Abstract
In this ongoing theme of coronavirus disease 2019 (COVID-19) pandemic, highly sensitive analytical testing platforms are extremely necessary to detect SARS-CoV-2 RNA and antiviral antibodies. To limit the viral spread, prompt and precise diagnosis is crucial to facilitate treatment and ensure effective isolation. Accurate detection of antibodies (IgG and IgM) is imperative to understand the prevalence of SARS-CoV-2 in public and to inspect the proportion of immune individuals. In this review, we demonstrate and evaluate some tests that have been used commonly to detect SARS-CoV-2. These include nucleic acid and serological tests for the detection of SARS-CoV-2 RNA and specific antibodies in infected people. Moreover, the vitality of biosensing technologies emphasizing on optical and electrochemical biosensors toward the detection of SARS-CoV-2 has also been discussed here. The early diagnosis of COVID-19 based on detection of reactive oxygen species overproduction because of virus-induced dysfunctioning of lung cells has also been highlighted.
Keywords: Pandemic, SARS-CoV-2, COVID-19, Diagnostics, Emerging techniques, Biosensors
Graphical abstract
Introduction
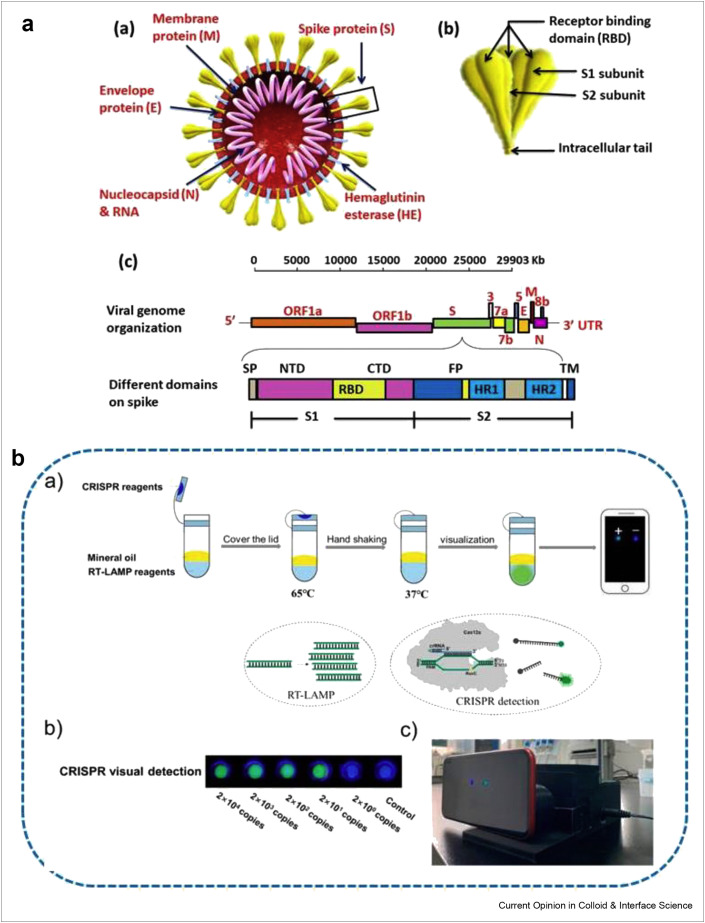
Recently, coronavirus disease 2019 (COVID-19) has caused a world health disaster and socioeconomic adversity by infecting millions and killing thousands of people. This pandemic is reported to have appeared as a result of a novel mutation of coronavirus as the seventh member of corona family outbreaking in December 2019 in Wuhan, China [1]. Subsequently, it was noticed that this novel coronavirus possessed 96% genome sequence similarity with SARSr-CoV-RaTG13 (bat SARS) and 79.5% with SARS-CoV [2]. The single-stranded RNA β-coronavirus [3] with Nidovirales order belongs to the Coronaviridae family [4] and comprises of nucleocapsid, spike (S), membrane (M), and envelope (E) proteins as shown in Figure 1 a. The receptor binding domain (RBD) in spike-protein shares the angiotensin-converting enzyme 2 (ACE2) of human cells as a receptor [5]. The spike proteins are strongly attracted toward ACE2, which defines the virus-host array to RBD and ensures the viral fusion with host cell membrane [6].
Figure 1.
The detailed genomic representation of SARS-CoV-2 and its mobile phone detection (a) Structural illustration of SARS-CoV-2 (a) structural image of SARS-CoV-2, containing four different types of structural proteins (b) is the inset of key features of spike protein, representing the receptor binding domain, S1 and S2 regions, whereas (c) shows the full-length viral genome (29,903 nucleotides long) single-stranded positive RNA sequence. The leader sequence at the 5′ end and poly A tail ends at the 3′, contains structural proteins E,M,N,S along with different accessory genes (ORF 1a, 1b, 3a, 6, 7a, 7b, and 8b). The open reading frame ORF1a/b ‘the replicase’ is responsible for the coding of polymerase enzymes for viral RNA synthesis as well as non-structural proteins. The S1 subunit of spike protein consist of N-terminal domain (NTD), receptor binding domain (RBD) and C-terminal domain, whereas the S2 subunit comprises heptad repeat 1and 2, which plays a critical role in viral entry (b) The flow chart diagram describing the whole experimental process and principle for contamination-free visual detection method to detect different concentrations of RNA. Reprinted with permission from Ref. [56].
As of today, in the absence of any recommended vaccine so far, swift and reliable diagnostic methods are of great significance for the diagnosis of symptomatic as well as asymptomatic COVID-19 cases [7]. The quick and reliable treatment decisions and quarantine strategies to slow down the spread of this infectious disease are all based on the diagnostics [8]. Some conventional testing methodologies like thoracic imaging, portable chest X-ray, flexible bronchoscopy, and computed tomography (CT) scan have been used for the preliminary diagnosis of infection. The quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) based testing [9] is regarded as the most popular test these days. Although these testing techniques are of great value but time consumption, tedious sample preparation and the need of expertise put them at the back. The infection rate and contagiousness of SARS-CoV-2 is much higher compared with other SARS infections. Therefore, rapid and point-of-care diagnostic methods are highly desirable to overcome the limitations of conventional techniques.
For rapid and point-of-care detection, lateral flow assay (LFA), loop-mediated isothermal amplification (LAMP) method, reverse transcription-LAMP (RT-LAMP) approach, clustered regularly interspaced short palindromic repeats (CRISPR) technology [10], and SHERLOCK [11] detection methods are generally implemented these days. Moreover, enzyme-linked immunosorbent assay (ELISA), LFAs, and microfluidic devices are the point-of-care testing methods under consideration. The technology behind testing is based on biosensing; either in the form of undeviating detection of biochemical and biotic molecules from the living sources or it is biomarker detection from bionics. The direct binding of analyte with target molecules and biochemical reactions are two benchmark spectacles of biosensing approaches. However, easy handling, small sample size, real time analyte tracing, and flexibility to detect the target analyte of interest within seconds are the other major advantages of biosensing platforms, which can circumvent the bottlenecks of other techniques [12]. It is also worth mentioning that electrochemical biosensors are reliable analytical techniques with the potential of being modest to sample absorbance or turbidity for the detection of contagious disease.
Herein, we outline the SARS-CoV-2 detection techniques with their pros and cons. We also present recent advances in standard testing tools and emerging biosensing platforms as portable devices for the timely detection of SARS-CoV-2 to prevent the spread of the infection. We also aim to offer a new perspective on the biosensor-based technologies for the rapid and reliable detection of SARS-CoV-2 to carry out the early diagnosis of COVID-19. This review concludes with a short discussion regarding current challenges and future perspectives.
Current detection approaches for SARS-CoV-2
The most commonly used techniques for COVID-19 diagnostics are nucleic acid test and CT scan. The procedure of identification and targeting the virus is considered more suitable when molecular approaches are used in comparison to syndromic or CT scan methods. These molecular approaches are developed after analyzing the proteomic and genomic structure of the microbes. The first genomic research on SARS-CoV-2 was done using metagenomic RNA sequence, which is an efficient way of sequencing different genomes [13].
Designing nucleic acid tests
The RT-qPCR is a version qPCR technique, which is unequivocally developed for RNA detection. So, it is being used for SARS-CoV-2 detection because it directly tests for the existence of virus RNA. RT-qPCR is regarded as the standard approach for COVID-19 identification. This approach is sufficiently reliable and can complete the results in few hours [14]. The commercially available kits use RT-qPCR techniques which detect more than two target regions and consequently the detection sensitivity is enhanced. This technique amplifies even a small viral genetic material in the testing samples. It works on two consecutive reactions; firstly, it converts the virus RNA to small complementary DNA sequence (cDNA) for special identification of paired sequencing on viral RNA. Secondly, cDNA is amplified through PCR using gene-specified primers and fluorescent-labeled hydrolysis probes. The DNA produced in first step is used in second step, where it is multiplied through repeated thermal cycling and finally the virus is detected using quantitative qPCR machine. To date, numerous RT-qPCR kits have been established for the SARS-CoV-2 detection by targeting the structural proteins and accessory genes (nucleocapsid (N), spike (S) protein (RdRP), envelope (E), or ORF1b, ORF8 genes) as biomarkers for SARS-CoV-2 detection. The reverse transcription of SARS-CoV-2 RNA into cDNA takes place in RT-qPCR test case where the specific cDNA zone is further amplified to complete the process [15,16]. Zambon's research group conducted a research on alignment and investigation of several SARS-associated genome sequencing to develop the primers and probes [17]. The E, N proteins, and RNAP (RNA polymerase) genes are amid three of discovered genes at present. First two of the three genes are found to have high analytical sensitivity, whereas the third one offers feeble sensitivity for detection. The RT-qPCR tests are carried out both in single-step and double-step procedure. The single-step assay exhibits rapid response and good reproducibility, and the whole process of reverse transcription and PCR amplification takes place in one step. However, this procedure finds it difficult to optimize reverse transcription and amplification based on the simultaneous occurrence of both steps. In comparison, double-step assay shows high sensitivity but requires more time as the reactions are completed in separate tubes. Although the two-step assay is highly sensitive and flexible, one-step assay is favored for the detection of SARS-CoV-2 because of its speed, simplicity, limited sample requirement, minimum counter time, as well as less cross contamination during the RT and real-time polymerase reaction periods [18,19]. Xiang's research group presented DNA nanoscaffold-based SARS-CoV-2 detection to diagnose COVID-19 [20]. Recently, Lubke's research group designed RNA extraction free protocol based on RT-qPCR for the detection of SARS-CoV-2 where the E-gene was targeted as novel primer [21]. Without dilution and heat inactivated, 5 μl respiratory samples were used. The assay was validated using 91 respiratory samples, where 81.3% samples were detected positive. It is concluded that direct RT-qPCR is a well-acknowledged alternative to conventional RNA RT-qPCR for analyzing fresh samples. Li et al. has studied 610 patients who were diagnosed with COVID-19 according to CT scan diagnosis, and reported the higher rate of false-negative RT-PCR results for SARS-CoV-2 detection [22]. The variations in the results by RT-PCR detection might be possible due to the low viral load in the test samples at early stages of infection, prolonged RNA conversion, and transmutations in the probes target regions [23]. PCR assays are not sensitive enough to differentiate between symptomatic and asymptomatic patients [24] and susceptible to few generalizations, which limit their applications such as:
-
1.
