Abstract
Hepatocellular carcinoma (HCC) is a representative primary liver cancer caused by long-term and repetitive liver injury. Surgical resection is generally selected as the radical cure treatment. Because the early recurrence of HCC after resection is associated with low overall survival, the prediction of recurrence after resection is clinically important. However, the pathological characteristics of the early recurrence of HCC have not yet been elucidated. We attempted to predict the early recurrence of HCC after resection based on digital pathologic images of hematoxylin and eosin-stained specimens and machine learning applying a support vector machine (SVM). The 158 HCC patients meeting the Milan criteria who underwent surgical resection were included in this study. The patients were categorized into three groups: Group I, patients with HCC recurrence within 1 year after resection (16 for training and 23 for test); Group II, patients with HCC recurrence between 1 and 2 years after resection (22 and 28); and Group III, patients with no HCC recurrence within 4 years after resection (31 and 38). The SVM-based prediction method separated the three groups with 89.9% (80/89) accuracy. Prediction of Groups I was consistent for all cases, while Group II was predicted to be Group III in one case, and Group III was predicted to be Group II in 8 cases. The use of digital pathology and machine learning could be used for highly accurate prediction of HCC recurrence after surgical resection, especially that for early recurrence. Currently, in most cases after HCC resection, regular blood tests and diagnostic imaging are used for follow-up observation; however, the use of digital pathology coupled with machine learning offers potential as a method for objective postoprative follow-up observation.
Subject terms: Cancer imaging, Liver cancer
Introduction
Hepatocellular carcinoma (HCC) accounts for ~90% of primary liver cancers [1]. The annual recurrence rate of HCC after surgical resection is ≥10% and reaches 70–80% after 5 years [2–5]. Therefore, it is clinically important to identify patients at high risk of HCC recurrence after curative surgical resection. Currently, there are no useful postoperative recurrence markers; thus, measurement of levels of tumor markers, such as alpha-fetoprotein and des-gamma-carboxy prothrombin, and periodic imaging tests, such as computed tomography and magnetic resonance imaging, are used to identify HCC recurrence. Previous studies have shown that early recurrence of HCC after resection is associated with low overall survival [6, 7]. Early recurrence after resection is also associated with tumor size, number of tumors, and portal vein invasion [8, 9]. However, pathological evaluation for early recurrence of HCC has not been described.
In digital pathology, machine learning (ML) approaches have been applied to a variety of image processing and classification tasks, with diagnosis methods reported for breast [10] and prostate [11] cancers. Image processing approaches are currently being used to evaluate the spatial arrangement and architecture of different types of tissue elements to predict clinical outcomes. Several examples of prognosis prediction methods using ML approaches based on pathological information have been described, including the relationship between the spatial arrangement of clusters of tumor-infiltrating lymphocyte and prognosis of non-small-cell lung carcinoma [12], prediction of overall survival for breast cancer [13], and the relationship between nuclear features of the stromal and the epithelial compartments and prediction of human papillomavirus-positive oropharyngeal cancer [14]. The application of ML to predict HCC recurrence include assessment of miRNA expression in exosomes [15] and CpG methylation signatures [16]. HCC diagnosis based on circulating miRNA information has also been reported [17]. However, to our knowledge, there are no reports of using images for HCC diagnosis and recurrence prediction.
Here, we applied support vector machine (SVM) methods to digital pathologic images of HE-stained specimens from resected tissues to predict the early recurrence of HCC after resection.
Materials and methods
Sample information
We included a total of 158 patients meeting the Milan criteria who underwent hepatic resection for HCC as curative treatment at Yamaguchi University (100 cases), Ogaki Municipal Hospital (47 cases), and Tokyo Medical University (11 cases). All cases used were primary HCC. Previously, our group revealed that the pattern of recurrence of HCC after surgical resection varied from year to year. In other words, recurrence within 2 years are mostly residual recurrences, and in 2–4 years, it was residual recurrence and/or multicentric (MC) carcinogenesis, and it was MC carcinogenesis after 4 years. Moreover, recurrence rate after resection is gradually decreasing with each passage [18]. The patients were categorized into three groups. Group I (39 cases) included patients with HCC recurrence within 1 year after hepatic resection. Group II (50 cases) included patients with HCC recurrence between 1 and 2 years after resection. Group III (69 patients) included patients without HCC recurrence at 4 years after resection (Table 1). To analyze recurrence prediction, we randomly selected 69 cases (16, 22, and 31 from Groups I–III, respectively) as training set; the remaining 89 cases (23, 28, and 38 from Groups I–III, respectively) were the model test set. The study was performed according to the principles of the Declaration of Helsinki and was approved by the ethical committees of Yamaguchi University, Ogaki Municipal Hospital, and Tokyo Medical University (SH4140).
Table 1.
