Abstract
LncRNA HCP5 has been confirmed to play crucial roles in many types of cancers. However, the role of lncRNA HCP5 in regulating the occurrence and development of gastric cancer (GC) remains unknown. In the current study, we aimed to investigate the precise effects of lncRNA HCP5 on cell proliferation, migration and invasion and molecular mechanisms in gastric cancer. Using RT-qPCR analysis, we found that lncRNA HCP5 was differentially expressed in GC cell lines. CCK-8, wound healing and transwell assay indicated that the proliferation, migration and invasion of gastric cancer cells were inhibited by downregulation of lncRNA HCP5 and lncRNA HCP5 overexpression exhibited the opposite effects in gastric cancer cells. Mechanistically, RNA binding protein immunoprecipitation and dual luciferase reporter assay confirmed the interaction between lncRNA HCP5 and DDX21. The effects of lncRNA HCP5 overexpression the proliferation, migration and invasion of GC cells were partly rescued by DDX21 silencing. Taken together, downregulation of lncRNA HCP5 exerted inhibitory effects on GC cell proliferation, migration and invasion through modulation of DDX21 expression, demonstrating the function of lncRNA HCP5 and DDX21 in GC progression.
Keywords: LncRNA HCP5, DDX21, Gastric cancer
Introduction
Gastric cancer as the common digestive malignancy is the third leading cause of cancer associated death (Eichelberger et al. 2015). Wherein, the mortality of gastric cancer in China accounts for over thirty percent of the cases across the world (Chen et al. 2016). As it is known to all, conventional treatments for gastric cancer consist of chemotherapy, radiotherapy, surgical excision and so forth, while their effects are far from satisfaction (Chen et al. 2020; Endo et al. 2019; Yang et al. 2020; Yu et al. 2019). Furthermore, poor prognosis of gastric cancer and low five-year survival rate especially for advanced gastric cancer make it less possible to control cancer progression. Therefore, it is of great significance to explore the effective biomarkers for the diagnosis of gastric cancer that can also serve as therapeutic targets and investigate the underlying molecular mechanism.
LncRNAs emerge as the promising functional molecules for the diagnosis and therapiesI of cancers (Fattahi et al. 2020; Luzón-Toro et al. 2019). LncRNAs transcribed by RNA polymerase II are RNAs with more than 200 nucleotides in length. LncRNAs, which were considered as transcription noise previously, have been confirmed to regulate gene expression at transcriptional, post-transcriptional and epigenetic levels (Gong et al. 2015; Goyal et al. 2017; Qin et al. 2016). LncRNAs were confirmed to be closely related to gastric cancer and aberrantly expressed in cancer cells. LncRNA LINC01419 was reported to be up-regulated in gastric cancer and downregulation of lncRNA LINC01419 exerted protective effects against gastric cancer (Wang et al. 2019b). LINC00565 was dysregulated and contributed to the progression of gastric cancer (Hu et al. 2020).
LncRNA HCP5 was reported to be aberrantly expressed in many types of cancers. LncRNA HCP5 was downregulated in skin cutaneous melanoma and lncRNA HCP5 overexpression exerted anti-cancer effects against skin cutaneous melanoma (Wei et al. 2019). LncRNA HCP5 was increased in triple negative breast cancer and contributed to the progression of triple negative breast cancer (Lin et al. 2019). LncRNA HCP5 was up-regulated in cervical cancer and exerted promoting effects on cervical cancer (Yu et al. 2018). As outlined, aberrant lncRNA HCP5 expression in these types of cancers suggested that lncRNA HCP5 may serve as a broad-spectrum tumor marker and a therapeutic target for cancers.
DEAD cassette helicase 21 (DDX21), functions as the co-regulator of transcription, was aberrantly expressed in many cancers including breast cancer, colorectal cancer, gastric cancer and so forth (Cao et al. 2018; Jung et al. 2011; Zhang et al. 2018). DDX21 is reported to be up-regulated in gastric cancer, contributing to the development of gastric cancer via promotion of cell proliferation (Cao et al. 2018).
Herein, we studied the biological functions of lncRNA HCP5 in gastric cancer and revealed its regulatory relationships with DDX21. Starbase predicted that DDX21 targeted lncRNA HCP5 and we analyzed that lncRNA HCP5 positively regulated DDX21 expression in gastric cancer. It was found that lncRNA HCP5 influenced the progression of gastric cancer through regulating DDX21 expression, suggesting that lncRNA HCP5 and DDX21 could be considered as potential targets for GC treatment.
