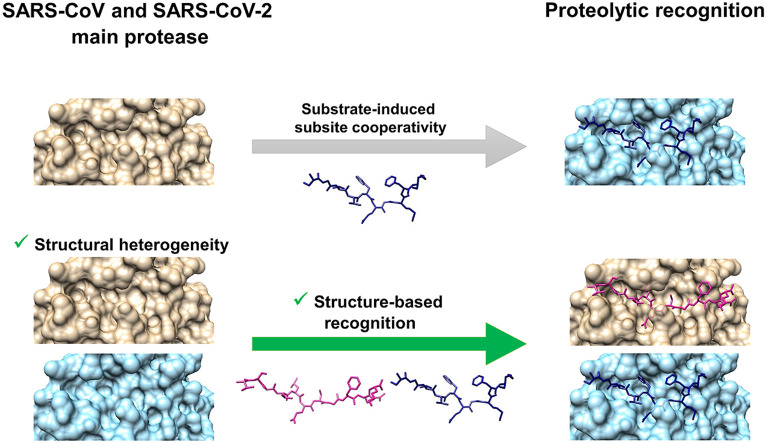
Abstract
The main protease (Mpro) of SARS-CoV and SARS-CoV-2 is a key enzyme in viral replication and a promising target for the development of antiviral therapeutics. The understanding of this protein is based on a number of observations derived from earlier x-ray structures, which mostly consider substrates or ligands as the main reason behind modulation of the active site. This lead to the concept of substrate-induced subsite cooperativity as an initial attempt to explain the dual binding specificity of this enzyme in recognizing the cleavage sequences at its N- and C-termini, which are important processing steps in obtaining the mature protease. The presented hypothesis proposes that structural heterogeneity is a property of the enzyme, independent of the presence of a substrate or ligand. Indeed, the analysis of Mpro structures of SARS-CoV and SARS-CoV-2 reveals a conformational diversity for the catalytically competent state in ligand-free structures. Variation in the binding site appears to result from flexibility at residues lining the S1 subpocket and segments incorporating methionine 49 and glutamine 189. The structural evidence introduces “structure-based recognition” as a new paradigm in substrate proteolysis by Mpro. In this concept, the binding space in subpockets of the enzyme varies in a non-cooperative manner, causing distinct conformations, which recognize and process different cleavage sites, as the N- and C-termini. Insights into the recognition basis of the protease provide explanation to the ordered processing of cleavage sites. The hypothesis expands the conformational space of the enzyme and consequently opportunities for drug development and repurposing efforts.
Keywords: SARS-CoV, SARS-CoV-2, Conformational selection, Viral protease, Antiviral drugs
Graphical abstract
1. Introduction
The coronavirus main protease (Mpro, also called 3CLpro) is a cysteine protease, which contributes to the processing of the viral polyproteins, derived from the translation of the viral RNA, at 11 cleavage sites [1]. A number of studies suggest that the catalytic activity of Mpro is associated mostly with dimer structures through a proximity between the N-terminal residues of one protomer and the S1 subpocket of the other protomer, mediated by an interaction between Glu166 and Ser1∗ (Fig. S2). The arrangement shapes the S1 subpocket and maintains the oxyanion loop in a catalytically competent state (Fig. S3) [2]. Relative to the peptide substrate, the active site is divided into subpockets (S and S’), where residues (P and P’) of a substrate or ligand can fit [3].
Among the cleavage sites processed by Mpro are its N- and C-termini, which are important for the release of the mature protease [1]. At these two sites, the binding specificity was proposed by x-ray structures to vary depending on substrate-induced subsite cooperativity [4]. The presented study hypothesizes an alternate mechanism, where structural heterogeneity of this protein, independent of the presence of the substrate or ligand, is the main determinant for proteolytic recognition by Mpro. To support this hypothesis, and in order to gain an understanding of the interplay between structure and function in this target, an analysis of x-ray structures is carried out and interpreted within the context of in-solution biochemical and structural results of relevance to the proteolytic recognition mechanism. The strategy has been previously applied for the study of conformations of the flaviviral protease, a viral protease from another RNA virus [5].
2. Material and methods
2.1. X-ray structures of SARS-CoV and SARS-CoV-2 main protease
Table 1.