Clinical background.
| Total | Group I <1 year |
Group II 1 < years < 2 |
Group III >4 years |
||
|---|---|---|---|---|---|
| SVM model training patients | |||||
| Cases | 69 | 16 | 22 | 31 | |
| ROI | HCC | 1369 | 219 | 558 | 592 |
| non-HCC | 738 | 136 | 273 | 329 | |
| Total | 2107 | 355 | 831 | 921 | |
| Nuclei | HCC | 979,086 | 130,178 | 413,665 | 435,243 |
| Normal | 328,463 | 57,437 | 120,702 | 150,324 | |
| Total | 1,307,549 | 187,615 | 534,367 | 585,567 | |
| Gender | M | 48 | 11 | 16 | 21 |
| F | 21 | 5 | 6 | 10 | |
| Age | Average | 65.4 | 65.8 | 67.0 | 64.0 |
| MAX | 80 | 75 | 80 | 80 | |
| MIN | 40 | 42 | 54 | 40 | |
| Virus | HBV | 19 | 5 | 6 | 8 |
| HCV | 36 | 8 | 13 | 15 | |
| NBNC | 14 | 3 | 3 | 8 | |
| Background | LC | 34 | 8 | 11 | 15 |
| CH | 32 | 8 | 11 | 13 | |
| Normal | 3 | 0 | 0 | 3 | |
| Size (mm) | Average | 27.8 | 28.3 | 26.5 | 28.4 |
| MAX | 48.0 | 48.0 | 43.0 | 47.0 | |
| MIN | 9.0 | 20.0 | 9.0 | 10.0 | |
| Nodule | Average | 1.3 | 1.7 | 1.1 | 1.2 |
| MAX | 3 | 3 | 2 | 3 | |
| MIN | 1 | 1 | 1 | 1 | |
| SVM model test patients | |||||
| Cases | 89 | 23 | 28 | 38 | |
| ROI | HCC | 1346 | 434 | 324 | 588 |
| non-HCC | 680 | 189 | 168 | 323 | |
| Total | 2026 | 623 | 492 | 911 | |
| Nuclei | HCC | 886,900 | 317,855 | 185,672 | 383,373 |
| Normal | 251,376 | 66,243 | 59,148 | 125,985 | |
| Total | 1,138,276 | 384,098 | 244,820 | 509,358 | |
| Gender | M | 67 | 17 | 23 | 27 |
| F | 22 | 6 | 5 | 11 | |
| Age | Average | 67.8 | 68.2 | 71.3 | 65.1 |
| MAX | 83 | 79 | 82 | 83 | |
| MIN | 28 | 47 | 46 | 28 | |
| Virus | HBV | 12 | 2 | 0 | 10 |
| HCV | 53 | 16 | 17 | 20 | |
| NBNC | 24 | 5 | 11 | 8 | |
| Background | LC | 53 | 16 | 14 | 23 |
| CH | 32 | 7 | 12 | 13 | |
| NON | 4 | 0 | 2 | 2 | |
| Size (mm) | Average | 26.3 | 38.5 | 27.7 | 24.2 |
| MAX | 45.0 | 45.0 | 45.0 | 45.0 | |
| MIN | 10.0 | 15.0 | 14.0 | 10.0 | |
| Nodule | Average | 1.3 | 1.7 | 1.4 | 1.1 |
| MAX | 3 | 3 | 3 | 2 | |
| MIN | 1 | 1 | 1 | 1 | |
HCC HCC area, non-HCC surrounding non-HCC area, ROI used ROI number, Nuclei calculated number of nuclei, NBNC neither HBV nor HCV infection.
Region of interest (ROI) selection and image size
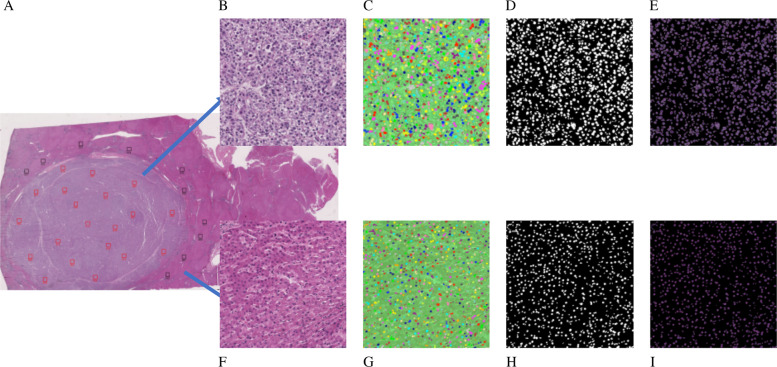
Formalin-fixed, paraffin-embedded HE-stained slides were scanned using a whole slide image (WSI) scanner (NanoZoomer-RS; Hamamatsu Photonics, Hamamatsu, Japan) at ×20 image magnification. Images acquired by WSI were in ndpi (scanner original) format with an average size of 1 GB and converted to tiff format for parsing due to memory limitations in the Windows operating. Under low-magnification WSI, 20 and 10 ROIs per sample were selected in HCC and surrounding non-HCC areas, respectively. ROIs with strong necrosis, low tumor cell content, and high blood cell aggregation in the selected images were excluded from the analysis. The analysis included a total of 1369 ROIs from 69 training set images and 1346 ROIs from 89 test set images from HCC areas, as well as 738 ROIs from 69 training sets images and 680 ROIs from 89 test set images from non-HCC areas (Fig. 1 and Supplementary Fig. 1).
