Abstract
In this study, we showed that a codon optimized version of the spike (S) protein of SARS-CoV-2 can migrate to the cell membrane. However, efficient production of Moloney murine leukemia (MLV) infectious viral particles was only achieved with stable expression of a shorter S version in C-terminal (ΔS) in MLV Gag-pol expressing cells. As compared to transient transfections, this platform generated viruses with a 1000-fold higher titer. ΔS was 15-times more efficiently incorporated into VLPs as compared to S, and that was not due to steric interference between the cytoplasmic tail and the MLV capsid, as similar differences were also observed with extracellular vesicles. The amount of ΔS incorporated into VLPs released from producer cells was high and estimated at 1.25 μg/mL S2 equivalent (S is comprised of S1 and S2). The resulting VLPs could potentially be used alone or as a boost of other immunization strategies for COVID-19.
Keywords: COVID-19, Coronavirus, SARS-CoV-2, VLP, Virus-like particle, Retrovirus, Moloney, Vaccine
1. Introduction
A cluster of severe pneumonia cases emerged in Wuhan in the Chinese province of Hubei in December 2019 and has quickly become a worldwide pandemic. A new virus was later identified as the etiological agent: the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the condition was named coronavirus disease 2019 (COVID-19) by the World Health Organization (Zhou et al., 2020; Zhu et al., 2020b). As of today, January 7th 2021, 88 million people have been infected, and 1.9 million deaths have been recorded (gisaid.org), but these numbers are probably well underestimated. In addition to its severe health threat, COVID-19 has profound socioeconomic consequences (Nicola et al., 2020).
SARS-CoV-2 is the seventh human coronavirus that has been identified so far. HCoV-NL63, HCoV-229E, HCoV-OC43 and HKU1 strains are constantly present in the human population and cause mild common-cold symptoms (Fung and Liu, 2019). The other two, SARS-CoV and the Middle East respiratory syndrome (MERS)-CoV, are similar to SARS-CoV-2 in that they are pathogenic causing acute respiratory disease (Cui et al., 2019). Two epidemics were caused by SARS-CoV and MERS-CoV, respectively: SARS that originated in China in 2002 and MERS which emerged 10 years later in the Middle East. These two viruses did not spread widely as only 8096 cases were reported for SARS-CoV and 2494 for MERS, but they had an exceedingly high mortality rate (9–35 %) as compared to SARS-CoV-2 (Petersen et al., 2020). There have been no new cases of SARS-CoV reported since 2004 although MERS is still endemic in the Middle East (Docea et al., 2020). The three pathogenic coronaviruses are zoonotic and have emerged from bats with dromedary camels, palm civet and most likely pangolin being the intermediary host for MERS-CoV, SARS-CoV and SARS-CoV-2, respectively (Corman et al., 2014; Lam et al., 2020; Lau et al., 2020; Li et al., 2005; Reusken et al., 2016; Zhang et al., 2020). Coronaviruses are single-stranded positive-sense RNA viruses that are composed of four structural proteins: spike (S), nucleocapsid, envelope and membrane (Fung and Liu, 2019). The S protein that is about 180 kDa assembles as a trimer at the virus surface. It is composed of two subunits S1 and S2 that are responsible for the virus attachment and fusion, respectively. MERS uses dipeptidyl peptidase 4 as its receptor, while SARS-CoV and SARS-CoV-2 share the same receptor for entering cells: the angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Letko et al., 2020; Li et al., 2003; Ou et al., 2020; Raj et al., 2013).
Efforts are being made to identify candidate neutralizing antibodies (Nabs) that could block the interaction of SARS-CoV-2 S with its receptor and that could be used for treating infected patients (Marovich et al., 2020; Renn et al., 2020; Smith et al., 2020). Several vaccine strategies for COVID-19 are also intensively pursued, with S protein being the major target (Amanat and Krammer, 2020; Dong et al., 2020; Moore and Klasse, 2020; Moreno-Fierros et al., 2020). These vaccines are produced from different platforms: RNA, DNA, recombinant proteins, viral vector-based or virus-like particles (VLPs), and live attenuated and inactivated viruses (Amanat and Krammer, 2020; Dong et al., 2020; Moore and Klasse, 2020; Moreno-Fierros et al., 2020; Xu et al., 2020).
Vaccines made from RNA, DNA or proteins are usually easier to manufacture than those that are virus-derived, but it is generally accepted that vaccines made of the original virus (attenuated) or from VLPs induce a better immune response (Mohsen et al., 2017). This is an important point to consider as a COVID-19 vaccine ideally should induce high-titer Nabs for a long-lasting period of time.
Results obtained in animals and in humans have shown that both humoral and cellular immune responses can be obtained with different vaccine strategies, and that Nab titers achieved by vaccination in humans were comparable to those measured in the serum of COVID-19 convalescent individuals (Erasmus et al., 2020; Folegatti et al., 2020; Gao et al., 2020; Guebre-Xabier et al., 2020; Jackson et al., 2020; Keech et al., 2020; Logunov et al., 2020; Mercado et al., 2020; Moore and Klasse, 2020; Mulligan et al., 2020; Polack et al., 2020; Ravichandran et al., 2020; Ren et al., 2020; Smith et al., 2020; Walls et al., 2020; Wang et al., 2020; Xia et al., 2021; Yu et al., 2020; Zhu et al., 2020a). Thanks to world global efforts, some vaccines are reaching the commercialization phase (Ledford, 2020; Ledford et al., 2020).
VLPs are produced by the assembly of viral proteins that do not contain genetic material, and that are then unable to replicate. VLPs are advantageous for their immunostimulatory activity: they are highly recognized by antigen-presenting cells and the repetitive arrangement of antigens on their surface is capable of inducing both innate and adaptive immune responses with a high level of Nabs (Mohsen et al., 2017). VLP-based vaccines have already been successfully developed for Human Papilloma Virus, Hepatitis B, E, and A Virus and influenza virus (Mohsen et al., 2017).
