Abstract
Bacterial RNA has recently emerged as an immune-stimulating factor during viral infection. The immune response in an organism is directly related to the progression of virus infections. Lactic acid bacteria in particular have anticancer, bioprotective, and antiallergic effects by modulating immunity. Here, we aimed to demonstrate the effect of bacterial RNA on in vitro production of IL-12, a proinflammatory cytokine, and on in vivo activity against influenza A virus (IFV) infection. Oral administration of heat-killed Enterococcus faecalis KH2 (KH2) or Lactobacillus plantarum SNK12 (SNK) in IFV-infected mice suppressed viral replication and stimulated production of virus-specific antibodies. However, ribonuclease-treated KH2 or SNK abrogated the effect, reducing IL-12 production in vitro and anti-IFV effects in vivo. Taken together, KH2 or SNK showed antiviral effects in vivo when administered orally, and the RNAs of KH2 and SNK play a part in these effects, despite the phylogenetic differences between the bacteria.
Keywords: viral infection, influenza virus, lactic acid bacteria, E. faecalis, L. plantarum, bacterial RNA
INTRODUCTION
Lactic acid bacteria (LAB) intake has several major health benefits, such as improvement of fecal microbiota [1,2,3] and antibacterial [4, 5], anti-allergy [6, 7], antitumor [8, 9], and antiviral effects [10,11,12]. Bacterial cell walls [13] and extracellular polysaccharides produced by bacteria [14] have been reported as factors affecting the immune stimulation and biological defense provided by LAB, but no active component has yet been clearly elucidated. In this study, we aimed to identify immunomodulators of LAB and evaluate their effects on influenza virus infection in a mouse model. First, we focused on the RNA of LAB because Staphylococcus aureus DSM20231 23S rRNA has been reported to stimulate toll-like receptor (TLR) 13 and produce various cytokines [15]. Furthermore, a sequence containing 13 nucleotides near the active site of 23S rRNA ribozyme, which catalyzes peptide bond synthesis, was necessary and sufficient to trigger TLR13-dependent interleukin (IL)-1β production [16]. It has also been reported that the RNAs of other bacteria, including Enterococcus faecalis EC-12, Lactobacillus gasseri JCM5344, Bifidobacterium breve JCM1192 [17], Pediococcus acidilactici strain K15, Lactobacillus plantarum ATCC14197T, Lactobacillus pentosus ATCC8041T, and Lactococcus lactis subsp. lactis ATCC19435 [18], influence IL-12 production. That is, RNase A treatment of heat-killed bacteria significantly decreased the IL-12 production of human peripheral blood mononuclear cells. IL-12 production induced by bacterial RNA was reduced by a treatment with siRNA against TLR8, suggesting that the recognition of bacterial RNA was mediated by TLR8. In addition, IL-12 is a proinflammatory cytokine produced by dendritic cells, macrophages, and B cells [19, 20] that have immunomodulatory effects, such as antitumor and antiviral effects. These reports also suggest that bacterial RNA influences immune stimulation and that the health benefits of LAB via the immune system are affected by bacterial RNA. We selected the same bacterial species as reported in previous studies [17, 18] from the species we use in our studies and tested whether RNase treatment would affect IL-12 production in the species we have been studying. Although it is a classical method, we evaluated the production of IL-12 in bacteria using mouse splenocytes and examined the influence of RNase treatment [21, 22].
Influenza continues to be a serious infectious disease worldwide. On average, influenza viruses infect 5–15% of the global population annually, resulting in approximately 500,000 deaths each year [23]. Influenza viruses belong to the family Orthomyxoviridae and are classified into four different types: A, B, C, and D [24, 25]. Among these viruses, influenza A viruses exhibit a broad host spectrum, including mammalians and birds, and they can quickly mutate into highly pathogenic strains [26, 27]. The two principal clinical approaches for combating influenza are antiviral drugs and vaccines. Among the antivirals, neuraminidase inhibitors are commonly used for treatment of influenza. However, drug-resistant viruses have been reported clinically [28, 29]. Therefore, the importance of prophylaxis by vaccination has been increasing. However, vaccination has several limitations and problems that need to be resolved, such as egg adaptation [30], antigen mismatching [31], and insufficient antibody responses by intradermal or intramuscular injection. Therefore, we believe that the countermeasures for influenza should involve not only vaccines and drugs but also enhancement of host defense functions. LAB have been reported to have an anti-influenza effect [32], and we hoped that this study would reveal the active components of LAB in host defense against this virus. In the present study, we investigated two strains of heat-killed LAB, E. faecalis KH2 (KH2) and L. plantarum SNK12 (SNK), to confirm the effects of KH2 and SNK on viral load and antiviral antibody production in mice infected with influenza virus. By comparing the effects of ribonuclease-treated and untreated LAB, we also tested whether LAB RNA is necessary for the antiviral effects.
MATERIALS AND METHODS
Preparation of heat-killed bacteria and ribonuclease treatment
E. faecalis KH2 (International Patent Organism Depositary in Japan number, NITE P-14444; GenBank Accession number, AB534553) and L. plantarum SNK12 (International Patent Organism Depositary in Japan number, NITE P-1445; GenBank Accession number, AB715330) were stored at Bio-Lab Co., Ltd. All LAB were grown aerobically overnight at 37°C in MRS broth (Difco, Detroit, MI, USA) and washed with distilled water, followed by centrifugation at 10,000 × g for 3 min. The bacterial suspension in distilled water (20–30 mg [wet bacteria weight]/mL) was heated at 105°C for 30 min using an autoclave (HV-25IILB, Hirayama Manufacturing Corp., Saitama, Japan).