Fig. 1. Image analysis process.
Whole slide image (WSI) scans of hepatocellular carcinoma (HCC) and surrounding non-HCC area (a), region of interest (ROI) selection for HCC (b), and non-HCC (f), nuclei selection by Ilastik for HCC (c) and non-HCC (g), mask image for HCC (d) and non-HCC (h), and analysis images for HCC (e) and non-HCC (i).
Nuclei segmentation on ROI images
The nuclei <80 pixels were removed as potential nucleus fragments by ilastik software (http://ilastik.org/). Each ROI was 2048 × 2048 pixels (at ×40 resolution), corresponding to a tissue area of 0.25 mm2. A total of 970,986 and 886,900 nuclei in these ROIs were analyzed in the training and test sets for the HCC area, respectively, and 328,463 and 251,376 nuclei in the ROIs in the training and test sets in the non-HCC areas.
Quantitative nucleus and ROI feature measurement
The morphological features of the segmented nuclei were analyzed using Cellprofiler (https://cellprofiler.org) and Cell Feature Level Co-occurrence Matrix [19]. Each ROI contained 300–2000 nuclei, with 903 ROI features based on nuclei information: for example, average and standard deviation, heterogeneity, and morphological features for nucleus size, contour line length, orientation, roundness and intra-nucleus texture (chromatin pattern) entropy, variance, second angular moment, etc.
Analysis of nuclear features and their rate of postoperative early recurrence
Nuclei (9907/565,421, 1.7%) in Groups I and III of the training set were randomly selected and an SVM model was created using information from 81 nuclei features generated from CellProfiler output (http://cellprofiler-manual.s3.amazonaws.com/CellProfiler-3.0.0/modules/measurement.html). On ROI-based SVM discrimination, we listed highly contributed features top 20 on Supplementary Table 1. The features having high weight value were contributed strongly for discrimination.
Analysis of the morphological features of ROIs
The prediction of recurrence analysis was performed with both linear and radial basis function (RBF) kernel SVM methods (e1071 library on R system) (https://cran.r-project.org/web/packages/e1071/index.html). We used the training data to create the SVM model, which was applied to the test patient data set. As the SVM with a linear kernel showed higher prediction accuracy than the SVM with an RBF kernel, we reported results based on the former SVM.
Results
Nuclei features-based analysis results
First, we analyzed proportion of nuclei belonging to Group I with CellProfiler outputted nucleus features. In Group I cases, 16 of the 23 cases were >20%. In contrast, in 22 of 38 cases in Group III, <10% of nuclei of the nuclei were characterized by early recurrence (Table 2).
Table 2.
Group classification based on nuclear information.
| Percentage of nuclei with Group I characters | ||||||
|---|---|---|---|---|---|---|
| >30% | 20–29% | 10–19% | <10% | Total | ||
| Truth | Group I | 8 | 8 | 5 | 2 | 23 |
| Group II | 4 | 5 | 8 | 11 | 28 | |
| Group III | 0 | 2 | 14 | 22 | 38 | |
| Total | 12 | 15 | 27 | 35 | 89 | |
ROI-based analysis results
Classification of the ROI of the HCC area into three groups using SVM model training (linear kernel) showed an accuracy of 99.8% (Table 3a). The ROI of the non-HCC area was then classified into three groups using SVM, with a probability of 100% (Table 3b). When the classification formula created using the training set was verified using the test set, the probabilities of correct classification of the ROIs in the HCC and non-HCC areas were 80.6% and 68.1%, respectively (Table 3c, d).
Table 3.
Region of interest (ROI)-based support vector machine (SVM) prediction of hepatocellular carcinoma (HCC) recurrence.
| Prediction | |||||
|---|---|---|---|---|---|
| Group I | Group II | Group III | Total | ||
| (a) HCC area Training set result (Accuracy: 99.8%) | |||||
| Truth | Group I | 219 | 0 | 0 | 219 |
| Group II | 0 | 556 | 2 | 558 | |
| Group III | 0 | 1 | 591 | 592 | |
| Total | 219 | 557 | 593 | 1369 | |
| (b) Non-HCC area Training set result (Accuracy: 100.0%) | |||||
| Truth | Group I | 136 | 0 | 0 | 136 |
| Group II | 0 | 273 | 0 | 273 | |
| Group III | 0 | 0 | 329 | 329 | |
| Total | 136 | 273 | 329 | 738 | |
| (c) HCC area Test set prediction result (Accuracy: 80.6%) | |||||
| Truth | Group I | 337 | 46 | 61 | 444 |
| Group II | 16 | 359 | 70 | 445 | |
| Group III | 35 | 58 | 495 | 588 | |
| Total | 388 | 463 | 626 | 1477 | |
| (d) Non-HCC area Test set prediction result (Accuracy: 68.1%) | |||||
| Truth | Group I | 131 | 32 | 26 | 189 |
| Group II | 15 | 164 | 61 | 240 | |
| Group III | 19 | 70 | 180 | 269 | |
| Total | 165 | 266 | 267 | 698 | |
In addition, the information on ROIs contained in the HCC or non-HCC areas were summed, and the accuracy of the classification between the three groups was verified on a case-by-case rather than an ROI basis. The group to which the maximum number of ROIs belonged was the group to which the case belonged. The accuracies for HCC and non-HCC areas were 88.8% and 64.0%, respectively (Table 4a, b).