The difficulty of developing COVID-19 vaccines in a short period of time is compounded by the major hurdle of creating mass production capacity to deliver the final product for the entire world population. In this study, we have engineered and characterized a Moloney murine leukemia virus (MLV) VLP platform that has the potential for large-scale production of a COVID-19 vaccine.
2. Results
2.1. The SARS-CoV-2 S protein migrates to the cell surface
The production of an MLV-derived VLP COVID-19 vaccine requires the presence of the carried immunogenic molecule (in our case the S protein) at the surface of the producer cell. As coronaviruses assemble at the ER-Golgi intermediate compartment (Fung and Liu, 2019), we had first to verify if the S protein could migrate efficiently to the cell surface. A full-length, codon-optimized S gene as well as a shorter version in which the last 3′ 57 nucleotides are lacking (ΔS) were cloned into an expression vector. The rationale for the construction of the latter is based on the presence of an endoplasmic reticulum retention signal in the cytoplasmic tail of the coronaviruses S protein, and previous reports that a 19-amino acid C-terminal deletion of SARS-CoV S increases the production of MLV or vesicular stomatitis virus (VSV) infectious particles (Fukushi et al., 2005; Giroglou et al., 2004; Howard et al., 2008; Lontok et al., 2004; Petit et al., 2005; Sadasivan et al., 2017; Ujike et al., 2016). After transfection in 293 cells, both S versions were detected by FACS with a very similar intensity at the cell surface, and there was no staining with untransfected cells or cells transfected with a gibbon ape leukemia virus (Galv) envelope plasmid control (Fig. 1 ). These results indicated that S was able to efficiently migrate to the cell membrane and that, in these experimental conditions, the endoplasmic reticulum retention signal did not affect its localization.
Fig. 1.
Expression of S protein at the surface of 293 cells. FACS analysis of untransfected and transiently transfected cells with plasmids encoding the Galv envelope, the full-length S protein, and the ΔS version. S was detected with an anti-S1 antibody. The results shown are representative of three independent experiments.
2.2. Inefficient transient production of infectious recombinant MLV viruses pseudotyped with the SARS-CoV-2 S protein
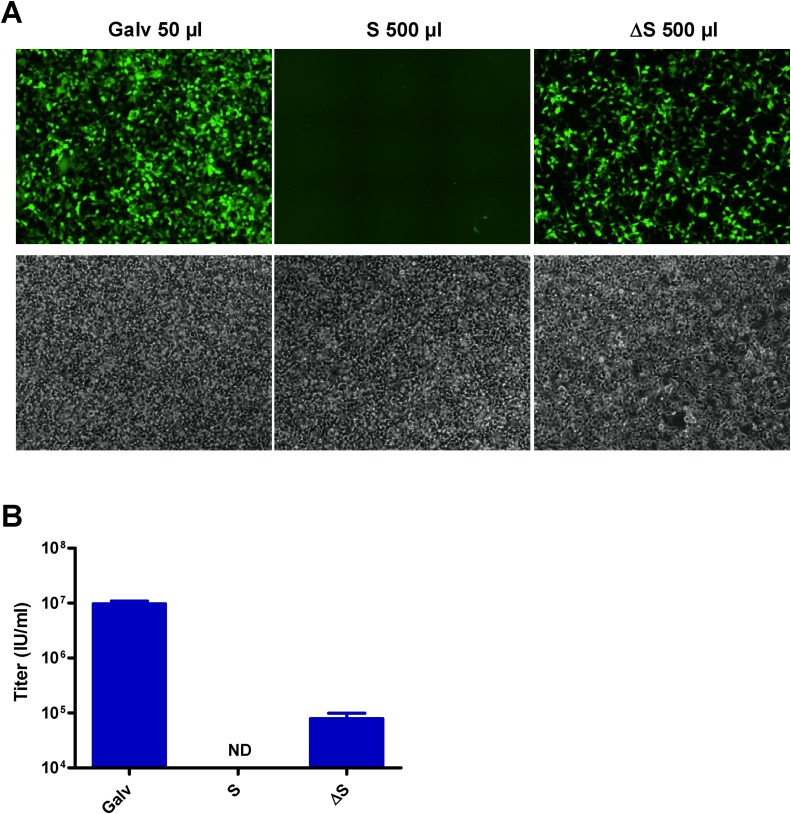
The production of VLPs pseudotyped with S or ΔS (VLP-S) was next assessed by generating GFP retroviral vectors in transient transfections. Titers were measured by FACS analysis on 293-ACE2 cells, a cell line generated by stable transfection that is 52 % positive for ACE2 (Fig. 2 A). Titers of 3.2 × 107 infectious units (IU)/mL and 1.5 × 106 IU/mL were obtained for VSV-G- and the Galv-pseudotyped viruses, although titers of S and ΔS-pseudotyped viruses were below the detection limit of 104 IU/mL (Fig. 2B). Only few GFP cells could be observed by fluorescence microscopy after infection with the ΔS-pseudotyped virus and there were none when the S-pseudotyped vector was used (Fig. 2C). Thus, these results indicated that the transient production was extremely inefficient for generating VLP-S, even with ΔS.
Fig. 2.
Transduction efficiency of different GFP pseudotyped vectors produced in transient transfections. (A) Expression of ACE2 at the surface of 293-ACE2 cells measured by FACS analysis with an anti-ACE2 antibody. The dark grey histogram represents parental 293 cells. The results shown are representative of two independent experiments. Two days after infection of 293-ACE2 cells, titers of VSV-G-, Galv-, S- and ΔS-pseudotyped vectors were (B) measured by FACS analysis or (C) evaluated by fluorescence microscopy. Values presented are the mean ± SD of three independent experiments. Titers of S- and ΔS-pseudotyped viruses were too low to be detected by FACS analysis. Fluorescent and bright-field pictures are displayed. The envelope pseudotype and the volume used for infection are indicated. ND: not determined.