Ribonuclease treatment was performed with RNase A (Invitrogen, Tokyo, Japan). RNase A was added to heat-killed KH2 and SNK suspended in distilled water at a final concentration of 10 µg/mL. After 120 min of incubation at 37°C, the ribonuclease-treated bacteria were washed with distilled water and resuspended in distilled water. The ribonuclease-treated KH2 and SNK were designated as R-KH2 and R-SNK, respectively.
To determine the RNA content of the bacteria, the solutions of the bacteria (n=3) with and without ribonuclease treatment were centrifuged at 10,000 × g for 3 min, and the bacteria pellets were collected. Next, 100 µL of distilled water was added, 100 mg of 0.1-mm zirconia beads (BioSpec Products, Bartlesville, OK, USA) were added, the bacteria were crushed in a Micro SmashTM MS-100 (TOMY, Tokyo, Japan). RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the RNA concentration was measured using a NanoDrop One (Thermo Fisher Scientific, Waltham, MA, USA).
IL-12 production by mouse splenocytes
The bacterial suspension was added at a final concentration of 1 µg/mL (culture medium, RPMI1640, Wako, Osaka, Japan) to 6 wells of a 96-well cell culture plate seeded with mouse splenocytes collected from BALB/c mice (8 to 9 weeks old) obtained from CLEA Japan (Tokyo, Japan).
The mixtures of mouse cells and bacteria were cultured in a humidified 5% CO2 incubator at 37°C. After incubation for 24 hr, the culture supernatants of the mixtures were collected to measure the concentration of IL-12 by enzyme-linked immunosorbent assay (ELISA). The reagents used in the ELISA were the primary antibody (purified anti-mouse IL-12 [p70] antibody, BioLegend Inc., San Diego, CA, USA), secondary antibody (Biotin anti-mouse IL-12/IL-23 p40 antibody; BioLegend), blocking reagent (Block Ace Powder, KAC Co., Ltd., Kyoto, Japan), capture antibody (HRP Avidin, BioLegend), substrate (tetramethylbenzidine, Sigma-Aldrich, St. Louis, MO, USA), and standard (Recombinant Mouse IL-12 [p70] [ELISA Std.], BioLegend), and the IL-12 levels were measured by sandwich ELISA method [33].
Animal experiments
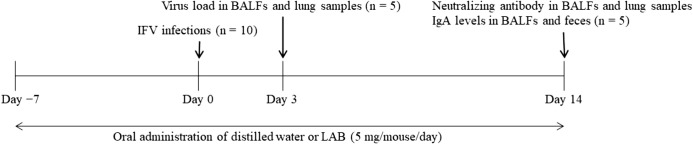
Female specific pathogen-free BALB/c mice (5–6 weeks old, 16–18 g) were obtained from Japan SLC (Shizuoka, Japan). All experiments were conducted in accordance with the animal experimentation guidelines of Chubu University and permitted by the Animal Care Committee of Chubu University (Permission number: 3010057). No side effects due to drug administration were detected throughout the experiments. Mice were intranasally infected with influenza A virus (A/NWS/33, H1N1 subtype) [34] at 2 × 104 plaque-forming units (PFU)/50 µL per mouse (n=10) on day 0. KH2 and SNK (5 mg/mouse/day; contents of RNAs in 5 mg of KH2, R-KH2, SNK, and R-SNK: 543.7 ± 17.5 ng, 54.9 ± 5.8 ng, 397.8 ± 5.8 ng, and 40.7 ± 2.3 ng, respectively) with or without ribonuclease treatment were suspended in distilled water. Oseltamivir phosphate (OSL; 0.2 mg/mouse/day) was used as a positive control for antiviral effect and was also dissolved in distilled water. Each of KH2, R-KH2, SNK, R-SNK, and OSL was given by oral administration twice per day from 7 days before virus inoculation until 14 days after inoculation. Control mice were administered orally with vehicle (distilled water) alone. As IFV infection causes a reduction in body weight [35, 36], mice in each treatment group were weighed daily for 14 days beginning on the day of IFV inoculation (designated day 0). Lung samples and bronchoalveolar lavage fluid (BALF) were collected from each group on days 3 and 14, and blood and fecal samples were collected on day 14 (Fig. 1). Lung samples were sonicated for 10 sec after the addition of 10 µL phosphate-buffered saline (PBS) per 1 mg of lung tissue and centrifuged at 10,000 × g for 30 min to separate the supernatants, which were stored at −80°C. BALFs were collected by four washes with 0.8 mL of ice-cold PBS via a tracheal cannula and centrifuged at 1,500 rpm for 10 min; supernatants were stored at −80°C. Blood samples were centrifuged at 3,000 rpm for 10 min, and the sera were stored at −20°C. Fecal extracts were prepared by adding PBS at 10 µL per mg of feces. The amount of virus in the lung and BALF samples collected on day 3 post-infection were quantified by plaque assays on Madin–Darby canine kidney (MDCK) cell monolayers. Sera and BALFs were subjected to neutralizing antibody titer assays using a 50% plaque reduction method, as described previously [37, 38], and BALFs and fecal extracts were assessed for mucosal IgA levels by ELISA.
Fig. 1.
Experimental procedure of influenza virus infection.
