Abstract
Objective
To provide an automatic method for segmentation and diameter measurement of type B aortic dissection (TBAD).
Materials and Methods
Aortic computed tomography angiographic images from 139 patients with TBAD were consecutively collected. We implemented a deep learning method based on a three-dimensional (3D) deep convolutional neural (CNN) network, which realizes automatic segmentation and measurement of the entire aorta (EA), true lumen (TL), and false lumen (FL). The accuracy, stability, and measurement time were compared between deep learning and manual methods. The intra- and inter-observer reproducibility of the manual method was also evaluated.
Results
The mean dice coefficient scores were 0.958, 0.961, and 0.932 for EA, TL, and FL, respectively. There was a linear relationship between the reference standard and measurement by the manual and deep learning method (r = 0.964 and 0.991, respectively). The average measurement error of the deep learning method was less than that of the manual method (EA, 1.64% vs. 4.13%; TL, 2.46% vs. 11.67%; FL, 2.50% vs. 8.02%). Bland-Altman plots revealed that the deviations of the diameters between the deep learning method and the reference standard were −0.042 mm (−3.412 to 3.330 mm), −0.376 mm (−3.328 to 2.577 mm), and 0.026 mm (−3.040 to 3.092 mm) for EA, TL, and FL, respectively. For the manual method, the corresponding deviations were −0.166 mm (−1.419 to 1.086 mm), −0.050 mm (−0.970 to 1.070 mm), and −0.085 mm (−1.010 to 0.084 mm). Intra- and inter-observer differences were found in measurements with the manual method, but not with the deep learning method. The measurement time with the deep learning method was markedly shorter than with the manual method (21.7 ± 1.1 vs. 82.5 ± 16.1 minutes, p < 0.001).
Conclusion
The performance of efficient segmentation and diameter measurement of TBADs based on the 3D deep CNN was both accurate and stable. This method is promising for evaluating aortic morphology automatically and alleviating the workload of radiologists in the near future.
Keywords: Aortic dissection; Tomography, X-ray computed; Deep learning
INTRODUCTION
In patients with suspected aortic dissection, computed tomography (CT) is the preferred imaging modality due to its widespread availability, and rapid image acquisition and processing compared to other imaging modalities, as well as excellent sensitivity of 95% (1,2,3,4). As stated in the 2014 European Society of Cardiology guidelines on the diagnosis and treatment of aortic diseases, multiplanar reformation images based on CT play an important complementary role in confirming diagnosis, determining the extent of involvement, and measuring the vascular diameter specifically and precisely, including the entire aorta (EA), true lumen (TL), and false lumen (FL) (1,2,3,4,5). Multiplanar reformation includes centerline analysis involving calculation of the geometric vessel center, and allows for measurement of the diameter along its course (6). For type B aortic dissection (TBAD), it is important to measure vascular diameters accurately. In patients with uncomplicated TBAD, conservative treatment is recommended (1), and accurate diameter measurements are needed to monitor lumen morphology during follow-up. In patients with complicated TBAD, thoracic endovascular aortic repair is recommended (1), and it is important to choose the appropriate size of stent by accurate assessment of the proximal and distal landing zones in order to reduce postoperative complications, such as displacement of stent, retrograde type A aortic dissection, and endoleaks (6,7). The guidelines require that accurate diameter measurements should be made perpendicular to the axis of flow in the aorta. However, due to restrictions in the positioning of X-ray tubes, only anteroposterior to left anterior oblique 60° projections can be obtained during endovascular repair, which makes it difficult to measure the true diameter. Instead, true measurements can be achieved from CT images through multiplanar reformation, since this method presents planes perpendicular to the course of the aorta.
At present, the vascular diameter on CT images is measured manually, which is subjective, time-consuming, and introduces inter- and intra-observer variations (8). Medical image-processing with the help of artificial intelligence may lead to automated measurement without these limitations. Deep learning (DL) is a powerful new machine-learning tool that has made breakthroughs in disease detection (9). DL has been applied across many medical tomographic imaging areas, inspired by numerous achievements in image processing (10,11). Various methods have been developed for vessel segmentation, and convolutional neural network (CNN) is a typical method for image recognition, processing, and segmentation that has shown promising results (12,13,14,15,16,17). Although previous research has mainly focused on two-dimensional (2D) images, with the development of DL, DL has been applied to three-dimensional (3D) images in recent years. Automatic segmentation of the aorta and detection of abdominal aortic aneurysms have been reported using a modified 3D U-Net combined with ellipse fitting. Recently, a serial multi-task CNN model has provided accurate and efficient segmentation of TBADs (18). However, automatic measurement of the aortic diameter has not yet been explored.
In this study, we implemented a DL algorithm to segment TBADs, process 3D images, establish the centerline, and measure the diameter automatically. Our hypothesis was that aortic diameter could be measured more accurately, reliably, and efficiently using a DL method than with the manual method. The success of this technique could reduce the work of radiologists in aortic measurement and establish a foundation for the application of DL in TBAD image-processing and diagnosis.