A list of all structures of the main protease of SARS-CoV and SARS-CoV-2 retrieved from the protein data bank (PDB) until June 2020.
| Virus | Structure type | PDB code | References |
|---|---|---|---|
| SARS-CoV-2 | Ligand-free | 6Y2E | [2] |
| 6WQF | [6] | ||
| 6WTM | [7] | ||
| 6M03, 6M2Q, 6YB7, 6Y84, 7BRO | None | ||
| Protein-ligand complex |
6Y2F, 6Y2G | [2] | |
| 6LU7, 7BQY | [8] | ||
| 6LZE, 6M0K | [9] | ||
| 7BUY | [10] | ||
| 6WTJ, 6WTK | [7] | ||
| 6M2N, 6W63, 6WNP, 6WTT, 6YNQ, 6YT8, 6XA4, 6XB0, 6XB1, 6XB2, 6XBG, 6XBH, 6XBI, 6XFN, 6XG2, 6Z2E, 7BRP, 5RG0, group of structures with title panDDA (115 structures) |
None |
||
| SARS-CoV | Ligand-free | 1UJ1, 1UK2, 1UK3 | [11] |
| 1Z1I | [12] | ||
| 2C3S | [13] | ||
| 2DUC | [4] | ||
| 2H2Z | [14] | ||
| 3EBN | [15] | ||
| 3VB3 | [16] | ||
| 2A5A | [17] | ||
| 2GT7, 2GT8 | [18] | ||
| 2BX4 | [19] | ||
| 3IWM | [20] | ||
| 1Q2W | None | ||
| Ligand-free mutant | 1Z1J | [12] | |
| 2QCY | [21] | ||
| 3AW1 | [22] | ||
| 3FZD | [23] | ||
| 4HI3 | [24] | ||
| 3F9G, 3F9E, 3F9F, 3F9H | [25] | ||
| 3EA7, 3EA8, 3EA9, 3EAJ, 3M3S, 3M3T, 3M3V, 3E91 | None | ||
| Protein-ligand complex |
1UK4 | [11] | |
| 2V6N, 2VJ1 | [26] | ||
| 2Z3C, 2Z3D, 2Z3E | [27] | ||
| 1WOF, 2AMD, 2AMQ, 2D2D | [28] | ||
| 2A5K, 2A5I | [17] | ||
| 2ALV | [29] | ||
| 2GTB | [18] | ||
| 2GX4 | [30] | ||
| 2GZ7, 2GZ8, 2GZ9 | [31] | ||
| 2HOB | [14] | ||
| 2OP9 | [32] | ||
| 2QIQ | [33] | ||
| 2Z9G, 2Z9J, 2Z9K, 2Z9L, 2Z94 | [34] | ||
| 2ZU4, 2ZU5 | [35] | ||
| 3ATW, 3AVZ, 3AW0 | [22] | ||
| 3D62 | [36] | ||
| 3SN8, 3SNA, 3SNB, 3SNC, 3SND, 3SNE | [37] | ||
| 3V3M | [38] | ||
| 3VB4, 3VB5, 3BV6, 3BV7 | [16] | ||
| 4MDS | [39] | ||
| 4TWW, 4TWY, 4WY3 | [40] | ||
| 3SZN, 3TIT, 3TIU, 3TNS, 3TNT, 5C5N, 5C5O, 5N5O, 5N19, 6Y7M |
None |
||
| Mutant-complex with C-terminus autoprocessing site | 5B6O | [4] | |
| Mutant-complex with N-terminus autoprocessing site | 2Q6G | [41] |
2.2. Assessment of protein structures
Computational analysis of protein conformations was conducted as previously described [2], using UCSF Chimera version 1.13.1 [42]. Briefly, structural alignment was performed with the “Match Maker” tool in Chimera using default settings. PDB files with dimeric protein structures were split to individual protomers and each protomer was included in the analysis. Default settings were used for visualization.
3. Analysis design and considerations
SARS-CoV-2 Mpro shares 96% sequence similarity with, the more extensively studied, SARS-CoV Mpro (Fig. S1). This high degree of conservation allows exploring the conformational space using all x-ray structures from both viruses, with a total of 241 structures deposited in the PDB until June 2020 (See Table 1). Catalytic competence is determined based on the orientation of the backbone NH groups for residues 143 and 145 in the oxyanion hole [11]. Root-mean-square deviation (RMSD) values are included relative to the average resolution of representative compared structures. Muramatsu et al. reported the first attempt to interpret biochemical results of dual specificity for processing of the N-terminal and C-terminal cleavage sites through an analysis limited to only two ligand-complexed structures [4]. This maturation step is important as in the dimer form, both the N- and C-termini of one protomer are oriented towards the active site of the other protomer, and thus can affect access of ligands or substrates for recognition and processing. In contrast to the analysis design by Muramatsu et al., the current study is conducted on a more comprehensive scope without disregarding ligand-free crystal structures, to pinpoint the basis for the molecular recognition by Mpro.
In-solution structural and biochemical studies of an enzyme often provide more relevant mechanistic insight. For the flaviviral protease, for example, the NMR structures (PDB code: 2M9P and 2M9Q) [43] show distinct conformations compared to those observed in x-ray structures.