2.3. ΔS-pseudotyped MLV recombinant viral particles are efficiently released from stable producer cells
We have shown that stable retrovirus packaging cell lines can generate Galv-pseudotyped vectors with at least 10-fold higher titers as compared to transient transfection productions (Ghani et al., 2009). We then hypothesized that S or ΔS stably expressed in 293GP cells (293 cells that express MLV Gag-pol) could be a better system to produce VLP-S. Stable populations of 293GP cells expressing S and ΔS were then generated by transfection. In these cells, S and ΔS were able to localize at the cell surface at even higher levels than what we found in transient transfection (Fig. 3 A). A GFP retroviral vector was then introduced in these cells by infection (Fig. 3B), and titers of GFP viruses released by these new producers were measured after infecting 293-ACE2 cells. Only few GFP positive cells could be detected by fluorescence microscopy after infection of 293-ACE2 cells with the S-pseudotyped vector, but a very high percentage of fluorescent cells was observed after infection with the ΔS virus. A high number of GFP positive cells was seen with the Galv virus diluted 10-times as compared to the two other vectors (Fig. 4 A). Titers of 1.6 × 107 IU/mL and 105 IU/mL were measured for the Galv and ΔS-pseudotyped viruses, respectively, and the S-pseudotyped vector titer was below the detection limit of 104 IU/mL, as expected (Fig. 4B). We could conclude that the production of recombinant viral particles was robust from stable producers expressing ΔS and inefficient with the S full-length version of SARS-CoV-2.
Fig. 3.
Characterization of stable VLPs producer cells. (A) S expression was measured by FACS analysis of 293GP, 293GP-S and 293GP-ΔS cells with an anti-S1 antibody. (B) GFP fluorescence of 293GP-Galv/GFP, 293GP-S/GFP and 293GP-ΔS/GFP measured by FACS analysis. The results shown are representative of three independent experiments.
Fig. 4.
Transduction efficiency of GFP pseudotyped vectors released from stable producers. (A) Fluorescent and bright-field pictures are displayed. The envelope pseudotype and the volume used for infection are indicated. (B) Titers of Galv-, S- and ΔS-pseudotyped vectors produced from stable producers were measured by FACS analysis two days after infection. Values presented are the mean ± SD of three independent experiments. ND: not determined.
2.4. High amounts of SARS-CoV-2 ΔS protein are released from stable producer cells
A VLP-derived SARS-CoV-2 vaccine will be a viable option if sufficient amounts of S protein are incorporated at the surface of the released particles. Western blots were performed with an anti-S2 antibody to evaluate the quantity of S protein into VLPs produced in transient transfections and from stable producers. Two bands were detected around 90 KDa that are most likely two glycosylated forms of S2. The uncleaved S protein migrated around 180 kDa, and two other bands above 250 kDa were also detected in the ΔS samples that had more intense signals. These bands could be dimeric and trimeric forms of S as it has been suggested (Ou et al., 2020). The amount of S2 detected in transient transfections or released from stable producers was much higher with the truncated version of S than with the full-length molecule (Fig. 5 A). A 4- and a 15-fold difference was found with the transient and the stable production systems, respectively (Fig. 5B), although there was less than a 1.5-fold difference between S and ΔS in cellular extracts (Fig. 5C). More ΔS was also released as compared to the full-length protein in the supernatants of stably transfected 293-ΔS cells, however the amount of ΔS detected was 4-to-5 times lower than the one released from the 293GP-ΔS cells. The amount of S2 equivalent present in the supernatant of 293GP-ΔS cells was high and evaluated at 1.25 μg/mL using the IgG-S2 standard (Fig. 5A). In this blot, MLV viral particles produced in transient transfection or from stable producers were detected with an antibody against p30 (Fig. 5A). In conclusion, our results indicated that the production of S is very efficient in stable producers but only with the truncated version of S.
Fig. 5.
Quantification of S and ΔS incorporated into VLPs. (A) Western blot analysis from concentrated supernatants of 293GP and 293 cells using anti-S2 and anti-p30 antibodies. Different amounts of Fc-tagged S2 were also loaded on the gel to quantify S2 in VLPs. (B) Differences between S and ΔS incorporation into VLPs. All the bands detected by the anti-S2 antibody in S- and ΔS-containing samples were quantified and normalized with the signal obtained for MLV p30. Values presented are the mean ± SD of three independent experiments analyzed twice in Western blot. (C) Western blot analysis of SARS-CoV-2 S protein in cellular extracts. Signals for S2, S, and multimeric forms of S were detected with the anti-S2 antibody. The Gag precursor pr65 was detected with the anti-p30 antibody, and β-tubulin was assessed for loading comparison.
2.5. SARS-CoV2 ΔS protein is preferentially incorporated into MLV VLPs versus extracellular vesicles
As stable transfected 293 cells were capable of releasing S or ΔS, we decided to further characterize the supernatants of the 293GP-ΔS. We used an iodixanol velocity gradient to discriminate VLPs from extracellular vesicles (EVs) as iodixanol solution has a neutral pH and an osmolarity that renders it suitable for the separation of viral particles and EVs (Ford et al., 1994). Furthermore, this technique has been used in the past to successfully separate human immunodeficiency viruses (HIV) from EVs (Cantin et al., 2008; Dettenhofer and Yu, 1999). Western blots with anti-S2 and anti-p30 antibodies were performed on the collected gradient fractions of 293-ΔS and 293GP-ΔS supernatants. S2 was detected in the top fractions from the 293-ΔS supernatant but there were none in the last 3 bottom fractions (Fig. 6 A). A similar detection pattern was observed in the top fractions of the supernatant from 293GP-ΔS, but the majority of S2 came from the 2 bottom fractions in which a band corresponding to the uncleaved S protein was also detected. The p30 signal was present in these two fractions, which indicated that the majority of ΔS released from 293GP-ΔS cells was incorporated into VLPs (Fig. 6B).