MATERIALS AND METHODS
Dataset
This retrospective study was approved by the Institutional Review Board with a waiver of informed consent. Patients diagnosed with TBAD according to aortic computed tomography angiography (CTA) from December 1, 2016 to June 30, 2019 were consecutively included. Patients with thrombus in the FL were excluded (n = 89); 13 patients were excluded due to aortic rupture or poor image quality; 21 were excluded as the TL was compressed to a thin line with a thickness less than 3 mm; and another 18 were excluded because their supra-arch branches had abnormal origins. Finally, 139 patients were included. Using a random number table, the data were randomly divided into training, validation, and testing sets at an approximate ratio of 7:1:2. Aortic CTA was performed with various multidetector-row (64-slice to 256- slice) scanners across different years, including the Discovery CT750 HD scanner (GE Healthcare), Revolution CT (GE Healthcare), SOMATOM Definition (Siemens Healthineers), SOMATOM Definition Flash (Siemens Healthineers), or the Brilliance iCT (Philips Healthcare). Spiral CT scans were performed from the level of the thoracic inlet to the pubic symphysis, and reconstructed with slices of 0.625, 0.75, 1, or 1.5 mm. A tube voltage of 120 kV was used for patients with a body mass index (BMI) > 30 kg/m2, 100 kV for a BMI 20–30 kg/m2, and 80 kV for a BMI < 20 kg/m2. The X-ray tube current was adjusted automatically. The characteristics of patients and CTA parameters are listed in Table 1.
Table 1. Characteristics of Patients and CTA Parameters.
| Overall (n = 139) | Training Set (n = 99) | Validation Set (n = 15) | Testing Set (n = 25) | |
|---|---|---|---|---|
| Patient characteristics | ||||
| Male* | 111 (79.9) | 78 (78.8) | 12 (80.0) | 21 (84.0) |
| Age (years)† | 48.7 ± 8.7 | 48.3 ± 8.2 | 47.6 ± 7.1 | 50.1 ± 9.7 |
| CTA Parameters | ||||
| Slice thickness (mm)† | 0.855 ± 0.023 | 0.782 ± 0.019 | 0.891 ± 0.025 | 0.875 ± 0.014 |
| CT scanners* | ||||
| GE Discovery CT | 23 (16.5) | 18 (18.2) | 2 (13.3) | 3 (12.0) |
| GE Revolution CT | 22 (15.8) | 15 (15.2) | 3 (20.0) | 4 (16.0) |
| Siemens Definition CT | 9 (6.5) | 5 (5.1) | 2 (13.3) | 2 (8.0) |
| Siemens Definition Flash CT | 31 (22.3) | 22 (22.2) | 3 (20.0) | 6 (24.0) |
| Philips iCT | 54 (38.8) | 39 (39.4) | 5 (33.3) | 10 (40.0) |
*Data are number of patients and data in parentheses are percentages, †Data are mean ± standard deviation. CT = computed tomography, CTA = computed tomography angiography
Training, Validation, and Testing Sets
The training set included 99 patients to train the model, and 15 patients were included in the validation set to estimate the performance of the model. The images for training and validation were labeled manually by experts via Mimics (Materialise).
Additional aortic CTA data from 25 TBAD patients were collected to demonstrate the segmentation and measurement performance of the model. In terms of verifying the segmentation accuracy of the model, the segmentation results were compared with the ground truth, which was the manual annotation of aortic lumen by an experienced radiologist. Four radiologists (R1–R4) with > 3 years of experience in vascular image post-processing and diagnosis were invited to measure. All the diameters were measured based on multiplanar reformation, which is considered the most accurate and reliable, as well as having the lowest inter-observer variability (2,3,19). The reference standard was the average value of measurements from R1 and R2. If the difference between the two radiologists was > 10% ([R1–R2]/R > 10%), a third radiologist (R3) was invited to reach a final consensus. The first radiologist measured each diameter twice, and the measurements were performed after an interval of at least 2 weeks to assess intra-observer variability. Additionally, the first measurements performed by R1 and the measurements performed by R2 were compared to evaluate the inter-observer variability of manual measurements. To access the accuracy and consistency of manual and DL methods, the measurements from R4 and the DL method were each compared to the reference standard.
Data Preprocessing
First, all images were denoised with a non-local mean threshold algorithm. Second, a region of interest detection algorithm was used to target the aortic lumen. Considering the huge diversity of data, the CTA images were resampled for normalization regarding the voxel size and value before training.