Table 4.
Case-based prediction of hepatocellular carcinoma (HCC) recurrence.
| Prediction | |||||
|---|---|---|---|---|---|
| Group I | Group II | Group III | Total | ||
| (a) Prediction used in HCC area information (Accuracy: 88.8%) | |||||
| Truth | Group I | 19 | 0 | 4 | 23 |
| Group II | 1 | 25 | 2 | 28 | |
| Group III | 0 | 3 | 35 | 38 | |
| Total | 20 | 28 | 41 | 89 | |
| (b) Prediction used in non-HCC area information (Accuracy: 64.0%) | |||||
| Truth | Group I | 13 | 7 | 1 | 21 |
| Group II | 1 | 17 | 9 | 27 | |
| Group III | 3 | 10 | 25 | 38 | |
| Total | 17 | 34 | 35 | 86 | |
Aggregated case-based prediction results
Finally, three integrated SVM models; ROI of HCC and non-HCC area based SVM, and nuclei features based SVM, were used for the prediction of HCC recurrence.
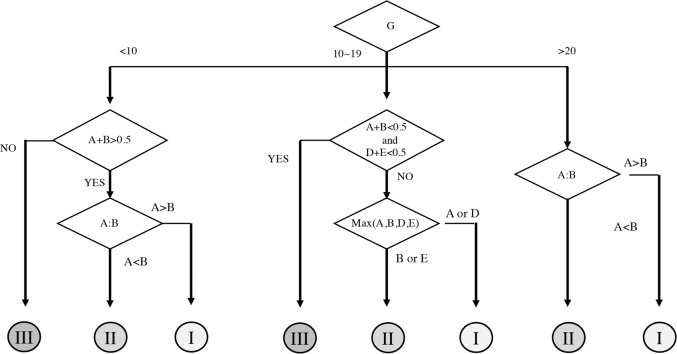
The values of A, B, and C were calculated as the average of the probabilities for ROIs in the HCC areas predicted to be Groups I, II, and III, respectively. The values of D, E, and F were also calculated as the averages of the probabilities of ROIs in the non-HCC areas predicted to be Groups I, II, and III, respectively. At the nuclei feature base, G was defined as the percentage of case nuclei in Group I. The prediction algorithm is shown in Fig. 2.
Fig. 2. Algorithm for prediction using three support vector machine (SVM) models.
The average values of the probabilities predicted to be Groups I, II, and III in the region of interest (ROI) of the HCC area were set as A, B, and C, respectively, whereas the average values of the probabilities predicted to be Groups I, II, and III in the ROI in the non-HCC area were set as D, E, and F, respectively. The predicted probability of a nucleus belonging to Group I was set as G.
(1) If the value of G was ≥20%, it was assumed that the case was Group I or II. Next, if comparisons of the ROI values of the HCC area showed A > B, the case was categorized as Group I; similarly, if A < B, the case was categorized as Group II.
For example, Case 1 had a G value of 34.8, which is >20%. Next, since A was 0.92 and B was 0.08, A > B; thus, Case 1 was predicted to belong to Group I. Case 25 had a G value of 35.2, also >20%. As A was 0.01 and B was 0.70, this case was predicted to belong to Group II because A < B.
(2) If the G value was 10–19% and A + B and D + E were ≤0.5, the case was predicted to be in Group III. If A + B and D + E were not <0.5, the values of A, B, D, and E were compared. If the value of A or D was larger than the other values, the case was predicted to be in Group I; if the value of B or E was larger, the case was predicted to belong to Group II.
For example, Case 35 had a G value of 17.8. The values of A + B and D + E were 0.93 and 0.98, respectively, both of which were >0.5. Of A, B, D, and E, E was the largest, at 0.98; therefore, the case was predicted to belong to Group II. Similarly, the G value for Case 65 was 18.8. Since A + B was 0.47 and D + E was 0.18, both <0.5, the case was predicted to belong to Group III.
(3) When the value of G was ≤10, the case was predicted to belong to Group I when A + B was >0.5 and A > B, and Group II when A < B, and Group III if A + B < 0.5. For example, Case 41 had a G value of 4.0. A + B was >0.5 and A (0.02) was <B (0.75). Therefore, Case 41 was predicted to belong to the Group II.
