Abstract
The recent outbreak of coronavirus disease 2019 (COVID-19) by SARS-CoV-2 has led to uptodate 24.3 M cases and 0.8 M deaths. It is thus in urgent need to rationalize potential therapeutic targets against the progression of diseases. An effective, feasible way is to use the pre-existing ΔORF6 mutant of SARS-CoV as a surrogate for SARS-CoV-2, since both lack the moiety responsible for interferon antagonistic effects. By analyzing temporal profiles of upregulated genes in ΔORF6-infected Calu-3 cells, we prioritized 55 genes and 238 ligands to reposition currently available medications for COVID-19 therapy. Eight of them are already in clinical trials, including dexamethasone, ritonavir, baricitinib, tofacitinib, naproxen, budesonide, ciclesonide and formoterol. We also pinpointed 16 drug groups from the Anatomical Therapeutic Chemical classification system, with the potential to mitigate symptoms of SARS-CoV-2 infection and thus to be repositioned for COVID-19 therapy.
Keywords: SARS-CoV-2, COVID-19, Transcriptomic and network analysis, Drug repositioning
1. Introduction
The COVID-19 pandemic, which has led to huge economic losses by the Great Lockdown, is an ongoing global threat caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. As of August 2020, ~24.3 M cases with ~828 k deaths have been reported across 188 countries and territories, leading to an ~3.4% mortality rate, 34 times higher than that of H1N1. Unfortunately, no drug or vaccine has been approved to treat human coronaviruses, and new interventions are likely to take months to years to develop. In this regard, rationalizing semiempirical or empirical strategies to protect currently hospitalized victims (~8.4 M uptodate) from death is urgent and necessary. The SARS-CoV-2 is linked to the severe acute respiratory syndrome coronavirus (SARS-CoV, which led to 774 deaths out of 8096 infected individuals ~16 years ago) and the Middle East respiratory syndrome coronavirus (MERS-CoV, which led to 858 deaths out of 2494 cases ~8 years ago) in that they share a high degree of genomic similarity (~80% between SARS-CoV and SARS-CoV-2, e.g.) [2]. As such, one plausible way to rationalize empirical strategies in a timely manner is to use available SARS-CoV/MERS-CoV data and perform fast, effective in silico analyses, thereby identifying candidate remedies from currently available medications.
In addition to genomics, SARS-CoV-2 and SARS-CoV share a certain degree of similiarity. It has been shown that SARS-CoV-2 shares similar transmission mechanisms and pathogenic sequelae with SARS-CoV. First, both viruses are group B coronaviruses utilizing the same receptor, angiotensin-converting enzyme 2 (ACE2), to bind to the host cell surface [3]. Second, the pathology of SARS-CoV-infected lung shows features of acute injury and fibrinous organizing pneumonia patterns [4], which were also found in SARS-CoV-2 infected lung, including diffusive acute alveolar injury with edema, hyaline membrane formation, proteinaceous exudate, focal reactive hyperplasia, immune cell infiltration, and intra-alveolar fibroblastic plug formation [5]. The consequence is the respiratory failure, the leading cause for the mortality, as no gas exchange can be performed in such foamy, fibroblastic alveolar space. Intervening in the disease progression requires an explicit molecular cellular understanding of the pathogenesis, which is currently limited. What has been noted is the involvement of highly inflammatory responses that produce enormous chemokines implicated in cytokine storm [6,7]. Futhermore, a key difference between SARS-CoV and SARS-CoV-2 is a truncation of the open reading frame (ORF) 3b, ORF6, in SARS-CoV-2, which affects the viral antagonistic effect toward interferon response [8]. These lines of evidence suggest that it is plausible to use available SARS-CoV data as a surrogate for devising the empirtical strategy and that immune modulation might be involved in such strategy. Fortunately, a mutant of SARS-CoV with the truncation in ORF6 has been found and used to infect a human lung cell line, Calu-3, for empirical study. Here, we took advantage of these preexisting data and applied bioinformatic analysis on SARS-CoV-infected cells (as the reference) and SARS-CoV ΔORF6-infected cells (as the surrogate to mimic SARS-CoV-2 infection) to identify medications that can be repurposed for COVID-19 therapy.
2. Materials and method
2.1. Expression data
Transcriptomes of Calu-3 cells infected by infectious clone-derived (ic)SARS CoV and icSARS ΔORF6 mutant (GSE33267) were queried using GEOquery. The downloaded data had already been normalized. Probe-level expression values (n = 30,772) were log2-transformed and were aggregated to become gene-level expression values (n = 18,113) by maximum value. Gene symbols were further checked and corrected using the HGNChelper package.
2.2. Identification of differentially expressed genes
Differentially expressed tests were conducted on the time-series microarray data (ΔORF6-infected cells vs. control cells or SARS-infected cells vs. control cells) based on the following post-treatment time-points: 0, 3, 7, 12, 24, 30, 36, 48, 54, 60 and 72 h. To ensure the reliability of identified DEGs, we conducted three different methods, and defined differential expressed genes (DEGs) as genes that showed consensus (both on significance and effect direction) across methods. The three methods used different ways of normalization (normexp method and quantile normalization for background correction and robust multichip average (RMA) for method 1 and 3; normexp method and quantile normalization for background correction, RMA and between-sample normalization for method 2), different ways of gene summarization (maximum value aggregation for methods 1 and 3; Tukey Biweight estimator for method 2), and different ways of identifying significant genes across groups, as following: Method (1): Limma method. By assuming smooth expression changes over time, we fitted a regression spline with effective degrees of freedom of five to capture the temporal trends, and then compared the differences in curves between groups (∆ORF6 vs. mock and wild-type vs. mock). Then for each gene, a linear model that implemented in limma [9] was fitted and an empirical Bayes moderated F-statistic was calculated for testing the change in expression level over the time points. Five coefficients corresponded to differences in two curves were retrieved from the fitted model and then averaged to produce a mean coefficient value. To control the false positive rate, raw p-values were adjusted using the Benjamini-Hochberg —(BH) method. Significant genes (out of 18,113 genes) of the method (1) were defined as genes with BH-adjusted p-value <0.05. Up or downregulated DEGs were defined by the average coefficient of > 0 or < 0, respectively. Method (2): Rnits method. In this method, probe expression values were further undergone between sample normalization and collapsed into gene-level using a robust Tukey Biweight estimator, which performs penalization on outlier values. Next, significant genes were identified by testing whether each gene expression trajectory from the treatment group (∆ORF6 or wild-type) and mock (control) group are the realizations of the same underlying basis B-spline curve [10]. The significant genes (out of 18,109 genes) of the method (2) were defined as genes that passed a BH-adjusted p-value threshold of 0.05. Up or downregulated significant genes were defined by ratio statistic of > 0 or < 0, respectively. Method (3): timecourse (tc) method. This method was implemented in timecourse package, which calculated a statistic for each gene using multivariate empirical Bayes approach. This method incorporated the correlation coefficient information across genes to diminish false-positivity and false-negativity. The significance of each gene was calculated by F-statistic that derived from the statistic. The significant genes (from total 18,113 genes) of the method (3) were defined as genes that passed a Bonferroni-adjusted p-value threshold of 0.05. Since this method didn't output information regarding the up or downregulation of significant genes, we thus calculated the logarithmic-transformed multiple of change (logFC) as , ∀j∈∆ORF6 or wild-type-infected cells and ∀i∈control cells, of each gene from normalized expression data. Finally, we defined DEGs as genes that were identified as significant genes in the above three methods and with the same effect direction.