DL Framework
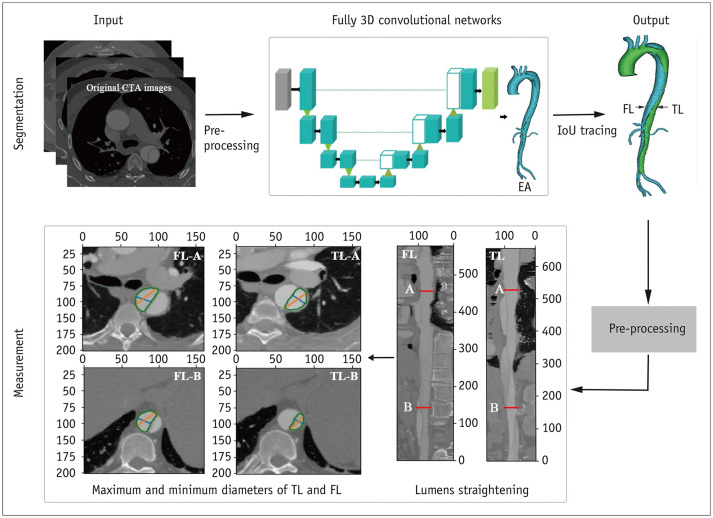
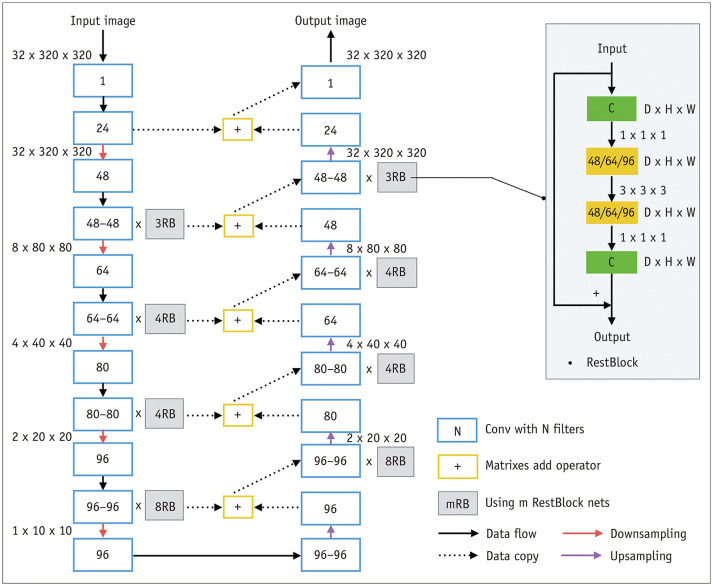
The framework of our deep CNN is illustrated in Figure 1. An end-to-end connected 3D U-Net architecture was used for EA segmentation, and intersection over union (IoU) tracing was used to identify the TL and FL. The U-Net model was a full CNN model with 125 layers, as shown in Figure 2. This pipeline combined a U-Net with DenseNet and an IoU tracing model. The U-Net used a network structure that included down-sampling and up-sampling processes. The down-sampling path had a filter size of 3 × 3 × 32 and stride 2 in each convolution layer. All convolution layers were processed with batch normalization, Rectified Linear Units, and same-padding. The final EA segmentation result from the U-Net model was obtained by assigning each pixel to the class with the highest probability. The IoU tracing method was applied to recognize the membrane, divide the EA into two lumens, and judge the TL and FL. After automatic segmentation of the EA, TL, and FL, a traversal algorithm was used to measure the diameter.
Fig. 1. Overview of the applied framework.
CTA = computed tomography angiography, FL = false lumen, IoU = intersection over union, TL = true lumen, 3D = three-dimensional
Fig. 2. Framework of convolutional neural network.
D = depth, H = height, W = width
Model Training
The network was implemented in PyTorch (version 1.0) on a machine with 125G memory, Interl CPU, and two NVIDIA GeForce GTX1080 Ti GPUs. Training stopped when the loss value stopped decreasing in the validation set after 30 steps. An adaptive learning rate was adopted with an initial setting value of 0.01, and the learning rate was reduced to 0.95 times the previous value every 80 epochs. The SoftMax activation function was used in the last layer in all tasks to identify the aortic branches. Before feeding into the networks, the CTA images were pre-processed and re-sized to 128 × 128 × 128, and data augmentation by rotation and flipping were applied. The hyper parameters α, β, and γ were set to 1, 3, and 6 respectively.
Quantitative Evaluation with Parameter Measurement
Eight measurement positions were chosen, including those recommended in the guidelines (1). The positions were proximal to the innominate artery origin (P1), proximal to the left common carotid artery origin (P2), proximal to the left subclavian artery origin (P3), distal to the left subclavian artery origin (P4), at the level of the diaphragm (P5), at the superior border of the celiac axis origin (P6), at the superior border of the lower renal aorta origin (P7), and proximal to the aortic bifurcation (P8). If there were two renal arteries on one side, the larger one was the target. The diameters of 32 parameters were assessed, from the inner to inner lumen wall, excluding calcium, as follows: The maximum and minimum diameter of the EA at P1–P8, and the maximum and minimum diameter of the TL and FL at P5–P8. If the TBAD did not involve P5–P8, the maximum and minimum diameters of the TL and FL were recorded as 0 mm. In the DL method, we applied a traversal algorithm to measure the diameter after the automatic segmentation and IoU tracing methods. At the specified level, the center point on the centerline was selected, the lumen was rotated to obtain different diameters, and the maximum and minimum values were selected as the maximum and minimum diameters. In the manual method, the diameters were measured with an image post-processing workstation (Advantage Workstation Release 4.6 software, GE Healthcare). The window width and level were set to 700/200 and individually adjusted if necessary. The diameters at each position were calculated based on the following steps: 1) The centerlines of the EA, TL, and the FL were each extracted and smoothed. The centerlines of the EA, TL, and the FL were acquired by placing seed points into the lumen of the ascending aorta and the iliac artery, the proximal and distal TL, and the proximal and distal FL. The centerline was manually adjusted if it was not in the center of the vessel. 2) The corresponding straightened images were generated according to each of their centerlines. 3) Each branch was located based on the centerline to determine the specified measurement position. 4) The maximum and minimum diameters of the EA, TL, and the FL auto-recognized by the workstation were recorded. The time required, including for centerline preparation and diameter measurement, was recorded for each patient.
