s) responsible for distinct profiles of seroconversion is necessary to be able to fully understand false-negative results.
Time since symptoms onset is self-reported which might introduce variability taking into consideration the diversity of clinical manifestations of COVID-19 and of symptom perception by each individual. However, we consider that it introduces less variability than time since disease diagnosis by RT-qPCR, which is performed for some patients before symptoms onset and for others several days after.
Despite variability in the overall sensitivity among the assayed tests, the PPV in COVID-19 hospitalised patients when assuming ≥50% seroprevalence is >87.5%, except for Cellex. However, using serological tests as a seroprevalence and surveillance tool when the estimated prevalence is low e.g., 3%, which is the estimated seroprevalence for the Portuguese population (Rodrigues, 2020), is more challenging due to the observed variation. Still, some of the tests are promising in low prevalence scenarios. For the semiquantitative tests, that can only be used in a laboratory, Euroimmun presented the best PPV (mainly IgG) which was expected due to its high specificity. For the rapid diagnostic tests, the combined IgG and IgM from Liming and Render has 100% PPV. The high NPV of these serological tests in low prevalence scenarios reinforces their suitability for use in the general population if the objective is to detect the presence of SARS CoV-2 antibodies in seroprevalence studies. However, for use in seroprevalence studies during a pandemic phase, a highly specific test is required to assure an acceptable PPV in a low seroprevalence population, as well as a reasonable sensitivity. Euroimmun IgG (ELISA) and the Liming IgG and IgM combination (LFIA) meet these criteria. Both tests have 100% PPV and an overall specificity close to 75% on a 3% seroprevalence scenario meaning that all the positive tests will be specific for SARS CoV-2 antibodies and 75% of subjects with Ig against SARS-CoV-2 will be detected (Table 4).
Our results highlight that Ig levels in a hospitalised COVID-19 population are associated with disease severity. Our data suggest a delay in Ig detection in patients with more severe disease presentation; at <10 days since symptoms onset they present relatively lower levels of Ig (IgA, IgM and IgG) compared to patients with non-severe clinical presentation. To our knowledge this has not previously been reported; some studies actually found no variability in Ig production between patients with different clinical presentation in the initial phase of the disease. As has been previously reported (Chen et al., 2020, Gudbjartsson et al., 2020a, Liu et al., 2020b, Wang et al., 2020, Zhao et al., 2020), we observed that patients with severe clinical presentation show higher relative amounts of Ig at later periods since symptoms onset (>21 days). A recent study showed that higher amounts of Ig in patients with a severe clinical presentation are associated with higher neutralising capability (Wang et al., 2020). The association of Ig production with disease severity is of utmost relevance in clinical settings and needs to be further explored in a larger cohort of patients and evaluated longitudinally from the earliest perceived onset of symptoms through the long-term course of the disease.
Our results are based on measurements performed in a specific population of hospitalised COVID-19 patients. Asymptomatic and non-hospitalised COVID-19 patients were not analysed, for this reason, extrapolation of these data to other COVID-19 settings or the general population should be made only cautiously. However, the results provide the basis for an informed selection of a serological test to be assayed and applied in a larger population setting to evaluate the potential value of each test as a complement for COVID-19 diagnosis and to understand the dynamics of Ig production on infection with SARS-CoV-2 or on vaccination.
Data availability
Data will be available from the corresponding author upon justified request.
Funding
This work was funded by National funds, through the Foundation for Science and Technology (FCT) R4COVID (596694995), POCI-01-0145-FEDER-016428, UIDB/50026/2020 and UIDP/50026/2020; and by the projects NORTE-01-0145-FEDER-000013 and NORTE-01-0145-FEDER-000023, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). CN, SR and NV are junior researchers under the scope of the FCT Transitional Rule DL57/2016. JC-G is supported by an FCT PhD grant, in the context the Doctoral Program in Aging and Chronic Diseases (PhDOC; PD/BD/137433/2018); CSS is supported by an FCT PhD grant, in the context of the Doctoral Program in Applied Health Sciences (PD/BDE/142976/2018).