2.3. Construction of co-expression clusters
ΔORF6-infected cell expression data were summarized to Gencode v33 [11] and the median-centering before network analysis. Gene clusters were identified using weighted correlation network analysis (WGCNA) [12]. A weighted signed network was constructed using a thresholding power of 10, which is the smallest value to get a scale-free r 2 fit of 0.85. The pairwise gene-to-gene topological overlap (TO) matrix was then calculated. The pre-clusters were identified by applying a top-down dynamic tree cut method (module gene size > 200 and deepSplit parameter 2), and pre-clusters were defined as branches of the hierarchical cluster tree. Pre-clusters with pairwise cluster eigengene (PC1) correlation values (r) > 0.85 were further merged into clusters.
2.4. Cluster enrichment test for DEGs
Enrichment test for DEGs across multiple time points in co-expressed clusters was conducted using two-tailed Fisher's exact test. The odds ratios (ORs) were calculated. We defined OR > 1 and p-value < 0.05 as significant overrepresentation (enrichment).
2.5. Transcription factor binding site (TFBS) analysis
Depending on the initial data (either PWM information or target genes of a TF) retrieved from databases, two different strategies were adopted as follows: (1) sequence-based enrichment test. Given a set a gene with size N G and several candidate transcription factors (TFs) with known position weight matrix (PWM) or motif, we want to identify whether the TFs' binding motif was enriched in the promoter sequence of genes in the given gene set. To do this, we scanned the promoter region (2000 basepair (bp) upstream from the transcription start site (TSS)) for the N G genes. We retrieved TFs' human PWM information from MotifDb package. For each PWM, a threshold-free log-normal distribution was calculated by fitting 500 bp chunks of 2000 bp promoters from all human genes (hg19). The PWM enrichment test of each TF was performed using PWMEnrich package. We further conducted Bonferroni correction to reduce false positivity, and (2) gene-based permutation test. Given a set of gene with size N G and several candidate TFs with known target genes from database(s) (such as ChIP-seq results from the ENCODE), we conducted gene-based permutation test by counting the overlapped genes of TF-regulated genes to the given gene set and performed permutation test to assess the enrichment of overlapped count number as following: For each TF, we counted the overlapped target number (n) in the given gene set with size N. Next, we randomly selected N genes from the background gene set and then counted the overlapped number of targets of this TFs. This process was repeated for 99,999 times to produce a null distribution n 1 , …, n 99999 of target number for the given TFs. We then calculated the single-tailed p-value as (no. of n, n 1 , …, n 99999 ≥ n)/(size of n, n 1 , …, n 99999).
2.6. Gene ontology and REACTOME pathway analysis
Genes in co-expressed clusters were characterized using Gene Ontology (GO) biological processes (BPs) and REACTOME pathways using clusterProfiler [13] and ReactomePA [14], respectively. Q-values were calculated for multiple testing corrections. The top 20 enriched terms were selected for visualization.
2.7. Identification of targetable genes
Targetable genes were defined as genes with it (protein) products that could be targeted by specific ligands (drugs) from the IUPHAR/BPS Guide to PHARMACOLOGY [15]. Further data sanitization steps including (i) limited only drugs with target species of human; (ii) ligands that annotated as non-endogenous agents; (iii) ligands with clear and explicit “action” and “type” annotations; and (iii) ligands with explicit target gene (symbol). Finally, the drug-gene pairs were collapsed across time points and further visualized using circlize, VennDiagram, RColorBrewer, and igraph packages.
2.8. Permutation test for the ATC groups of drugs
The Anatomical Therapeutic Chemical (ATC) classification of each ligand was annotated from the DrugBank database [16]. Given the size of N G for the final targetable genes, we randomly selected N G genes from the targetable gene list and mapped the ligands that may target the genes. Next, we summarized the number of drugs that belong to a specific ATC group. We repeated this process for n = 99,999 times and calculated the probability of obtaining a number of drugs that equal to or greater than original data in the particular ATC group. We further computing BH-adjusted p-values for multiple testing corrections and selected a BH-adjusted p-value of 0.1 as a threshold of significance.
2.9. Statistical analyses
In this study, we used R (http://www.r-project.org/ and http://cran.r-project.org/) or Bioconductor (http://www.bioconductor.org/) to perform all statistical association tests, enrichment analyses, and visualization tasks.
3. Results
3.1. Characterization of icSARS ΔORF6 infection-related genes
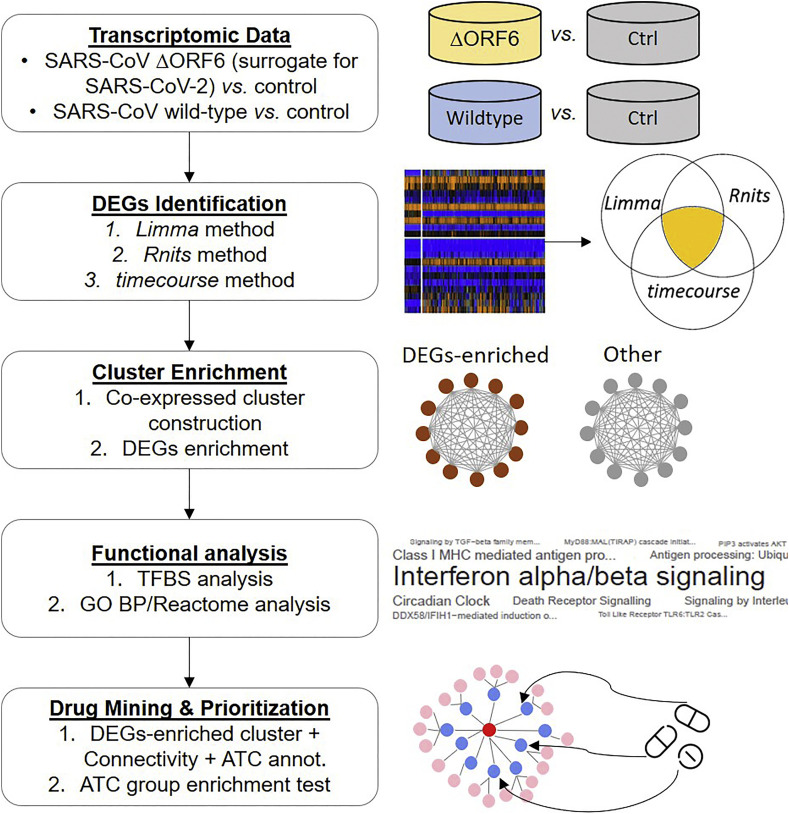
In contrast to SARS-CoV, SARS-CoV-2 shows ablated antagonism to interferon (IFN) due to an amino acid truncation in the viral ORF6 gene [8]. We therefore leveraged a previously published dataset (GSE33267) [17] that contains time-dependent transcriptomic profiling of infectious clone-derived (ic)SARS-CoV ΔORF6 (henceforth called “ΔORF6”) in human Calu-3 cells. Since ΔORF6 is a mutant form of SARS-CoV that cannot express ORF6, it is reasonable to use ΔORF6 as a surrogate for SARS-CoV-2 (which also contains truncated ORF6) to reveal its pathophysiology and explore drug repositioning (Fig. 1 ).
Fig. 1.
Flowchart of the approach in this study. Transcriptomic-driven network discovers pathophysiology of human coronavirus (HCoV) and contribute to drug repositioning for SARS-CoV-2 infection.