However, very few NMR studies were conducted on the main protease of SARS-CoV [[44], [45], [46], [47]] and SARS-CoV-2 [7]. For SARS-CoV, the separate C- and N-domains of Mpro were deposited with PDB code 2LIZ, or biological magnetic resonance bank (BMRB) codes 17911 and 17251. For SARS-CoV-2, NMR signal assignment for Mpro is available under BMRB code 50262. As opposed to other viral proteases, such as the flaviviral protease [5], NMR studies were not combined to the use of tags (lanthanide tags for pseudocontact shifts or tert-butyl groups for high signal intensity) for the protein or the ligand to increase the technique sensitivity in tracking conformational dynamics. The equilibrium between the monomeric and dimeric forms of Mpro is suggested to play a role in the understanding of the catalytic mechanism of this enzyme [48]. However, a high variability in monomer-dimer equilibrium dissociation constants was reported for Mpro between different research groups and measurement conditions [48,49]. In some instances, the values are one or more orders of magnitude higher than the common concentration range (submicromolar to nanomolar) used in biochemical assays, where ligand recognition is noticed and assessed. Advances in NMR and monomer-dimer studies of Mpro have been reviewed [47,48], and will be addressed within the proteolytic recognition focus of this work and the outcomes of the performed analysis of crystal structures.
4. Outcomes of structural analysis
4.1. Structural heterogeneity in Mpro
A number of hypotheses govern the understanding of SARS-CoV and SARS-CoV-2 Mpro: First, the Glu166-Ser1∗ interaction is needed to achieve catalytic activity, second, the number of catalytically competent protomers within the dimer structure varies in a pH-dependent manner [19], and third, depending on the ligand steric demand, binding can trigger via induced-fit a change of the inactive state into active state and a modulation of the protein structure in the binding site [18,26,39]. As such, only dimeric structures of Mpro that display the Glu166-Ser1∗ interaction are used as starting point for structure-based design and optimization or repurposing of inhibitors [2,9].
The presented analysis provides structural insights, which contradict the above mentioned concepts for this enzyme and shows that deposited structures are not identical. The study hypothesizes a more general understanding of the coronavirus Mpro based on conformational heterogeneity.
Representative structures with distinct conformations are depicted in Fig. 1 : 6Y2E, 6Y2F, 6Y2G, 6M0K, 7BRO for SARS-CoV-2, and 1UK3, 2GT7, 2GTB, 2C3S, 2Q6G, 5B6O, 2VJ1, 4MDS, 2DUC for SARS-CoV. The following points are observed:
-
A)
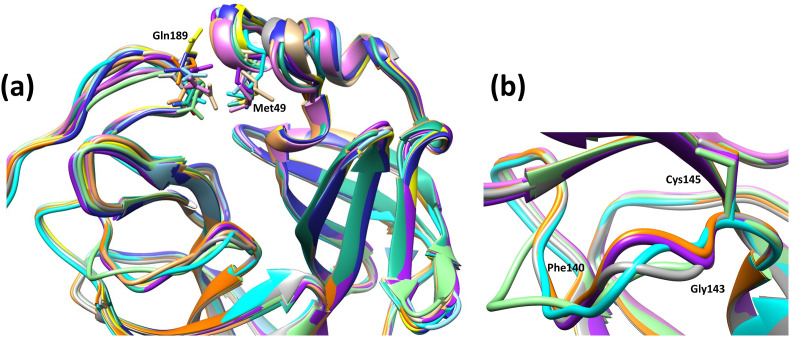
The flexibility in Gly143 affects the orientation of its backbone NH group, as seen in 1UJ1, 2VJ1 and 2DUC, and hence the switch of the oxyanion loop from inactive to active state (Figs. S2, S4a, and S5). Modest variations are noticed for segment 136–138 in 2DUC, and for residues 139–142 of the catalytically competent protomers in structures 6Y2E, 6Y2G, and 2GT7 in comparison to 1UK3 or 2VJ1 (Fig. 2a and b).
-
B)
The number of active protomers in a dimer structure is pH-independent. 1UJ1 displays one active protomer in the dimer, while in 2GT7 both protomers are in the active state, despite the fact that both structures are crystallized under the same pH and the same space group, which contradicts earlier hypothesis about the influence of pH on the number of active protomers. However, pH is reported to induce variation of the conformation of protomers in crystal structures obtained at different pH [19] or temperature [6]. The pH-related structural variations are proposed to be caused by changes in the protonation states of His163 and His172.
-
C)
Mpro is catalytically active even in the absence of the Glu166-Ser1∗ proximity. 2VJ1 depicts a dimer in the absence of the Glu166-Ser1∗ interaction, where the N-terminus points away from the S1 subpocket, as in an inactive enzyme, but the oxyanion hole retains catalytic competence in one protomer (Fig. S4). The construct used for this structure is a SARS-CoV Mpro mutant with a Ser1 deletion, which displays a degree of catalytic activity in biochemical assays [26]. The difference in the orientation of the N-terminus and the oxyanion loop conformation results in a modulation of the space available for ligand binding at the dimer interface and in the vicinity of the catalytic center in 6Y2G vs 2VJ1.