Fig. 6.
Incorporation of SARS-CoV-2 ΔS into MLV VLPs. Western blot analysis with antibodies against S2 and p30 on collected fractions separated with an iodixanol velocity gradient of (A) 293-ΔS and (B) 293GP-ΔS supernatants. The arrow below the blot indicates the density gradient. The results shown are representative of two independent experiments.
3. Discussion
Immunization will be the best preventive strategy to address the current COVID-19 pandemic, although therapeutic alternatives cannot be neglected as a vaccine that would trigger long-term protection is not a certainty (Amanat and Krammer, 2020; Moore and Klasse, 2020; Morris, 2020). Yet results from preclinical and clinical studies are encouraging and has started to lead to commercialization approval (Erasmus et al., 2020; Folegatti et al., 2020; Gao et al., 2020; Guebre-Xabier et al., 2020; Jackson et al., 2020; Keech et al., 2020; Logunov et al., 2020; Mercado et al., 2020; Moore and Klasse, 2020; Mulligan et al., 2020; Polack et al., 2020; Ravichandran et al., 2020; Ren et al., 2020; Smith et al., 2020; Walls et al., 2020; Wang et al., 2020; Xia et al., 2021; Yu et al., 2020; Zhu et al., 2020a). How efficient and how long Nabs will be present in vaccinated people remains an open question that will only be answered with time (Moore and Klasse, 2020). Also, antibody-dependent enhancement will have to be carefully monitored in vaccinated people as it is a side effect that cannot be underestimated with coronaviruses (Amanat and Krammer, 2020; Moore and Klasse, 2020; Morris, 2020). One other major challenge ahead is the capacity to mass produce COVID-19 vaccines. In this study, we have established and characterized a new MLV-derived VLP platform that could be used for the production of a COVID-19 vaccine.
The efficient pseudotyping of MLV particles with S is a prerequisite to establish a robust VLP platform. Studies on SARS-CoV and more recently on SARS-CoV-2 have shown that the codon optimization of S and the deletion of the ER retention signal located in the cytoplasmic tail are modifications that increase the pseudotyping of MLV, HIV, simian immunodeficiency virus and VSV viral vectors (Fukushi et al., 2005; Giroglou et al., 2004; Kobinger et al., 2007; Moore et al., 2004). The codon optimization enhances the level of S expression, but the role of the ER retention signal is less clear. Indeed, it was recently reported that the localization of SARS-CoV-2 S at the cell surface was not improved after disrupting the ER retention signal by missense mutations (Crawford et al., 2020). In this study, we showed that S could be detected at the cell surface at a similar level to that achieved by ΔS in transiently transfected cells as well as in stable producers (Fig. 1, Fig. 3A), a finding that has also been reported for SARS-CoV S expressed in transient transfections (Giroglou et al., 2004; Moore et al., 2004). These results indicate that S can bypass its natural localization and efficiently migrates to the cell surface when it is overexpressed.
Despite similar amounts of S and ΔS at the cellular membrane, the truncated version was more efficiently incorporated into MLV viral particles. Four- and 15-fold differences were obtained with VLPs produced in transient transfection experiments and from stable producers, respectively (Fig. 5B). The hypothesis that has been proposed for SARS−CoV and SARS−CoV-2 is that the 19 amino-acid deletion in the S cytoplasmic tail facilitates the pseudotyping by decreasing the steric interference with the retroviral matrix proteins (Crawford et al., 2020; Johnson et al., 2020; Moore et al., 2004; Schmidt et al., 2020). Our results invalidate this hypothesis as more ΔS was also found in the supernatant of 293 transfected cells that did not express MLV Gag-Pol (Fig. 5A). Parental 293 and 293GP cells release EVs that can incorporate ΔS more efficiently than S (Figs. 5A and 6). VLPs and EVs are very similar in composition, and it has been postulated that they use similar pathways for vesicle trafficking (Gould et al., 2003; Nolte-’t Hoen et al., 2016). So, unlike S, ΔS was efficiently incorporated into VLPs or EVs, like for example tetraspanins or endosomal markers that are equally found in both particle types (Gould et al., 2003; Nolte-’t Hoen et al., 2016).
EVs released from 293GP-ΔS contain less than 10 % of the total ΔS protein, and they would not need to be removed from vaccine preparations as they could be as good immunogens as VLPs. It was even reported that EVs containing the S protein of SARS-CoV could induce high levels of Nabs (Kuate et al., 2007).
Titers of recombinant GFP retroviruses released from stable producers were at least a 1000-fold higher with ΔS versus S despite a 15-fold difference in the amount of the two proteins incorporated at the surface of VLPs (Figs. 4A and 5 B). Our results suggest that recombinant viruses become fully infectious when a certain threshold of S protein is incorporated at their surface.
Recombinant GFP or luciferase retroviruses pseudotyped with S are commonly used to measure the activity of Nabs present in serum of infected or vaccinated people (Crawford et al., 2020; Jackson et al., 2020; Johnson et al., 2020; Mercado et al., 2020; Ou et al., 2020; Schmidt et al., 2020). These reagents are convenient, as unlike SARS-CoV-2 they can be manipulated in a BSL-2 laboratory. The robust production system with the 293 GP-ΔS cell line could be highly valuable to evaluate the presence of Nabs in large cohorts.