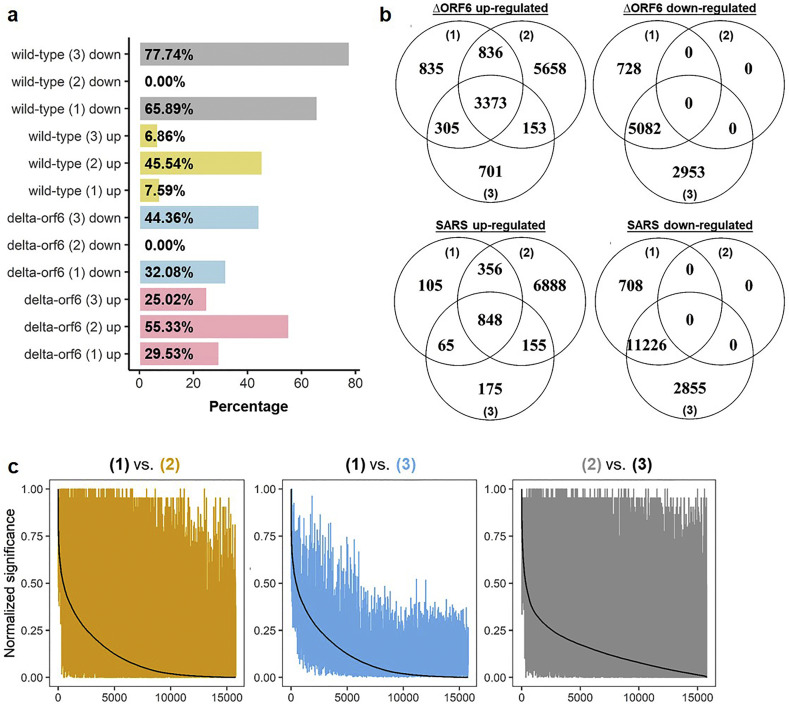
To identify genes that respond to ΔORF6 infection in Calu-3 cells (ΔORF6 infection-related genes), we analyzed the differential expression of genes by comparing ΔORF6-infected cells (n = 33; 3 replicates for 11 different time points including 0, 3, 7, 12, 24, 30, 36, 48, 54, 60 and 72 h) to control cells (n = 33; 3 replicates for 11 different time points) by applying three complementary methods (i.e. method (1), (2) and (3) as described in “Materials and Methods” section) that used different strategies to identify differentially expressed genes (DEGs) of time-series expression data. In parallel, we conducted similar tests on wild-type icSARS-CoV-infected Calu-3 cells (henceforth called “wild-type” or “SARS”) for comparison (Fig. 2a). We revealed a wide range of percentages of up or downregulated DEGs (0% to 77.74%), suggesting different sensitivity and specificity across methods.
Fig. 2.
Up-regulated differentially expressed genes (DEGs) in ΔORF6- and SARS-infected Calu-3 cells. (A) Barplot showing the percentage (x-axis) of identified significant genes in methods (1), (2) and (3) (y-axis). The results of ΔORF6 (delta-orf6) and SARS (wild-type) were further classified by the effect of direction, i.e. upregulation or downregulation. The numbers in parenthesis indicate methods. (B) Venn diagrams showing the number of intersected up or downregulated significant genes in ΔORF6- and SARS-infected cells across three methods. (C) Pairwise comparison of three methods by using normalized significance values of genes. The numbers in parenthesis indicate methods. The color of the lines is corresponding to the color of numbers in the main title. In each comparison, one method (black) is selected and we sort the genes (x-axes) according to its normalized significance values (y-axes). The normalized significant values of genes from the other method (brown/blue/grey) are then compared with the black line, with smaller bandwidth indicates higher outcome similarity between methods. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For significant genes detected by each method, we defined DEGs as genes identified by all three methods and with the same effect of direction (upregulated and downregulated) across methods (Fig. 2b). Therefore, we detected no down-regulated DEGs in this study. Especially, we detected 3373 and 848 upregulated DEGs in ΔORF6-infected cells and SARS-infected cells, respectively. However, none of the ΔORF6 group or SARS group shows consensus results regarding downregulated genes.
Significance values of genes were further transformed into 0–1, and the outcome similarity of three methods was quantified using Pearson's product-moment correlation coefficients (r P). We observed moderate-to-high correlations (r P = 0.7 and 0.69 in ∆ORF6-infected cells and SARS-infected cells, respectively; Fig. S1) between the results of methods (1) and (3); while the results from the method (2) deviated from that of methods (1) and (3) (r P ranged from 0.25–0.60; Figs. 2c and S2).
To investigate the effect of ∆ORF6 or SARS infection on cytokinome profiles, we assessed the significance of 121 cytokines/chemokines (ligands) and 144 corresponding receptor genes (in total 265 cytokines/chemokines-related genes, a.k.a. cytokinome genes; Fig. S3) from the results of the differentially expressed test across multiple time points. We quantified the number (and percentage) of ligands or receptors. The p-values were calculated by using a one-tailed binomial test (with a “greater” alternative hypothesis and confidence level of 95%) by comparing it to the background gene list (excluding cytokinome genes). False positivity was controlled by using Bonferroni correction. As a result, we found that the numbers of ligand genes (cytokines/chemokines) were significantly higher than the background genes (Fig. S4). However, we didn't identify any significance in receptor genes. These data suggested that ∆ORF6 or SARS infection has a great effect on the cytokine (and chemokine) levels.
3.2. Time-dependent co-expression networks of icSARS ΔORF6-induced genes
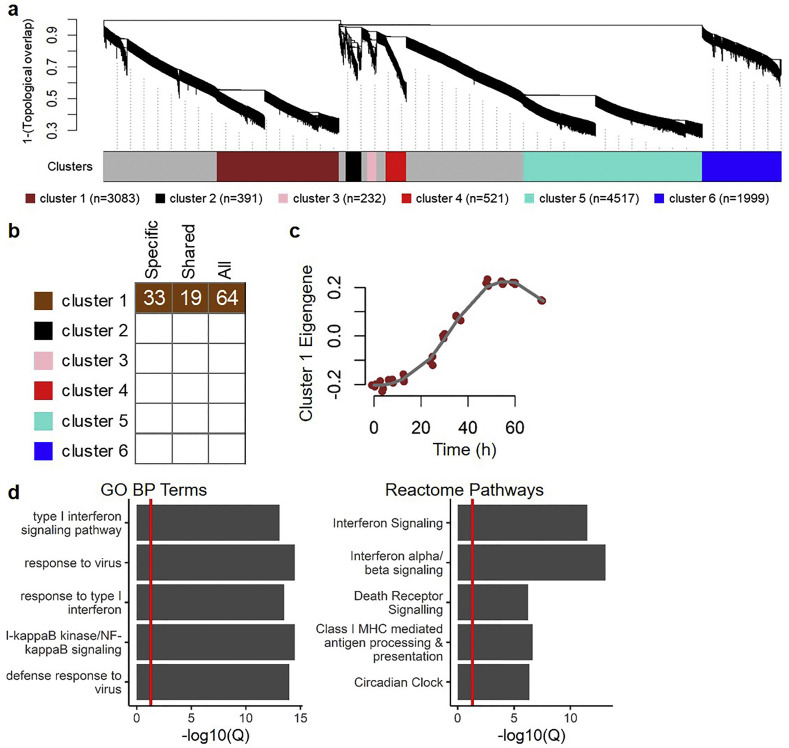
To depict the co-expression dynamics of ΔORF6 infection-related genes in human lung cells after viral infection, we constructed time-dependent co-expression networks from the GSE33267. Post-infection transcriptomic profiles of ΔORF6-infected Calu-3 cells (from 0 to 72 h, across 11 time-points, each with 3 replicates) were found to be partitioned into 6 clusters based on pairwise correlations between gene expression values; the number of genes in each cluster ranged from 232 to 4517 (Fig. 3a). In the following steps, we categorized the significantly upregulated DEGs in ΔORF6-infected cells into two groups: (i) specifically in ΔORF6-infected cells (denoted by “ΔORF6-specific” or “specific”; which containing 2613 genes) and shared in both ΔORF6- and wild-type-infected cells (denoted by “shared”; which containing 760 genes).
Fig. 3.