The small countryside hospitals are not well equipped to carry out PCR tests. Lack of expertise and infrastructure limit this facility.
-
2.
There is a possibility of false-negative results because the appropriate sampling time for nasopharyngeal swab is still vague for the patients on ventilators [25].
-
3.
A tedious procedure for sample preparation is required, which includes cell lysis and nucleic acid purification. Also, the extractions kits are less than the ramping up demand of tests [26].
-
4.
During transportation, the denaturation of sample may occur, which leads to false-negative results [27].
-
5.
In case of fully recovered patients, PCR testing can even result false-positive outcome detecting genetic material of inactive SARS-CoV-2, which is actually not reinfection. The genetic material never dies and can persist in the body for several days to months, so it can be detected after months of recovery [28].
-
6.
The nucleic acid amplification is used in PCR testing, where distinct primers and probes for every target are needed that consequently limit PCR's flexibility of scaling up for other nucleic acids in an easy and prompt way.
-
7.
If mutations of SARS-CoV-2 (which are likely to be) are undergoing, then it may be a problem for same target. Thus, continuous monitoring of viral genetic material is mandatory to warrant the accuracy of test [29].
No doubt qPCR and RT-qPCR are evolving day by day with the number of advantages but these few considerations are needed to be improved in RT-qPCR to make quick, more sensitive, and especially cost-effective detection method.
CT scan method
In case of inadequate availability of these kits and false-negative results of RT-qPCR, a noninvasive CT scan technique has also been used in clinical diagnostics of COVID-19 [30]. The characteristics of these images are closely related to stage of infection when the symptoms are shown. At the early-stage infection, transparent, localized lesions were confined to the subpleural area of one or both lungs of patients after the appearance of symptoms [31]. When bilateral and peripheral ground-glass denseness and amalgamation of the lungs are appeared, then these are the characteristic symbols of COVID-19. With the progression of COVID-19 infection, the lungs consolidation increases in addition to crazy paving pattern. The aforementioned outcomes have revealed that CT scan method showing better false-negative rates is more sensitive in comparison with RT-qPCR but the initial level screening with CT scan is restricted. The major drawback of CT scan method in COVID-19 diagnosis is the lack of specificity because of the overlap of lungs images with other pneumonia. Moreover, the CT scan diagnoses the patients at advanced stage of infection [32].
Antibody detection
The reliable diagnosis of COVID-19 is carried out using protein tests such as antigens and antibodies [33]. The saliva test may vary with passage of time after the symptoms appear but in comparison, antibodies persist for a long time, which enable large span for indirect SARS-CoV-2 detection [34]. The commonly used methods for antibody detection are LFAs and ELISA. In LFA, the sample is deposited onto sample pad of cassette-like device and moved through strip by capillary action. The antibodies labeled with Au NPs bind with target molecules present in sample at the first line. Owing to continuously moving sample, Au-labeled antibodies are bound by capture antibodies existing in lines. The excessive Au-labeled antibodies are captured at the control line while moving through the strip. If the target molecules are not present in sample solution, Au-labeled antibodies are surely captured at control line, which makes control line valid for test. For IgG or IgM-type antibodies or both, there should be one, two, or three stripes in display window [35]. The average test time of 10–30 min places it in the rapid diagnostic methods. It provides only qualitative analysis but has the advantages of easy handling and cost effectiveness and allows direct assessment of ongoing infection as anti-CoV antibodies are used instead of immobilized viral antigen [36]. Recently, Kong's group has developed highly sensitive, fast, easy to use, and on-site immunoassay, which can complete the assay in 15 min to detect SARS-CoV-2 antibodies and antigens at the same time. The antibodies from human serum samples and SARS-CoV-2 antigens from nasal swab were detected successfully from 26 COVID-19 infected and 28 healthy individuals using this integrated method [37]. Moreover, Feng's research group has constructed a simple operated point-of-care lateral flow immunoassay to simultaneously detect antibodies (IgG, IgM) developed against SARS-CoV-2 in blood samples of affected people at various phases of infection. The achieved sensitivity and selectivity of this testing method were 86.6% and 90.6%, respectively [38]. The assay for simultaneous detection of both IgG–IgM possesses better usability and sensitivity in comparison with single IgG and IgM tests. Convalescent serum containing large quantity of neutralization IgG is extremely helpful in curing COVID-19 patients. Tan's et al. has successfully demonstrated an ELISA method to detect anti-SARS-CoV-2 S1 IgG from human serum using just 8 μl sample [39]. The practicability of this method has been verified in detecting anti-SARS-CoV-2 S1 IgG in 16 convalescent SARS-CoV-2–infected persons.
The immunoglobulin IgG and IgM have been detected from serum sample of infected individuals using ELISA where nucleocapsid protein of SARS-CoV-2 is used. The typical result time of ELISA is 60 min to 5 h either the test is quantitative or qualitative. ELISA being fast, flexible, robotics, high throughput with variable sensitive range, and ability to test multiple samples is specifically suitable as point-of-care purpose [40,41]. In ELISA test, human serum sample is added to the well where the recombinant viral antigen is already coated. The bindings occur between antibodies and target antigen present in sample and then washing is done several times to make sure the complete removal of unbound substrate. Another solution comprising labeled antibodies is mixed and binding will occur if the antibodies of interest are present in the sample. After washing again to remove unbound substrate, horseradish peroxidase is used as color changing reaction to confirm the binding of target antibodies. Using a spectrometer, color change is read and concentration of antibodies is measured [42]. It has been noticed that the antibodies are developed in SARS-CoV-2–infected people after around 14 days of onset of symptoms. However, some studies depict the development of IgG and IgM antibodies after five days of SARS-CoV-2 infection [43]. The level of antibodies in patients may go on increasing after five days of symptoms appear. The antibodies were also found in respiratory fluids, blood and fecal specimens.
LFAs are preferred over ELISA tests because these tests can be performed at home without the need of any expert. ELISA is the analytical technique, which can only be carried out in laboratories with the help of skilled personnel and high protocol as well as it requires different steps, long turnaround time and special instruments. Owing to these limitations, the recent studies are more focusing to enhance the sensitivity and selectivity of LFAs. Therefore, signal amplification and multiplexed detection methods are under development to achieve increased sensitivity, selectivity, and detection throughput. The development in serological tests for SARS-CoV-2 antibody faces the issues of being cross-reactive with other family members of coronavirus. Okba observed the cross-reactivity among S proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV. But there was no cross-reactivity for S1 subunit of MERS-CoV spike protein. The most conserved subunit among all coronaviruses is S2, which can increase the suitability of S1 subunit for serological tests [44].
Antigen detection
Antigen tests can be considered more reliable compared with antibodies tests because antigens are target specific and introduce antibodies [45]. Both the LFAs and ELISA methods can be used for the detection of antigens. Kim's research group developed a novel detection method where they used ACE2 receptor to detect SARS-CoV-2 spike 1 (S1) protein [46]. The receptor is further paired with commercially available antibodies and this pair was captured and detected using lateral flow immunoassay. There was no cross-reactivity with other coronaviruses showing a detection limit of 1.86 × 105 copies/mL in clinical samples. The fluorescent immunochromatographic LFA was designed by Diao et al. to detect nucleocapsid (N) protein of SARS-CoV-2 [36]. The anti-N mouse antibodies and anti-rabbit IgG were used to generate test and control line, respectively. The conventional Au NPs were replaced by anti-N rabbit IgG marked with carboxylate-modified polystyrene Europium (III) chelate microparticles. The samples were consisting of urine and nasopharyngeal swab. The assay demonstrated 68% sensitivity and 100% selectivity in comparison with nucleic acid test. These are the early researches and it will take much time to be available in markets for practical use irrespective of their superb sensitivity and specificity. LFAs and ELISA are well established methods and antigen testing is the need of the hours but there is just one Sofia 2 SARS Antigen Test Kit available commercially so far to detect SARS-CoV-2. It works on the principle of immunofluorescence strip technology for the detection of N protein of SARS-CoV-2 and SARS-CoV that mean the kit is unable to make differentiation between these two viruses. The kit showed clinical sensitivity of 80% and selectivity of 100% for 47 positive and 96 negative individuals [47]. Following the failures of kits, WHO discouraged the medical staff to use rapid commercial assays to detect SARS-CoV-2. There are two companies “QUIDEL as well as Avacta and Cytiva,” which are working for the development of trustworthy antigen test kits. The targets for these two kits are N protein, and SARS-CoV-2 viral antigens or S glycoproteins [48].