Mice in the control or LAB groups were administered distilled water or LAB (5 mg/day in two doses per day) during the study period (day −7 to 14). Mice were intranasally infected with IFV on day 0. On day 3 after IFV infection, five mice from each group were sacrificed to quantify virus loads in BALFs and lungs. The remaining five mice were sacrificed for measurement of neutralizing antibody and IgA levels on day 14. BALF: bronchoalveolar lavage fluid; IFV: influenza A virus; LAB: lactic acid bacteria.
Statistical analysis
The effects of the drugs were analyzed by one-way analysis of variance, and correction for multiple comparisons was done by Tukey’s multiple comparison test. A p value of <0.05 was considered to be significant.
RESULTS
Comparison of RNA concentration with and without ribonuclease treatment
The RNA contents were 108.74 ± 3.50 ng/mg for KH2, 10.98 ± 1.16 ng/mg for R-KH2, 79.55 ± 1.16 ng/mg for SNK, and 8.13 ± 0.47 ng/mg for R-SNK. Ribonuclease treatment reduced the quantity of RNA to 1/10.
Effects of ribonuclease-treated KH2 and SNK on IL-12 production in mouse splenocytes
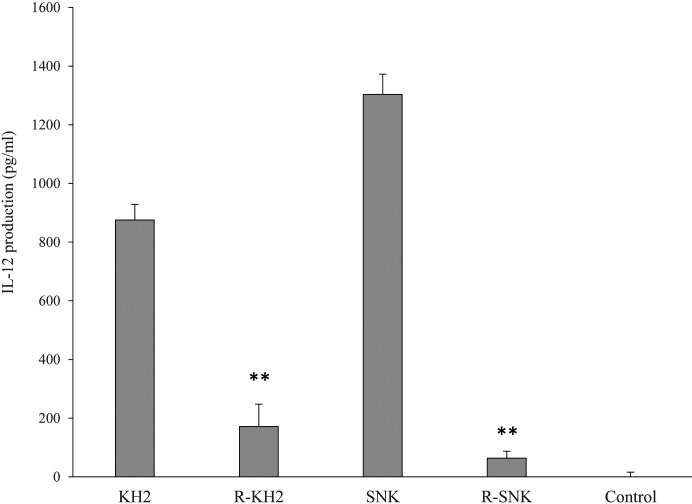
Ribonuclease treatment of both KH2 and SNK markedly reduced the levels of IL-12 produced by mouse splenocytes (p<0.01) (Fig. 2). The reduction in IL-12 production was more pronounced in the SNK strain than in the KH2 strain.
Fig. 2.
Effect of ribonuclease treatment of LAB on IL-12 production in mouse splenocytes.
Heat-killed E. faecalis KH2 and L. plantarum SNK12 were treated with or without ribonuclease and co-cultured with mouse splenocytes for 24 hr. IL-12 protein concentration in the culture supernatant was measured by enzyme-linked immunosorbent assay (ELISA). KH2: non-treated E. faecalis KH2; R-KH2: ribonuclease-treated E. faecalis KH2; LAB: lactic acid bacteria; SNK: non-treated L. plantarum SNK12; R-SNK: ribonuclease-treated L. plantarum SNK 12; control: culture medium only. Each value is presented as the mean ± SD. n=6. **p<0.01 vs. untreated for each LAB.
Effects of ribonuclease-treated KH2 and SNK on IVF infection in mice
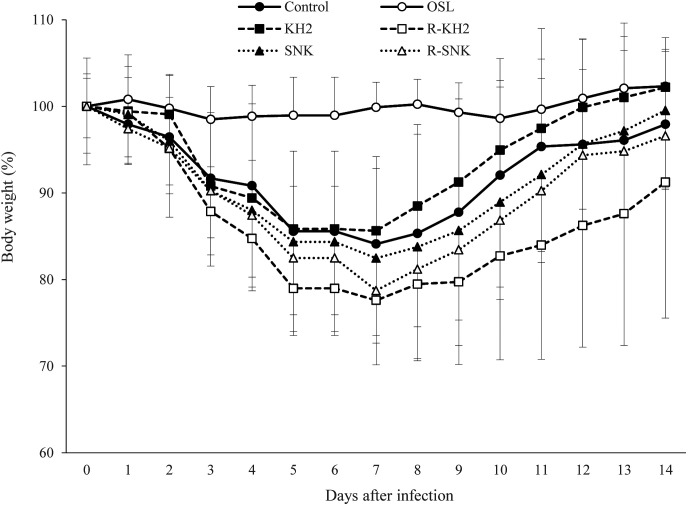
The effects of ribonuclease-treated (R) or untreated KH2 and SNK on the change in body weight of mice infected with IFV were examined (Fig. 3). The control group without KH2 or SNK showed approximately 16% loss of body weight on day 7 following IFV infection. The KH2, R-KH2, SNK, and R-SNK groups showed approximately 14%, 22%, 17%, and 21% losses, respectively, on day 7 post-infection. Although no significant difference was observed between the ribonuclease-treated and untreated groups, KH2 and SNK slightly suppressed weight loss more so than R-KH2 and R-SNK. Thereafter, mice of these groups gradually gained body weight, and the mice in the KH2 group returned to their pre-infection body weight levels on day 14 post-infection.
Fig. 3.
Body weight changes of mice infected with the IFV.
IFV-infected mice were orally administered distilled water (control, filled circle), 0.2 mg/day of oseltamivir (OSL, white circle), 5 mg/day of bacteria (KH2, untreated E. faecalis KH2, filled square; R-KH2, ribonuclease-treated E. faecalis KH2, white square; SNK, untreated L. plantarum SNK12, filled triangle; R-SNK, ribonuclease-treated L. plantarum SNK12, white triangle) from 7 days prior to virus infection to 14 days post-infection. Body weights are relative to those on the day of viral infection (day 0), which was set as 100%. Each value is presented as the mean ± SD. n=5. IFV: influenza A virus.