Cluster identification and enrichment for up-regulated differentially expressed genes (DEGs) in ΔORF6-infected Calu-3 cells. (A) Network analysis dendrogram showing co-expressed clusters of post-infected ΔORF6-infected Calu-3 cells' expression profiles across multiple time points. The size of each cluster (n = gene number) is illustrated in the parenthesis. (B) For each cluster, enrichment tests for up-regulated DEGs (ΔORF6-specific only, denoted as “Specific”; wild-type-sharing, denoted as “Shared”; and all up-regulated DEGs upon ΔORF6 treatment, denoted as “All”) are performed. This heat map shows the results of enrichment tests, with significant enrichments colored in corresponding to the color of the enriched module. Clusters that are not significant at a particular time point are in white. The numbers indicate the odds ratios (ORs) of enrichment tests. (C) Characterization of temporal dynamics of cluster 1. Each colored point indicates the principal component 1 (PC1; a.k.a. eigengene; y-axis) of genes in cluster 1 at different time points (x-axis). The post-infection temporal expression trajectory (grey line) is fitted by using a locally least-squares fitting algorithm. (D) The top 5 significant (all Q-values < 0.05) gene ontology (GO) biological process (BP) terms and Reactome pathways, with red vertical line indicates a Q-value of 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We posited that additional mapping of ΔORF6 infection-related DEGs in an identified cluster should be observed if the particular cluster reflects a pathophysiological process in ΔORF6-infected Calu-3 cells. To determine the convergence of identified co-expressed clusters of ΔORF6 infection-related genes, we conducted enrichment tests for ΔORF6-specific and shared DEGs using an over-representation analysis (ORA). Among up-regulated DEGs (both ΔORF6-specific and shared genes) in response to ΔORF6 infection, significant enrichment was detected for clusters 1 (Fig. 3b). We next depicted the temporal trajectory of cluster 1 by calculating the first principal component (PC1; i.e., cluster eigengene) and assessing the value of eigengene across time. Eigengene can be viewed as a summarized value of gene expression in the specific cluster. In particular, genes in cluster 1 were up-regulated and sustained at approximately 30 to 72 h upon ΔORF6 infection (Fig. 3c). In details, the eigengene of cluster 1 from ΔORF6-infected cells gradually increased from 0 to 54 h post-infection, and then slightly dropped from 60 to 72 h.
3.3. Biological meanings of cluster enriched for upregulated ΔORF6 infection-related DEGs
To characterize the key factor(s) in up-regulated DEGs, we hypothesized that if there exists a key factor that causes the up-regulation and sustainability of genes from approximately 30 to 72 h after ∆ORF6 infection, the key factor gene(s) should be also continuously expressed, and exert its/their regulatory functions. Since key factors should be able to regulate—turn on and off—genes' expression (upregulation), we thence focused on transcription factor(s) (TFs). Therefore, we derived a sequence-based enrichment test (please see “Materials and Methods” section) to identify TFs that may regulate the expression of ∆ORF6-specific or shared (with wild-type/SARS) DEGs. As a result, we identified 57 and 18 TFs that passed the significance threshold in ∆ORF6-specific and shared groups, respectively (Fig. S5). To validate our findings in ∆ORF6-specific up-regulated DEGs, we leveraged the chromatin immunoprecipitation sequencing (ChIP-seq) data from the Encyclopedia of DNA Elements (ENCODE) database. Of the 10 available TFs data in ENCODE, only the FOSL1 was overlapped with the 57 identified TFs. We, therefore, adopted the FOSL1 ChIP-seq results for validation. Given the information on regulatory interactions regarding FOSL1, we conducted a gene-based permutation test (please see “Materials and Methods” section) to assess the enrichment of (∆ORF6-specific) up-regulated DEGs in FOSL1 targets. As a result, we successfully replicated the significance of enrichment regarding FOSL1 using ENCODE ChIP-seq evidence (p-value <0.00001; Fig. S6).
We next sought to determine whether correlated genes in cluster 1 share biological functions or regulatory mechanisms. The top 20 most significantly enriched Reactome pathways and Gene Ontology (GO) biological process (BP) annotation terms suggested that the genes in cluster 1 were implicated in cellular defensive response to the virus, which includes interferon (especially, type I) signaling pathways (Fig. 3d).
3.4. Discovery of candidate pharmacological ligands (drugs) that potentially oppose SARS-CoV-2 infection
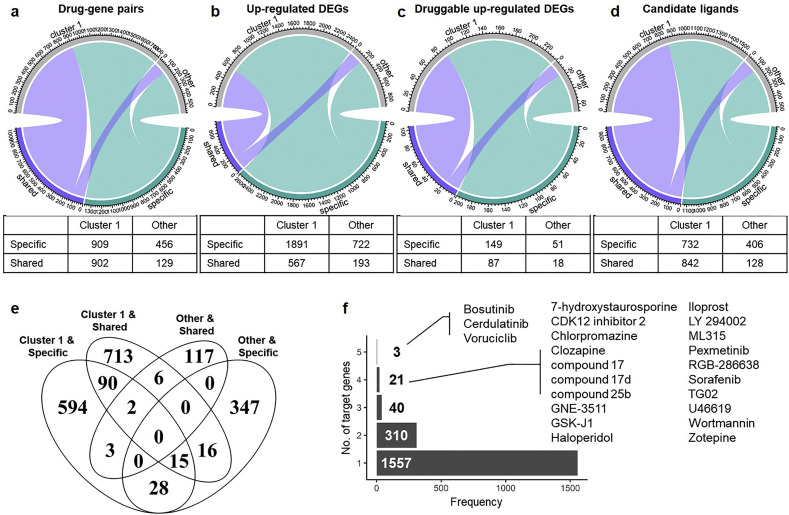
Based on the fact that SARS-CoV-2 possesses a truncated ORF6 gene, which is critical for viral sensitivity to type I IFN and may influence host immune response [17], we utilized ΔORF6 as a model for SARS-CoV-2 to compare specific and/or shared biological functions of DEGs in human lung cancer cell line infected with wild-type or ΔORF6 SARS-CoV. Specific and shared genes induced in ΔORF6-infected cells were of interest for our analysis, as both groups may provide clues for drug repositioning. By restricting our study to upregulated genes responsive to ΔORF6 infection, we were able to pinpoint candidate drugs that may inhibit the gene products. DEGs upregulated at different time-points were classified into two cluster categories (i.e., “Cluster 1” and “Other”). Together, DEGs that were specifically induced by ΔORF6 (denoted as “Specific”) or up-regulated in both ΔORF6 and SARS treatment (denoted as “Shared”) were subjected to drug repositioning analysis using the drug (ligand)-gene pairing information from the IUPHAR PHARMACOLOGICAL database. In total, 2396 drug-gene pairs (305 unique targetable genes and 1931 unique ligands) were identified, with 1365 (56.97%) pairs from the “Specific” group and 1031 (43.03%) from the “Shared” group (Fig. 4a). Among them, 1811 (75.58%) pairs were from “Cluster 1” and the remaining 585 (24.42%) pairs were from “Other”. Therefore, we identified a substantial number of candidate drug-gene pairs in “Cluster 1” and “Other” groups. The identified 3373 up-regulated genes were considered “targetable” or “druggable” and may be potentially useful to guide repositioning of drugs for the treatment of SARS-CoV-2 (Fig. 4b).
Fig. 4.
Drug repositioning based on up-regulated DEGs from ΔORF6-infected cells. Chord diagrams (top) and tables (bottom) showing the relationship between ΔORF6-specific (“specific”) and wild-type-sharing (“shared”) to cluster categories (“cluster 1” or “other”) of (A) drug-gene pairs; (B) up-regulated DEGs; (C) intersected genes between up-regulated DEGs and targetable (druggable) genes; and (D) candidate ligands for repurposing. (E) Venn diagram showing the intersected number of candidate ligands between ligands from ΔORF6-specific (“Specific”) or wild-type-sharing (“Shared”) and cluster categories (“Cluster 1” or “Other”). (F) Bar plot showing the number of targeting DEGs (x-axis) of candidate ligands. The numbers of ligands that may target 1– 5 up-regulated DEGs are counted (y-axis).