Mass production will be a major challenge with all types of SARS-CoV-2 vaccine that are being developed as the entire worldwide population will have to be vaccinated. Based on the results of a nanoparticle vaccine containing S, whose 5 and 25 μg doses triggered a high level of Nabs in people (Keech et al., 2020), we assume that a vaccine derived from the VLP platform described in this study could be efficient with similar or lower amounts of S per dose. The yield of VLPs produced from the 293 GP-ΔS cells could be increased if a high producer clone is selected instead of a bulk population, and if cells are cultured in bioreactors in fed-batch or perfusion modes. The average titer of gene therapy vectors produced with a derivative of the 293GP cell line was increased by 5.6-fold in bioreactor versus a 10-layer cell factory, and the total vector yield was increased by 13.1-fold (Wang et al., 2015). Mutations of the furin cleavage site located between S1 and S2 and the D614G variant that became more prevalent in the infected population could increase the amount of S incorporated into VLPs (Korber et al., 2020).
A very concise review that compared the first results of different COVID-19 vaccines concluded that the most immunogenic ones were made with recombinant proteins (Moore and Klasse, 2020). These results emphasize the importance of the platform developed in this study because VLPs present the antigen in a protein format that seems more potent for vaccination than the protein alone. Indeed, MLV VLPs displaying the human cytomegalovirus glycoprotein B antigen could trigger 10-times more Nabs in mice than the protein alone using the same amount of antigen (Kirchmeier et al., 2014). Finally, VLP-ΔS could be used as a boost for other types of vaccine like measle virus- and adenovirus-based recombinant vectors. These combinations were highly potent for triggering Nabs against hepatitis C proteins in mice and macaques (Garrone et al., 2011).
In conclusion, we have developed and characterized a new VLP platform that can efficiently incorporate the S protein from SARS-CoV-2, and that has the potential to produce a pan-coronavirus vaccine. The next logical step is to validate this vaccine in experimental animals and in humans thereafter.
4. Materials and methods
4.1. Plasmids
The expression plasmid pMD2ACE2iPuror containing the human angiotensin-converting enzyme (ACE2) cDNA used to generate ACE2 positive cells was constructed as follows: the ACE2 PmeI cDNA fragment obtained from the plasmid hACE2 (Addgene; #1786) was cloned in pMD2iPuror opened in EcoRV.
The SARS-CoV-2 S gene from the Wuhan-Hu-1 isolate (GenBank: MN908947.3) was codon optimized (Genscript, Township, NJ) and cloned in pMD2iPuror in EcoRI/XhoI. A shorter version with a 19-codon deletion in C-terminal (ΔS) was also constructed in a similar way. These sequences will be made available upon request.
The pMD2GPiZeor, pMD2.GalviPuror and pMD2.G plasmids that encode MLV Gag-pol, and the Galv and VSV-G envelopes have been described elsewhere (Ghani et al., 2007). RetroVec is a retroviral vector plasmid containing the GFP gene under the control of the 5′ long terminal repeat sequence and that was derived from GFP3 (Qiao et al., 2002).
4.2. Cell lines
293GP, 293 cells (ATCC, CRL-11,268), and their derivatives expressing the ACE2 receptor (293-ACE2), S (293GP–S and 293-S), DeltaS (293GP-ΔS and 293-ΔS) and the Galv envelope (293GP-Galv) were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Wisent, Saint-Jean-Baptiste, Canada) supplemented with 10 % fetal calf serum (Life Technologies, Grand island, NY) and antibiotics (Wisent). Bulk populations of 293-ACE2, 293-S, 293-ΔS, 293GP–S, 293GP-ΔS and 293GP-Galv were established by transfection using the calcium phosphate procedure. Briefly, subconfluent 293 or 293GP cells plated in 10-cm dishes were transfected with 20 μg of the pMD2 plasmids expressing ACE2, S, ΔS or Galv. Two days later, cells were selected in puromycin for a period of 10 days (0.5 μg/mL). Bulk populations of 293GP–S/GFP, 293GP-ΔS/GFP and 293GP-Galv/GFP were generated by infections at a high multiplicity of infection with the RetroVec GFP recombinant virus. The virus was pseudotyped with VSV-G and produced by transient transfection in 293 cells as described in the next paragraph. The 3 derived cell lines were at least 86 % GFP positive (Fig. 3B).
4.3. Virus productions and infections
The production of GFP retroviral vectors was generated by transient transfection of 293 cells. One day prior transfection, 3 × 106 cells were plated in 60-mm dishes. 293 cells were transfected for 4 h by the calcium phosphate procedure with 1 μg of envelope expression plasmids (pMD2.G, pMD2.GalviPuror, pMD2.SiPuror or pMD2.ΔSiPuror), 4 μg of Gag-pol expression plasmid (pMD2GPiZeor) and 5 μg of RetroVec plasmid. Two days later, 2.5 mL of viral supernatants were harvested and frozen at -80 °C. Recombinant viruses from stable 293GP–S/GFP, 293GP-ΔS/GFP and 293GP-Galv/GFP cells were also produced similarly in 60-mm dishes.
Transduction efficiency of GFP vectors was determined by scoring fluorescent-positive target cells. 293-ACE2 cells were inoculated at a density of 2 × 105 cells per well in 24-well plates. The next day, the medium from each well was replaced with different volume of viral supernatants containing 8 μg/mL polybrene. Two days later, cells were trypsinized and analyzed by flow cytometry. Vector titers were calculated using the following formula (N x P) x 2/(V x D). N = Cell number on the day of infection; P = percentage of fluorescent-positive cells determined by flow cytometry; V is the viral volume applied; and D is the virus dilution factor. Titers were calculated when the percentage of fluorescent-positive cells was comprised between 2–20 %. Alternatively, GFP positive cells were assessed under a fluorescent microscope. The 3 × 3 mosaic images of GFP and transmitted light were acquired with a Nikon TI-E inverted microscope with a PlanApo VC 20 × 0.75 NA objective using a Hamamatsu Orca-ER CCD camera. Acquisition and stitching were performed with the Nikon NIS Elements 5.02 software program. The fluorescence intensity of infected cells displayed in Fig. 4A were scanned using the Fiji software to evaluate the difference in viral titers (Schindelin et al., 2012).