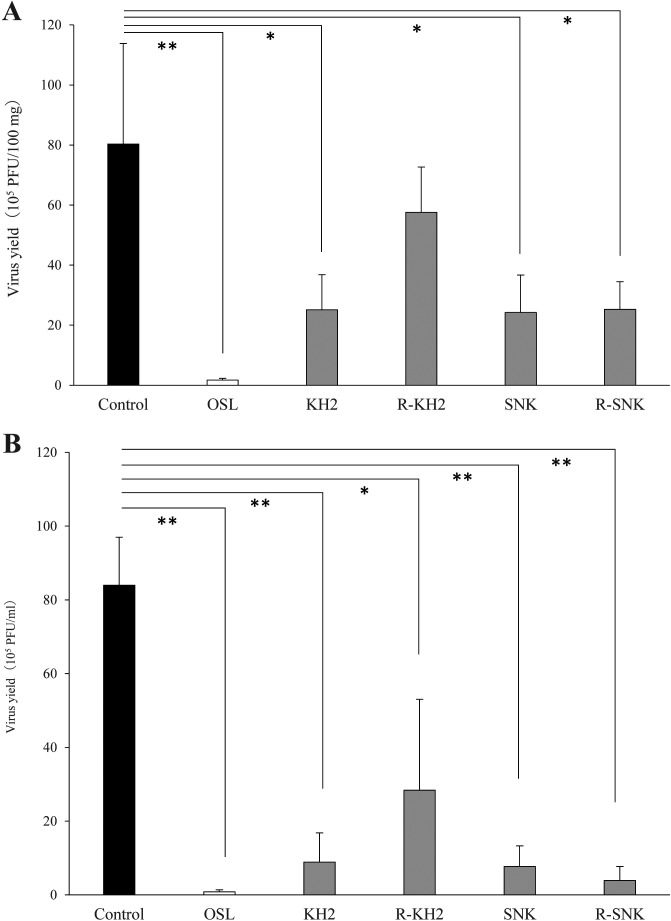
The virus yields in the lungs and BALFs of IFV-infected mice on day 3 post-infection are shown in Fig. 4A and Fig. 4B, respectively. Oral administration of the ribonuclease-treated or untreated forms of KH2 and SNK significantly reduced the virus load in the lungs compared with the control group (p<0.05), except for the lung samples of R-KH2, which showed no significant difference. A similar tendency was observed in BALF samples. Virus loads in the OSL group were markedly low, as shown in Fig. 4.
Fig. 4.
Effect of LAB administration on virus load in the mice.
Virus yield in BALFs (A) and lung samples (B) were measured by a plaque assay on day 3 post-infection. Each value is presented as the mean ± SD. n=5. **p<0.01; *p<0.05. BALF: bronchoalveolar lavage fluid; KH2: non-treated E. faecalis KH2; R-KH2: ribonuclease-treated E. faecalis KH2; LAB: lactic acid bacteria; OSL: oseltamivir; PFU: plaque-forming units; SNK: non-treated L. plantarum SNK12; R-SNK: ribonuclease-treated L. plantarum SNK12.
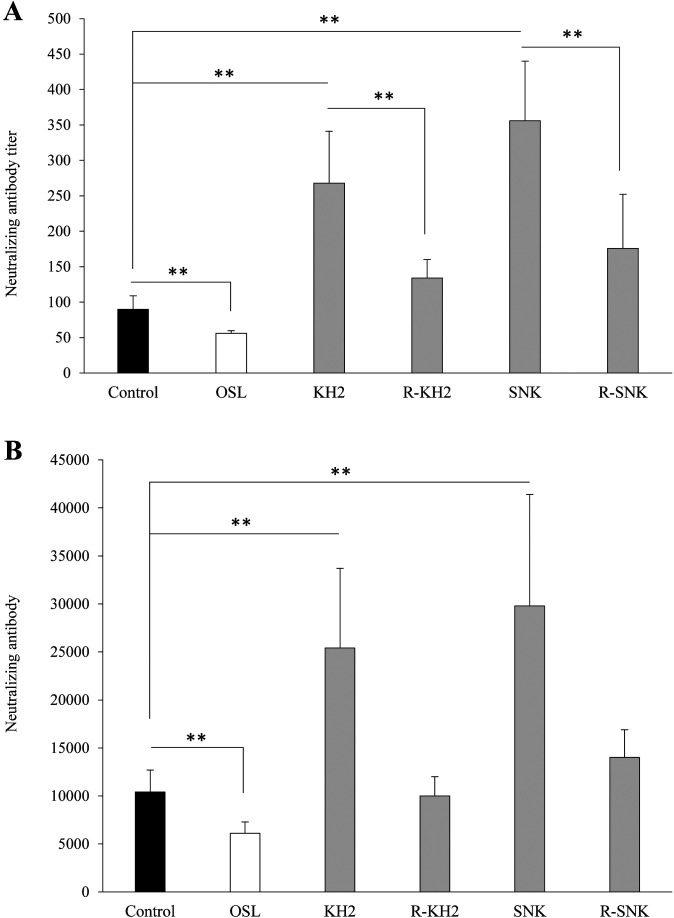
Figure 5 shows the effects of the ribonuclease-treated and untreated forms of KH2 and SNK on the neutralizing antibody response to IFV in BALFs (Fig. 5A) and sera (Fig. 5B) at day 14 post-infection. The antibody titers of BALFs and sera in the mice administered with untreated KH2 or SNK were significantly high as compared with those obtained in the control group (p<0.01). By contrast, antibody titers in the ribonuclease-treated KH2 and SNK groups were almost equivalent to those of the control group, but those in BALF samples were decreased significantly by ribonuclease treatment (p<0.05) (Fig. 5A). OSL group titers were significantly lower than those of the control group (p<0.01) in both BALF and serum samples.