From the gene perspective, for the initial 3373 up-regulated DEGs, we consequently identified 305 unique targetable (druggable) genes by intersecting the DEGs with druggable genes from the database, with 200 (65.57%) genes were “Specific” and 105 (34.43%) genes were “Shared” (Fig. 4c). Moreover, 236 (77.38%) genes were from “Cluster 1” and 69 (22.62%) genes were from “Other”. From the ligand perspective, we ascertained in total 1931 unique candidate ligands (Fig. 4d), with some duplicated ligands been identified in different categories due to multiple target genes (Fig. 4e). We thus investigated the number of multiple gene targets of each ligand and found that most of the ligands (1557 out of 1931, 80.63%) may target unique up-regulated DEGs (Fig. 4f). However, 374 (19.37%) candidate ligands have multiple targets.
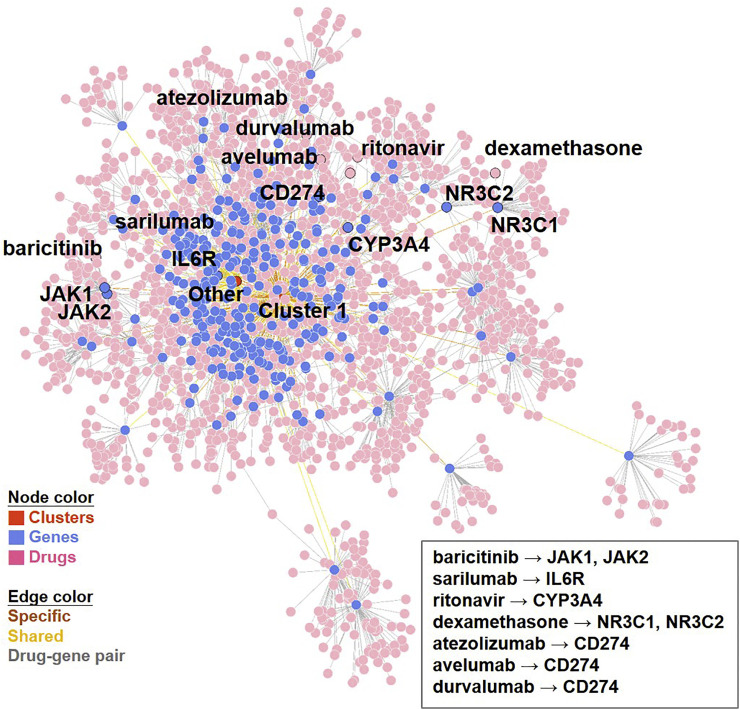
Since McCreary et al. suggested several putative pharmacotherapeutic options for COVID-19 based on current clinical evidence (up to March 28, 2020) [18], we sought to verify our primary findings by examining the candidate drugs identified and discussed in the study. Among previously identified drug candidates, our analysis successfully identified baricitinib (targeting JAK1 and JAK2), sarilumab (targeting IL6R) and ritonavir (targeting CYP3A4) as potential drugs for the treatment of SARS-CoV-2 infection (Fig. 5 ). Since the upstream (IL6R) and downstream (JAK1/2) genes in IL6 signaling pathway were up-regulated, we thus checked the significance of IL6 gene. Indeed, IL6 has successfully been identified as up-regulated DEGs (i.e., passed the significant threshold of three methods, Fig. S7). After prioritization, we found two drugs (siltuximab and sirukumab) as candidate ligands for the treatment COVID-19. Furthermore, we also detected dexamethasone (targeting gene product of NC3R1 and NC3R2) as a candidate ligands for SARS-CoV-2, which was proved to reduce mortality in COVID-19 patients who received respiratory support [19]. These results suggested that our pipeline could be empirically supported by current therapeutic options.
Fig. 5.
The network of (total) 1931 drug-gene pairs. The total identified drug-gene pairing network is displayed by using the Fruchterman-Reingold (FR) force-directed algorithm. In addition to dexamethasone (targeting gene product of NR3C1 and NR3C2), the drugs (and corresponding genes) that have been discussed in the review article of EK McCreary et al.21 and immune checkpoint inhibitors (ICIs) targeting PD-L1 (gene product of CD274) and their targeting genes are illustrated.
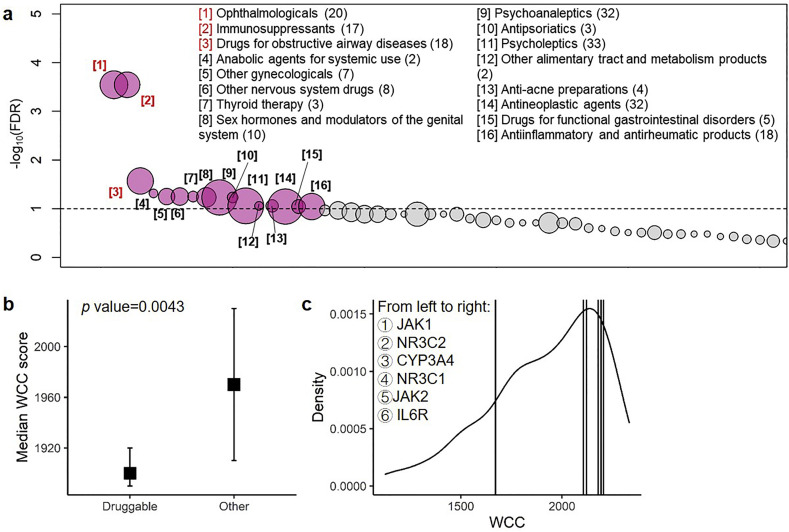
We reasoned that our transcriptomic-driven drug discovery approach may provide a novel perspective regarding the pharmacological mechanisms of drugs that could be considered for repositioning to SARS-CoV-2 infection. To assess the pharmacological actions of the candidate ligands, pharmacological and chemical properties of drugs were annotated using the Anatomical Therapeutic Chemical (ATC) classification code. As a result, we identified 345 (17.87% of 1931) ligands with available ATC code which may target 92 druggable genes. A permutation test was conducted to assess the enrichment of specific ATC groups for the 92 unique targetable genes and revealed significant enrichment in 16 out of 57 (28.07%) ATC groups (FDR < 0.1; Fig. 6a). Especially, the top 3 most significantly enriched ATC groups with FDR < 0.05 were “ophthalmologicals” (FDR = 0.00057), “immunosuppressants” (FDR = 0.00086), and “drugs for obstructive airway diseases” (FDR = 0.0268).
Fig. 6.
Enrichment in the Anatomical Therapeutic Chemical (ATC) classification system and gene-based prioritization using within-cluster connectivity (WCC). (A) Enrichment of the drugs' Anatomical Therapeutic Chemical (ATC) classification groups from 92 targetable genes. Scatterplot showing the permutation results of drugs' ATC classification groups by randomly sampling 92 targetable genes for 99,999 times. The nodes representing ATC groups (x-axis) that pass the significant threshold (grey dashed horizontal line, i.e., false discovery rate (FDR) = 0.1; y-axis) are colored in red, while the remaining are colored in grey. The 16 significant ATC groups are listed, with the number in parenthesis indicates the number of drugs that may target the 92 genes and number in bracket indicates the rank of significance. The top 3 ATC groups (i.e. “Ophthalmologicals”, “Immunosuppressants” and “Drugs for obstructive airway diseases”) that passed FDR < 0.05 were highlighted using red color. (B) Box plot showing the distribution of WCC scores (y-axis) of druggable genes and the remaining genes (x-axis). (C) The distribution of WCC (x-axis) across genes in Cluster 1. The vertical lines indicate the genes that are targeted by dexamethasone or potential ligands discussed in EK McCreary et al. [18]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Since “immunosuppressants” was identified as an enriched ATC term, implying the roles of immune-related functions for drug repositioning to COVID-19. Therefore, we investigated whether other immune-relevant molecules could be exploited for drug repositioning in SARS-CoV-2 infection, which exemplified by immune checkpoint markers, such as PDCD1 and HAVCR2. However, in this study, differentially expressed tests and following drug repositioning analyses were derived from virus-infected human Calu-3 cells. Since T cells data was not being included in our research, we could not analyze the differential expression pattern of PDCD1 and HAVCR2, which mainly be expressed on T cells. We thus investigated the DEG results of CD274 and PDCD1LG2 genes due to the following reasons: (1) CD274 and PDCD1LG2 were druggable by immune checkpoint inhibitors (ICIs) and (2) CD274 and PDCD1LG2 could bind to PDCD1 gene product to trigger T-cell exhaustion. We found a significant up-regulation of CD274 (but not for PDCD1LG2) as DEG, and thus identified atezolizumab, avelumab, and durvalumab (targeting gene product of CD274) as candidate ligands.