Point-of-care testing
Point-of-care testing is also much imperative to diagnose SARS-CoV-2–infected persons without sending them to hospitals. In pandemic, the most important is the mass testing to identify the persons with viral exposure for quarantine and treatment strategies. When the viral infection continues to spread at exponential rate, then RT-PCR and immunoassays become limited because of the turnaround time. Simple design, noncomplicated mechanism, and sample collection to result readout by anyone to everyone without any expertise are the key features of point-of-care testing, which is important to control the viral spread. LFAs are the point-of-care testing methods, which are being developed for the detection of SARS-CoV-2. The development in such point-of-care devices is the need-of-the-hour that anyone, anywhere with minor to moderate symptom, or even asymptomatic individuals can get the result within minutes to confirm whether they are positive or negative for SARS-CoV-2 [49]. It is reported that currently, four point-of-care setting devices are being used under Food and Drug Administration- Emergency Use Authorization (FDA-EUA) approval that we have enlisted below with their specific characteristics [50].
-
•
ID NOW COVID-19 test depends on the isothermal nucleic acid amplification to target the RdPR gene at specific site using 4–6 primer and a polymerase at constant temperature. It has great advantage to produce large amount of nucleic acid duplicates in a very short time with excellent sensitivity of 125 copies/mL. Moreover, the result time is approximately 13 min from upper respiratory samples [51].
-
•
Mesa Biotech's Accula SARS-CoV-2 test target is N gene and it is basically the combination of two techniques, RT-PCR plus lateral flow immunoassay. With the analytical sensitivity of 200 copies/mL and it approximately provides result in 30 min [50].
-
•
Xpert Xpress SARS-CoV-2 test offers an additional pledge in SARS-CoV-2 detection targeting more the one gene. It targets the N and E genes and gives the turnout in 40 min. With the analytical sensitivity of 140 copies/mL and it provides selectivity of 100% [50].
-
•
Cue COVID-19 test relies on isothermal amplification to target N gene of SARS-CoV-2 and gives turnout approximately in 20 min. It shows 100% selectivity. Cue test offers the portable and easy to use analytical podium connected with cell phone, which is advantageous to access the health condition at fingerprint making it more promising point-of-care testing approach compared with aforementioned three methods [50].
Developing techniques for SARS-CoV-2 detection
The imperative need of time is, to develop nucleic acid and protein-based testing for diagnosing COVID-19 to compensate the RT-qPCR drawbacks. Fascinatingly, an international company has used the combination of PCR with LFA technique as an alternate method to detect SARS-CoV-2 within the exhaled breath condensate. Moreover, isothermal amplification of genetic material is an alternative approach to PCR for developing point-of-care devices to carry out the quick detection of nucleic acid.
Viral gene detection
Several organizations are striving to implement the isothermal nucleic acid amplification techniques in the detection of SARS-CoV-2. The isothermal amplification approaches are the best options that can be operated at a single temperature without particular laboratory apparatus providing sensitive results [52]. The helicase-dependent amplification, LAMP, and RT-LAMP approaches turned out to be established to detect SARS-CoV-2 and are more specific, sensitive, and reaction efficient [53]. The LAMP technique is more sensitive and specific but it uses high number of primers. In comparison, RT-LAMP just uses 4 to 6 primers and DNA polymerase to target genome at 6 positions. Currently, RT-LAMP has been used to detect SARS-CoV-2 in less than 13 min via reverse of transcription of virus RNA to cDNA. The assay using RT-LAMP technique was developed, which targeted Nsp3 for SARS-CoV-2 detection and showed a detection limit of 100 copies/reaction [54]. Baek et al. designed RT-LAMP primer sets to pick out the N gene of viral RNA, which showed comparable results with RT-PCR displaying Limit of Detection (LOD) of 100 copies of RNA [55]. The test displayed no cross reactivity to other coronaviruses in the detection of SARS-CoV-2 viral RNAs. Chen's group introduced contamination-free naked-eye detection of SARS-CoV-2 using RT-LAMP approach as shown in Figure 1b [56]. The portable 3D printing instrument and smartphone were used and the created fluorescence was seen by naked eyes without using some devoted tools. Tanner’s group used DNA-binding dye for calorimetric detection. It is reported that at the level of 480 RNA copies in cell lysates, colorimetric LAMP can recognize the viral RNA without any interference and can be the alternative of RT-PCR being fast and simple detection method. Moreover, high selectivity, sensitivity, and only requiring heating system and optical inspection make it more favorable for SARS-CoV-2 detection. Zhu et al. developed an approach by integrating multiplex RT-LAMP with nanoparticle-based LFA to detect SARS-CoV-2 through ORF1ab and N gene detection [57]. The SARS-CoV-2 detection is possible in less than 13 min but the one sample/run and the optimizations of primers and reaction conditions are the major limitations associated with LAMP techniques [58]. These isothermal amplification systems may also be multiplexed at the amplification and/or readout stages. The process of multiplexing is done using organic fluorescent molecules as beads for barcoding. Furthermore, visual detection approaches are also being carried out using some dyes, which use intrinsic by-products of widespread DNA synthesis. Fascinatingly, CRISPR-Cas12 lateral flow test was used to achieve enhanced sensitivity, simplified result read, and reduced detection time for RT-LAMP method [59]. Researchers are devoting much efforts in using collateral cleavage activity of Cas nucleases (e.g., Cas12a, Cas12b, and Cas13a) to establish point-of-care viral RNA detection methods. Hou et al. achieved enhanced sensitivity with a detection limit of 7.5 copies per reaction in 40 min using RPA and CRISPR-Cas13–mediated enzymatic signal amplification [60]. A comparative study on clinical evaluation methods demonstrated that CRISPR-Cas13–based tests possess more detection capacity compared with RT-PCR methods. To make the detection process further easy, a compact dual CRISPR-Cas12a (AIOD-CRISPR) assay has been developed, where single-reaction system is used to incubate all components and high sensitivity as well as specificity has been achieved for RNA detection avoiding the need of separate preamplification steps [10]. The detection limit of 4.6 copies/reaction has been achieved for SARS-CoV-2 N gene in 40 min. Recently, Wang's group designed an assay by combining RT-LAMP with Cas12a cleavage and named it as opvCRISPR to detect SARS-CoV-2 visually [61]. This assay was validated with 50 clinical specimens of SARS-CoV-2–infected people. The RT-LAMP amplicon triggered collateral activity against single-stranded DNA reporters of activated Cas12a, which consequently increased the detection sensitivity and made the outcomes to be observed by naked eye. Moreover, SHERLOCK detection methods for SARS-CoV-2 are also used based on nucleic acid testing technique. In this approach, the targeted virus RNA is reverse transcribed to cDNA where isothermal amplification takes place by reverse polymerase. Further the amplified products are transcribed back into RNA [62].
Biosensing techniques for SARS-CoV-2 detection
The biosensors are emerging area of analytical chemistry. The biosensing systems are able to provide quantitative analysis and measurements without demanding extra processing steps or reagents [63,64]. The biosensor-based technologies offer alternative approach to standard PCR tests, which are not only sensitive but also helpful in diagnostics and therapeutic assessments. Biosensors have the capability to detect pathogens in numerous environments without requiring tedious sample preparing steps [65,66]. Several biosensors with diverse biorecognition elements (enzyme, antibody, deoxyribonucleic acid, cell, or microorganisms) and transducers (such as mechanical and optical transducers) have been extensively applied in the detection of pathogens [67]. Labels can also be named as reporters which are molecular species including organic dyes or quantum dots which bind to the target, either in a direct way or via a biorecognition element to enable the detection [68,69]. The biosensors are now trending for SARS-CoV-2 detection, specifically the CRISPR technology–based biosensing systems such as CRISPR-Chip to electrically detect genetic mutations [70] and electrochemical CRISPR-biosensing platforms for microRNA analysis [71]. These approaches are easily adapted to any nucleic acid and can be able to report the mutations of SARS-CoV-2 in timely way. Moreover, successful differentiation in similar gene sequences with superb sensitivity has been recently reported by using combination of plasmonic photothermal (PPT) effect and localized surface plasmon resonance. Here, we have summarized the recently used several analytical approaches (see Table 1 ) for the detection of SARS-CoV-2.
Table 1.