Fig. 5.
Effect of LAB administration on the neutralizing antibody titer against IFV in the mice.
The titer of the virus-neutralizing antibody is presented as the reciprocal of the dilution of BALFs (A) and sera (B) that reduced the plaque number to a level below 50% of that seen in the virus control. Each value is presented as the mean ± SD. n=5. **p<0.01; *p<0.05. BALF: bronchoalveolar lavage fluid; IFV: influenza A virus; KH2: non-treated E. faecalis KH2; R-KH2: ribonuclease-treated E. faecalis KH2; LAB: lactic acid bacteria; OSL: oseltamivir; SNK: non-treated L. plantarum SNK12; R-SNK: ribonuclease-treated L. plantarum SNK12.
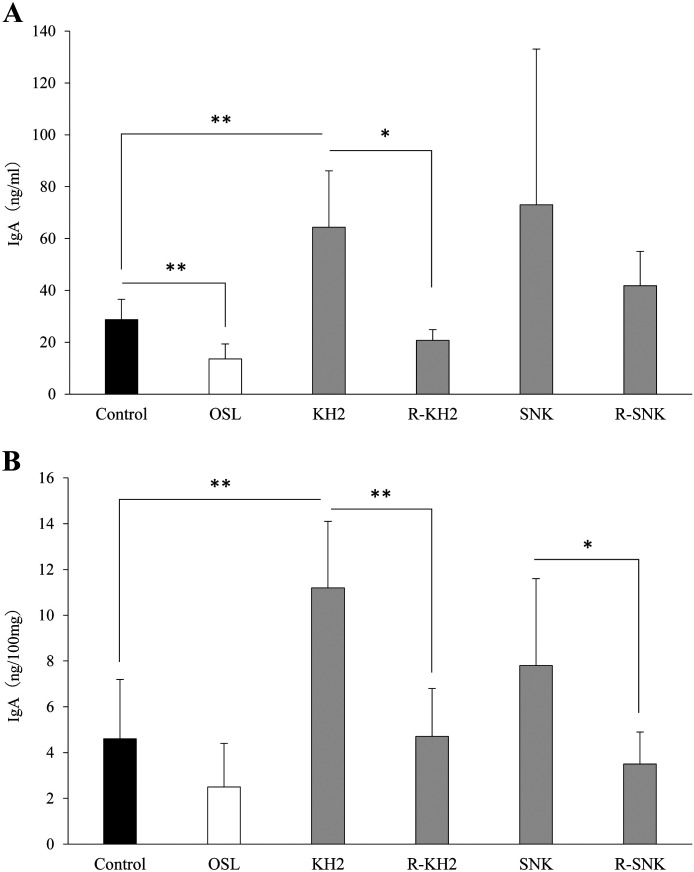
To elucidate whether ribonuclease treatment of KH2 and SNK stimulates the local immune response in mice, the levels of IFV-specific IgA in BALFs and feces were determined at day 14 post-infection (Fig. 6A and Fig. 6B). IgA production in the KH2 group was significantly increased (p<0.01). The IgA levels of the ribonuclease-treated KH2 group were almost equivalent to those of the control group and significantly lower than those of the KH2 group in the BALFs (p<0.05) and feces (p<0.01). For the SNK and R-SNK groups, no significant difference in IgA levels was observed in the BALF samples, whereas R-SNK showed significantly reduced IgA production in the feces (p<0.05). By contrast, marked suppression of IgA production was observed in the OSL group as compared with the control group (p<0.01) in BALF samples.
Fig. 6.
Effect of LAB administration on the production of IFV-specific IgA in mice.
The IFV-specific IgA levels in BALFs (A) and feces (B) were determined by ELISA. Each value is presented as the mean ± SD. n=5. **p<0.01; *p<0.05. BALF: bronchoalveolar lavage fluid; IFV: influenza A virus; KH2: non-treated E. faecalis KH2; R-KH2: ribonuclease-treated E. faecalis KH2; LAB: lactic acid bacteria; OSL: oseltamivir; SNK: non-treated L. plantarum SNK12; R-SNK: ribonuclease-treated L. plantarum SNK12.
DISCUSSION
To validate the immune-related active components of KH2 and SNK, IL-12 production by splenocytes and a mouse IFV infection model were used. The results showed that oral administration of KH2 or SNK produced an anti-IFV effect. In addition, the RNAs of KH2 and SNK were degraded by ribonuclease, which markedly reduced IL-12 production in splenocytes and had an impact on the anti-IFV effects. The RNAs of KH2 and SNK were suggested to be important factors for the anti-IFV effects based on the relationship between IL-12 production and anti-IFV effects. Although the cell wall has been reported to be an important factor in IL-12 production [13, 39], the present study found that RNA in bacteria is also a major factor affecting IL-12 production, though it should be noted that the validation method differed between this study and previous studies. The cell wall of L. plantarum and Streptococcus mutans strongly induces IL-12 production via TLR2 and TLR4 signaling in DCs and macrophages [13, 39], whereas KH2 and SNK may be signaling differently due to reduced IL-12 production by RNA degradation. Since bacteria have been reported to produce IL-12 via TLR3, 7, and 8 [17, 18], KH2 and SNK may also be signaling via TLR3, 7, and 8.