Materials and methods
Cell culture and transfection
GES-1, AGS and FU97 cells were purchased from Cobioer (Nanjing, China). MKN-45 and MGC-803 cells were purchased from Mingzhou company (Ningbo, China). GES-1, AGS, MKN-45, MGC-803 and FU97 cells at a density of 1.0 × 103 cells/well were seeded in 96-well cell culture plates, and cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen, CA, USA) supplemented with penicillin (100 U/mL), streptomycin (100 mg/mL) as well as 10% Fetal Bovine Serum (FBS; Gibco, NY, USA) in an incubator with 5% CO2 atmosphere at 37 °C.
For cell transfection, shRNA-HCP5-1, shRNA-HCP5-2, shRNA-DDX21-1, shRNA-DDX21-2 and its negative control (shRNA-NC) were all obtained from GenePharma (Shanghai, China). The transfection of the above products was performed using Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA) for 48 h. PcDNA HCP5 and its negative control (pcDNA) were purchased from Genecopoeia (Rockville, MD, USA). The transfection of pcDNA HCP5 and pcDNA was performed by EndoFectin Lenti transfection reagent (Genecopoeia, MD, USA) according to the manufacturer’s instructions. The related sequences were presented as following: shRNA-NC (5′-TTCTCCGAACGTGTCACGT-3′), shRNA-HCP5-1 (5′-GGCTCAACTCACAAGAAAC-3′), shRNA-HCP5-2 (GGUUGGUCACCUAAAGAAAUA), shRNA-DDX21-1: (CGGGAAGGACTTAATTGCA), shRNA-DDX21-2: (GGACGCACTATCATCTTTT).
Real-time quantitative polymerase chain reaction (RT-qPCR) assay
Trizol reagent (Beyotime, Shanghai, China) was used to extract the total RNA according to the protocols of the manufacturer. TaKaRa Reverse Transcription Kit (TaKaRa, Dalian, China) was applied to synthesise cDNA. Then, qRT-PCR assay was performed on a Bio-Rad Real-Time PCR System (Bio-Rad, CA, USA) under the following conditions: firstly, 95 °C for 5 min, then thirty-five cycles of 95 °C for 10 s, 60 °C for 20 s, 70 °C for 10 s. The primers were presented as follows: HCP5: CAGCCTGAGAGAAGTAGGGC (Forward), TCAGTCGCATTTCCAGGTAATTT (Reverse); DDX21, 5′-AATGTTGCTGCACGTGGGTTAG-3′ (Forward) and 5′-CCCGGATCGATGAATGTAGGAC-3′ (Reverse); NGAL: 5′-CCTCCCTGAAAACCACATCGT-3′ (Forward), 5′-TGTGCACTCAGCCGTCGATA-3 (Reverse); MMP7: 5’-TGTAAAACGACGGCCAGT-3’ (Forward), 5′-CAGGAAACAGCTATGACC-3′ (Reverse). MMP13 5′-TCCTGATGTGGGTGAATACAATG-3′ (Forward); 5′-GCCATCGTGAAGTCTGGTAAAAT-3′ (Reverse). The gene expression level was determined by using the 2−ΔΔCt method.
CCK-8 assay
Cells (5.0 × 103 cells/well) after transfection were cultured in a 96-well cell culture plate in an incubator with 5% CO2 atmosphere at 37 °C for 24 h. Then, 10 µl CCK-8 solution (Beyotime, Shanghai, China) was added into each well and cells after treatment were cultured for 2 h. The cell viability was determined by detecting the absorbance at 450 nm under Spectrometer Varioskan® Flash (Thermo Fisher Scientific, MA, USA).
Wound healing assay
Wound healing assay was performed to evaluate the ability of cell migration. For wound healing assay, cells after transfection were cultured in RPMI 1640 supplemented with 10% FBS and grown to 90% confluence. Then, the scratch was made on cells using a pipette tip. Subsequently, the culture medium was changed to serum-free RPMI 1640 Cell migration was evaluated by the ratio of scratch wound area at 24 h over 0 h.
Transwell assay
Transwell assay was performed to evaluate the ability of cell invasion. For transwell assay, cells at a density of 1 × 105 cells/mL after transfection were maintained in serum-free medium and added in the upper chamber that was coated with Matrigel (Corning Inc, NY, USA). Then, the medium containing 20% FBS was added into the lower chamber. After 36 h incubation, 0.05% crystal violet (Sigma, MO, USA) was used to stain the migrated cells in the lower chamber. The migrated cells were photographed in 5 randomly selected fields under microscope (magnification 100×; Leica, Wetzlar, Germany).