Recently used several analytical approaches to detect SARS-CoV-2.
| Test type | Institute | LOD | Ref |
|---|---|---|---|
| Virus blood culture and high throughput sequence of whole genome | Wuhan Institute of Virology (WIV) | N/A | [72] |
| RT-PCR | Berlin Institute of Virology (BIV) | 3.9 copies and 3.6 copies | [73] |
| High-resolution CT scan | HUST | N/A | [74] |
| Dual CRISPR Cas12a test | University of Connecticut Health Center | 1.2 DNA copies and 4.6 RNA copies | [10] |
| Fast IgM and IgG combined Ab testing kit | Guangzhou Medical University | N/A | [38] |
| Closed tube one-stage LAMP3 | University of Pennsylvania | 70 copies/reaction | [75] |
| Closed tube two-stage RAMP4 assay | University of Pennsylvania | 7 copies/reaction | [75] |
| RNA-based paper LFA5 PoC-based LAMP assay | National Tsinghua University | N/A | [76] |
| ELISA6 and GICA for combined IgG-IgM | Wuhan University | N/A | [49] |
| FTO-based Ab sensor | National-Institute of Animal-Biotechnology | 10 fM | [77] |
| SPCE-based Ab biosensor | National-Institute of Animal-Biotechnology | 10 fM | [77] |
Optical biosensor
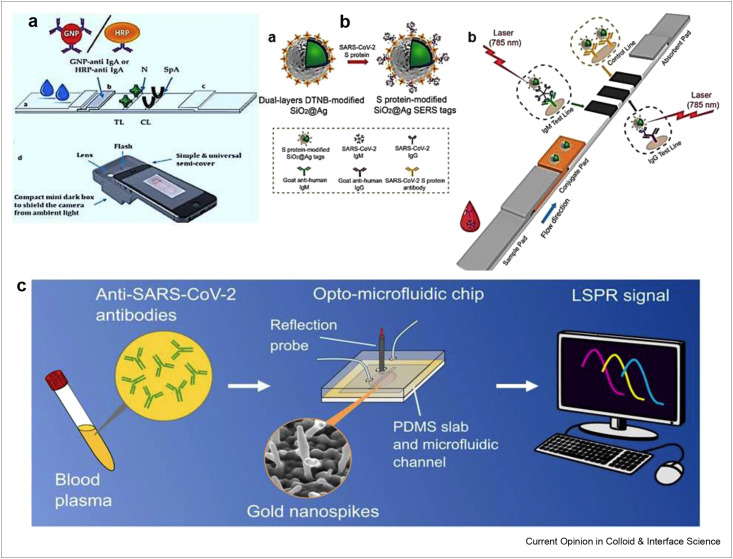
To timely and efficiently diagnose COVID-19 infection, the detection of immunoglobulin A (IgA) specific to SARS-CoV-2 is highly crucial to complement the assays used in the detection of IgM and IgG. Anfossi's research group has designed a dual functioning optical/chemiluminescence immunosensor for the detection of IgA from serum and saliva samples as presented in Figure 2 a [78]. The recombinant nucleocapsid antigen was used to capture SARS-CoV-2 antibodies present in sample in infected people. The assay was successfully used in the analysis of 25 serum and 9 saliva samples taken from infected and/or recovered persons with excellent sensitivity. This is a noninvasive immunosensor, which can be helpful to monitor early immune responses to COVID-19. Moreover, surface-enhanced Raman scattering–based lateral flow immunoassay (SERS-LFIA) has been developed by Liu et al. to detect IgM and IgG simultaneously as shown in Figure 2b [79]. To fabricate SERS, tags modified with dual-layers of Raman dye, SiO2 core was completely coated with Ag shell (SiO2@Ag), which showed superb SERS signal, decent monodispersity, and excellent stability. The anti-human antibodies were immobilized which captured SiO2@Ag-spike S protein anti-SARS-CoV-2 IgM/IgG complexes. The as proposed SERS-LFIA biosensor showed a detection limit of 800 times higher than Au NPs based LFIA. The direct detection of S protein has been achieved using nanoplasmonic resonance sensors with a detection limit of 370 vp/mL in 15 min [80]. The assay was very economical showing good performance under clinical environment. Recently, surface plasmon resonance (SPR) sensing approach has been used to detect nucleocapsid antibodies in serum sample, which were produced in response to SARS-CoV-2 infection [81]. Here, after activating EDC-NHS AffiCoat surface, the binding of nucleocapsid protein of SARS-CoV-2 (rN) on the surface of the SPR chip was achieved and ethanolamine was used for the passivation of remaining activated sites. This method is quantitative, portable and good enough to pave the way for point-of-care assays and label-free detection of antibodies. The SPR sensor had the capability to detect anti-SARS-CoV-2 antibodies at nano-molar level after coating with peptide monolayer and functionalizing with SARS-CoV-2 nucleocapsid recombinant protein. The whole assay was conducted on portable SPR instrument without diluting the serum sample. This method is good enough to pave the way for point-of-care and label-free testing of antibodies. Another SPR biosensor has been developed based on pig sera-derived anti-SARS-CoV-2 antibodies [82]. Pig-sera were used to purify SARS-CoV-2 antibodies against nucleocapsidprotein (NP). The assay presented a detection limit of 1.02 pM for the detection of SARS-CoV-2. More recently, Shen's group detected antibodies against SARS-CoV-2 spike protein using Au nanospikes in opto-microfluidic sensing platform as shown in Figure 2c [83]. The opto-microfluidic sensor was developed based on the principle of localized surface plasmon resonance (LSPR), and Au nanospikes were electrodeposited for the detection of antibodies against SARS-CoV-2 spike protein 1 μL of human plasma. The label-free microfluidic sensor showed a detection limit of ∼0.5 pM and completed the assay in ∼30 min. The wavelength peak shift of Au nanospikes is measured, which is then correlated with the amount of target antibodies. This platform is very cheap, easy-to-use, fast, and offers point-of-care diagnostics. Furthermore, circle-to-circle amplification and optomagnetic analysis have been used to quantify SARS-CoV-2 RdRp coding sequence [84]. The proposed optomagnetic approach was successfully used to detect SARS-CoV-2 cDNA with a detection limit of 0.4 fM. In comparison to previous literature on circle-to-circle–based sensors, this method showed subfemtomolar level detection, reduced the time consumption and labor work. These biosensors possess the advantages to execute fast naked-eye diagnosis of viral infection with real-time analysis and without requiring many reagents. Consequently, preferable method of choice is the used of optical biosensors. They can detect analyte with very low quantity, high sensitivity and specificity as well as detect the variants of viral strains.
Figure 2.
The use of optical biosensors in the detection of SARS-CoV-2 (a) Schematic representation of smartphone reader used for optical immunosensor in the detection of IgA. Reprinted with permission from Ref. [78]. (b) Preparation and operating-principle of (a) SARS-CoV-2 S protein coupled SiO2@Ag SERS tags and (b) simultaneous detection of anti-SARS-CoV-2 antibodies. Reprinted with permission from Ref. [79]. (c) Schematic representation of localized surface plasmon resonance–based opto-microfluidic sensor to detect SARS-CoV-2 antibodies. Reprinted with permission from Ref. [83].
Dual-functional LSPR and PPT biosensor
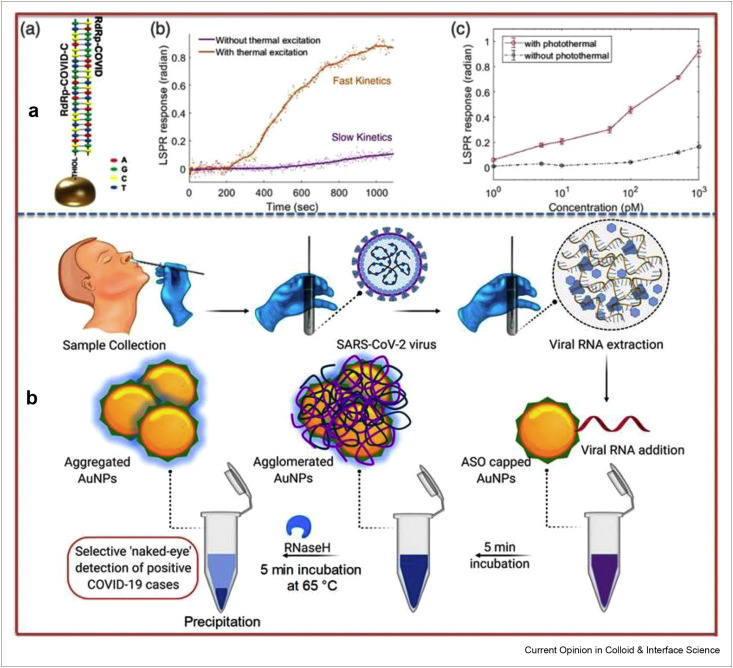
The biosensors are considered most trustworthy diagnosis approaches as an alternate way out to clinical analysis, real-time and nonstop monitoring to promote epidemic prevention and control [85,86]. It is well-documented that huge optical cross sections are shown by plasmonic nanoparticles and abundant heat energy is produced as a result of nonradiative relaxing process of absorbed light [87]. More recently, Wang's research group fabricated a dual-functionalized plasmonic biosensor using PPT effect in combination with localized surface plasmon resonance (LSPR) sensing transducer for sensitive and promising diagnosis of SARS-CoV-2 in clinical specimens [88]. The schematic drawing of hybridization, real-time hybridization of RdRp-COVID and its cDNA sequence, and RdRp-COVID sequence detection at various concentrations have been shown in Figure 3 a. The nucleic acid hybridization process has been used in the detection of selected sequences of virus with super sensitivity where two-dimensional Au nanoislands combined with cDNA receptors perform as biosensing platform. To achieve improved performance, the same Au nanoislands chip is used to generate plasmonic heat when illuminated. Furthermore, the localized PPT heat has the capability to increase the in-situ hybridization temperature, which in turn enables the perfect differentiation of two same gene sequences. The proposed LSPR biosensor has successfully demonstrated superb sensitivity in the selective determination of SARS-CoV-2 sequences showing low detection limit of 0.22 pM and has allowed accurate determination of particular target in mixed genes. This research gains insight into thermoplasmonic enhancement and envisions the practical application of LSPR biosensor in nucleic acid testing and viral infection diagnosis. Moreover, the integration of these biosensors with microfluidic systems may increase overall sensitivity of device helping in miniaturization process [89].
Figure 3.
Dual-functional plasmonic biosensing and naked-eye detection of SARS-CoV-2 (a) LSPR biosensing. (a) Schematic drawing of the hybridization and inhibited hybridization, (b) real-time hybridization of RdRp-COVID and its cDNA sequence, (c) RdRp-COVID sequence detection at various concentrations. Reprinted with permission from Ref. [88]. (b) Scheme illustrating the naked-eye detection of SARS-CoV-2 using ASO-capped AuNPs. Reprinted with permission from Ref. [91].