Fig. 1.
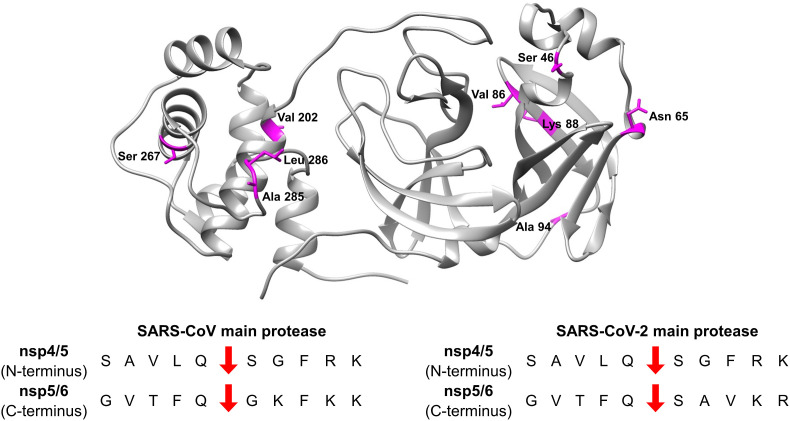
Mapping of residues of the main protease of SARS-CoV-2, which differ from SARS-CoV. Structure 6Y2E is depicted in ribbon form in grey, non-conserved residues are shown in stick form in magenta. Cleavage sequences at the N- and C-termini of the main protease (NSP5) are shown. (Sequence alignment and a complete list of cleavage sites are provided in the Supporting Information, as Fig. S1 and Table S1, respectively).
Fig. 2.
Structural heterogeneity of the main protease of SARS-CoV and SARS-CoV-2. (a) Overlay of representative ligand-free structures and complexes of SARS-CoV and SARS-CoV-2 main protease in ribbon form to show variation in the segments containing Met49 and Gln189. Color code: 6Y2E in light purple, 6Y2F in light blue, 6Y2G in purple, 6M0K in blue, 7BRO in dark cyan, 1UK3 in cyan, 2C3S in light orange, 2GT7 in orange, 2GTB in tan, 4MDS in yellow, 2VJ1 in grey, 2DUC in light green. Structures 6Y2E, 7BRO, 2C3S, 1UK3, 2GT7, and 2DUC are ligand-free structures. (b) Zoomed-in view of the backbone flexibility in Gly143 at the oxyanion loop. Same color code is used as in section (a).
It is thus generally noted that apart from the inactive conformation, different conformational states show catalytic competence for Mpro, with respect to the number of active protomers, the conformation for the residues lining the S1 subpocket, in addition to flexibility in other conserved segments of the protein such residues 45–52 and 189–196 (Fig. 2 a and b). Accordingly, the structural evidence demonstrates the presence of different conformational populations for this enzyme.
4.2. Proteolytic recognition: substrate-induced subsite cooperativity or structure-based recognition?
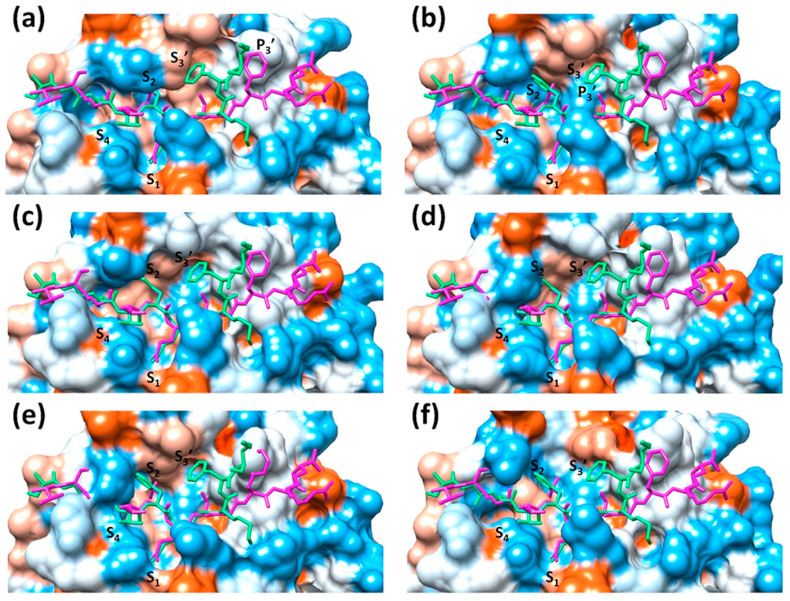
Substrate-induced subsite cooperativity (Fig. 3 a and b) is suggested between S2 and S3’ based on structures 2Q6G and 5B6O [4]. The phenomenon of subsite cooperativity is known to influence specificity in other proteases [50]. For Mpro, when the P2 leucine of the N-terminus cleavage site binds to S2, the S3’ subsite is not formed. However, the more bulky P2 phenylalanine in the C-terminus processing site induces a conformational change in S2, forming thus the S3’ subsite, where the P3’ residue fits. The alternate fitting of the N- and C-termini in the enzyme active site in 2Q6G and 5B6O, respectively, provides an interpretation for the dual specificity in autoprocessing observed biochemically in-solution. The performed structural analysis contradicts this mechanism by identifying conformations of the ligand-free protein depicting the S3’ binding space (Fig. 3c-f). Variation of the active site in S2 and S3’ does not occur in an exclusively cooperative manner but originates instead from conformational dynamics of the protein at Gln189 and Met49, and the segments incorporating them. This study introduces “structure-based recognition” as a paradigm to understand ligand binding in Mpro and hypothesizes that conformational heterogeneity is the main determinant for its proteolytic recognition. In this model, substrates or cleavage site sequences are recognized differently, based on the fitting of their residues into the active site of distinct conformations of the enzyme. Further support is provided by an analysis of inhibitor-protein complexes, to identify the ligand binding mechanism.