TLR3, which recognizes double-stranded RNA, and TLR7, which recognizes single-stranded RNA (TLR8 in humans), are also receptors that recognize viruses [40, 41]. This suggests that the anti-IFV effects of KH2 and SNK are influenced by RNA. The degradation of the RNAs of KH2 and SNK reduced the antiviral effect, suggesting that the RNAs of KH2 and SNK acted similarly to viral RNAs. Therefore, we would like to analyze the entire genome of KH2 or SNK to determine whether there are any sequences similar to those possessed by influenza viruses. In contrast, our observations that LAB reduced virus loads and increased antiviral antibodies were similar to those reported from previous studies on the anti-influenza virus effects of other LAB strains [42,43,44,45,46]. Because many reports on the anti-influenza effects of bacteria are conducted with probiotic strains and the heat-killed bacteria used in this study exerted similar effects to those of live bacteria, the viability of bacteria is not related to their immune-mediated antiviral effects. The mechanism of action is the uptake of bacteria from M cells in the intestinal tract and phagocytosis of them by DCs and macrophages, stimulating immunity [47]. Bacteria endocytosed by M cells have been reported to be transported to immunocompetent cells and to induce an immune response systemically or in the immune system [47, 48]. Since we confirmed the uptake of KH2 from the intestinal Peyer’s patch, we believe that this is a similar mechanism. However, it is unclear whether the mechanism is the same as that of other LAB, so we will use KH2 and SNK in future research to investigate their movement after transport from the Peyer’s patch and to analyze the influence in each tissue. In addition, it was interesting that the serum and BALF neutralizing antibody titers, BALF and fecal IgA, were significantly lower in the ribonuclease-treated KH2 and SNK intake groups after 14 days of IFV infection (Figs. 5 and 6). The R-KH2 group showed a trend in viral load than the KH2 group, but there was no difference between the SNK and R-SNK groups after 3 days of IFV infection (Fig. 4). Ribonuclease treatment also had a slight influence on weight change, with the R-KH2 group losing weight compared with the KH2 group, although not significantly; the R-SNK group also showed a slight loss of weight compared with the SNK group, although not as much as the R-KH2 group (Fig. 3). This difference may be related to the immune response to the viral infection. During virus infection, inflammatory cytokines such as antiviral type I interferon are produced, and the innate immune system plays an important role in viral control [49]. The major receptors involved in the recognition of a virus during innate immunity are TLRs, retinoic acid-inducible gene-1 like receptor, and nucleotide-binding oligomerization domain-containing protein 2 [50,51,52], and it is possible that the effects of the RNA of KH2 at the initial stages of infection (Fig. 4) affected the abovementioned receptors. In the future, we would like to use KH2 RNA to analyze the receptors involved in innate immunity. By contrast, SNK had little influence on the initial viral suppression effect on nuclease treatment (Fig. 4), as it has a different immune activation pathway from KH2. The titers of neutralizing antibody and IgA production at day 14 post-infection were significantly reduced by ribonuclease treatment (Figs. 5 and 6), suggesting that the RNAs of KH2 and SNK affect acquired rather than innate immunity.
In conclusion, herein we showed that both orally administered KH2 and SNK have potent profiles as influenza therapeutic agents involved in protection against IFV infection, inhibition of viral replication, and increased immune response. Furthermore, the RNAs of KH2 and SNK were shown to be active components and were suggested to affect the acquired immunity. We would also like to compare the quality and quantity of RNAs in the future by analyzing the RNAs of KH2 and SNK by RNA-seq. Based on the results obtained herein, we elucidated at least one mechanism of the protective effects of KH2 and SNK against virus infection. Future studies will search for more effective LAB species and explore not only the effect but also the mechanism through comparisons with live bacteria. Because the threat from viral infections still exists, the need for safer and more effective immunomodulators remains pressing. Among them, LAB are generally accepted as safe functional foods; we therefore hope that the widespread use of LAB species such as the KH2 and SNK used in this study will reduce the risk of viral infections.
Acknowledgments
We thank Ms. Kazumi Shimizu (Non-Profit Organization, The Japanese Association of Clinical Research on Supplements) and Yuriko Namatame (Bio-Lab Co., Ltd.) for skillful technical assistance and valuable discussions. The authors would like to thank Enago (www.enago.jp) for the English language review.