Microfluidic colorimetric biosensor
In last decades, microfluidic devices have been used for the contagious ailments like HIV, SARS-CoV, and others, which target the biomarkers in less than 5 min consuming only 6 μL of blood plasma sample [90]. For COVID-19 and other infectious viral diseases, microfluidic devices are raising the panorama of swift, low cost, and point-of-care diagnostic to avoid further transmission of virus. Moreover, microfluidic devices fulfill the “ASSURED” (affordable, selective, sensitive, user-friendly, rapid and robust, equipment-free, and deliverable) criteria for biosensing and point-of-care testing having the benefits of high precision, transportability, quick response, high reproducibility, less reagent usage, easily applied, and high throughput parallel processing. These biosensors offer multiplexing, miniaturization, and capability of integration with various techniques. Conversely, sample preparation and analysis, sample’s matrix effect reduction, and full automation in microfluidic devices are still challenging.
Colorimetric biosensors present the exciting properties including straightforwardness, rapid response, accurateness, and cost-effectiveness, and produce signals visible to naked-eye for detection of viral infectious diseases. The thiol-modified antisense oligonucleotides (ASOs) capped Au NPs (Au-ASOs) have been used as colorimetric biosensing platform to detect SARS-CoV-2 by Pan's research group [91]. The schematic illustration of Au-ASOs–based biosensor for naked-eye detection of SARS-CoV-2 has been shown in Figure 3b. The Au-ASOs–based assay enabled the specific detection of N-gene from nasal swab to detect SARS-CoV-2 infection in 10 min. The detection mechanism involved the agglomeration of Au-ASOs architecture with target SARS-CoV-2 RNA, which led to red-shift. The cleavage of RNA strands from RNA–DNA hybrid was done by adding RNaseH and finally the precipitations in solution owing additional agglomeration of Au NPs were visible to naked-eye. The biosensor showed a detection limit of 0.18 ng/μL for SARS-CoV-2 RNA with no cross-reactivity with MERS-CoV viral RNA. Moreover, Kumar et al. developed another similar colorimetric-based biosensing system for the detection of RdRp gene of SARS-CoV-2 form nasopharyngeal sample in less than 30 min [92]. The biosensor demonstrated low detection limit of 0.5 ng for SARS-CoV-2 RNA and no cross-reactivity toward cervical DNA sample, which was acquired from human papillomavirus-infected women. This biosensing platform may facilitate the mass screening during pandemic management in a cost-effective manner. The simultaneous detection of immunoglobulin IgM and IgG for SARS-CoV-2 has been carried out using Au NPs–based immunoassay [38]. The assay detected antibodies from human blood sample in 15 min with a clinical detection sensitivity and selectivity of 88.66% and 90.63%, respectively.
Electrochemical biosensing systems
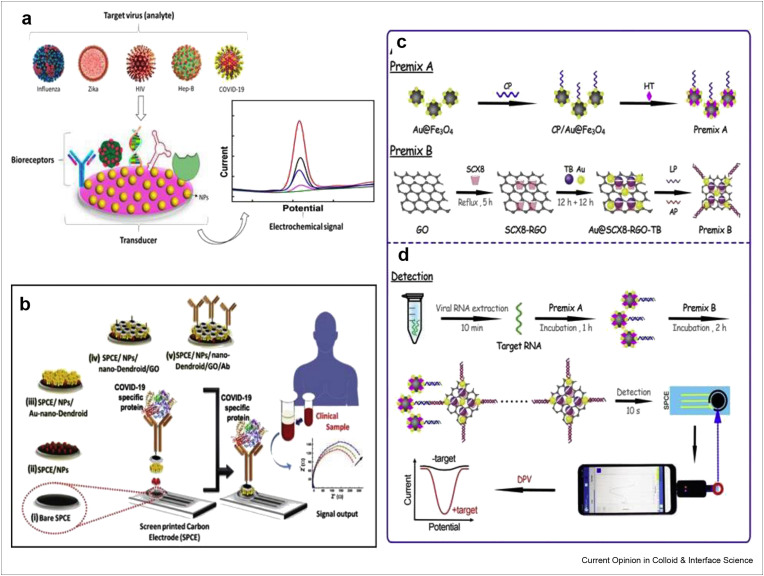
Moreover, electrochemical biosensors have attracted immense attention of the researchers because of their capabilities to offer sophisticated ways to interface at molecular level, DNA recognition and signal transduction elements and are economical requiring low volume and power for DNA detection [93,94]. The electrochemical biosensing podium to detect different viruses including SARS-CoV-2 has been described in Figure 4 a [95]. A hybridization-based genosensor was developed on a 100 nm sputtered gold film to detect 30-mer sequence, which is distinctive to SARS [96]. The alkaline phosphatase (AP) was attached with AP-conjugated streptavidin via eminent streptavidin–biotin binding on electrochemical nanoarchitectured screen printed carbon electrode and was used as a genosensor for electrochemical detection of SARS-associated coronavirus [97]. Here, the Au NPs act as immobilization surface through thiol–gold interaction and transduction surface as well. This enzymatic electrochemical biosensor demonstrated a linear range between 2.5 and 50 pmol/L with low LOD of 2.5 pmol/L in the detection of targeted DNA sequence. In this work, the usability of Au NPs immobilized surfaces has considerably improved the electrochemical sensing performance. Fung's research group developed a biosensor based on piezoelectric quartz crystal (PQC)-aptamer coupled with paramagnetic nanoparticle for specific detection of SCV helicase protein created by SARS-CoV replication. The fabricated aptamer-PQC biosensor showed low detection limit of 3.5 ng/mL having reproducibility of 6.8% (%RSD, n = 3) and good recoveries of 102% and 119% [98].
Figure 4.
The vitality of electrochemical sensing techniques to carry out SARS-CoV-2 detection (a) Electrochemical biosensing podiums to detect different viruses including SARS-CoV-2. Reprinted with permission from Ref. [95]. (b) Scheme representing the concept of SPCE/NPs/nano-Dendroids/GO/Ab probe fabrication to diagnose COVID-19. Reprinted with permission from Ref. [100], Schematic illustration of (c) Preparation of premix (d) Use of smartphone in SARS-CoV-2 detection using the electrochemical biosensing platform. Reprinted with permission from Ref. [104].
Recently, Pan's research group fabricated Au NPs capped with specific antisense oligonucleotides (ssDNA) based electrochemical biosensor chip, which enabled digital detection of SARS-CoV-2 nucleocapsid phosphoprotein (N-gene) [99]. The paper-based electrochemical system was used to immobilize sensing probes to construct nucleic acid testing device. The as-fabricated biosensor outperformed, exhibiting a sensitivity of 231 (copies μL−1)−1 and detection limit of 6.9 copies/μL when applied to test samples obtained from vero cells infected with SARS-CoV-2 as well as patients' specimens. The biosensor showed 100% accuracy, sensitivity, and selectivity when the results of 22 COVID-19 positive patients and 26 healthy individuals acquired by the proposed sensor with standard RT-PCR diagnostic kit. The sensor was also feasible during genomic mutation of SARS-CoV-2 because design of ssDNA probes which concurrently targeted two regions of SARS-CoV-2.
The scheme representing the concept of SPCE/NPs/nano-Dendroids/GO/Ab probe fabrication to diagnose COVID-19 has been shown in Figure 4b [100]. The label-free electrochemical impedimetric sensor was prepared by Rashed et al. to rapidly detect SARS-CoV-2 antibodies [101]. The sensor showed 100% accurate performance when results displayed by this sensor for clinical samples of COVID-19 negative and COVID-19 positive persons were compared with ELISA method. Moreover, Kim's research group has designed field-effect transistor (FET)-based biosensor in the detection of SARS-CoV-2 from clinical specimens [102]. Graphene nanosheets modified FET was coated with specific antibody against SARS-CoV-2. The as-constructed FET biosensor has shown excellent sensing capabilities in the detection of SARS-CoV-2 in culture medium and clinical samples with detection limits of 1.6 × 101 pfu/mL and 2.42 × 102 copies/Ml, respectively. The detection of S and N proteins from saliva samples has also been carried out using an electrochemical immunosensor based on magnetic beads as support of immunological chain and antibody with AP as immunological label [103]. The sensor depicted low detection limits of 19 ng/mL and 8 ng/mL for S and N proteins in buffer solution and untreated saliva, respectively. In this assay, saliva sample is used, which yields high sensitivity compared with nasopharyngeal sample owing to self-sampling of saliva. Magnetic bead–based sensor offers the possibility of loading enhanced amount to capture antibody because of high surface area. More recently, Li's research group has introduced an electrochemical sensing platform using smartphone for the detection of SARS-CoV-2 from infected patients as shown in Figure 4c and d [104]. Using supersandwich-type recognition approach, electrochemical smartphone was able to detect RNA of SARS-CoV-2 without nucleic acid amplification and reverse transcription. The detection capability of this biosensor (85.5% and 46.2%) was higher than RT-PCR (56.5% and 7.7%) when practically applied in to detect RNA of SARS-CoV-2 from 88 RNA extracts of 25 confirmed patients and 8 recovered individuals, respectively. The measured LOD was 200 copies/mL and only two copies (10 μL)/assay of SARS-CoV-2 were needed. The different biosensing platforms for virus detection have been presented in Table 2 . These biosensors not only demonstrate the very low detection limit, enhanced sensitivity and selectivity in detecting infectious agents without sample pretreatment but also produce rapid and cost-effective results in comparison with RT-PCR detection methods. Rapid testing, ease of handling, and miniaturization are also offered by electrochemical biosensors. The integration of theses biosensors with microfluidic platforms, and multiplexing can easily be achieved, which can further increase the sensitivity of device.
Table 2.