With this algorithm, these models showed an accuracy of 89.9% (80/89) (Table 5). Twenty-four cases were classified as Group I, of which 23 were really group I and the remaining one was Group II. Of the 35 cases predicted to be in Group II, 27 were actually Group II; the remaining 8 cases were Group III. Thirty cases predicted to be Group III were actually in Group III.
Table 5.
Results of hepatocellular carcinoma (HCC) recurrence prediction by integrating three types of support vector machine (SVM).
| A | B | C | D | E | F | G | |||
|---|---|---|---|---|---|---|---|---|---|
| HCC Area ROIs | non-HCC area ROIs | ||||||||
| Test case No. | Group | Group I Probability | Group II Probability | Group III Probability | Group I Probability | Group II Probability | Group III Probability | Nuclei percentage | Prediction Group |
| 1 | I | 0.92 | 0.08 | 0.00 | 0.99 | 0.00 | 0.00 | 34.80 | I |
| 6 | 1 | 0.65 | 0.06 | 0.29 | 0.78 | 0.00 | 0.22 | 31.50 | 1 |
| 9 | 1 | 0.49 | 0.02 | 0.49 | 0.33 | 0.00 | 0.67 | 28.20 | 1 |
| 13 | 1 | 0.57 | 0.42 | 0.02 | 0.12 | 0.77 | 0.11 | 28.00 | 1 |
| 17 | 1 | 0.55 | 0.10 | 0.35 | 0.20 | 0.76 | 0.04 | 71.90 | 1 |
| 18 | 1 | 0.50 | 0.25 | 0.25 | NI | NI | NI | 49.50 | 1 |
| 20 | 1 | 0.42 | 0.16 | 0.43 | 0.23 | 0.77 | 0.00 | 24.80 | 1 |
| 105 | I | 0.59 | 0.17 | 0.25 | 0.74 | 0.10 | 0.16 | 26.66 | I |
| 106 | I | 0.18 | 0.15 | 0.67 | 0.21 | 0.42 | 0.37 | 31.81 | I |
| 107 | I | 0.48 | 0.39 | 0.13 | 0.37 | 0.45 | 0.18 | 5.36 | I |
| 108 | I | 0.88 | 0.02 | 0.10 | 0.63 | 0.17 | 0.19 | 41.65 | I |
| 109 | I | 0.48 | 0.11 | 0.41 | 0.43 | 0.06 | 0.51 | 8.52 | I |
| 110 | I | 0.55 | 0.07 | 0.38 | 0.74 | 0.11 | 0.15 | 10.22 | I |
| 111 | I | 0.64 | 0.19 | 0.17 | 0.61 | 0.17 | 0.21 | 23.99 | I |
| 113 | I | 0.24 | 0.22 | 0.53 | 0.48 | 0.30 | 0.22 | 14.25 | I |
| 114 | I | 0.43 | 0.37 | 0.20 | 0.53 | 0.19 | 0.27 | 23.70 | I |
| 115 | I | 0.47 | 0.29 | 0.23 | NI | NI | NI | 42.02 | I |
| 116 | I | 0.47 | 0.44 | 0.09 | 0.41 | 0.25 | 0.34 | 61.27 | I |
| 118 | I | 0.94 | 0.04 | 0.02 | 0.64 | 0.11 | 0.25 | 10.07 | I |
| 119 | I | 0.41 | 0.26 | 0.33 | 0.73 | 0.18 | 0.09 | 25.78 | I |
| 120 | I | 0.33 | 0.21 | 0.43 | 0.22 | 0.37 | 0.41 | 21.69 | I |
| 123 | I | 0.57 | 0.21 | 0.22 | 0.63 | 0.15 | 0.22 | 12.14 | I |
| 124 | I | 0.45 | 0.41 | 0.14 | 0.41 | 0.48 | 0.11 | 13.86 | I |
| 25 | II | 0.01 | 0.70 | 0.29 | 0.42 | 0.15 | 0.43 | 35.20 | II |
| 32 | II | 0.00 | 0.71 | 0.29 | 0.14 | 0.18 | 0.68 | 20.90 | II |
| 34 | II | 0.00 | 0.20 | 0.80 | 0.00 | 0.71 | 0.29 | 22.90 | II |
| 35 | II | 0.11 | 0.82 | 0.08 | 0.00 | 0.98 | 0.02 | 17.80 | II |
| 38 | II | 0.01 | 0.28 | 0.71 | 0.01 | 0.63 | 0.36 | 10.60 | II |
| 41 | II | 0.02 | 0.75 | 0.23 | 0.03 | 0.44 | 0.53 | 4.00 | II |
| 42 | II | 0.01 | 0.89 | 0.10 | NI | NI | NI | 13.40 | II |
| 46 | II | 0.00 | 0.68 | 0.32 | 0.01 | 0.16 | 0.83 | 21.30 | II |
| 50 | II | 0.01 | 0.69 | 0.31 | 0.00 | 0.03 | 0.97 | 9.50 | II |
| 52 | II | 0.00 | 0.56 | 0.44 | 0.08 | 0.62 | 0.30 | 5.00 | II |
| 54 | II | 0.04 | 0.71 | 0.25 | 0.00 | 0.89 | 0.11 | 4.60 | II |
| 125 | II | 0.02 | 0.83 | 0.15 | 0.05 | 0.35 | 0.60 | 6.20 | II |
| 126 | II | 0.38 | 0.33 | 0.29 | 0.10 | 0.81 | 0.09 | 19.99 | I |
| 127 | II | 0.16 | 0.57 | 0.27 | 0.11 | 0.49 | 0.40 | 23.90 | II |
| 128 | II | 0.04 | 0.87 | 0.09 | 0.15 | 0.56 | 0.29 | 39.23 | II |