Statistical Analysis
Continuous data are reported as means ± standard deviations. Segmentation performance was evaluated with the dice coefficient score (DCS). Kolmogorov-Smirnov tests were used to assess the distribution of the data. Measurements and times between the DL and manual methods were compared, and the intra- and inter-observer reproducibility of the manual method was assessed. Paired t test (for variables normally distributed) or Wilcoxon signed rank test (for variables not normally distributed) was applied for these comparisons. All p values were two-sided, and a p value < 0.05 was considered statistically significant. Two-way mixed intra-class correlation (ICC) analyses were used to assess the consistency of R1, R2, and R4, and an ICC coefficient of > 0.8 was considered an excellent agreement. Linear correlation graphs and Bland-Altman plots were used to show the differences between reference standards and measurements from the two methods. Correlation coefficients were calculated. All statistical analyses were performed using SPSS version 22.0 (IBM Corp.).
RESULTS
Accuracy of Segmentation by DL
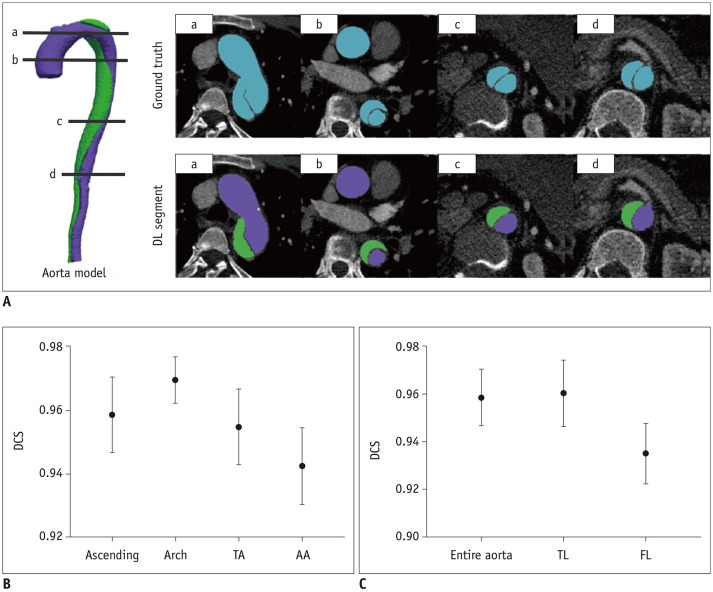
The DCSs of the EA, TL, and the FL for 25 cases are shown in Figure 3. The segmentation results of DL corresponded well with ground truth. The corresponding distributions of the DCS values for different aortic regions and lumens are shown in Figure 3B and C. The model worked well in the testing process, reaching mean DCS values of 0.958, 0.961, and 0.932 for EA, TL, and FL segmentation, respectively (Fig. 3C), which was significantly higher than that reported in a previous study (18). In order to further evaluate the performance of aorta segmentation in difference regions, we separated the aorta into the ascending aorta, aortic arch, descending aorta, and the abdominal aorta (Fig. 3B). The results showed that the DCS was high in each region, while the segmentation performance was comparatively poor in the abdominal aorta.
Fig. 3. Automatic segmentation results with the DL method.