Different biosensing platforms for virus detection.
| Biosensor | Pathogen | Detection target | Limit of detection | Linear range | Ref. |
|---|---|---|---|---|---|
| EC | SARS-CoV-2 | S or N protein | 19 ng/mL and 8 ng/mL | – | [103] |
| EC | SARS-CoV-2 | Antibodies | – | – | [101] |
| EC | SARS-CoV-2 | N-gene | 6.9 copies/μL | 585.4 to 5.854 × 107 copies/μL |
[99] |
| Cell-based | SARS-CoV-2 | Antigen | 1 fgmL1- | 10 fg and 1 μg mL1- | [105] |
| Optofluidic | SARS-CoV-2 | Antibody | 0.5 pM | 1- | [83] |
| Nanoplasmonic | SARS-CoV-2 | Virus particles | 370 vp/mL | 0 to 107 vp/mL | [80] |
| SPR | SARS-CoV-2 | Antibody | 1.02 pM | 2–1000 pM | [82] |
| LSPR | SARS-CoV-2 | RNA | 1 pM | 1 nM to 1 μM | [106] |
| Lateral flow optical/chemiluminescence | SARS-CoV-2 | Serum IgA | – | – | [78] |
| SERS-LFIA | SARS-CoV-2 | Antibody | 1.28 × 107-fold dilution | – | [79] |
| Lateral flow | SARS-CoV-2 | RNA | 12 copies/reaction | – | [57] |
| Liquid crystal | SARS-CoV-2 | RNA | – | – | [107] |
| Optomagnetic | SARS-CoV-2 | RdRp | 0.4 fM | 0.1–10 fM | [84] |
Viral-induced reactive oxygen species detection based on EC sensor
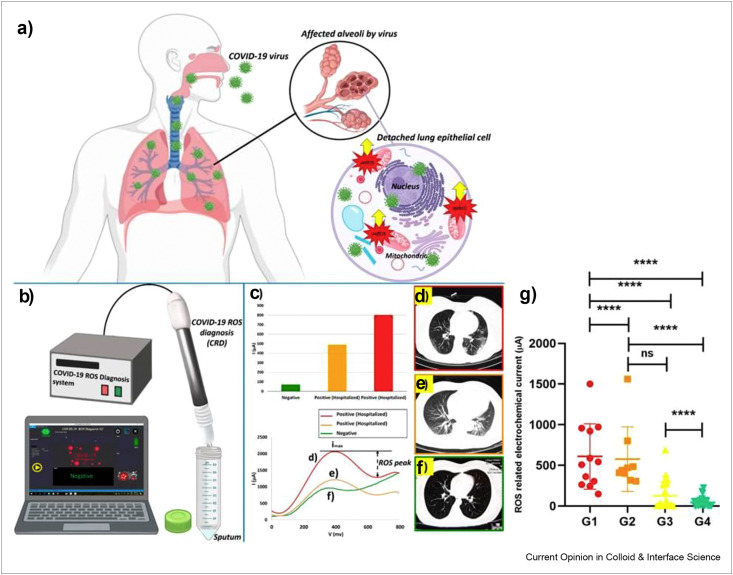
The biomarker-based detection of COVID-19 without using viral RNA, antigen, antibodies, and whole virus particles for detection, may be an interesting strategy in pandemic. It is well-documented that the existence of viral RNA is acknowledged to initiate the activation of NLRP3 inflammasome through RNA-modulating proteins and to trigger the generation of reactive oxygen species (ROS) [108]. Moreover, Zika virus has also been found to induce oxidative stress, which ultimately lowers antioxidant enzyme activities. During in-vitro and in-vivo studies, it has been confirmed that the infection caused by Zika virus significantly enhances ROS production and lipid peroxidation products, whereas decreases superoxide dismutase and catalase activities [109]. Previously, in case of SARS-CoV, Li's group investigated that the ROS level was noticeably augmented in SARS-CoV 3C-like protease expressing cells. The authors confirmed the role of SARS-CoV in virus-induced cell apoptosis in Vero-E6 cells [110]. It is very crucial to carry out early diagnosis of COVID-19 pneumonia to avoid care consumption and mortalities at mass level. Considering the early diagnosis, Miripour et al. presented a diagnostic method based on early traces of overproduced mitochondrial ROS in lungs due to viral-infected lung epithelium [111]. The recent investigation on COVID-19 diagnostic mechanism has been carried out monitoring overproduction of early traces of mitochondrial ROS as lung cells' dysfunctions triggered by SARS-CoV-2. The scheme of side effects of virus in lungs producing mitochondrial ROS which promote viral replication has been shown in Figure 5 a. The electrochemical sensor for the detection of ROS consisting of three steel needle electrodes with working electrode modified by multiwalled carbon nanotube (MWCNT), reference electrode and counter electrode in a triangular form with 3 mm distance from each other, comparison among DPV responses of confirmed and healthy persons and their corresponding CT scan images have been displayed in Figure 5b–f. The practical application of proposed electrochemical sensor in differentiating patient samples by measuring the produced current under sweeping potential range of −0.8 to 0.8 V and with scan rate of 100 mV/s has also been presented in Figure 5g. So far, this is the first report on construction of electrochemical sensing platform, which can specifically detect concentration of ROS in sputum specimens to screen the individuals with or without COVID-19 infection. The sensor can work with a volume of less than 500 μl. The MWCNT-decorated electrode showed a sensitivity of 97% when applied in ROS detection in sputum samples of more than 140 healthy and confirmed cases and compared the results with clinical diagnostics. The sensor displayed the diagnosis results in less than 30 s and it can be a supreme assistant in rapid screening of patients. However, the authentication on specificity of clinical analysis would be needed further. Though, this type of portable, light weight, and sensitive biosensing platform is extremely needed to execute rapid screening of patients for further medical checkup to decrease the workload on front line medical workers during pandemic.
Figure 5.
The ROS-based detection of COVID-19 using an electrochemical biosensor consisting of three needle electrodes modified with functionalized multiwalled carbon nanotubes (a) Scheme representing the side effects of virus in lungs producing mitochondrial ROS which promote viral replication. (b) Electrochemical detection of ROS. (c) Comparison among different fresh sputum samples from patients and their corresponding CV. (d–f) CT-scan images of infected patients and healthy individual. (g) G1; hospitalized in ICU (n = 25), G2; hospitalized without need to ICU care (n = 36), G3; PCR-positive nonhospitalized (n = 45), G4; PCR-negative healthy controls (n = 36). Reprinted with permission from Ref. [111].
Comparison among various detection approaches
The detailed comparison among various detection approaches has been discussed with their distinguish characteristics as given in Table 3 . There are different detection methods such as immunofluorescence approaches, cell culture–based methods, molecular assays, and electrochemical biosensing–based approaches.
Table 3.
Comparing the pros and cons of conventional and electrochemical biosensing techniques.
| Techniques | Significant points | Drawbacks | Ref. |
|---|---|---|---|
| Immunofluorescence (IF) approaches | Highly specific and sensitive. | Sensitivity is less as compared to cell culture–based methods, as well as, being highly specific cannot be used for all types of viruses, exhibit poor sensitivity against some viral particles. Moreover, need expertise. | [112] |
| Cell culture–based methods | Sensitivity is higher as compared to most antigen testing methods. Specific viral particles can be isolated even from the mixed culture medium. Facilitate the Antiviral, serotype as well as epidemiological studies. | Long incubation period and need of expertise are major disadvantages related to cell culturing methods. | [113] |
| Molecular assays | Sensitivity and specificity are good enough, turnout time is less in real-time analysis, even appropriately can detect the viral particles that cannot be cultured by cell culture methods. | These assays need highly specific primer and probes, need skills to be intramural use, as well as are expensive and most of them can only be done in research laboratories. In case of mixed infections, chances of false results are higher. Moreover, FDA-approved kits are not available for all types of viruses. | [114] |
| Electrochemical biosensors | Portable biosensors facilitate the in-house healthcare services without the need of highly trained personals. Being highly cost effective, with frequent response, less result turnout time, small sampling size, as well as admirable detection limits biosensors are key technologies in health care systems. | Sensitivity toward the samples matrix effect and poor stability are the bottleneck of biosensing technology. | [115,116] |
The issues need to be addressed
No doubt, there are lots of successful advancements in detection and diagnostic assays which are effectively contributing to control the SARS-CoV-2 spread worldwide but some unmet problems still need to be solved for better control and prevention of COVID-19. Especially in the case of asymptomatic, presymptomatic, and the patients with less viral load, the point-of-care devices that work without RNA extraction, met the accuracy problem for the sensitivity and detection of SARS-CoV-2. The long detection time also does not fit into the fast and on-site conductive screening of COVID-19 suspected individuals [117]. In case of COVID-19, it has been observed clinically that most of the infected persons have low viral load as well as less expression of nucleocapsid protein in infected cells, which ultimately produce a less nucleocapsid concentration in body fluids comparing to other CoV viruses. This thing by putting the question marks on the accuracy of the antigen and viral particle detection, suggested the development of ultrasensitive and POC detection technologies for the fast and in-situ sensing of antigen in suspected cases for the early diagnostics purposes [118].
Furthermore, low seroconversion is the main reason of trace antibody level (anti SP-antibody) in most of the COVID-19 patients, specifically the level of anti-RBD and anti-NP-antibodies is less at the start of infection, which results in the false results during the examination. During 5–6 days of incubation period, due to the trace level of targeted SARS-CoV-2 antibodies, there are more chances that infected patients could be neglected by the commercially available kits. Ultimately, these presymptomatic and asymptomatic persons will be the source of virus spread to their close contacts [119]. To solve these unmet problems, in addition to smarter strategies, there is an urgent need to develop the ultrasensitive and cost-effective detection technologies for the accurate detection of antigen, antibodies, and integration of RNA release through preconcentration, amplification, and detection in portable microfluidic-based chips.