| 129 | II | 0.16 | 0.51 | 0.33 | 0.30 | 0.57 | 0.13 | 42.34 | II |
| 130 | II | 0.02 | 0.85 | 0.13 | 0.13 | 0.39 | 0.48 | 7.70 | II |
| 131 | II | 0.03 | 0.69 | 0.28 | 0.06 | 0.53 | 0.41 | 7.96 | II |
| 132 | II | 0.08 | 0.61 | 0.31 | 0.10 | 0.66 | 0.25 | 4.21 | II |
| 133 | II | 0.04 | 0.55 | 0.41 | 0.05 | 0.52 | 0.43 | 17.26 | II |
| 134 | II | 0.22 | 0.63 | 0.15 | 0.24 | 0.62 | 0.14 | 6.95 | II |
| 135 | II | 0.21 | 0.61 | 0.18 | 0.44 | 0.35 | 0.21 | 12.94 | II |
| 136 | II | 0.09 | 0.56 | 0.35 | 0.09 | 0.56 | 0.34 | 15.91 | II |
| 137 | II | 0.02 | 0.52 | 0.46 | 0.19 | 0.52 | 0.29 | 21.87 | II |
| 138 | II | 0.04 | 0.50 | 0.46 | 0.07 | 0.46 | 0.47 | 3.48 | II |
| 139 | II | 0.00 | 0.47 | 0.53 | 0.02 | 0.75 | 0.24 | 19.06 | II |
| 141 | II | 0.06 | 0.58 | 0.37 | 0.11 | 0.31 | 0.58 | 5.51 | II |
| 142 | II | 0.12 | 0.79 | 0.09 | 0.08 | 0.39 | 0.53 | 40.96 | II |
| 58 | III | 0.13 | 0.47 | 0.40 | 0.61 | 0.16 | 0.23 | 25.80 | II |
| 61 | III | 0.00 | 0.00 | 1.00 | 0.03 | 0.03 | 0.94 | 8.60 | III |
| 65 | III | 0.47 | 0.00 | 0.53 | 0.14 | 0.04 | 0.83 | 18.80 | III |
| 66 | III | 0.01 | 0.00 | 0.99 | 0.01 | 0.01 | 0.98 | 9.10 | III |
| 71 | III | 0.02 | 0.35 | 0.43 | 0.11 | 0.87 | 0.02 | 13.60 | II |
| 77 | III | 0.19 | 0.03 | 0.78 | 0.02 | 0.03 | 0.98 | 8.20 | III |
| 78 | III | 0.00 | 0.12 | 0.88 | 0.02 | 0.64 | 0.34 | 5.70 | III |
| 82 | III | 0.11 | 0.12 | 0.77 | 0.00 | 0.78 | 0.22 | 7.00 | III |
| 84 | III | 0.06 | 0.27 | 0.67 | 0.00 | 0.37 | 0.63 | 3.70 | III |
| 86 | III | 0.00 | 0.31 | 0.68 | 0.00 | 0.04 | 0.96 | 26.70 | II |
| 88 | III | 0.02 | 0.90 | 0.07 | 0.01 | 0.91 | 0.08 | 7.50 | II |
| 90 | III | 0.01 | 0.07 | 0.92 | 0.00 | 0.65 | 0.35 | 2.70 | III |
| 91 | III | 0.00 | 0.04 | 0.96 | 0.01 | 0.60 | 0.40 | 6.20 | III |
| 92 | III | 0.16 | 0.20 | 0.64 | 0.00 | 0.76 | 0.24 | 12.00 | II |
| 95 | III | 0.00 | 0.08 | 0.92 | 0.01 | 0.59 | 0.49 | 14.40 | III |
| 97 | III | 0.00 | 0.06 | 0.94 | 0.03 | 0.60 | 0.37 | 3.90 | III |
| 102 | III | 0.00 | 0.27 | 0.73 | 0.01 | 0.02 | 0.97 | 6.60 | III |
| 143 | III | 0.25 | 0.33 | 0.41 | 0.44 | 0.13 | 0.43 | 11.21 | II |
| 144 | III | 0.04 | 0.18 | 0.78 | 0.11 | 0.32 | 0.56 | 8.44 | III |
| 145 | III | 0.04 | 0.29 | 0.67 | 0.04 | 0.45 | 0.50 | 10.02 | III |
| 146 | III | 0.02 | 0.47 | 0.51 | 0.11 | 0.37 | 0.52 | 7.48 | III |
| 147 | III | 0.01 | 0.63 | 0.36 | 0.06 | 0.47 | 0.47 | 6.05 | II |
| 148 | III | 0.02 | 0.28 | 0.70 | 0.09 | 0.19 | 0.35 | 13.92 | III |
| 149 | III | 0.09 | 0.21 | 0.71 | 0.16 | 0.24 | 0.49 | 13.05 | III |
| 150 | III | 0.04 | 0.03 | 0.93 | 0.25 | 0.20 | 0.55 | 7.26 | III |
| 151 | III | 0.08 | 0.22 | 0.69 | 0.13 | 0.32 | 0.55 | 5.70 | III |
| 153 | III | 0.07 | 0.26 | 0.66 | 0.06 | 0.34 | 0.60 | 14.54 | III |
| 154 | III | 0.19 | 0.27 | 0.54 | 0.14 | 0.37 | 0.48 | 4.56 | III |
| 155 | III | 0.07 | 0.09 | 0.83 | 0.08 | 0.30 | 0.61 | 19.68 | III |
| 158 | III | 0.01 | 0.38 | 0.61 | 0.21 | 0.23 | 0.56 | 8.43 | III |
| 160 | III | 0.27 | 0.15 | 0.58 | 0.37 | 0.10 | 0.52 | 5.69 | III |
| 161 | III | 0.04 | 0.23 | 0.73 | 0.12 | 0.19 | 0.69 | 7.67 | III |
| 162 | III | 0.01 | 0.27 | 0.72 | 0.01 | 0.35 | 0.64 | 1.46 | III |
| 163 | III | 0.20 | 0.24 | 0.56 | 0.03 | 0.33 | 0.63 | 12.86 | III |
| 164 | III | 0.02 | 0.19 | 0.79 | 0.18 | 0.26 | 0.56 | 5.51 | III |
| 165 | III | 0.01 | 0.05 | 0.94 | 0.03 | 0.25 | 0.72 | 11.14 | III |