3.5. Prioritization of candidate ligand-gene pairs for drug repositioning to SARS-CoV-2 infection
Given the substantial number of identified drugs with the potential to be repositioned for treatment of SARS-CoV-2 infection, we utilized several drug-based (a.k.a. ligand-based) and gene-based criteria to prioritize our candidate drug-gene pairs. For six identified clusters, we first restricted the results to “Cluster 1” genes based on following reasons (Fig. S8): (1) known candidate targetable genes (JAK1, JAK2, CYP3A4, IL6R, NC3R1 and NC3R2) were restricted in Cluster 1; (2) In compared to Cluster 1 (18% ~ 61%, approximately), Clusters 2–6 showed a lower level of intersection to the ∆ORF6-infected DEGs (0% ~ 11%, approximately); (3) For Cluster 1, a number of 236 druggable genes were found. However, for Clusters 2–6, only 0–1 druggable gene was identified; (4) Similar results were inspected while examining the number of drug-gene pairs and number of candidate ligands. In light of the underrepresentation of ∆ORF6-infected DEGs and candidate druggable genes in Clusters 2–6, Cluster 1 was focused for drug repositioning, and resulting in 236 (77.38%) unique genes and 1467 (75.97%) unique ligands.
We then leveraged a network (cluster) property, so-called within-cluster connectivity (WCC; i.e. the sum of weights connecting to the gene) to perform gene-based prioritization. We compared the distributions of WCC scores between druggable genes and the remaining genes and found that WCC scores of druggable genes were significantly lower than that of the remaining genes in Cluster 1 (asymptotic general independence test Z value = −2.85; p-value = 0.0043; Fig. 6b). This result indicated that genes with lower connectivity (non-hub) tend to be developed as a pharmacotherapeutic target. We therefore selected the genes with a WCC score of ≤ 2177.606, which was the WCC score of NR3C1 (targeted by dexamethasone), and resulting in 188 druggable genes (Fig. 6c). These 188 genes corresponded to 1165 candidate ligands. Notably, JAK2 and IL6R were filtered in this step due to higher WCC scores.
Additionally, in consideration of some candidate ligands from IUPHAR PHARMACOLOGY database were under investigation and thus may not be suitable for drug repositioning, we performed ligand-based prioritization by limiting our candidate drugs to those with ATC annotation. Finally, 55 druggable genes and 238 candidate ligands were discovered for drug repositioning to SARS-CoV-2 infection (Fig. S9).
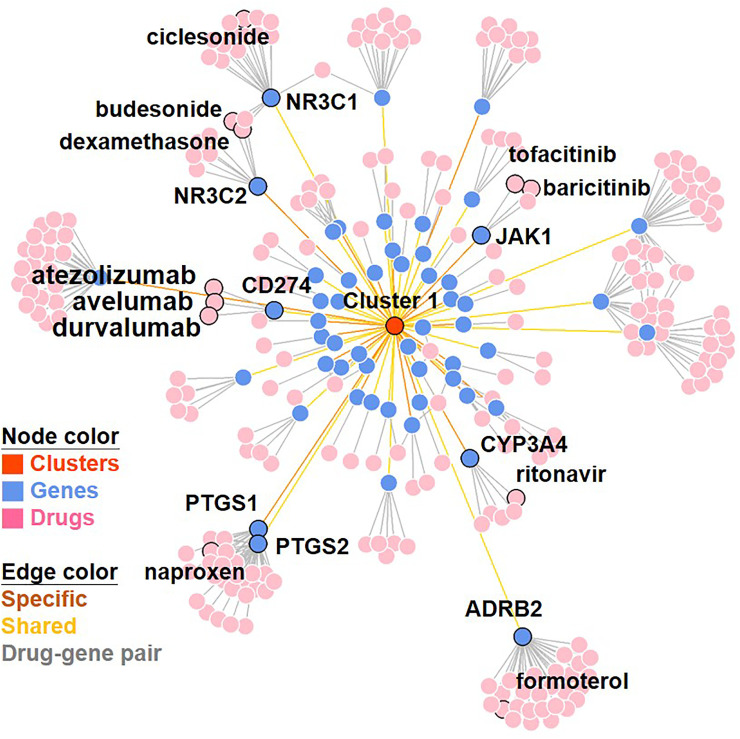
To validate our findings, we next asked whether some of the candidate drugs for repositioning to SARS-CoV-2 infection identified from our study are in ongoing clinical trials for COVID-19. Indeed, our analysis identified seven drugs (including ritonavir, baricitinib, tofacitinib, naproxen, budesonide, ciclesonide and formoterol) in ongoing clinical trials. These seven drugs target 7 genes found in our study, including CYP3A4, JAK1, PTGS1, PTGS2, NR3C1, NR3C2 and ADRB2. It is noteworthy that dexamethasone was also been identified as candidate ligands, which targets PTGS1 and PTGS2 genes (Fig. 7 ).
Fig. 7.
The network of (prioritized) 296 drug-gene pairs. The final prioritized drug-gene pairing network is displayed by using the Fruchterman-Reingold (FR) force-directed algorithm. The drugs (and corresponding genes) that belonged to immune checkpoint inhibitors (ICIs), that had undergoing clinical trials and that have been discussed in the review article of EK McCreary et al. [18] and it's targeting gene(s) are illustrated.
4. Discussion
With the pandemic threat of COVID-19 as it currently stands and the lack of time to develop new medical intervention for over millions of hospitalized victims, devising an empirical yet rationably effective therapeutic strategy is in urgent need. Unfortunately, no preclinical omic-level data from SARS-CoV-2-infected human tissues, especially for lung transcriptomics, are currently not available, posing substantial challenges for drug development or repositioning. A feasible, appealing approach is to use pre-existing data that closely resembled SARS-CoV-2-infection for bioinformatics analyses, thereby identifying candidates of currently available medications to be repositioned for SARS-CoV-2 therapy. In this study, we performed such task by analyzing the gene expression profile of a human lung cell line, Calu-3, infected with SARS-CoV ΔORF6, a mutant closely resembling SARS-CoV-2. The rationale for using the mutant as a surrogate is justified by the fact that both the mutant and SARS-CoV-2 lack the antagonistic effect toward interferon response due to a truncation in ORF6. Our method of transcriptomics-driven drug discovery ultimately provided a candidate list of marketed drugs (approved for other indications) to be used in empirical clinical therapy for the urgent need of SARS-CoV-2 infection.