Fig. 3.
Proteolytic recognition based on substrate-induced subsite cooperativity, or structural heterogeneity and structure-based recognition. (a–f) Overlay of the N-terminal processing site in magenta and the C-terminal processing site in green in stick form to the solvent accessible surface of different structures: (a) 2Q6G, SARS-CoV, H41A mutant cocrystallized with the N-terminal site; (b) 5B6O, SARS-CoV C145A cocrystallized with the C-terminal site; (c) 6Y2E, SARS-CoV-2, ligand-free structure; (d) 7BRO, SARS-CoV-2, ligand-free structure; (e) 2C3S, SARS-CoV, ligand-free structure; (f) 2GT7, SARS-CoV, ligand-free structure. (a) and (b) represent the basis behind the concept of substrate-induced subsite cooperativity between S2 and S3’, while (c–f) oppose this mechanism by showing that variations in the active site occur in the absence of a ligand due to conformational heterogeneity and thus substrate processing depends on structure-based recognition. The binding spaces in S2 and S3’ does not vary in an exclusively cooperative manner. The solvent accessible surface is colored according to hydrophobicity, where blue is hydrophilic and orange is hydrophobic.
While the Muramatsu et al. work is considered as a starting step to assess the reason behind altered specificity for Mpro, the difference in the mechanistic insight compared to the current hypothesis can be explained by certain considerations. First, Muramatsu et al. limited their analysis to two structures, which excluded the high diversity from crystallized ligand-free Mpro. The highest measured RMSD is 2.4 Å in S2 at residue 189, and in S3’ at residues 46–47 and 49, where the average resolution of the compared structures is in range of 2.2–2.5 Å. It is however true that the fit of the P3’ residue is quite different between 2Q6G and 5B6O, with an RMSD of 6.8 Å, and in fact the orientation of the P3’ amino acid in 5B6O cannot be accommodated in 2Q6G due to steric clash with amino acids lining the S3’ subpocket. Ligand-free structures, such as 1UK3 or 2GT7 and 6Y2E, show variations with RMSD values of 3.1 Å in S2 at residue 189, 4.9 Å in S3’ at residues 46–47, and 3.9 Å with residue 49. The biochemical evidence supporting the subsite-induced cooperativity was assessed under one testing condition, which may not represent the entire conformational diversity of this protein. To lean further support to this point, a comparison of the recognition properties of different synthesized substrates by Mpro depicts a degree of variation in substrate processing for example in the pH profile curve. This includes the shape of the curve being steep [19,[51], [52], [53]] or flat [54] as the pH increases from 6 to 8, and the peak for substrate cleavage. The pH optima is observed at pH 6.5, [54] 7, [19] 7.5, [53,55] or 8 [51,52]. Difference in results can be interpreted based on the use and type of fluorophores, or on structure-based recognition caused by pH-dependent conformational changes in Mpro. In fact, a change in the pH profile is reported with mutation near the active site that influences dimerization compared to the wild-type enzyme [56]. Furthermore, monomer to dimer studies measured distinct equilibrium dissociation constants under conditions involving alteration in pH or salt concentration [56]. An influence of these factors on the distribution of conformation populations in-solution is also known for other viral proteases [5]. This in turn can have an impact on the proteolytic processing of substrates recognized by certain conformations, rather than an overall effect on the catalytic competence of the enzyme. It is expected that changes in assay conditions and the inclusion of additives or detergents can possibly alter molecular recognition by affecting conformational states, or even by direct interaction of buffer components with the binding site.