Western blot analysis
Total protein extraction was performed using RIPA lysis buffer containing proteinase inhibitors (Beyotime, Shanghai, China). Total proteins were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, CA, USA). Later, we used 5% skim milk to block the membranes. Subsequently, the primary antibody against MMP7 (#ab205525, Abcam, MA, USA), MMP13 (#ab219620, Abcam, MA, USA), DDX21 (#ab181870, Abcam, MA, USA) and GAPDH (#ab128915, Abcam, MA, USA) were incubated with the membranes overnight at 4 °C, respectively. After that, the membranes were incubated with HRP-conjugated antibody for 1 h at room temperature. Enhanced chemiluminescence detection system (Amersham International, Buckinghamshire, UK) was used to obtain the bands and Image Lab software (Bio-Rad, CA, USA) was used to quantify the protein expression levels.
RNA binding protein immunoprecipitation assay
Starbase (http://starbase.sysu.edu.cn) was used to predict the target gene of lncRNA HCP5. Imprint RNA immunoprecipitation kit (Sigma-Aldrich, MO, USA) was used to further verify the target binding between lncRNA HCP5 and target gene, according to the instructions of the manufacturer. Briefly, the cells were lysed in RIPA lysis buffer (Solarbio, Beijing, China) to obtain the cell lysates. Then the anti-IgG (#ab133470, Abcam, MA, USA) and anti-DDX21 antibody (ab244458, Abcam, MA, USA) were incubated with the cell lysates at 4 °C. RT-qPCR was used to analyze the purified RNA.
Dual luciferase reporter assay
DDX21 sequences containing the wild-type (WT) or mutated-type (MUT) lncRNA HCP5 binding sites were respectively inserted into the pGL3 luciferase reporter vector (Promega, WI, USA). Next, cells were co-transfected with pcDNA HCP5 along with DDX21-WT or DDX21-MUT reporter vector using Lipofectamine 2000. At 48 h after transfection, luciferase activity was assessed using the Dual Luciferase Reporter Assay system (Promega, WI, USA) and normalized to Renilla luciferase activity..
Statistical analysis
The data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test using GraphPad Prism Software (GraphPad Prism, Inc., CA, USA) and presented as mean ± standard deviation (SD). P < 0.05 represented a statistically significant difference.
Results
LncRNA HCP5 was highly expressed in gastric cancer cells
We primarily evaluated lncRNA HCP5 levels in varieties of gastric cancer cells. LncRNA HCP5 levels were higher in various GC cells (AGS, MKN-45, MGC-803 and FU97) than that in gastric epithelial cells (GES-1). Besides, MGC-803 cells presented the most obvious upregulation of lncRNA HCP5 and MGC-803 cells were chosen for the subsequent experiments (Fig. 1). All these findings indicated that lncRNA HCP5 may play an important role in gastric cancer.
Fig. 1.