REFERENCES
- 1.Lidbeck A, Gustafsson JA, Nord CE. 1987. Impact of Lactobacillus acidophilus supplements on the human oropharyngeal and intestinal microflora. Scand J Infect Dis 19: 531–537. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara S, Seto Y, Kimura A, Hashiba H. 2001. Establishment of orally-administered Lactobacillus gasseri SBT2055SR in the gastrointestinal tract of humans and its influence on intestinal microflora and metabolism. J Appl Microbiol 90: 343–352. [DOI] [PubMed] [Google Scholar]
- 3.Unno T, Choi JH, Hur HG, Sadowsky MJ, Ahn YT, Huh CS, Kim GB, Cha CJ. 2015. Changes in human gut microbiota influenced by probiotic fermented milk ingestion. J Dairy Sci 98: 3568–3576. [DOI] [PubMed] [Google Scholar]
- 4.Reid G, Bruce AW, McGroarty JA, Cheng KJ, Costerton JW. 1990. Is there a role for lactobacilli in prevention of urogenital and intestinal infections? Clin Microbiol Rev 3: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahara T, Shimizu K, Nomoto K, Hamabata T, Ozawa A, Takeda Y. 2004. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect Immun 72: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross ML, Stevenson LM, Gill HS. 2001. Anti-allergy properties of fermented foods: an important immunoregulatory mechanism of lactic acid bacteria? Int Immunopharmacol 1: 891–901. [DOI] [PubMed] [Google Scholar]
- 7.Inoue R, Nishio A, Fukushima Y, Ushida K. 2007. Oral treatment with probiotic Lactobacillus johnsonii NCC533 (La1) for a specific part of the weaning period prevents the development of atopic dermatitis induced after maturation in model mice, NC/Nga. Br J Dermatol 156: 499–509. [DOI] [PubMed] [Google Scholar]
- 8.Tsukahara T, Nakamura SI, Romero-Pèrez GA, Ohwaki M, Yanagisawa T, Kan T. 2018. Stimulation of murine cell-mediated immunity by dietary administration of a cell preparation of Enterococcus faecalis strain KH-2 and its possible activity against tumour development in mice. Biosci Microbiota Food Health 37: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagi A, Matsuzaki T, Sato M, Nomoto K, Morotomi M, Yokokura T. 2001. Enhancement of natural killer cytotoxicity delayed murine carcinogenesis by a probiotic microorganism. Carcinogenesis 22: 599–605. [DOI] [PubMed] [Google Scholar]
- 10.Goto H, Sagitani A, Ashida N, Kato S, Hirota T, Shinoda T, Yamamoto N. 2013. Anti-influenza virus effects of both live and non-live Lactobacillus acidophilus L-92 accompanied by the activation of innate immunity. Br J Nutr 110: 1810–1818. [DOI] [PubMed] [Google Scholar]
- 11.Kumova OK, Fike AJ, Thayer JL, Nguyen LT, Mell JC, Pascasio J, Stairiker C, Leon LG, Katsikis PD, Carey AJ. 2019. Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLoS Pathog 15: e1008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae JY, Kim JI, Park S, Yoo K, Kim IH, Joo W, Ryu BH, Park MS, Lee I, Park MS. 2018. Effects of Lactobacillus plantarum and Leuconostoc mesenteroides probiotics on human seasonal and Avian influenza viruses. J Microbiol Biotechnol 28: 893–901. [DOI] [PubMed] [Google Scholar]
- 13.Ren C, Zhang Q, de Haan BJ, Zhang H, Faas MM, de Vos P. 2016. Identification of TLR2/TLR6 signalling lactic acid bacteria for supporting immune regulation. Sci Rep 6: 34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino S, Ikegami S, Kano H, Sashihara T, Sugano H, Horiuchi H, Saito T, Oda M. 2006. Immunomodulatory effects of polysaccharides produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci 89: 2873–2881. [DOI] [PubMed] [Google Scholar]
- 15.Oldenburg M, Krüger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, Koedel U, Akira S, Kawai T, Buer J, Wagner H, Bauer S, Hochrein H, Kirschning CJ. 2012. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337: 1111–1115. [DOI] [PubMed] [Google Scholar]
- 16.Li XD, Chen ZJ. 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife 1: e00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishibayashi R, Inoue R, Harada Y, Watanabe T, Makioka Y, Ushida K. 2015. RNA of Enterococcus faecalis strain EC-12 is a major component inducing Interleukin-12 production from human monocytic cells. PLoS One 10: e0129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima T, Ikari N, Watanabe Y, Kubota Y, Yoshio S, Kanto T, Motohashi S, Shimojo N, Tsuji NM. 2018. Double-stranded RNA derived from lactic acid bacteria augments Th1 Immunity via interferon-β from human dendritic cells. Front Immunol 9: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdi K, Singh NJ, Spooner E, Kessler BM, Radaev S, Lantz L, Xiao TS, Matzinger P, Sun PD, Ploegh HL. 2014. Free IL-12p40 monomer is a polyfunctional adaptor for generating novel IL-12-like heterodimers extracellularly. J Immunol 192: 6028–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trinchieri G. 1995. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol 13: 251–276. [DOI] [PubMed] [Google Scholar]
- 21.Sashihara T, Sueki N, Ikegami S. 2006. An analysis of the effectiveness of heat-killed lactic acid bacteria in alleviating allergic diseases. J Dairy Sci 89: 2846–2855. [DOI] [PubMed] [Google Scholar]
- 22.Ongol MP, Iguchi T, Tanaka M, Sone T, Ikeda H, Asano K, Nishimura T. 2008. Potential of selected strains of lactic acid bacteria to induce a Th1 immune profile. Biosci Biotechnol Biochem 72: 2847–2857. [DOI] [PubMed] [Google Scholar]
- 23.Stöhr K. 2002. Influenza—WHO cares. Lancet Infect Dis 2: 517. [DOI] [PubMed] [Google Scholar]
- 24.Manuguerra JC, Hannoun C. 1997. [Influenza: interspecies transmissions and viral rearrangement]. Bull Acad Natl Med 181: 421–430 (in French). [PubMed] [Google Scholar]
- 25.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9: e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459: 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med 360: 2616–2625. [DOI] [PubMed] [Google Scholar]