A. Four levels (a, b, c, d) of ground truth and DL segmentation of aorta. B. DCSs for the Ascending, Arch, DA, and AA in the test set. C. DCSs for the entire aorta, TL and FL in the test set. (B, C) are the corresponding mean and error bar plot, displaying the distribution of DCS in test set based on minimum, mean, and maximum values. AA = abdominal aorta, Arch = aortic arch, Ascending = ascending aorta, DA = descending aorta, DCS = dice coefficient score, DL = deep learning
Accuracy and Reliability of Manual and DL Methods
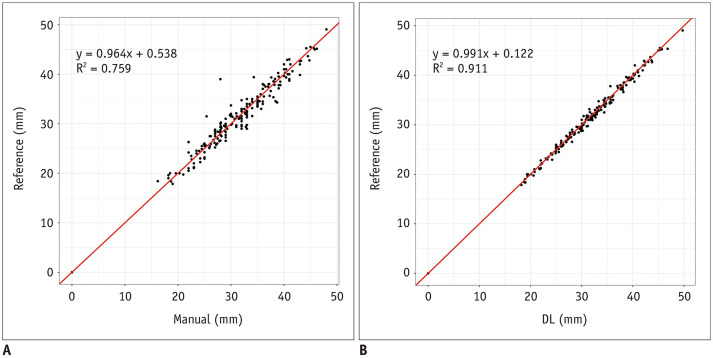
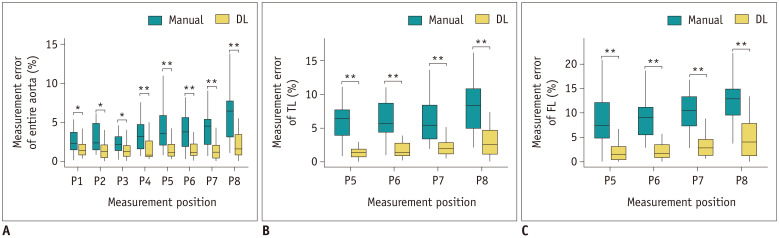
The diameters of the EA, TL, and the FL were calculated at each of the 8 positions using the two methods. The ICC coefficient was 0.976 (95% confidence interval, 0.973–0.979) (p < 0.001) between R1, R2, and R4, demonstrating an excellent consistency in the measurements of these three observers. We found linear relationships between reference and measurements from manual and DL methods, with Pearson's correlation coefficients of 0.964 and 0.991, and coefficients of determination of 0.759 and 0.911, respectively (Fig. 4). The measurement errors with the DL method were much fewer than with manual method at all positions in each lumen (Fig. 5). The DL model maintained high accuracy at all positions in the EA, TL, and the FL, while the performance of the manual method in the TL and FL was not as good as in the EA.
Fig. 4. Linear relationships between reference and measurements from manual and DL methods.
A. Correlation between measurements from manual method and reference standard (p < 0.001, r = 0.964, R2 = 0.759). B. Correlation between measurements from DL method and reference standard (p < 0.001, r = 0.991, R2 = 0.911). The correlation between measurements from DL method and reference standard was better than that from manual method. R = Pearson's correlation coefficient, R2 = coefficient of determination
Fig. 5. Measurement performance for the entire aorta, TL, and FL in 25 cases with DL and manual methods.
A. Measurement error for the entire aorta at eight levels. B. Measurement error for the TL at four levels. C. Measurement error for the FL at four levels. In the box plots, the boxes indicate the median and interquartile range, while the error bars represent the minimum and maximum values. Significance is labeled as follows: *p < 0.05, **p < 0.01. P1 = innominate artery origin, P2 = proximal to the left common carotid artery origin, P3 = proximal to the left subclavian artery origin, P4 = distal to the left subclavian artery origin, P5 = at the level of the diaphragm, P6 = at the superior border of the celiac axis origin, P7 = at the superior border of the lower renal aorta origin, P8 = proximal to the aortic bifurcation
The detailed measurements are listed in Table 2. The average measurement error was the mean error from the reference standard in all 25 cases at each measurement position. It was clear that the average DL measurement error at each position was fewer than that observed with the manual measurement. For the EA, TL, and the FL, the average measurement errors at all positions with the DL method were 1.64, 2.46, and 2.50 mm, while those with manual method were 4.13, 11.67, and 8.02 mm, respectively. The performance for the EA was better than that for the TL and FL, regardless of the method used, and the differences were greater in the manual measurements. There were statistically significant differences in the measurements between the two methods, which indicated that the measurements using the DL method were more accurate than those using the manual method.
Table 2. Measurement Errors of Deep Learning and Manual Methods, and Differences between the Methods.