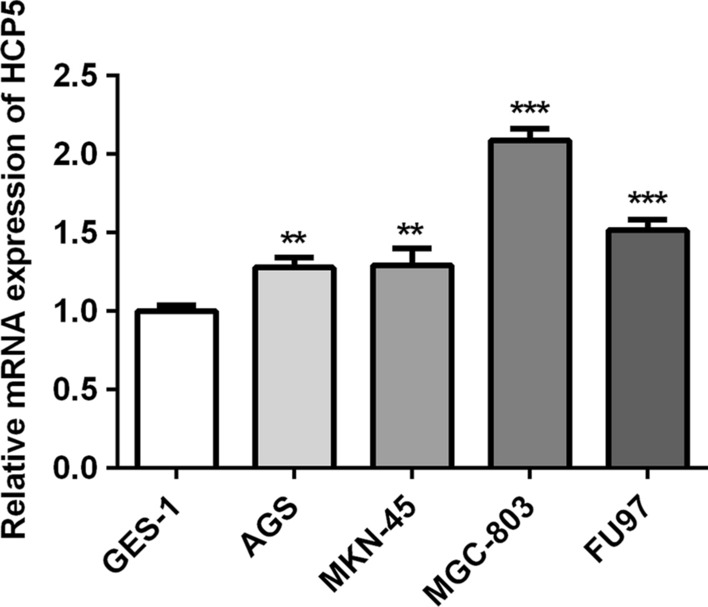
The expression of lncRNA HCP5 in different gastric cancer cells (AGS, MKN-45, MGC-803 and FU97) and gastric epithelial cells (GES-1). ***P < 0.001, **P < 0.01 vs. GES-1
Downregulation of lncRNA HCP5 exhibited inhibitory effects on the proliferation of MGC-803 cells
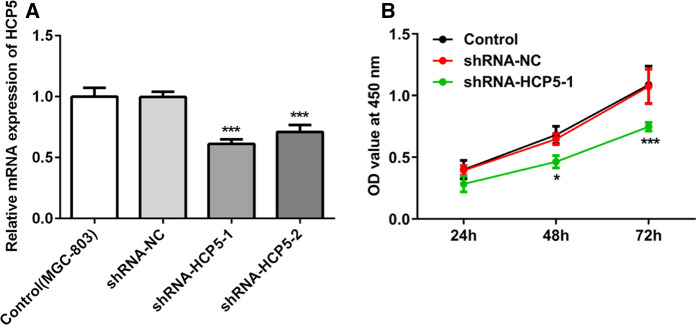
We further evaluated the effects of lncRNA HCP5 on the proliferation of MGC-803 cells. As shown by the results above, lncRNA HCP5 level was relatively higher in MGC-803 cells. Hence, the effect of lncRNA HCP5 downregulation was investigated herein. LncRNA HCP5 level was more significantly downregulated by shRNA-HCP5-1 than that by shRNA-HCP5-2. Therefore, shRNA-HCP5-1 was used for downregulation of lncRNA HCP5 in the following experiment (Fig. 2a). A significant decrease in MGC-803 cell proliferation was observed in shRNA-HCP5-1 group in contrast to control group and shRNA-NC group, confirming that downregulation of lncRNA HCP5 had inhibitory effects on the proliferation of MGC-803 cells (Figs. 2b).
Fig. 2.
The effects of lncRNA HCP5 downregulation on the proliferation of MGC-803 cells. a LncRNA HCP5 levels in MGC-803 cells after transfection with shRNA-HCP5-1 or shRNA-HCP5-2; b Cell viability assessed by CCK-8 assay. *** P < 0.001, *P < 0.05 vs. shRNA-NC
Downregulation of lncRNA HCP5 exhibited inhibitory effects on the migration and invasion of MGC-803 cells
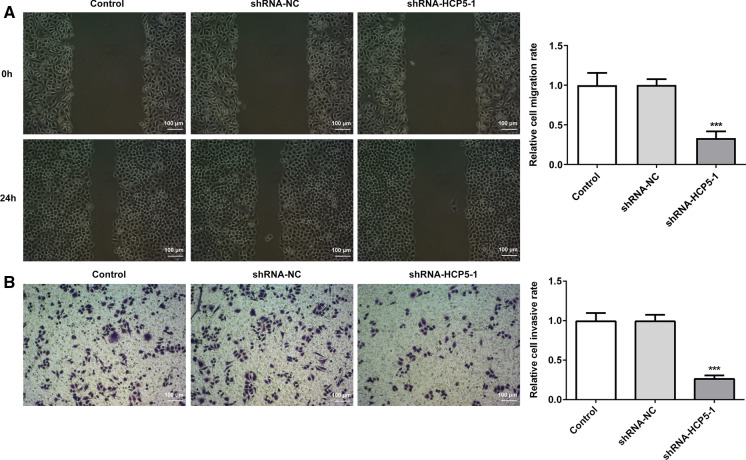
The effects of lncRNA HCP5 downregulation on the migration and invasion of MGC-803 cells were examined by wound healing assay and transwell assay, respectively. Both the migration ability (Fig. 3a) and the invasion ability (Fig. 3b) were significantly decreased in shRNA-HCP5-1 group than that in shRNA-NC group and control group, demonstrating that downregulation of lncRNA HCP5 exerted inhibitory effects on the invasion and migration of MGC-803 cells. To further confirm the authenticity of these findings, the levels of MMP7 and MMP13, which are key proteins related to migration and invasion (Gao et al. 2019; Li and Cao 2019; Liu et al. 2020; Wang et al. 2019a, b), were assessed herein (Fig. 4). MMP7 and MMP13 expression were remarkably decreased in shRNA-HCP5-1 group in comparison with the control group and shRNA-NC group, further confirming that the invasion and migration abilities of MGC-803 cells were obviously inhibited by downregulation of lncRNA HCP5 (Fig. 4).