4.4. Protein analysis
The presence of S at the surface of 293 cells was assessed in transient transfections. Subconfluent cells in 6-well plates were transfected for 4 h with 5 μg of S or ΔS plasmids by the calcium phosphate procedure, and 24 h later, the media was replaced with serum-free media (SFM) BalanCD HEK293 (Fujifilm Irvine Scientific, Santa Ana, CA). The next day, cells were detached without trypsin by gently pipetting up and down the medium on top of the cells. A human chimeric anti-S1 antibody (1:200; Genscript) followed by an Alexa647-conjugated goat anti-human IgG (1;400; Jackson Laboratories) were successively incubated with cells for labelling. The fixable viability stain 450 (BD Biosciences, San Jose, CA, USA) was used to exclude dead cells. The presence of S was then analyzed by flow cytometry with a BD FACSAria II (BD Biosciences). Cells transfected with a Galv expression plasmid were used as control. The presence of stably expressed S at the cell surface of 293GP–S and 293GP-ΔS was similarly analyzed by flow cytometry.
The presence of ACE2 at the surface of 293-ACE2 cells was also checked by FACS. Detached cells were labelled with a mouse anti-ACE2 antibody (1:200; R&D Systems, Minneapolis, MN) followed by an Alexa488 goat anti-mouse (1:1,000; Invitrogen, Carlsbad, CA).
The presence of S released in the supernatant of transiently transfected 293GP cells was analyzed by Western blot. Subconfluent cells plated in 60 mm were transfected for 4 h with 5 μg of envelope expression plasmids and 5 μg of the GFP retroviral plasmid. One day later, the media was replaced with 2.5 mL of SFM that was then harvested the following day. Supernatants were concentrated 10-fold with a 30 kDa Amicon centrifugal unit (Millipore Sigma, Oakville, Canada) and were stored at −80 °C until use. The GFP fluorescence evaluated under a microscope at the time of harvest was very similar among the different transfected plates.
Supernatants from confluent 293GP–S, 293GP-ΔS, 293-S and 293-ΔS cells were also harvested and concentrated from 60-mm dishes.
Cell pellets of 1 × 106 cells were resuspended in 100 μL RIPA lysis buffer containing a protease inhibitor cocktail (Roche). Samples were centrifuged for 5 min to remove cell debris and stored at −20 °C until use for Western blot analysis.
Samples of 20 μL were incubated 5 min at 95 °C in loading buffer containing 1 % SDS and 2.5 % β-mercaptoethanol, and run on a 10 % SDS-polyacrylamide gel (4 % stacking), followed by transfer onto nitrocellulose membranes (GE Healthcare, Chicago, IL). Immunoblotting was performed with a rabbit polyclonal antibody anti-S2 (1:400; SinoBiological, Beijing, China), a rat monoclonal antibody anti-MLV p30 produced from the hybridoma R187 (1:2,000; American Type Culture Collection, Manassas, VA) and a mouse monoclonal antibody anti-β-tubulin (1:1,000; Sigma). Blots were then incubated with secondary antibodies IRDylight680 goat anti-rat IgG (1:10,000; Invitrogen) and IRDye 800CW anti-rabbit IgG (1:10,000; Li-Cor Biosciences, Lincoln, NE), and analyzed with the Odyssey Infrared Imaging System (Li-Cor Biosciences). Serial dilutions of known amounts of C-terminally Fc-tagged S2 (BioVendor, Brno, Czech Republic) were used for quantification.
4.5. Velocity gradient
Thirty mL of supernatant from confluent 150-mm dishes of 293GP-ΔS and 293-ΔS cells were harvested and filtered through a 0.45 μ membrane, and concentrated by ultracentrifugation for 90 min at 100,000 x g in a AH629 rotor. Pellets containing virions and EVs were resuspended in 1 mL PBS containing a protease cocktail inhibitor (Roche, Laval, Canada) during 2 h at 4 °C. The resuspended vesicles were layered onto a 6–18 % Optiprep™ 11-step discontinuous velocity gradient (Stemcell Technologies, Vancouver, Canada), and centrifuged for 90 min at 176,000 x g in a SW40Ti rotor as previously described (Cantin et al., 2008; Dettenhofer and Yu, 1999). Fractions of approximately 800 μL were collected from the bottom after puncturing the wall of the centrifuge tube with a gauge needle, and 20 μL of each sample were analyzed by Western blot.
CRediT authorship contribution statement
Sylvie Roy: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing - review & editing. Karim Ghani: Data curation, Investigation, Formal analysis, Methodology, Validation, Writing - review & editing. Pedro O. de Campos-Lima: Formal analysis, Methodology, Validation, Writing - review & editing. Manuel Caruso: Data curation, Formal analysis, Investigation, Methodology, Funding acquisition, Validation, Supervision, Writing - original draft.
Declaration of Competing Interest
K.G, P.O. de C-L and M.C. are co-founders and shareholders of BioVec Pharma. M.C. is an author of a patent application covering the VLP platform presented in this study.
Acknowledgments
The authors thank Carl Saint-Pierre for his technical assistance in microscopy. This work was supported by BioVec Pharma, Canada.