| Levels | EA AME (%) | TL AME (%) | FL AME (%) | DL vs. M | |||||
|---|---|---|---|---|---|---|---|---|---|
| DL | M | DL | M | DL | M | EA P | TL P | FL P | |
| P1 | 1.56 | 2.75 | - | - | - | - | 0.021 | - | - |
| P2 | 1.63 | 4.04 | - | - | - | - | 0.050 | - | - |
| P3 | 1.49 | 2.96 | - | - | - | - | 0.035 | - | - |
| P4 | 1.72 | 3.57 | - | - | - | - | < 0.001 | - | - |
| P5 | 1.73 | 4.37 | 2.43 | 11.32 | 2.02 | 7.59 | 0.001 | < 0.001 | < 0.001 |
| P6 | 1.61 | 4.81 | 2.19 | 10.61 | 2.19 | 5.61 | 0.001 | < 0.001 | < 0.001 |
| P7 | 1.40 | 4.65 | 3.17 | 12.34 | 2.44 | 7.55 | < 0.001 | < 0.001 | < 0.001 |
| P8 | 1.98 | 5.85 | 2.03 | 12.41 | 3.36 | 11.33 | < 0.001 | < 0.001 | < 0.001 |
| AVE | 1.64 | 4.13 | 2.46 | 11.67 | 2.50 | 8.02 | - | - | - |
The average measurement error of the diameter of EA, TL, and FL at each measurement position in 25 cases. AME = average measurement error, AVE = average measurement error at all evaluated positions, DL = measurement by deep learning method, EA = entire aorta, FL = false lumen, M = measurement by manual method, P1 = innominate artery origin, P2 = proximal to the left common carotid artery origin, P3 = proximal to the left subclavian artery origin, P4 = distal to the left subclavian artery origin, P5 = at the level of the diaphragm, P6 = at the superior border of the celiac axis origin, P7 = at the superior border of the lower renal aorta origin, P8 = proximal to the aortic bifurcation, TL = true lumen
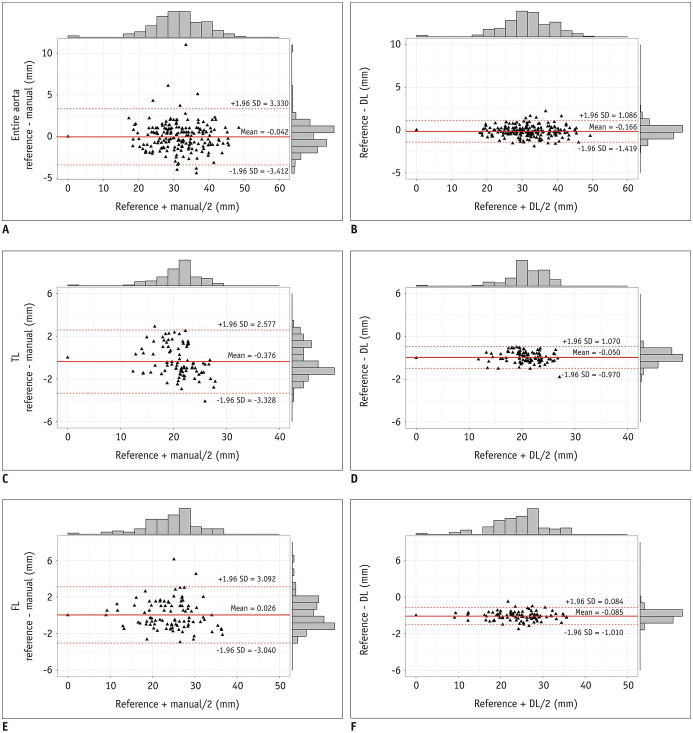
Bland-Altman plots revealed the deviations of the diameters between the reference standard and the manual or DL method. The mean difference in the diameters between the manual method and reference standard was −0.042 mm (−3.412 to 3.330 mm) for the EA, −0.376 mm (−3.328 to 2.577 mm) for the TL, and 0.026 mm (−3.040 to 3.092 mm) for the FL. The mean difference in the diameters between the DL method and the reference standard was −0.166 mm (−1.419 to 1.086 mm) for the EA, −0.050 mm (−0.970 to 1.070 mm) for the TL, and −0.085 mm (−1.010 to 0.084 mm) for the FL. We found that the diameters measured with the DL method were considerably more consistent with the reference standard than those measured with the manual method (Fig. 6).
Fig. 6. Bland-Altman plots for diameter measurements.
Bland-Altman plots for the entire aorta (A, B), TL (C, D), and FL (E, F) for deviations of diameters between manual method and reference standard (A, C, E), as well as DL method and reference standard (B, D, F) at each measurement position in the 25 cases.
Intra- and Inter-Observer Reproducibility of Diameter Measurements
The diameters measured by the manual method were assessed to evaluate reliability. The intra-observer reproducibility of the EA diameter was good (p = 0.889), while the differences between the two measurements for the TL and FL diameters were statistically significant (p = 0.013 and 0.018, respectively) (Table 3). For inter-observer reproducibility, the difference in the EA diameter between the two observers was statistically significant (p < 0.001), while there was no statistical difference in the TL and FL. There were no intra- and inter-observer differences in the DL method.
Table 3. Intra- and Inter-Observer Differences in Diameter with Manual Method.
| Method | Mean Values | Paired Differences | P | 95% Limits of Agreement |
|---|---|---|---|---|
| Intra-observer | ||||
| EA (n = 400) | 29.78 ± 7.69 vs. 29.79 ± 7.75 | −0.011 ± 1.974 | 0.889 | −0.171–0.148 |
| TL (n = 200) | 16.64 ± 6.27 vs. 16.16 ± 5.79 | 0.482 ± 2.721 | 0.013 | 0.103–0.861 |
| FL (n = 200) | 18.96 ± 8.07 vs. 19.29 ± 7.86 | −0.340 ± 2.004 | 0.018 | −0.619– −0.601 |
| Inter-observer | ||||
| EA (n = 400) | 29.79 ± 7.75 vs. 29.43 ± 7.59 | 0.364 ± 1.821 | < 0.001 | 0.217–0.511 |
| TL (n = 200) | 16.16 ± 5.79 vs. 16.09 ± 6.25 | 0.071 ± 2.729 | 0.713 | −0.310–0.452 |
| FL (n = 200) | 19.29 ± 7.86 vs. 19.14 ± 8.05 | 0.153 ± 1.819 | 0.237 | −0.101–0.406 |
Values are reported as mean ± standard deviation.