- 28.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98: 174–185. [DOI] [PubMed] [Google Scholar]
- 29.Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A, Lim W, Meijer A, Penn C, Tashiro M, Uyeki TM, Zambon M, WHO Consultation on Pandemic Influenza A (H1N1) 2009 Virus Resistance to Antivirals2012. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12: 240–248. [DOI] [PubMed] [Google Scholar]
- 30.Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, Fonseca K, Winter AL, Gubbay JB, Krajden M, Petric M, Charest H, Bastien N, Kwindt TL, Mahmud SM, Van Caeseele P, Li Y. 2014. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One 9: e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNeil SA, Andrew MK, Ye L, Haguinet F, Hatchette TF, ElSherif M, LeBlanc J, Ambrose A, McGeer A, McElhaney JE, Loeb M, MacKinnon-Cameron D, Sharma R, Dos Santos G, Shinde V, Investigators of the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network (CIRN).2015. Interim estimates of 2014/15 influenza vaccine effectiveness in preventing laboratory-confirmed influenza-related hospitalisation from the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network, January 2015. Euro Surveill 20: 21024. [DOI] [PubMed] [Google Scholar]
- 32.Iwabuchi N, Yonezawa S, Odamaki T, Yaeshima T, Iwatsuki K, Xiao JZ. 2012. Immunomodulating and anti-infective effects of a novel strain of Lactobacillus paracasei that strongly induces interleukin-12. FEMS Immunol Med Microbiol 66: 230–239. [DOI] [PubMed] [Google Scholar]
- 33.Kragstrup TW, Vorup-Jensen T, Deleuran B, Hvid M. 2013. A simple set of validation steps identifies and removes false results in a sandwich enzyme-linked immunosorbent assay caused by anti-animal IgG antibodies in plasma from arthritis patients. Springerplus 2: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govorkova EA, Fang HB, Tan M, Webster RG. 2004. Neuraminidase inhibitor-rimantadine combinations exert additive and synergistic anti-influenza virus effects in MDCK cells. Antimicrob Agents Chemother 48: 4855–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinoshita E, Hayashi K, Katayama H, Hayashi T, Obata A. 2012. Anti-influenza virus effects of elderberry juice and its fractions. Biosci Biotechnol Biochem 76: 1633–1638. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi K, Narutaki K, Nagaoka Y, Hayashi T, Uesato S. 2010. Therapeutic effect of arctiin and arctigenin in immunocompetent and immunocompromised mice infected with influenza A virus. Biol Pharm Bull 33: 1199–1205. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki K, Hayashi K, Lee JB, Kurosaki F, Hayashi T. 2015. Characterization of a novel mutation in NS1 protein of influenza A virus induced by a chemical substance for the attenuation of pathogenicity. PLoS One 10: e0121205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terasawa M, Hayashi K, Lee JB, Nishiura K, Matsuda K, Hayashi T, Kawahara T. 2020. Anti-influenza A virus activity of rhamnan sulfate from green algae Monostroma nitidum in mice with normal and compromised immunity. Mar Drugs 18: E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Wang D. 2014. Streptococcus mutans wall-associated protein A promotes TLR4-induced dendritic cell maturation. Scand J Immunol 80: 121–126. [DOI] [PubMed] [Google Scholar]
- 40.Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat Rev Immunol 4: 499–511. [DOI] [PubMed] [Google Scholar]
- 41.Akira S. 2003. Toll-like receptor signaling. J Biol Chem 278: 38105–38108. [DOI] [PubMed] [Google Scholar]
- 42.Kechaou N, Chain F, Gratadoux JJ, Blugeon S, Bertho N, Chevalier C, Le Goffic R, Courau S, Molimard P, Chatel JM, Langella P, Bermúdez-Humarán LG. 2013. Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl Environ Microbiol 79: 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee YN, Youn HN, Kwon JH, Lee DH, Park JK, Yuk SS, Erdene-Ochir TO, Kim KT, Lee JB, Park SY, Choi IS, Song CS. 2013. Sublingual administration of Lactobacillus rhamnosus affects respiratory immune responses and facilitates protection against influenza virus infection in mice. Antiviral Res 98: 284–290. [DOI] [PubMed] [Google Scholar]
- 44.Youn HN, Lee DH, Lee YN, Park JK, Yuk SS, Yang SY, Lee HJ, Woo SH, Kim HM, Lee JB, Park SY, Choi IS, Song CS. 2012. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res 93: 138–143. [DOI] [PubMed] [Google Scholar]
- 45.Hori T, Kiyoshima J, Shida K, Yasui H. 2001. Effect of intranasal administration of Lactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin Diagn Lab Immunol 8: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harata G, He F, Hiruta N, Kawase M, Kubota A, Hiramatsu M, Yausi H. 2010. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett Appl Microbiol 50: 597–602. [DOI] [PubMed] [Google Scholar]
- 47.Yanagihara S, Kato S, Ashida N, Yamamoto N. 2015. Lactobacillus acidophilus CP23 with weak immunomodulatory activity lacks anchoring structure for surface layer protein. J Biosci Bioeng 119: 521–525. [DOI] [PubMed] [Google Scholar]
- 48.Savidge TC, Smith MW, James PS, Aldred P. 1991. Salmonella-induced M-cell formation in germ-free mouse Peyer’s patch tissue. Am J Pathol 139: 177–184. [PMC free article] [PubMed] [Google Scholar]
- 49.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 50.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34: 637–650. [DOI] [PubMed] [Google Scholar]
- 51.Desmet CJ, Ishii KJ. 2012. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol 12: 479–491. [DOI] [PubMed] [Google Scholar]
- 52.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. 2009. Activation of innate immune antiviral responses by Nod2. Nat Immunol 10: 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]