One important consideration is the careful interpretation of results, especially when derived by mutations of the enzyme. A study reported a substrate-induced dimerization of R298A/L mutants of Mpro, however the same effect could not be observed for the wild-type enzyme [57]. The same study used high concentrations of both the substrate and the enzyme, beyond those normally used in biochemical assays on the isolated protein. While mutational studies can provide valuable mechanistic insight by pinpointing key residues involved in specific interactions, the used model protein can deviate significantly in its behavior from the wild-type enzyme.
4.3. Ligand binding: induced-fit or conformational selection?
The change in the oxyanion loop (residues 139–143) from inactive to active state between the two protomers of 1UJ1 (Figs. S2 and S4a) shows a backbone RMSD of 3.2 Å and 6.5 Å as overall RMSD. This conformational dynamic is in the range of the oxyanion hole activation for the West Nile virus (WNV) protease, PDB structures 2FP7 [58] and 5IDK [59], which displays a backbone and overall RMSD values in the range of 1.5–2.0 Å. Minor variation of the oxyanion loop in Mpro can be observed within the catalytically competent protomers (Fig. 2b), due to flexibility in Gly143. For example, the RMSD value for Asn142 is 3.5 Å for the catalytically competent protomers in 2VJ1 and 6Y2E, in comparison to an RMSD of 7.0 Å between the inactive and active protomers in 2U1J. This modestly alters the binding space in the S1 subpocket, however, a more significant influence on ligand fitting is possible, as discussed previously for the P3’ residue in section 4.2.
Structures 1UK3, 1UJ1, or 6Y2E in comparison to 1UK4, 2VJ1, 6Y2F, or 6Y2G show that, contrary to earlier interpretations in the field, the catalytic competence represented by the conformation of the oxyanion loop is independent of the presence of ligands or their steric demand (Figs. S2 and S5). As such, there is no steric threshold needed by the compound to target a catalytically competent state of this protein. In the conformational selection model, fine-tuning of the ligand-protein interactions can involve secondary induced-fit events. For SARS-CoV Mpro, inhibitor-induced modulation of the active site beyond the S1 subpocket is reported for example in 4MDS, however, it appears to originate instead from conformation dynamics of the protein, as observed in the analysis for segments incorporating Gln189 or Met49 (Fig. 1). As such, the structural evidence postulates a primary contribution for the conformational selection model [60,61] in ligand recognition for Mpro. The versatility of this mechanism makes it more suitable to depict the conformational and kinetic complexity of biological macromolecules [61].
The catalytic competence of Mpro is often mentioned to require dimerization, through a Glu166-Ser1∗ interaction between the N-terminal residues of one protomer or “N-finger” and the active site of the other protomer. This is supported by the fact that dimeric Mpro displays a much higher trans-cleavage of synthesized substrates in biochemical testing in comparison to the monomeric form. However, this general understanding does not explain a multitude of obtained results that show proteolytic cleavage despite deviation from this criterion. This is demonstrated by mutants of the enzyme that retain dimeric structures but lack catalytic activity and those that exist predominantly as monomers in-solution but still are able to process substrates in biochemical assays. For example, both the E166A mutant and the Ser1 deletion (2VJ1) constructs lack the Glu166-Ser1∗ interaction, the first is reported as a dimer in-solution [57], and the second shows catalytic competence [26]. The N214A mutant retains catalytic activity, despite being mostly a monomer in-solution [45]. Another case is the mutant enzymes E290R and R298E, these examples show N-terminal autoprocessing, while dimerization and trans-cleavage do not take place [62]. This is interpreted as a step in the maturation of Mpro through immature monomers forming intermediate structures, which are distinct from the mature dimeric Mpro, described previously.
NMR studies on separate N- and C-domains of the enzyme support a more complex relationship between dimerization and catalysis. Both domains show proper folding. The N-terminal domain, which harbors the catalytic machinery exists predominantly in-solution as a monomer and displays a degree of catalytic activity, while the C-terminal domain or extra domain alone is present mainly as a dimer [44]. The fact that mutations in the extra domain (from residue 200), which is distant from the active site, influence substrate catalysis, and possibly binding, aligns mostly with a mechanism dominated by conformational selection [61,63].
5. Impact of the hypothesis on the understanding of Mpro
5.1. Spatial and temporal order of cleavage events
Viral proteases, such as those from coronaviruses or flaviviruses, are machineries of complex nature, known to exert their function by cis- or trans-processing of a number of cleavage sites in the viral polyprotein according to a spatial and temporal order [5,[64], [65], [66]]. For SARS-CoV and SARS-CoV-2 Mpro, this complexity in the function of the protease was attributed to substrate-induced subsite cooperativity. This work proposes an alternate understanding of the molecular mechanism for proteolytic recognition by SARS-CoV and SARS-CoV-2 Mpro based on structural heterogeneity. In "structure-based recognition", the enzyme possesses different conformations independent of the presence of a substrate or ligand. The resulting variation in the binding site is then responsible for the fitting of different substrate sequences in alternate orientations, leading to modulation of the binding specificity. When this is connected to changes in the distribution of enzyme populations showing distinct conformations through the replication cycle, it could then result into the ordered processing of cleavage sites by the protease.