Fig. 3.
The effects of lncRNA HCP5 downregulation on the invasion and migration of MGC-803 cells. a The migration capacity evaluated by wound healing assay; b The invasion capacity evaluated by transwell assay. ***P < 0.001 vs. shRNA-NC
Fig. 4.
The effects of lncRNA HCP5 downregulation on MMP7 and MMP13 expression. a The levels of MMP7 and MMP13 evaluated by western blot; b The levels of MMP7 and MMP13 evaluated by RT-qPCR. ***P < 0.001, **P < 0.01, *P < 0.05 vs. shRNA-NC
LncRNA HCP5 directly targeted DDX21 and downregulation of lncRNA HCP5 inhibited DDX21 expression in gastric cancer cells
As predicted by Starbase, lncRNA HCP5 directly targeted DDX21 (Fig. 5a). In order to further verify the binding relationship between DDX21 and lncRNA HCP5, RNA binding protein immunoprecipitation assay and dual luciferase reporter assay were performed. Compared with the control (IgG) group, lncRNA HCP5 level in DDX21 group was greatly elevated, confirming that lncRNA HCP5 could bind to DDX21 (Fig. 5b). Following transfection with pcDNA HCP5, lncRNA HCP5 overexpression was verified by RT-qPCR assay (Fig. 5c). Furthermore, the relative luciferase activity in DDX21-WT + pcDNA HCP 5 group was the lowest among all the groups and the relative luciferase activity in the remaining groups remained unchanged, further confirming that DDX21 was the target of lncRNA HCP5 (Fig. 5d). In addition, downregulation of lncRNA HCP5 suppressed DDX21 expression, indicating that lncRNA HCP5 positively regulated DDX21 expression (Fig. 5e). All these results demonstrated that DDX21 was the direct target of lncRNA HCP5.
Fig. 5.
Target gene prediction and verification a The target gene of lncRNA HCP5 predicted by Starbase; b The binding relationship between lncRNA HCP5 and DDX21 verified by RNA binding protein immunoprecipitation assay ***P < 0.001 vs. IgG; c lncRNA HCP5 expression in MGC-803 cells after transfection with pcDNA HCP5 ***P < 0.001 vs. pcDNA ; d The binding relationship between lncRNA HCP5 and DDX21 verified by dual luciferase reporter assay **P < 0.01 vs. DDX21-MUT; e DDX21 expression in MGC-803 cells after transfection with shRNA-HCP5-1 *P < 0.05 vs. shRNA-NC
Overexpression of lncRNA HCP5 promoted cell proliferation and downregulation of DDX21 rescued the pro-proliferation effect of lncRNA HCP5 in MGC-803 cells
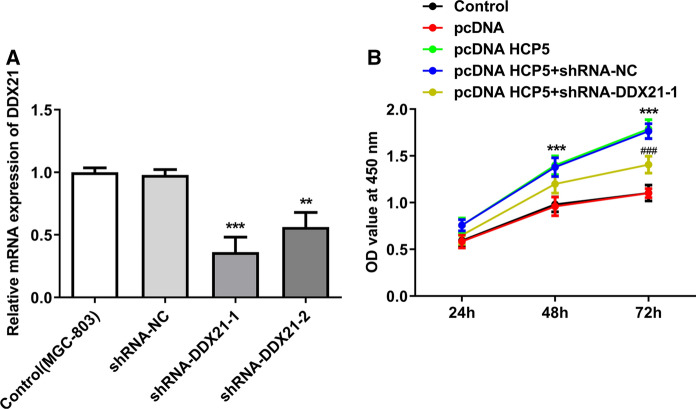
Following transfection with shRNA-DDX21-1 or shRNA-DDX21-2, DDX21 downregulation was verified by RT-qPCR assay. DDX21 level was more obviously downregulated by shRNA- DDX21-1 compared with shRNA- DDX21-2 (Fig. 6a). Hence, shRNA- DDX21-1 was selected to downregulate DDX21 expression. Following transfection with pcDNA HCP5, an obvious increase in cell proliferation was observed, demonstrating that overexpression of lncRNA HCP5 exerted pro-proliferation effect in MGC-803 cells. This pro-proliferation effect of lncRNA HCP5 overexpression was partly inhibited by downregulation of DDX21, indicating that the pro-proliferation effect of lncRNA HCP5 was realized via upregulation of DDX21 (Fig. 6b).