References
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin R., Diou J., Belanger D., Tremblay A.M., Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J. Immunol. Methods. 2008;338(1–2):21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88(19):11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., Pettie D., King N.P., Balazs A.B., Bloom J.D. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M., Yu X.F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 1999;73(2):1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docea A.O., Tsatsakis A., Albulescu D., Cristea O., Zlatian O., Vinceti M., Moschos S.A., Tsoukalas D., Goumenou M., Drakoulis N., Dumanov J.M., Tutelyan V.A., Onischenko G.G., Aschner M., Spandidos D.A., Calina D. A new threat from an old enemy: reemergence of coronavirus (Review) Int. J. Mol. Med. 2020;45(6):1631–1643. doi: 10.3892/ijmm.2020.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020;5(1):237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., Draves K.E., Randall S., Guerriero K.A., Duthie M.S., Carter D., Reed S.G., Hawman D.W., Feldmann H., Gale M., Jr., Veesler D., Berglund P., Fuller D.H. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12(555) doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Oxford C.V.T.G. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford T., Graham J., Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 1994;220(2):360–366. doi: 10.1006/abio.1994.1350. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Mizutani T., Saijo M., Matsuyama S., Miyajima N., Taguchi F., Itamura S., Kurane I., Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86(Pt 8):2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrone P., Fluckiger A.C., Mangeot P.E., Gauthier E., Dupeyrot-Lacas P., Mancip J., Cangialosi A., Du Chene I., LeGrand R., Mangeot I., Lavillette D., Bellier B., Cosset F.L., Tangy F., Klatzmann D., Dalba C. A prime-boost strategy using virus-like particles pseudotyped for HCV proteins triggers broadly neutralizing antibodies in macaques. Sci. Transl. Med. 2011;3(94):94ra71. doi: 10.1126/scitranslmed.3002330. [DOI] [PubMed] [Google Scholar]
- Ghani K., Cottin S., Kamen A., Caruso M. Generation of a high-titer packaging cell line for the production of retroviral vectors in suspension and serum-free media. Gene Ther. 2007;14(24):1705–1711. doi: 10.1038/sj.gt.3303039. [DOI] [PubMed] [Google Scholar]
- Ghani K., Wang X., de Campos-Lima P.O., Olszewska M., Kamen A., Riviere I., Caruso M. Efficient human hematopoietic cell transduction using RD114- and GALV-pseudotyped retroviral vectors produced in suspension and serum-free media. Hum. Gene Ther. 2009;20(9):966–974. doi: 10.1089/hum.2009.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroglou T., Cinatl J., Jr., Rabenau H., Drosten C., Schwalbe H., Doerr H.W., von Laer D. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 2004;78(17):9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Booth A.M., Hildreth J.E. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100(19):10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guebre-Xabier M., Patel N., Tian J.H., Zhou B., Maciejewski S., Lam K., Portnoff A.D., Massare M.J., Frieman M.B., Piedra P.A., Ellingsworth L., Glenn G., Smith G. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine. 2020;38(50):7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.W., Travanty E.A., Jeffers S.A., Smith M.K., Wennier S.T., Thackray L.B., Holmes K.V. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell-cell fusion. J. Virol. 2008;82(6):2883–2894. doi: 10.1128/JVI.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H., m, R.N.A.S.G An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized pseudotyping conditions for the SARS-COV-2 spike glycoprotein. J. Virol. 2020;94(21) doi: 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmeier M., Fluckiger A.C., Soare C., Bozic J., Ontsouka B., Ahmed T., Diress A., Pereira L., Schodel F., Plotkin S., Dalba C., Klatzmann D., Anderson D.E. Enveloped virus-like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity. Clin. Vaccine Immunol. 2014;21(2):174–180. doi: 10.1128/CVI.00662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger G.P., Limberis M.P., Somanathan S., Schumer G., Bell P., Wilson J.M. Human immunodeficiency viral vector pseudotyped with the spike envelope of severe acute respiratory syndrome coronavirus transduces human airway epithelial cells and dendritic cells. Hum. Gene Ther. 2007;18(5):413–422. doi: 10.1089/hum.2006.194. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuate S., Cinatl J., Doerr H.W., Uberla K. Exosomal vaccines containing the S protein of the SARS coronavirus induce high levels of neutralizing antibodies. Virology. 2007;362(1):26–37. doi: 10.1016/j.virol.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in malayan pangolins. Nature. 2020;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Luk H.K.H., Wong A.C.P., Li K.S.M., Zhu L., He Z., Fung J., Chan T.T.Y., Fung K.S.C., Woo P.C.Y. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect Dis. 2020;26(7):1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Moderna COVID vaccine becomes second to get US authorization. Nature. 2020 doi: 10.1038/d41586-020-03593-7. [DOI] [PubMed] [Google Scholar]
- Ledford H., Cyranoski D., Van Noorden R. The UK has approved a COVID vaccine - here’s what scientists now want to know. Nature. 2020;588(7837):205–206. doi: 10.1038/d41586-020-03441-8. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Lubenets N.L., Egorova D.A., Shmarov M.M., Nikitenko N.A., Morozova L.F., Smolyarchuk E.A., Kryukov E.V., Babira V.F., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396(10255):887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78(11):5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marovich M., Mascola J.R., Cohen M.S. Monoclonal antibodies for prevention and treatment of COVID-19. JAMA. 2020;324(2):131–132. doi: 10.1001/jama.2020.10245. [DOI] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., He X., Martinez D.R., Rutten L., Bos R., van Manen D., Vellinga J., Custers J., Langedijk J.P., Kwaks T., Bakkers M.J.G., Zuijdgeest D., Rosendahl Huber S.K., Atyeo C., Fischinger S., Burke J.S., Feldman J., Hauser B.M., Caradonna T.M., Bondzie E.A., Dagotto G., Gebre M.S., Hoffman E., Jacob-Dolan C., Kirilova M., Li Z., Lin Z., Mahrokhian S.H., Maxfield L.F., Nampanya F., Nityanandam R., Nkolola J.P., Patel S., Ventura J.D., Verrington K., Wan H., Pessaint L., Van Ry A., Blade K., Strasbaugh A., Cabus M., Brown R., Cook A., Zouantchangadou S., Teow E., Andersen H., Lewis M.G., Cai Y., Chen B., Schmidt A.G., Reeves R.K., Baric R.S., Lauffenburger D.A., Alter G., Stoffels P., Mammen M., Van Hoof J., Schuitemaker H., Barouch D.