Time for Analysis
The measurement time using the DL method was significantly shorter than that of the manual method (21.7 ± 1.1 [20.0 to 23.5] vs. 82.5 ± 16.1 [60 to 100] minutes for one case, p < 0.001). This finding demonstrated that the DL method is both time- and labor-saving.
DISCUSSION
Using the 3D multi-task deep CNN method, we successfully achieved automatic segmentation and measurement of TBADs. In our study, this method segmented TBAD into the EA, TL, and FL with high DCSs. Compared to the manual method, the DL method showed better performance with higher accuracy for the EA, TL, and the FL at all positions. Thus, automatic measurement is helpful to accurately judge the size changes of the vascular lumen during follow-up, and to reveal signs of surgery, including aortic expansion. The average measurement errors of the manual method were greater than the interval value between the different types of stent (2 mm), which could lead to the selection of an inappropriate stent size and increase the risk of adverse events, while the DL method had smaller measurement errors, conducive to selecting appropriate stents. In addition, better consistency and stability was observed between the DL method and the reference standard. Moreover, there were variations across different readers and times of measurement in the manual method, while the DL method was stable without any inter- and intra-observer difference. In addition, the DL method was more efficient in terms of time. The diameters of the EA, TL, and the FL were each measured after the centerlines were found. Although most commercially available software packages facilitate automatic centerline detection, in our clinical work, we found that the accuracy of the centerlines was poor. Careful manual adjustment is required to keep the centerline as close to the real situation as possible and to minimize deviations for each reader, which is why manual measurements take so long.
Automated methods for aorta segmentation have been investigated extensively; most are based on classical image-processing technologies (20,21,22), and rarely on artificial neural networks (23,24). With the prevalence of DL in medical image segmentation (25,26), deep CNN has been proposed to segment the aorta based on 2D images in small datasets (18). Recently, Cao et al. (18) applied a multi-task output CNN network for automatic TBAD segmentation, but only the accuracy of segmentation and volume measurement were assessed. Here, we used a 3D multi-task deep CNN in a unified framework based on a large database. To the best of our knowledge, this is the first application of 3D DL neural networks in TBAD to measure the vascular diameter, as well as to evaluate the accuracy and consistency of the model. With the DL method, the true aortic morphology was displayed due to it representing an accurate and stable measurement. Furthermore, the computerized approach was more efficient and labor-saving.
The current study has several limitations. First, the training and testing datasets were not large enough due to the low prevalence of TBAD. More data, especially for instances where there were various morphologies from multiple centers, would be helpful to further improve the robustness of the performance. Second, only TBAD was included, since for type A, replacement of the aorta by an artificial vessel is recommended and is of little importance to measure diameters. Furthermore, TBAD with thrombus in the FL was not included; we intend to focus on thrombus segmentation in a subsequent study.
In conclusion, the DL model was successfully applied to automatically segment the aorta and measure the diameter for TBAD. This model enabled image processing and aortic measurements to be implemented efficiently. With more accurate, reliable, and stable measurements, DL is likely to play an important role in guiding intervention and assessing aortic remodeling. The proposed 3D deep CNN method for automatic segmentation and measurement of TBAD will reduce the labor of radiologists in the near future.
Acknowledgments
The authors thank Dr. Iain Charles Bruce for help in editing the manuscript.