| 166 | III | 0.02 | 0.50 | 0.49 | 0.05 | 0.28 | 0.66 | 19.47 | II |
| 168 | III | 0.19 | 0.02 | 0.78 | 0.64 | 0.07 | 0.30 | 19.85 | III |
NI no information.
The prediction algorithm is created on training data set, one-third of ROIs removed as a validation set, SVM models created on the remaining data set. This process was repeated three times (Supplementary Table 2).
Prediction was performed using average value of probability of ROIs belonging to each case. The number of prediction of ROIs is shown in Supplementary Table 3.
Discussion
The recurrence rate of surgically resected HCC is high, and until now, no predictive method had been described. The results of the present study demonstrated the efficient prediction of recurrence after resection applying ML-based approach on pathological findings.
The pathological evaluation for early recurrence of HCC has not been reported. HCC patients are at high risk for MC tumors due to the strong carcinogenic background of the liver. Recurrence patterns after treatment for HCC are diverse, including MC and intrahepatic metastasis. Because the mode of development and clinical course differ, it is important to distinguish between the two; however, it is difficult to do so clinically or pathologically [20].
We performed classification using the tumor marker and histopathological information between the three groups, the accuracy of prediction using the tumor markers AFP and DCP was 44.8%, and that using the tumor and histopathological information was 53.8%, respectively (Supplementary Table 4). In addition, the significant difference between groups of each item used for these predictions was not demonstrated (Supplementary Table 5). Moreover, while the recurrence risk factors for liver cancer include the number of tumors, size, vascular invasion, distant metastasis, etc., the patients’ backgrounds in the present study were normalized by enrolling patients according to the Milan criteria.
The prediction of breast and prostate cancer recurrence using pathological findings has been reported. However, image-based prognosis prediction in these conditions is based on glandular structures and tumor invasion patterns in adenocarcinomas, whereas the structure and invasion pattern in HCC differ from those of adenocarcinoma. In addition to the analysis of ROI units of the HCC area, the prediction accuracy could be improved to 89.9% by combining nuclear information in HCC areas and ROIs of non-HCC areas. The results of the analysis showed that all cases predicted to recur within 1 year indeed recur within 1 year; similarly, all cases predicted to have no recurrence did not recur. Among the cases with an incorrect prediction, 1/28 in Group II and 8/38 in Group III did not have a recurrence although they were predicted to have a recurrence in 1–2 years. Clinically, it is useful to predict both: no recurrence with no false-negative results and early recurrence. Although, there is a limitation in our current prediction model due to the low number of cases used for analyses, and there is a possibility of overfitting, especially in non-HCC in ROI-based SVM prediction results. However, the results were considered clinically useful due to the synthesis of three SVM models. We are currently conducting a study with an increased number of cases.