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Moore J.P., Klasse P.J. COVID-19 vaccines: “Warp speed” needs mind melds, not warped minds. J. Virol. 2020;94(17) doi: 10.1128/JVI.01083-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78(19):10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fierros L., Garcia-Silva I., Rosales-Mendoza S. Development of SARS-CoV-2 vaccines: should we focus on mucosal immunity? Expert Opin. Biol. Ther. 2020;20(8):831–836. doi: 10.1080/14712598.2020.1767062. [DOI] [PubMed] [Google Scholar]
- Morris K.V. The improbability of the rapid development of a vaccine for SARS-CoV-2. Mol. Ther. 2020;28(7):1548–1549. doi: 10.1016/j.ymthe.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Nicola M., Alsafi Z., Sohrabi C., Kerwan A., Al-Jabir A., Iosifidis C., Agha M., Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-’t Hoen E., Cremer T., Gallo R.C., Margolis L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. U. S. A. 2016;113(33):9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341(2):215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C., Group C.C.T. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Roy V., Girard M.-H., Caruso M. High translation efficiency is mediated by the encephalomyocarditis virus IRES if the natural sequence surrounding the 11th AUG is retained. Hum. Gene Ther. 2002;13:881–887. doi: 10.1089/10430340252899046. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S., Coyle E.M., Klenow L., Tang J., Grubbs G., Liu S., Wang T., Golding H., Khurana S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020;12(550) doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Sun H., Gao G.F., Chen J., Sun S., Zhao R., Gao G., Hu Y., Zhao G., Chen Y., Jin X., Fang F., Chen J., Wang Q., Gong S., Gao W., Sun Y., Su J., He A., Cheng X., Li M., Xia C., Li M., Sun L. Recombinant SARS-CoV-2 spike S1-Fc fusion protein induced high levels of neutralizing responses in nonhuman primates. Vaccine. 2020;38(35):5653–5658. doi: 10.1016/j.vaccine.2020.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn A., Fu Y., Hu X., Hall M.D., Simeonov A. Fruitful neutralizing antibody pipeline brings hope to defeat SARS-Cov-2. Trends Pharmacol. Sci. 2020;41(11):815–829. doi: 10.1016/j.tips.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Raj V.S., Koopmans M.P., Haagmans B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016;16:55–62. doi: 10.1016/j.coviro.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivan J., Singh M., Sarma J.D. Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals. J. Biosci. 2017;42(2):231–244. doi: 10.1007/s12038-017-9676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Caskey M., Robbiani D.F., Nussenzweig M.C., Rice C.M., Hatziioannou T., Bieniasz P.D. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217(11) doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E.L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M.A., Wu Y., Amante D., Park D.H., Dia Y., Ali A.R., Zaidi F.I., Generotti A., Kim K.Y., Herring T.A., Reeder S., Andrade V.M., Buttigieg K., Zhao G., Wu J.M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A.S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J.S., Muthumani K., Wang B., Carroll M.W., Kim J.J., Boyer J., Kulp D.W., Humeau L., Weiner D.B., Broderick K.E. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016;97(8):1853–1864. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., Navarro M.J., Miranda M.C., Pettie D., Ravichandran R., Kraft J.C., Ogohara C., Palser A., Chalk S., Lee E.C., Guerriero K., Kepl E., Chow C.M., Sydeman C., Hodge E.A., Brown B., Fuller J.T., Dinnon K.H., 3rd, Gralinski L.E., Leist S.R., Gully K.L., Lewis T.B., Guttman M., Chu H.Y., Lee K.K., Fuller D.H., Baric R.S., Kellam P., Carter L., Pepper M., Sheahan T.P., Veesler D., King N.P. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183(5):1367–1382. doi: 10.1016/j.cell.2020.10.043. e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Olszewska M., Qu J., Wasielewska T., Bartido S., Hermetet G., Sadelain M., Riviere I. Large-scale clinical-grade retroviral vector production in a fixed-bed bioreactor. J Immunother. 2015;38(3):127–135. doi: 10.1097/CJI.0000000000000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G.F., Tan W., Yang X. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182(3):713–721. doi: 10.1016/j.cell.2020.06.008. e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., Huang W., Xu W., Huang B., Wang H., Wang W., Zhang W., Li N., Xie Z., Ding L., You W., Zhao Y., Yang X., Liu Y., Wang Q., Huang L., Yang Y., Xu G., Luo B., Wang W., Liu P., Guo W., Yang X. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21(1):39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Shi M., Li J., Song P., Li N. Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 2020;8:862. doi: 10.3389/fbioe.2020.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., Martinez D.R., Loos C., Atyeo C., Fischinger S., Burke J.S., Slein M.D., Chen Y., Zuiani A., Lelis F.J.N., Travers M., Habibi S., Pessaint L., Van Ry A., Blade K., Brown R., Cook A., Finneyfrock B., Dodson A., Teow E., Velasco J., Zahn R., Wegmann F., Bondzie E.A., Dagotto G., Gebre M.S., He X., Jacob-Dolan C., Kirilova M., Kordana N., Lin Z., Maxfield L.F., Nampanya F., Nityanandam R., Ventura J.D., Wan H., Cai Y., Chen B., Schmidt A.G., Wesemann D.R., Baric R.S., Alter G., Andersen H., Lewis M.G., Barouch D.H. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z., Jia S.Y., Wang B.S., Hu Y., Liu J.J., Zhang J., Qian X.A., Li Q., Pan H.X., Jiang H.D., Deng P., Gou J.B., Wang X.W., Wang X.H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]