Footnotes
This study was granted by the Ministry of Science and Technology of China, National key research and development project (2016YFC1300400) and Chinese Academy of Medical Sciences Innovation Project (No. 2016-I2M-1-011).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Erbel R, Aboyans V, Boileau C, Bossone E, Bartolomeo RD, Eggebrecht H, et al. 2014 ESC guidelines on the diagnosis and treatment of aortic diseases: document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The Task Force for the Diagnosis and Treatment of Aortic Diseases of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2873–2926. doi: 10.1093/eurheartj/ehu281. [DOI] [PubMed] [Google Scholar]
- 2.Sommer T, Fehske W, Holzknecht N, Smekal AV, Keller E, Lutterbey G, et al. Aortic dissection: a comparative study of diagnosis with spiral CT, multiplanar transesophageal echocardiography, and MR imaging. Radiology. 1996;199:347–352. doi: 10.1148/radiology.199.2.8668776. [DOI] [PubMed] [Google Scholar]
- 3.Kaji S, Akasaka T, Katayama M, Yamamuro A, Yamabe K, Tamita K, et al. Long-term prognosis of patients with type B aortic intramural hematoma. Circulation. 2003;108 Suppl 1:II307–II311. doi: 10.1161/01.cir.0000087425.86049.74. [DOI] [PubMed] [Google Scholar]
- 4.Novelline RA, Rhea JT, Rao PM, Stuk JL. Helical CT in emergency radiology. Radiology. 1999;213:321–339. doi: 10.1148/radiology.213.2.r99nv01321. [DOI] [PubMed] [Google Scholar]
- 5.Hagan PG, Nienaber CA, Isselbacher EM, Bruckman D, Karavite DJ, Russman PL, et al. The International Registry of Acute Aortic Dissection (IRAD): new insights into an old disease. JAMA. 2000;283:897–903. doi: 10.1001/jama.283.7.897. [DOI] [PubMed] [Google Scholar]
- 6.Rengier F, Wörz S, Godinez WJ, Schumacher H, Böckler D, Rohr K, et al. Development of in vivo quantitative geometric mapping of the aortic arch for advanced endovascular aortic repair: feasibility and preliminary results. J Vasc Interv Radiol. 2011;22:980–986. doi: 10.1016/j.jvir.2011.01.434. [DOI] [PubMed] [Google Scholar]
- 7.Sternbergh WC, 3rd, Money SR, Greenberg RK, Chuter TA, Zenith Investigators. Influence of endograft oversizing on device migration, endoleak, aneurysm shrinkage, and aortic neck dilation: results from the Zenith Multicenter Trial. J Vasc Surg. 2004;39:20–26. doi: 10.1016/j.jvs.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Müller-Eschner M, Rengier F, Partovi S, Weber TF, Kopp-Schneider A, Geisbüsch P, et al. Accuracy and variability of semiautomatic centerline analysis versus manual aortic measurement techniques for TEVAR. Eur J Vasc Endovasc Surg. 2013;45:241–247. doi: 10.1016/j.ejvs.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Tajik AJ. Machine learning for echocardiographic imaging: embarking on another incredible journey. J Am Coll Cardiol. 2016;68:2296–2298. doi: 10.1016/j.jacc.2016.09.915. [DOI] [PubMed] [Google Scholar]
- 10.Wang G. A perspective on deep imaging. IEEE Access. 2016;4:8914–8924. [Google Scholar]
- 11.Wang G, Kalra M, Orton CG. Machine learning will transform radiology significantly within the next 5 years. Med Phys. 2017;44:2041–2044. doi: 10.1002/mp.12204. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y, Chen Q, Wang W, Peng Y, Wang Q, Duan F, et al. A vessel active contour model for vascular segmentation. Biomed Res Int. 2014;2014:106490. doi: 10.1155/2014/106490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 14.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drozdzal M, Chartrand G, Vorontsov E, Shakeri M, Di Jorio L, Tang A, et al. Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal. 2018;44:1–13. doi: 10.1016/j.media.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016;35:1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- 17.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 18.Cao L, Shi R, Ge Y, Xing L, Zuo P, Jia Y, et al. Fully automatic segmentation of type B aortic dissection from CTA images enabled by deep learning. Eur J Radiol. 2019;121:108713. doi: 10.1016/j.ejrad.2019.108713. [DOI] [PubMed] [Google Scholar]
- 19.Rengier F, Weber TF, Partovi S, Müller-Eschner M, Böckler D, Kauczor HU, et al. Reliability of semiautomatic centerline analysis versus manual aortic measurement techniques for TEVAR among non-experts. Eur J Vasc Endovasc Surg. 2011;42:324–331. doi: 10.1016/j.ejvs.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Sedghi Gamechi Z, Bons LR, Giordano M, Bos D, Budde RPJ, Kofoed KF, et al. Automated 3D segmentation and diameter measurement of the thoracic aorta on non-contrast enhanced CT. Eur Radiol. 2019;29:4613–4623. doi: 10.1007/s00330-018-5931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrens T, Rohr K, Stiehl HS. Robust segmentation of tubular structures in 3-D medical images by parametric object detection and tracking. IEEE Trans Syst Man Cybern B Cybern. 2003;33:554–561. doi: 10.1109/TSMCB.2003.814305. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation From CT volumes. IEEE Trans Med Imaging. 2018;37:2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 23.Herment A, Kachenoura N, Lefort M, Bensalah M, Dogui A, Frouin F, et al. Automated segmentation of the aorta from phase contrast MR images: validation against expert tracing in healthy volunteers and in patients with a dilated aorta. J Magn Reson Imaging. 2010;31:881–888. doi: 10.1002/jmri.22124. [DOI] [PubMed] [Google Scholar]
- 24.Ben Ayed I, Wang M, Miles B, Garvin GJ. TRIC: trust region for invariant compactness and its application to abdominal aorta segmentation. Med Image Comput Comput Assist Interv. 2014;17(Pt 1):381–388. doi: 10.1007/978-3-319-10404-1_48. [DOI] [PubMed] [Google Scholar]
- 25.Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, et al. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46:e1–e36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham) 2019;6:014006. doi: 10.1117/1.JMI.6.1.014006. [DOI] [PMC free article] [PubMed] [Google Scholar]