Contribution
C.S-M., C.N., S.R., C.C., J.A.P., P.G.C. and M.C-N. conceptualized the study; C.S-M., C.N., S.R. and M.C-N. designed the experiments; J.C., A.R., M.F., H.S., O.P., A.C., C.C. and P.G.C. recruited the patients and collected the clinical data; C.S-M., C.N., S.R., J.C-G., C.S.S., N.V., P.B-S. and P.A-P. performed the experiments; C.S-M., C.N. and S.R. prepared the figures and performed the statistical analysis; J.A.P., P.G.C. and M.C-N. supervised the study; and all authors discussed the results and contributed to the final manuscript.
Conflict of interest
Getein kits were provided free of charge by the manufacturer. No other conflicts of interest reported.
Acknowledgments
The authors gratefully acknowledge Drs AC Braga and R Menezes (University of Minho) for discussions of the statistical properties of the data, Dr Qi Huan (Nanjing Tembusu New Material Research Institute) for assistance in obtaining the Medomics kits, Dr Lei Wu (INL) for assistance in obtaining the Getein kits, and Hao Tu (Overseas Business, Getein) for providing the Getein kits for this project free of charge. We are thankful to all study participants.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.01.038.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Alter G., Seder R. The power of antibody-based surveillance. N Engl J Med. 2020;383:1782–1784. doi: 10.1056/NEJMe2028079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caini S., Bellerba F., Corso F., Díaz-Basabe A., Natoli G., Paget J. Meta-analysis of diagnostic performance of serological tests for SARS-CoV-2 antibodies up to 25 April 2020 and public health implications. Euro Surveill. 2020;25(23):1–5. doi: 10.2807/1560-7917.ES.2020.25.23.2000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Pan Z., Yue S., Yu F., Zhang J., Yang Y. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther. 2020;5(1) doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Ph D, Helgason A., Ph D, Jonsson H., Ph D Early spread of SARS-Cov-2 in the Icelandic population. N Engl J Med. 2020;382(24):2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kontou P.I., Braliou G.G., Dimou N.L., Nikolopoulos G., Bagos P.G. Antibody tests in detecting SARS-CoV-2 infection. Diagnostics. 2020;10(5):319. doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos I., Gruson D., Kabamba B., Dahma H., Wijngaert S. Van Den, Reza S. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128(April):104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Muller M.A., LI W., Wang C., Geurtsvankessel C.H., Corman V.M. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osório N.S., Correia-Neves M. Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays. Lancet Infect Dis. 2020:1–2. doi: 10.1016/S1473-3099(20)30435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta. 2020;507(April):164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 2020;3099(20):1–5. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y., Chee R.S. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rb-Silva R., Nobrega C., Azevedo C., Athayde E., Canto-Gomes J., Ferreira I. Thymic fuction as a predictor of immune recovery in chronically HIV-infected patients initiating antiretreoviral therapy. Front Immunol. 2019;10:25. doi: 10.3389/fimmu.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.P. INSA-Instituto Nacional de Saúde Dr Ricardo Jorge; 2020. Relatório de apresentação dos resultados preliminares do primeiro inquérito serológico nacional COVID-19.http://www.insa.min-saude.pt/wp-content/uploads/2020/08/ISN_COVID19_Relatorio_06_08_2020.pdf [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenny S., Hoffman M.R. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2020. Prevalence. Available from: https://www.ncbi.nlm.nih.gov/books/NBK430867/ [Google Scholar]
- Walls A.C., Park Y., Tortorici M.A., Wall A., Mcguire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. 281-292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130(1):5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9(May):833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Nie S., Zhang Z., Zhang Z. Longitudinal change of severe acute respiratory syndrome coronavirus 2 antibodies in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):183–188. doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020:1–22. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available from the corresponding author upon justified request.