In conclusion, there has been no useful method to predict recurrence after HCC resection until now. We developed a recurrence prediction method based on ML by comprehensively using information on cancer tissue, peripheral non-cancerous tissue, and nuclei. While all cases are considered high risk after HCC resection, our method showed promise as a novel follow-up method to review the frequency of tests and determine the need for additional treatment.
Supplementary information
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoshiki Murakami, Email: yoshikim@tokyo-med.ac.jp.
Masahiko Kuroda, Email: kuroda@tokyo-med.ac.jp.
Supplementary information
The online version of this article (10.1038/s41379-020-00671-z) contains supplementary material, which is available to authorized users.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243:229–35. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah SA, Cleary SP, Wei AC, Yang I, Taylor BR, Hemming AW, et al. Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery. 2007;141:330–9. doi: 10.1016/j.surg.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359:2045–7. doi: 10.1056/NEJMe0807581. [DOI] [PubMed] [Google Scholar]
- 5.Shimada M, Takenaka K, Gion T, Fujiwara Y, Kajiyama K, Maeda T, et al. Prognosis of recurrent hepatocellular carcinoma: a 10-year surgical experience in Japan. Gastroenterology. 1996;111:720–6. doi: 10.1053/gast.1996.v111.pm8780578. [DOI] [PubMed] [Google Scholar]
- 6.Takeishi K, Maeda T, Tsujita E, Yamashita Y, Harada N, Itoh S, et al. Predictors of intrahepatic multiple recurrences after curative hepatectomy for hepatocellular carcinoma. Anticancer Res. 2015;35:3061–6. [PubMed] [Google Scholar]
- 7.Shah SA, Greig PD, Gallinger S, Cattral MS, Dixon E, Kim RD, et al. Factors associated with early recurrence after resection for hepatocellular carcinoma and outcomes. J Am Coll Surg. 2006;202:275–83. doi: 10.1016/j.jamcollsurg.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Shamai G, Binenbaum Y, Slossberg R, Duek I, Gil Z, Kimmel R, et al. Artificial intelligence algorithms to assess hormonal status from tissue microarrays in patients with breast cancer. JAMA Netw Open. 2019;2:e197700. doi: 10.1001/jamanetworkopen.2019.7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck-Krauss-Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019. 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed]
- 10.Veta M, Kornegoor R, Huisman A, Verschuur-Maes AH, Viergever MA, Pluim JP, et al. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol. 2012;25:1559–65. doi: 10.1038/modpathol.2012.126. [DOI] [PubMed] [Google Scholar]
- 11.Lee G, Sparks R, Ali S, Shih NN, Feldman MD, Spangler E, et al. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients. PLoS ONE. 2014;9:e97954. doi: 10.1371/journal.pone.0097954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2019;25:1526–34. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J R Soc Interface. 2015;12. 10.1098/rsif.2014.1153. [DOI] [PMC free article] [PubMed]
- 14.Ali S, Lewis J, Madabhushi A. Spatially aware cell cluster(spACC1) graphs: predicting outcome in oropharyngeal pl6+ tumors. Med Image Comput Comput Assist Inter. 2013;16(Pt 1):412–9. doi: 10.1007/978-3-642-40811-3_52. [DOI] [PubMed] [Google Scholar]
- 15.Itami-Matsumoto S, Hayakawa M, Uchida-Kobayashi S, Enomoto M, Tamori A, Mizuno K, et al. Circulating exosomal miRNA profiles predict the occurrence and recurrence of hepatocellular carcinoma in patients with direct-acting antiviral-induced sustained viral response. Biomedicines. 2019;7. 10.3390/biomedicines7040087. [DOI] [PMC free article] [PubMed]
- 16.Qiu J, Peng B, Tang Y, Qian Y, Guo P, Li M, et al. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: results from a multicenter study. J Clin Oncol. 2017;35:734–42. doi: 10.1200/JCO.2016.68.2153. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Kondo S, Matsuzaki J, Esaki M, Okusaka T, Shimada K, et al. Highly sensitive circulating microRNA panel for accurate detection of hepatocellular carcinoma in patients with liver disease. Hepatol Commun. 2020;4:284–97. doi: 10.1002/hep4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakon M, Umeshita K, Nagano H, Eguchi H, Kishimoto S, Miyamoto A, et al. Clinical significance of hepatic resection in hepatocellular carcinoma analysis by disease-free survival curves. Arch Surg. 2000;135:1456–9. doi: 10.1001/archsurg.135.12.1456. [DOI] [PubMed] [Google Scholar]
- 19.Saito A, Numata Y, Hamada T, Horisawa T, Cosatto E, Graf HP, et al. A novel method for morphological pleomorphism and heterogeneity quantitative measurement: named cell feature level co-occurrence matrix. J Pathol Inf. 2016;7:36. doi: 10.4103/2153-3539.189699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singal AK, Freeman DH, Jr., Anand BS. Meta-analysis: interferon improves outcomes following ablation or resection of hepatocellular carcinoma. Aliment Pharm Ther. 2010;32:851–8. doi: 10.1111/j.1365-2036.2010.04414.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.