Conclusion and future outlook
The ultrasensitive and specific diagnostic methods are necessary for the early detection of SARS-CoV-2 to expedite the treatment and isolation of infected individuals. In addition, diagnostic techniques are highly mandatory to empower medical staff with direct resources and to better combat with COVID-19. These days, CT scan, RT-PCR, and lateral flow immunoassays have been developed for the diagnosis of COVID-19. The nucleic acid testing methods and RT-PCR kits being sensitive and specific offer the best way to detect SARS-CoV-2, but the reduction in false negative results is needed to increase the applicability of RT-PCR test. To overcome the bottleneck of standard diagnostic tools, emerging diagnostic approaches with low cost, wide availability, rapid response, and superb reliability are very essential to identify and handle the viral spread. Here of, we have critically discussed the state-of-the-art biosensing strategies for SARS-CoV-2 detection. Biomarker-based diagnosis potential of biosensors will be the captivating approach in combination with clinical observations and risk factors to treat the patients according to the severity of their disease. Recently, biosensors have successfully detected SARS-CoV-2 from biological samples; therefore, the use of biosensing technologies is the best way to diagnose the disease at its earliest stage.
With the innovation of nanotechnology, the sensing performance of biosensor devices should be further improved in terms of low detection limits, specificity and reproducibility, making them more trustworthy for in-vitro and in-vivo diagnostics. To improve the accuracy, combined detection of various biomarkers with multiplex biosensors can be the alternative sensitive approach. To guarantee the fast measurements at the point-of-care, sensing devices should be inexpensive and easy to handle. More work is required to connect sensing devices through databases with medical staff to realize a decentralized healthcare, which would be an important tool for the diagnosis of emerging infectious disease and to determine the herd-immunity regions. Furthermore, colorimetric strips and smartphone-based biosensors that can target antibody or antigen possess great potential as home-used point-of-care testing. Moreover, rapid translation of laboratory research on biosensors into commercially practicable prototype by industry is the main challenge to bridge the gap between laboratory work and industrial needs. The wastewater-based epidemiology should be further explored to develop proficient analytical devices for accurate and fast detection of trace level of SARS-CoV-2 to determine the virus carrier regions. The society needs the massive defense to contain the enormous SARS-CoV-2 like viral attacks and we believe that the best way to deal with such situations is, the development of the highly sensitive, swift, easily accessible point-of-care testing kits, which can be used even by the common man.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research work was funded by National Key Research and Development Program of China under Grant 2017YFC1104402, China Postdoctoral Science Foundation (2016M602291), the initial research fund from Chinese Scholarship Council (CSC); and 3551 Project, Optics Valley of China.
This review comes from a themed issue on Hot Topic: COVID-19
Edited by Reinhard Miller and Libero Liggieri
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiol. 2020:1. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S. Another decade, another coronavirus. N Engl J Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchis-Gomar F., Lavie C.J., Perez Quilis C., Henry B.M., Lippi G. Angiotensin-converting enzyme 2 and antihypertensives (angiotensin receptor blockers and angiotensin-converting enzyme inhibitors) in coronavirus disease 2019. Mayo Clin Proc. 2020;95:1222–1230. doi: 10.1016/j.mayocp.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan W. j., Ni Z. y., Hu Y., Liang W. h., Ou C. q., He J. x., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 9.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., et al. Positive RT-PCR test results in patients recovered from COVID-19. Jama. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X., Yin K., Li Z., Liu C. All-in-One dual CRISPR-cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. BioRxiv. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F., Abudayyeh O.O., Gootenberg J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. CRISPR diagnostics. 2020;8 [Google Scholar]
- 12.Liu J., Chen X., Wang Q., Xiao M., Zhong D., Sun W., et al. Ultrasensitive monolayer MoS2 field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. 2019;19:1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- 13.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr : Clin Res Rev. 2020;14:241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green K, Winter A, Dickinson R, Graziadio S, Wolff R, Mallett S, et al.: What tests could potentially be used for the screening, diagnosis and monitoring of COVID-19 and what are their advantages.
- 15.Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 16.Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1560–7917. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniq. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 19.Bustin S. International University Line; San Diego. CA: 2004. AZ of quantitative PCR. [Google Scholar]
- 20.Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lübke N., Senff T., Scherger S., Hauka S., Andrée M., Adams O., et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol. 2020;130:104579. doi: 10.1016/j.jcv.2020.104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trisnawati I., Khair R.E., Puspitarani D.A., Fauzi A.R., Gunadi Prolonged nucleic acid conversion and false-negative RT-PCR results in patients with COVID-19: a case series. Annal Medic Surg. 2020;59 doi: 10.1016/j.amsu.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020;92:1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piras A., Rizzo D., Uzzau S., De Riu G., Rubino S., Bussu F. Inappropriate nasopharyngeal sampling for SARS-CoV-2 detection is a relevant cause of false-negative reports. Otolaryngol Head Neck Surg. 2020:163. doi: 10.1177/0194599820931793. [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Johnson M., Hill P., Gale B.K. Microfluidic sample preparation: cell lysis and nucleic acid purification. Integrat Biol. 2009;1:574–586. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- Qin Z., Peng R., Baravik I.K., Liu X. Fighting COVID-19: integrated micro- and nanosystems for viral infection diagnostics. Matter. 2020;3:628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes the micro and nanosystems against viral infections diagnostics.
- 28.An J., Liao X., Xiao T., Qian S., Yuan J., Ye H., et al. Clinical characteristics of the recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med. 2020;8:1084. doi: 10.21037/atm-20-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M., et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee E.Y., Ng M.Y., Khong P.L. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 2020;20:384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiol. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiol. 2020;12:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microb Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.To K.K.W., Tsang Ot Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soh J.H., Chan H.M., Ying J.Y. Strategies for developing sensitive and specific nanoparticle-based lateral flow assays as point-of-care diagnostic device. Nano Today. 2020;30:100831. [Google Scholar]
- 36.Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. MedRxiv. 2020 [Google Scholar]
- 37.Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., et al. Microfluidic immunoassays for sensitive and simultaneous detection of IgG/IgM/antigen of SARS-CoV-2 within 15 min. Anal Chem. 2020;92:9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan X., Krel M., Dolgov E., Park S., Li X., Wu W., et al. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens Bioelectron. 2020;169:112572. doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.FDA U. 2020. Cellex qSARS-CoV-2 IgG/IgM rapid test. [Google Scholar]
- 41.FDA U. 2020. Approval of cellex, inc. qSARS-CoV-2 IgG/IgM rapid test. [Google Scholar]
- 42.Lin D., Liu L., Zhang M., Hu Y., Yang Q., Guo J., et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur J Clin Microbiol Infect Dis. 2020;39:2271–2277. doi: 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cevik M., Bamford C., Ho A. COVID-19 pandemic–A focused review for clinicians. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O N.M., Muller M., Li W., Wang C., GeurtsvanKessel C., Corman V., et al. 2020. SARS-CoV-2 specific antibody responses in COVID-19 patients. [Google Scholar]
- 45.Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostica. 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim J., et al. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porte L., Legarraga P., Iruretagoyena M., Vollrath V., Pizarro G., Munita J.M., et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity. MedRxiv. 2020 2020.10.04.20206466. [Google Scholar]
- 48.Ron H.L. FDA U; 2020. Sofia 2 SARS antigen FIA.https://www.fda.gov/media/137885/download [Google Scholar]
- 49.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., et al. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19) MedRxiv. 2020 [Google Scholar]
- 50.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Zinger T., Inglima K., Woo K. m., Atie O., Yurasits L., et al. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. J Clin Microbiol. 2020;58:e01136–e01220. doi: 10.1128/JCM.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article highlighted the point-of-care testing platform for SARS-CoV-2.
- 52.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 2020.02.26.20028373. [Google Scholar]
- 54.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., et al. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J Molecul Diagnost. 2020 doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baek Y.H., Um J., Antigua K.J.C., Park J.-H., Kim Y., Oh S., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microb Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z., et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens Bioelectron. 2020;169:112642. doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]; The CRISPR-based technology for contamination-free visual detection of SAR-CoV-2 has been presented in this article.
- 57.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ashist S.K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nature biotechnol. 2020:1–5. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., et al. Development and evaluation of a CRISPR-based diagnostic for 2019-novel coronavirus. MedRxiv. 2020 [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., et al. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper proposed an ultrasensitive visual SARS-CoV-2 detection method integrated RT-LAMP and CRISPR/Cas cleavage in one-pot and totally avoided amplicon contamination.