Specifically, we prioritized 55 targetable genes that respond to ΔORF6 infection. These genes can be modulated by 238 candidate drugs that are currently available and hence can be repositioned for COVID-19 theraphy. The reliability of our analyses were justified by the following facts. First, we predicted several drugs including ritonavir, baricitinib, tofacitinib, naproxen, budesonide, ciclesonide and formoterol that are currently under clinical trials. Second, despite of an intensive discussion on the empirical use of several drugs including hydroxychloroquine/chloroquine, remdesivir, lopinavir and ritonavir, interleukin (IL)-1 and − 6 inhibitors, and Janus kinase (JAK) inhibitors, no consistent or conclusive evidence for the safety and efficacy is available, except for dexamethasone, which remarkably was on our list of candidates. As such, the analyses we provided support the idea that analyzing transcriptomic data from virus-infected cells can yield valuable information for potential treatments, which can further be leveraged to identify potential candidates for drug repositioning.
Without any preclinical data or clinical experiments, current therapeutic options on COVID-19 infection are lacking in solid evidence and empirically limited to antiviral agents and host modifiers. In light of recent studies and reviews on potential drugs for COVID-19 therapy [18,[20], [21], [22], [23]], our list of candidates contains unexpected drugs. These candidate drug categories can expand the scope of COVID-19 drug targets, open new avenues for future study, and include agents with solid track records in clinical treatment of infectious disease for empirical tests. For instance, we identified several repositioned candidates (e.g., ophthalmological agents, immunosuppressive agents, drugs for obstructive airway diseases, etc.), which have proven useful in treating other viral infections. Also identified were drugs for obstructive airway diseases (e.g. formoterol and ciclesonide) as candidate for repositioning to COVID-19. Notably, a previous in vitro study has shown that combination use of formoterol with budesonide (both were identified as candidate ligands) and glycopyrronium can inhibit the viral replication and cytokine production of HCoV-229E in human respiratory epithelial cells [24,25]. Besides, several studies have suggested that ciclesonide may block the replication of SARS-CoV-2 ribonucleic acid and thus reduce the virus-induced cytopathogenic effect in COVID-19 patients [26,27]. Thus, it appears reasonable to reposition and include drugs from other categories for COVID-19 treatment.
One intriguing feature in our prediction was the emergence of drugs for immune modulation. Yet several lines of molecular clinical evidence suggest that taking into account immune modulation might be necessary for COVID-19 therapy. First, the pathology of the SARS-CoV-2 infected lungs showed mixed features of acute alveolar injury and diffusive immune responses including immune infiltration, focal reactive hyperplasia, and intra-alveolar fibroblastic plug formation [5]. Second, the clinical severity of COVID-19 infection is linked to highly inflammatory responses that produce enormous chemokines implicated in a cytokine storm [6,7]. Third, a key difference between SARS-CoV and SARS-CoV-2 is a truncation of the open reading frame (ORF) 3b, ORF6, in SARS-CoV-2, which affects the viral antagonistic effect toward interferon response [8]. On our list, we found a significant enrichment in the category of “anti-inflammatory and antirheumatic products”, which may alleviate immune-inflammatory injury that is known to cause increased oxidative stress and organ failure in virus-infected patients [28,29]. Also identified was β2-adrenergic signaling (elicited by some drugs for obstructive airway diseases such as formoterol, salbutamol, etc.) to be critical in norepinephrine-mediated processes and in the seretion of TNFα and IL-10, which may relieve tissue-damaging inflammation (and thus exerts a protective effect to patients) during viral infection [30].
Most striking was CD274 (PD-L1) identified as druggable gene in this study, suggesting the plausibility of repurposing ICIs (immune checkpoint inhibitors) for COVID-19 therapy. We attribute this finding to the cell lines applied in current analysis, namely, Calu-3, a human lung cancer cell line. These drugs include atezolizumab, avelumab and durvalumab. The idea of implimenting ICIs for COVID-19 therapy is supported by a previous study showing that T-cell exhaustion markers (PDCD1 and HAVCR2) were overexpressed in severe cases of COVID-19 patients [31]. Two recent commentary articles also addressed the possibility of using ICIs to reinvigorate exhuausted T-cell in the early phase of COVID-19 infection, yet the discussion was mainly focused on cancer patients receiving ICI treatment [32,33]. Should ICIs be given during pandemic conditions, especially for cancer patients ? We note that drugs targeting immune checkpoints may lead to immune checkpoint inhibitor-related pneumonitis (ICI-P) at a relatively low occurrence rate (4–13%) [[34], [35], [36], [37]]. Given that immunostimulants such as ICIs may exacerbate the clinical symptoms (especially pneumonitis) of SARS-CoV-2 infection, repositioning of these drugs to COVID-19 should be evaluated with great caution. Moreover, it may be useful to consider combining ICIs with other drugs that mitigate unwanted ICI-induced inflammation, such as IL6 and JAK2 inhibitors [[38], [39], [40]]. Consistently, the potential of targeting IL6 signaling pathway for COVID-19 therapy has already been considered [18,41]. Of note, our analyses pinpointed several ligands targeting IL6, IL6R, JAK1 and JAK2, suggesting the possibility of using ICIs with IL6 signaling pathway blockers in a combination therapy.
For COVID-19, concerns have been raised on enhancing SARS-CoV-2 virulence by treatment of widely used drugs, such as angiotensin-converting enzyme 2 (ACE2) inhibitors and angiotensin-receptor blockers (ARBs) [[42], [43], [44], [45]]. Consistent with this concern, we did not detect significant enrichment for the category “agents acting on the renin-angiotensin system”. To maximize the accuracy of our drug repositioning predictions, we prioritized the initially identified 2396 drug-gene pairs into a list of 296 (12.35%) drug-gene pairs. The final list was restricted to (i) drugs with gene target(s) in “Cluster 1”, (ii) drugs with target gene(s) with lower WCC compared to dexamethasone, and (iii) drugs with an available ATC code. Excluding candidate drug-gene pairs that lack ATC annotations, we identified high-priority drug-gene pairs with likely disease relevance. Note that our analysis may be undermined by a lack of data from preclinical cases of SARS-CoV-2 infection and a lack of clinical specimens from COVID-19 patients. It is possible that the DEGs identified from the surrogate system may not fully reflect genes that respond to SARS-CoV-2 and promote disease progression in COVID-19 infection. Another potential caveat is that we did not take into account physical binding affinities in drug-gene interactions. Moreover, since our analyses were based on data from infected cells, we might not be able to identify candidate drugs that act on viral proteins or human gene products involved in the process of virus entry. For example, ACE2 and TMPRSS2 were identified as the entry factors for SARS-CoV-2 [46]. Among those genes we analyzed, TMPRSS2 was identified as a differentially-expressed gene, and TMPRSS2-upregulating variants were more frequently found in European and American populations [47]. However, we failed to detect the corresponding candidate ligand targeting TMPRSS2. Neither was ACE2 identified as a DEG in our study. Based on the ATC enrichment test, we indeed failed to identify enrichment of “Agents acting on the renin-angiotensin system” in the candidate drug list. Thus, it is likely that the renin-angiotensin system may play an important role in the early stage of infection but not associate with the ongoing pathogenesis after viral infection. Although our initial attempt was to predict drugs for empirical clinical use, we should point out that the clinical feasibility of using candidate drugs for repositioning was not accessed in our study. As such, comorbidities and potential drug-drug interactions arising from polypharmacy in COVID-19 patients should be carefully considered and investigated before repositioning specific drugs and regimens for COVID-19 therapy. Likewise, preclinical experiments are needed to demonstrate the potential efficacy of identified drugs, and preclinical evidence should be gathered before moving to clinical application.
5. Conclusions
In summary, our results reveal potential candidates for drug repositioning to SARS-CoV-2 therapy. The candidates we found include drugs from categories other than “antivirals for systemic use” (based on ATC code), suggesting that drugs with various indications should be considered in future studies. Given the scarcity of omics-level data from SARS-CoV-2 infected lung tissue, our surrogate transcriptomic-driven drug discovery approach has prioritized the list of potential candidate drugs to be experimentally examined in preclinical studies, to be considered for empirical clinical use when it is needed, and to open novel therapeutic avenues for the treatment of COVID-19.