Fig. 6.
Downregulation of DDX21 rescued the pro-proliferation effect of lncRNA HCP5 overexpression in MGC-803 cells. a DDX21 levels in MGC-803 cells after transfection with shRNA-DDX21-1 or shRNA-DDX21-2 ***P < 0.001, **P < 0.01 vs. shRNA-NC; b Cell viability assessed by CCK-8 assay ***P < 0.001 vs. pcDNA, ###P < 0.001 vs. pcDNA HCP5 + shRNA-NC
LncRNA HCP5 overexpression promoted cell migration and invasion and these effects were inhibited by downregulation of DDX21 in MGC-803 cells
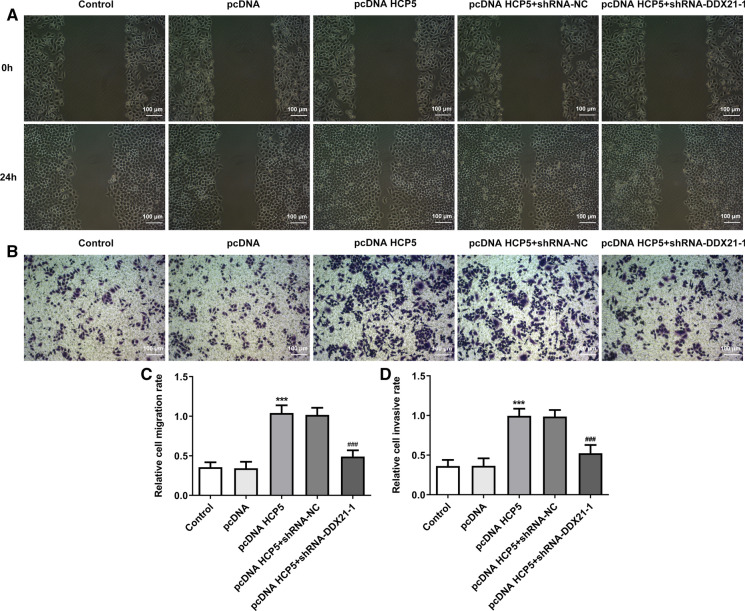
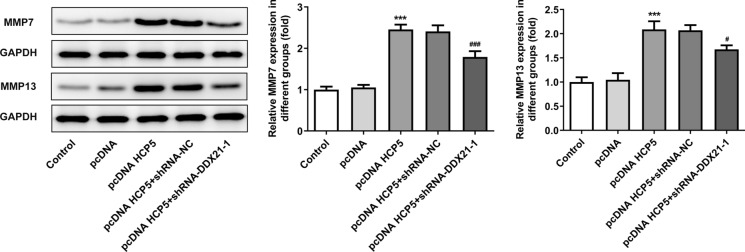
Overexpression of lncRNA HCP5 exerted promoting effects on migration and invasion in MGC-803 cells. Following transfection with shRNA- DDX21-1, the invasion and migration abilities of MGC-803 cells were greatly suppressed, confirming that the promoting effects of lncRNA HCP5 overexpression on migration and invasion were inhibited by downregulation of DDX21(Fig. 7a–d). MMP7 and MMP13 levels were examined as further evidence. Overexpression of lncRNA HCP5 upregulated MMP7 and MMP13 levels. Moreover, downregulation of DDX21 downregulated MMP7 and MMP13 levels and reversed the regulating effects of lncRNA HCP5 on MMP7 and MMP13 expression (Fig. 8). All the results are consistent and supported that the effects of lncRNA HCP5 overexpression was realized via up-regulation of DDX21.
Fig. 7.
Downregulation of DDX21 reversed the promoting effects of lncRNA HCP5 overexpression on the migration and invasion of MGC-803 cells. a, c The migration capacity evaluated by wound healing assay; b, d The invasion capacity evaluated by transwell assay. ***P < 0.001 vs. pcDNA, ###P < 0.001 vs. pcDNA HCP5 + shRNA-NC
Fig. 8.