- 62.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asif M., Aziz A., Dao A.Q., Hakeem A., Wang H., Dong S., et al. Real-time tracking of hydrogen peroxide secreted by live cells using MnO2 nanoparticles intercalated layered doubled hydroxide nanohybrids. Anal Chim Acta. 2015;898:34–41. doi: 10.1016/j.aca.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 64.Ashraf G., Asif M., Aziz A., Dao A.Q., Zhang T., Iftikhar T., et al. Facet-energy inspired metal oxide extended hexapods decorated with graphene quantum dots: sensitive detection of bisphenol A in live cells. Nanoscale. 2020;12:9014–9023. doi: 10.1039/c9nr10944g. [DOI] [PubMed] [Google Scholar]
- 65.Asif M., Liu H., Aziz A., Wang H., Wang Z., Ajmal M., et al. Core-shell iron oxide-layered double hydroxide: high electrochemical sensing performance of H2O2 biomarker in live cancer cells with plasma therapeutics. Biosens Bioelectron. 2017;97:352–359. doi: 10.1016/j.bios.2017.05.057. [DOI] [PubMed] [Google Scholar]
- 66.Asif M., Aziz A., Wang Z., Ashraf G., Wang J., Luo H., et al. Hierarchical CNTs@CuMn layered double hydroxide nanohybrid with enhanced electrochemical performance in H2S detection from live cells. Anal Chem. 2019;91:3912–3920. doi: 10.1021/acs.analchem.8b04685. [DOI] [PubMed] [Google Scholar]
- 67.Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., et al. The role of biosensors in coronavirus disease-2019 outbreak. Curr Opinion Electrochem. 2020;23:174. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aziz A., Asif M., Azeem M., Ashraf G., Wang Z., Xiao F., et al. Self-stacking of exfoliated charged nanosheets of LDHs and graphene as biosensor with real-time tracking of dopamine from live cells. Anal Chim Acta. 2019;1047:197–207. doi: 10.1016/j.aca.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Aziz A., Asif M., Ashraf G., Azeem M., Majeed I., Ajmal M., et al. Advancements in electrochemical sensing of hydrogen peroxide, glucose and dopamine by using 2D nanoarchitectures of layered double hydroxides or metal dichalcogenides. A review. Microchim Acta. 2019;186:671. doi: 10.1007/s00604-019-3776-z. [DOI] [PubMed] [Google Scholar]
- 70.Hajian R., Balderston S., Tran T., DeBoer T., Etienne J., Sandhu M., et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nature biomed Engineer. 2019;3:427. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruch R., Urban G.A., Dincer C. Unamplified gene sensing via Cas9 on graphene. Nature biomed Engineer. 2019;3:419–420. doi: 10.1038/s41551-019-0413-4. 427. 2019;3. [DOI] [PubMed] [Google Scholar]
- 72.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiol. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.El-Tholoth M., Bau H.H., Song J. 2020. A single and two-stage, closed-tube, molecular test for the 2019 Novel Coronavirus (COVID-19) at home, clinic, and points of entry. [Google Scholar]
- 76.Yang T., Wang Y.C., Shen C.F., Cheng C.M. Point-of-care RNA-based diagnostic device for COVID-19. Multidiscip Digit Publish Instit. 2020:165. doi: 10.3390/diagnostics10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. BioRxiv. 2020 [Google Scholar]; This manuscript sheds light on the ultrasensitive eCovSens EC devices for fast detection of SARS-CoV-2 via spike protein.
- 78.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., et al. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens Bioelectron. 2021;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sensor Actuator B Chem. 2020:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdelhadi D., Benjamin C., Maryam Hojjat J., Vincent T., Julien C., Keisean S., et al. 2020. A rapid and quantitative serum test for SARS-CoV-2 antibodies with portable surface plasmon resonance sensing. [Google Scholar]
- 82.Bong J.H., Kim T.H., Jung J., Lee S.J., Sung J.S., Lee C.K., et al. Pig sera-derived anti-SARS-CoV-2 antibodies in surface plasmon resonance biosensors. Biochip journal. 2020:1–11. doi: 10.1007/s13206-020-4404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Funari R., Chu K.-Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]; This explain the homogenous circle to circle amplification for SARS-CoV-2 detection.
- 85.Asif M., Haitao W., Shuang D., Aziz A., Zhang G., Xiao F., et al. Metal oxide intercalated layered double hydroxide nanosphere: ith enhanced electrocatalytic activity towards H2O2 for biological applications. Sensor Actuator B Chem. 2017;239:243–252. [Google Scholar]
- 86.Asif M., Aziz A., Wang H., Wang Z., Wang W., Ajmal M., et al. Superlattice stacking by hybridizing layered double hydroxide nanosheets with layers of reduced graphene oxide for electrochemical simultaneous determination of dopamine, uric acid and ascorbic acid. Mikrochim Acta. 2019;186:61. doi: 10.1007/s00604-018-3158-y. [DOI] [PubMed] [Google Scholar]
- 87.Jauffred L., Samadi A., Klingberg H., Bendix P.M., Oddershede L.B. Plasmonic heating of nanostructures. Chem Rev. 2019;119:8087–8130. doi: 10.1021/acs.chemrev.8b00738. [DOI] [PubMed] [Google Scholar]
- 88.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 89.Rampazzi S., Danese G., Leporati F., Marabelli F. A localized surface plasmon resonance-based portable instrument for quick on-site biomolecular detection. IEEE T INSTRUM MEAS. 2015;65:317–327. [Google Scholar]
- 90.Gray E.R., Turbé V., Lawson V.E., Page R.H., Cook Z.C., Ferns R.B., et al. Ultra-rapid, sensitive and specific digital diagnosis of HIV with a dual-channel SAW biosensor in a pilot clinical study. NPJ digital medicine. 2018;1:1–8. doi: 10.1038/s41746-018-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moitra P., Alafeef M., Dighe K., Frieman M., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020 doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar V., Mishra S., Sharma R., Agarwal J., Ghoshal U., Khanna T., et al. Development of RNA-based assay for rapid detection of SARS-CoV-2 in clinical samples. BioRxiv. 2020 doi: 10.1159/000522337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asif M., Aziz A., Ashraf G., Wang Z., Wang J., Azeem M., et al. Facet-inspired core-shell gold nanoislands on metal oxide octadecahedral heterostructures: high sensing performance toward sulfide in biotic fluids. ACS Appl Mater Interfaces. 2018;10:36675–36685. doi: 10.1021/acsami.8b12186. [DOI] [PubMed] [Google Scholar]
- 94.Asif M., Aziz A., Azeem M., Wang Z., Ashraf G., Xiao F., et al. A review on electrochemical biosensing platform based on layered double hydroxides for small molecule biomarkers determination. Adv Colloid Interface Sci. 2018;262:21–38. doi: 10.1016/j.cis.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 95.Khan M.Z.H., Hasan M.R., Hossain S.I., Ahommed M.S., Daizy M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: state of the art. Biosens Bioelectron. 2020;166:112431. doi: 10.1016/j.bios.2020.112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abad-Valle P., Fernandez-Abedul M.T., Costa-Garcia A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens Bioelectron. 2005;20:2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martínez-Paredes G., González-García M.B., Costa-García A. Genosensor for SARS virus detection based on gold nanostructured screen-printed carbon electrodes. Electroanalysis. 2009;21:379–385. [Google Scholar]
- 98.Albano DRB, Shum K, Tanner JA, Fung YS. Piezoelectric quartz crystal aptamer biosensor for detection and quantification of SARS CoV helicase protein.
- Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020 doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes the oligonucleotides-based EC biosensor for COVID-19 diagnostics.
- 100.Chandra P. Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sensor Intl. 2020;1:100019. doi: 10.1016/j.sintl.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., et al. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]; Excellent explanation about the rapid, label free detection of SARS-CoV-2 using commercially available equipment.
- 102.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 103.Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor Actuator B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ultrasensitive smartphone-based EC sensors for the detection of SARS-CoV-2.
- Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents the portable cell-based biosensor for the detection of SARS-CoV-2.
- Miti A., Thamm S., Muller P., Csaki A., Fritzsche W., Zuccheri G. A miRNA biosensor based on localized surface plasmon resonance enhanced by surface-bound hybridization chain reaction. Biosens Bioelectron. 2020;167:112465. doi: 10.1016/j.bios.2020.112465. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported the SPR resonance, as a promising substitute of traditional lab-based techniques for the detection and quantification of miRNAs.
- 107.Xu Y., Rather A.M., Song S., Fang J.-C., Dupont R.L., Kara U.I., et al. Ultrasensitive and selective detection of SARS-CoV-2 using thermotropic liquid crystals and image-based machine learning. Cell Report Physical Sci. 2020:100276. doi: 10.1016/j.xcrp.2020.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spel L., Martinon F. Detection of viruses by inflammasomes. Curr Opinion Virol. 2020;46:59–64. doi: 10.1016/j.coviro.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Almeida L.T., Ferraz A.C., da Silva Caetano C.C., da Silva Menegatto M.B., dos Santos Pereira Andrade A.C., Lima R.L.S., et al. Zika virus induces oxidative stress and decreases antioxidant enzyme activities in vitro and in vivo. Virus Res. 2020;286:198084. doi: 10.1016/j.virusres.2020.198084. [DOI] [PubMed] [Google Scholar]
- 110.Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., et al. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]; The new finding for real time tracing of reactive oxygen species for the effective detection of COVID-19 has been described in this paper.
- 112.Kumar P. Methods for rapid virus identification and quantification. Mater Method. 2013;3:207. [Google Scholar]
- 113.Li Q., Deng L., Kim J.-K., Zhu Y.Q., Holmes S.M., Perez-Page M., et al. Growth of carbon nanotubes on electrospun cellulose fibers for high performance supercapacitors. J Electrochem Soc. 2017;164:A3220. [Google Scholar]
- 114.Shieh W.J. Adv techniq diagn microbiol. Springer; 2018. Advanced pathology techniques for detecting emerging infectious disease pathogens; pp. 543–561. [Google Scholar]
- 115.Malecka K., Menon S., Palla G., Kumar K.G., Daniels M., Dehaen W., et al. Redox-active monolayers self-assembled on gold electrodes—effect of their structures on electrochemical parameters and DNA sensing ability. Molecules. 2020;25:607. doi: 10.3390/molecules25030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mathew M.R., Kumar K.G. Poly (amino hydroxy naphthalene sulphonic acid) modified glassy carbon electrode; an effective sensing platform for the simultaneous determination of xanthine and hypoxanthine. J Electrochem Soc. 2020;167 [Google Scholar]
- 117.Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol Hepatol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lv L., Xie X., Gong Q., Feng R., Guo X., Su B., et al. Transcriptional difference between SARS-COV-2 and other human coronaviruses revealed by sub-genomic RNA profiling. BioRxiv. 2020 [Google Scholar]
- 119.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Euro Surveill. 2020;25:2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]