5.2. Expanding opportunities for antiviral drugs against Mpro
Due to their pivotal role in the virus life cycle, viral proteases are generally promising targets for drug discovery, capable of achieving broad-spectrum activity against different viruses [67]. Structural heterogeneity and the conformational selection model have implications on the development of assays for hit identification against viral enzymes and on structure-based design as well as hit to lead optimization of antivirals. The latter takes place by expanding the conformational space in search for inhibitors in order to interfere with distinct or multiple conformational states or stages of activity of SARS-CoV and SARS-CoV-2 Mpro. A number of studies aiming at targeting SARS-CoV-2 main protease rely on structure-based strategies whether by virtual screening of compound libraries against an x-ray structure of Mpro or by fine tuning of known inhibitor scaffolds to optimize interactions with the active site of the enzyme. It is important to be aware that the outcome of such screenings and optimization efforts will vary depending on the starting structure used, due to inherent conformational heterogeneity of the enzyme. As such, the recommendation is to consider a conformational ensemble for the protein rather than an individual structure. The variation of the binding site in certain conformations of Mpro allows the accommodation of non-canonical and sterically demanding residues that can extend in adjacent subpockets as S2 and S3’, to modulate the properties of the designed inhibitors. It is possible that certain conformations of the same protein are more relevant for achieving antiviral activity, or that their inetraction requirements can allow a more favorable pharmacokinetic profile. As such, compounds acting against these states can have an accelerated preclinical development into antiviral drug candidates.
6. Conclusion
The present study introduces "structure-based recognition" as a new concept in the understanding of the main protease of SARS-CoV and SARS-CoV-2, where structural heterogeneity is the main determinant of proteolytic recognition of different cleavage sites. The spatial distribution in the active site of Mpro varies in a non-cooperative manner, independent of the presence of a substrate or ligand. The conformational diversity plays an important role in the molecular biology of this protein through modulation of the binding specificity at the N- and C-termini cleavage sites, which are implicated in the maturation of the protease. Identified hits from structure-based screening and optimization of inhibitors will vary depending on the used conformation of the enzyme.
Earlier work on the flaviviral protease by Behnam and Klein identified the conformational selection paradigm to be more relevant than the induced-fit theory to explain the complexity of this viral enzyme. The diversity of x-ray structures of the main protease of SARS-CoV and SARS-CoV-2 allows a step forward in the understanding of viral proteases, by demonstrating a direct impact on the proteolytic recognition. From a broader view of structural analysis, mechanistic insights about enzymes should be used with caution, when obtained from comparison of a small sample size of x-ray structures [68]. Instead, as demonstrated in this work, interpretations should be derived based on the entire conformational space of a protein.
Associated content
Supporting Information file with sequence alignment of the main protease of SARS-CoV and SARS-CoV-2, cleavage sites, and supplementary figures. Chimera session files for all figures presented in this manuscript.
Author contribution
MB performed the analysis, interpreted the results, and wrote the manuscript. The author read and approved the final manuscript.
Declaration of competing interest
The author declares no competing financial interest.
Acknowledgment
The author thankfully acknowledges valuable comments of Prof. Christian Klein, Heidelberg University, on aspects of the work related to drug discovery.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biochi.2021.01.010.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Ziebuhr J., Herold J., Siddell S.G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu T., et al. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behnam M.A.M., Klein C.D.P. Conformational selection in the flaviviral NS2B-NS3 protease. Biochimie. 2020;174:117–125. doi: 10.1016/j.biochi.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Kneller D.W., et al. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuong W., et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 9.Dai W., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Z., et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 11.Yang H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu M.-F., et al. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu T., et al. Structure of the SARS coronavirus main proteinase as an active C2 crystallographic dimer. Acta Crystallogr. F. 2005;61:964–966. doi: 10.1107/S1744309105033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue X., et al. Production of authentic SARS-CoV Mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong N., et al. C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer. Protein Sci. 2009;18:839–844. doi: 10.1002/pro.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuck C.-P., et al. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur. J. Med. Chem. 2013;59:1–6. doi: 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T.-W., et al. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J. Mol. Biol. 2005;353:1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T.-W., et al. Crystal structures reveal an induced-fit binding of a substrate-like aza-peptide epoxide to SARS coronavirus main peptidase. J. Mol. Biol. 2007;366:916–932. doi: 10.1016/j.jmb.2006.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan J., et al. pH-dependent conformational flexibility of the SARS-CoV main proteinase (Mpro) dimer: molecular dynamics simulations and multiple x-ray structure analyses. J. Mol. Biol. 2005;354:25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S., et al. Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease. Protein Cell. 2010;1:371–383. doi: 10.1007/s13238-010-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82:4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akaji K., et al. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J. Med. Chem. 2011;54:7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 23.Barrila J., Gabelli S.B., Bacha U., Amzel L.M., Freire E. Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CLpro. Biochemistry. 2010;49:4308–4317. doi: 10.1021/bi1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C.-G., et al. Mechanism for controlling the monomer-dimer conversion of SARS coronavirus main protease. Acta Crystallogr. D. 2013;69:747–755. doi: 10.1107/S0907444913001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu T., et al. Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology. 2009;388:324–334. doi: 10.1016/j.virol.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verschueren K.H.G., et al. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 2008;15:597–606. doi: 10.1016/j.chembiol.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin J., et al. A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase. J. Mol. Biol. 2007;371:1060–1074. doi: 10.1016/j.jmb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A.K., et al. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005;48:6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 30.Yang S., et al. Synthesis, crystal structure, structure−activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006;49:4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 31.Lu I.L., et al. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J. Med. Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 32.Goetz D.H., et al. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry. 2007;46:8744–8752. doi: 10.1021/bi0621415. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh A.K., et al. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007;17:5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C.-C., et al. Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors. FEBS Lett. 2007;581:5454–5458. doi: 10.1016/j.febslet.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C.-C., et al. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J. Biol. Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacha U., et al. Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CLpro. Chem. Biol. Drug Des. 2008;72:34–49. doi: 10.1111/j.1747-0285.2008.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L., et al. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antivir. Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs J., et al. Discovery, synthesis, and structure-based optimization of a series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turlington M., et al. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimamoto Y., et al. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23:876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue X., et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen E.F., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Gibbs A.C., Steele R., Liu G., Tounge B.A., Montelione G.T. Inhibitor bound dengue NS2B-NS3pro reveals multiple dynamic binding modes. Biochemistry. 2018;57:1591–1602. doi: 10.1021/acs.biochem.7b01127. [DOI] [PubMed] [Google Scholar]
- 44.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S., Zhong N., Ren X., Jin C., Xia B. 1H, 13C and 15N resonance assignments of SARS-CoV main protease N-terminal domain. Biomol. NMR Assign. 2011;5:143–145. doi: 10.1007/s12104-010-9287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim L., et al. Structurally- and dynamically-driven allostery of the chymotrypsin-like proteases of SARS, dengue and Zika viruses. Prog. Biophys. Mol. Biol. 2019;143:52–66. doi: 10.1016/j.pbiomolbio.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 49.Graziano V., McGrath W.J., Yang L., Mangel W.F. SARS CoV main proteinase: the monomer−dimer equilibrium dissociation constant. Biochemistry. 2006;45:14632–14641. doi: 10.1021/bi061746y. [DOI] [PubMed] [Google Scholar]
- 50.Ng N.M., Pike R.N., Boyd S.E. Subsite cooperativity in protease specificity. Biol. Chem. 2009;390:401–407. doi: 10.1515/BC.2009.065. [DOI] [PubMed] [Google Scholar]
- 51.Graziano V., McGrath W.J., DeGruccio A.M., Dunn J.J., Mangel W.F. Enzymatic activity of the SARS coronavirus main proteinase dimer. FEBS Lett. 2006;580:2577–2583. doi: 10.1016/j.febslet.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., et al. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta Pharmacol. Sin. 2005;26:99–106. doi: 10.1111/j.1745-7254.2005.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 54.Ma C., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou C.-Y., et al. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2004;43:14958–14970. doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- 57.Cheng S.-C., Chang G.-G., Chou C.-Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010;98:1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erbel P., et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 59.Nitsche C., et al. Peptide–boronic acid inhibitors of flaviviral proteases: medicinal chemistry and structural biology. J. Med. Chem. 2017;60:511–516. doi: 10.1021/acs.jmedchem.6b01021. [DOI] [PubMed] [Google Scholar]
- 60.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cera E.D. Mechanisms of ligand binding. Biophys. Rev. 2020;1 doi: 10.1063/5.0020997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S., Jonas F., Shen C., Hilgenfeld R. Liberation of SARS-CoV main protease from the viral polyprotein: N-terminal autocleavage does not depend on the mature dimerization mode. Protein Cell. 2010;1:59–74. doi: 10.1007/s13238-010-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weikl T.R., von Deuster C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 64.Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7–10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cahour A., Falgout B., Lai C.J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Constant D.A., Mateo R., Nagamine C.M., Kirkegaard K. Targeting intramolecular proteinase NS2B/3 cleavages for trans-dominant inhibition of dengue virus. Proc. Natl. Acad. Sci. U.S.A. 2018;115:10136. doi: 10.1073/pnas.1805195115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boldescu V., Behnam M.A.M., Vasilakis N., Klein C.D. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017;16:565–586. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DePristo M.A., de Bakker P.I., Blundell T.L. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure. 2004;12:831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.