Authors' Contributions
H.W.S.C., G.H.L, Y.O. and W.C.C. designed the study. H.W.S.C. and G.H.L. performed the bioinformatics. H.W.S.C., C.L.G., G.H.L, Y.O. and W.C.C. analyzed and discussed the data. H.W.S.C., G.H.L, K.Y.L., Y.O. and W.C.C. interpreted the data. H.W.S.C., C.L.G., G.H.L and W.C.C. wrote the paper.
Funding
This work was supported by grants from Ministry of Science and Technology, Taiwan (MOST109-2314-B-038-131 and MOST109-2628-B-038-012), and Taipei Medical University, Taiwan (12310-106079; Yusuke Nakamura Chair Professorship).
Declaration of Competing Interest
The authors declare that they have no known conflicts of interest associated with this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2020.12.041.
Appendix A. Supplementary data
Supplementary material
References
- 1.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 2.de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Chen L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature Medicine. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 8.Lokugamage K.G., Hage A., Vries M.d., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020;94:e01410–e01420. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005:4. doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 13.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 15.Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S., et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schafer A., Matzke M.M., Webb-Robertson B.J., Chang J., Luna M.L., Long C.E., et al. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCreary E.K., Pogue J.M. Coronavirus disease 2019 treatment: a review of early and emerging options. Open Forum Infect. Dis. 2020:7. doi: 10.1093/ofid/ofaa105. ofaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. medRxiv. 2020. Effect of dexamethasone in hospitalized patients with COVID-19: Preliminary report. [Google Scholar]
- 20.Simsek Yavuz S., Unal S. Antiviral treatment of COVID-19. Turk. J. Med. Sci. 2020;50:611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Ind. J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenkens M., de Wit H., Danser A.H., Esselink A.C., Horikx A., Ten Oever J., van de Veerdonk F., Kramers C. Medication and comedication in COVID-19 patients. Ned. Tijdschr. Geneeskd. 2020;164 [PubMed] [Google Scholar]
- 23.Some drugs for COVID-19 Med. Lett. Drugs Ther. 2020;62:49–50. [PubMed] [Google Scholar]
- 24.Halpin D.M.G., Singh D., Hadfield R.M. Inhaled corticosteroids and COVID-19: a systematic review and clinical perspective. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.01009-2020. 2001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaya M., Nishimura H., Deng X., Sugawara M., Watanabe O., Nomura K., Shimotai Y., Momma H., Ichinose M., Kawase T. Inhibitory effects of glycopyrronium, formoterol, and budesonide on coronavirus HCoV-229E replication and cytokine production by primary cultures of human nasal and tracheal epithelial cells. Respir. Investig. 2020;58:155–168. doi: 10.1016/j.resinv.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W., Shimojima M., Fukushi S. bioRxiv. 2020. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rainsford K.D. Influenza (“bird flu”), inflammation and anti-inflammatory/analgesic drugs. Inflammopharmacology. 2006;14:2–9. doi: 10.1007/s10787-006-0002-5. [DOI] [PubMed] [Google Scholar]
- 29.Garcia C.C., Guabiraba R., Soriani F.M., Teixeira M.M. The development of anti-inflammatory drugs for infectious diseases. Discov. Med. 2010;10:479–488. [PubMed] [Google Scholar]
- 30.Ağaç D., Estrada L.D., Maples R., Hooper L.V., Farrar J.D. The β2-adrenergic receptor controls inflammation by driving rapid IL-10 secretion. Brain Behav. Immun. 2018;74:176–185. doi: 10.1016/j.bbi.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., et al. medRxiv. 2020. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickles O.J., Lee L.Y.W., Starkey T., Freeman-Mills L., Olsson-Brown A., Cheng V., Hughes D.J., Lee A., Purshouse K., Middleton G. Immune checkpoint blockade: releasing the breaks or a protective barrier to COVID-19 severe acute respiratory syndrome? Br. J. Cancer. 2020;123:691–693. doi: 10.1038/s41416-020-0930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maio M., Hamid O., Larkin J., Covre A., Altomonte M., Calabro L., Vardhana S.A., Robert C., Ibrahim R., Anichini A., et al. Immune checkpoint inhibitors for cancer therapy in the COVID-19 era. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2020;26:4201–4205. doi: 10.1158/1078-0432.CCR-20-1657. [DOI] [PubMed] [Google Scholar]
- 34.Domagała-Kulawik J. Immune checkpoint inhibitors in non-small cell lung cancer - towards daily practice. Adv. Respir. Med. 2018;86 doi: 10.5603/ARM.2018.0022. [DOI] [PubMed] [Google Scholar]
- 35.Kalisz K.R., Ramaiya N.H., Laukamp K.R., Gupta A. Immune checkpoint inhibitor therapy-related pneumonitis: patterns and management. Radiographics. 2019;39:1923–1937. doi: 10.1148/rg.2019190036. [DOI] [PubMed] [Google Scholar]
- 36.Larsen B.T., Chae J.M., Dixit A.S., Hartman T.E., Peikert T., Roden A.C. Clinical and Histopathologic features of immune checkpoint inhibitor-related pneumonitis. Am. J. Surg. Pathol. 2019;43:1331–1340. doi: 10.1097/PAS.0000000000001298. [DOI] [PubMed] [Google Scholar]
- 37.Cho J.Y., Kim J., Lee J.S., Kim Y.J., Kim S.H., Lee Y.J., Cho Y.J., Yoon H.I., Lee J.H., Lee C.T., Park J.S. Characteristics, incidence, and risk factors of immune checkpoint inhibitor-related pneumonitis in patients with non-small cell lung cancer. Lung Cancer. 2018;125:150–156. doi: 10.1016/j.lungcan.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan R.J., Johnson D.B., Rini B.I., Neilan T.G., Lovly C.M., Moslehi J.J., Reynolds K.L. COVID-19 and immune checkpoint inhibitors: initial considerations. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Cosimo S., Malfettone A., Pérez-García J.M., Llombart-Cussac A., Miceli R., Curigliano G., Cortés J. Immune checkpoint inhibitors: a physiology-driven approach to the treatment of coronavirus disease 2019. Eur. J. Cancer. 2020;135:62–65. doi: 10.1016/j.ejca.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stroud C.R., Hegde A., Cherry C., Naqash A.R., Sharma N., Addepalli S., Cherukuri S., Parent T., Hardin J., Walker P. Tocilizumab for the management of immune mediated adverse events secondary to PD-1 blockade. J. Oncol. Pharm. Pract. 2019;25:551–557. doi: 10.1177/1078155217745144. [DOI] [PubMed] [Google Scholar]
- 41.Atal S., Fatima Z. IL-6 inhibitors in the treatment of serious COVID-19: a promising therapy? Pharm. Med. 2020:1–9. doi: 10.1007/s40290-020-00342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins J. Preventing a covid-19 pandemic. BMJ. 2020;368:m810. doi: 10.1136/bmj.m810. [DOI] [PubMed] [Google Scholar]
- 43.Esler M., Esler D. Can angiotensin receptor-blocking drugs perhaps be harmful in the COVID-19 pandemic? J. Hypertens. 2020;38:781–782. doi: 10.1097/HJH.0000000000002450. [DOI] [PubMed] [Google Scholar]
- 44.Diaz J.H. Hypothesis: angiotensin-converting enzyme inhibitors and angiotensin receptor blockers may increase the risk of severe COVID-19. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa041. taaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irham L.M., Chou W.H., Calkins M.J., Adikusuma W., Hsieh S.L., Chang W.C. Genetic variants that influence SARS-CoV-2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem Biophys Res Commun. 2020;529:263–269. doi: 10.1016/j.bbrc.2020.05.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material