Downregulation of DDX21 ablished the promoting effects of lncRNA HCP5 overexpression on MMP7 and MMP13 levels. The levels of MMP7 and MMP13 evaluated by western blot. ***P < 0.001 vs. pcDNA, #P < 0.05, ###P < 0.001 vs. pcDNA HCP5 + shRNA-NC
Discussion
As a progressive disease, gastric cancer has become one of the clinical problems that render us worrisome. Early diagnosis and timely treatment are extremely important for slowing lesion progression and prolonging lifetime. In the current research, we found that downregulation of lncRNA HCP5 inhibited the proliferation, migration and invasion of gastric cancer cells by modulation of DDX21 expression, indicating that lncRNA HCP5 may serve as an effective diagnosis biomarker and the therapeutic target.
In this study, lncRNA HCP5 level was greatly up-regulated in varieties of gastric cancer cells and it exhibited more significant up-regulation in MGC-803 cells in comparison with that in gastric epithelial cells, uncovering that lncRNA HCP5 may play a crucial role in gastric cancer. Increasing evidence has showed that aberrant expression of lncRNA may cause significant effects on the occurrence and development of several kinds of cancers. LINC01419 silencing was reported to exert protective effects against gastric cancer cell progression (Guo et al. 2014; Wang et al. 2019a, b). LINC00565 upregulation in gastric cancer was reported to contribute to gastric cancer progression via sponge of miR-665 (Hu et al. 2019). Aberrant expression of long non-coding RNA H19 in gastric cancer was confirmed to serve as a potential diagnostic biomarker in gastric cancer (Zhou et al. 2015).
In order to further explore the vital role of lncRNA HCP5 in gastric cancer, we investigated the precise effects of lncRNA HCP5 downregulation on proliferation in gastric cancer cells. Our data showed that the lncRNA HCP5 silencing remarkably inhibited the proliferation of MGC-803, demonstrating that downregulation of lncRNA HCP5 had anti-proliferation effects in gastric cancer. LncRNA HCP5 silencing may become a new strategy for GC therapy.
Increasing research has reported that the aberrant expression of lncRNAs has effects on cell migration and invasion in cancers. Overexpression of lncRNA MALAT1 contributed to the progression of gastric cancer by enhancing cell migration and invasion (Zhu et al. 2019). LncRNA GACAT1 has promoting effects on the invasion and migration of gastric cancer cells (Shi et al. 2018). LncRNA LUCAT1 was confirmed to enhance cell proliferation and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis (Chi et al. 2019). In the current study, migration and invasion were inhibited by downregulation of lncRNA HCP5. In addition, MMP7 and MMP13 were also down-regulated following lncRNA HCP5 downregulation. All these findings supported that downregulation of lncRNA HCP5 exerted anti-cancer effects by inhibiting cell proliferation, invasion and migration in gastric cancer.
As predicted by Starbase online tool, DDX21 was directly targeted by lncRNA HCP5. DDX21 was reported to be significantly up-regulated in gastric cancer in previous study (Cao et al. 2018). In this research, our data showed that lncRNA HCP5 was able to bind to DDX21 and lncRNA HCP5 silencing down-regulated DDX21 expression. We speculated that lncRNA HCP5 may promote the proliferation, migration and invasion of GC cells by binding DDX21.
Subsequently, we investigated the effects of lncRNA HCP5 overexpression and DDX21 silencing on gastric cancer. DDX21 had been confirmed to induce tumorigenesis by promoting AP-1 activity and rRNA processing (Zhang et al. 2014). Besides, DDX21 was reported to induce gastric cancer cell growth by promoting the protein levels of Cyclin D1 and CDK2 (Cao et al. 2018). Our data confirmed that the proliferation, migration and invasion of GC cells were all enhanced by lncRNA HCP5 overexpression and downregulation of lncRNA HCP5 presented opposite effects. The promoting effects of lncRNA HCP5 overexpression on GC cell proliferation, migration and invasion were distinctly reversed by DDX21 silencing, further confirming our speculation that lncRNA HCP5 acted on GC via up-regulating DDX21 expression. In other words, downregulation of lncRNA HCP5 inhibited GC cell proliferation, migration and invasion by silencing DDX21 expression.
Conclusions
In summary, our study was the first to investigate the role of lncRNA HCP5 in gastric cancer. The results indicated that lncRNA HCP5 is a novel potential oncogene in GC, and it is up-regulated in GC cells. LncRNA HCP5 can promote the proliferation, migration and invasion of GC cells. DDX21 is a target gene of lncRNA HCP5 and DDX21 mediates the function of lncRNA HCP5 in gastric cancer, implicating the potential application of lncRNA HCP5 in GC therapy.
Acknowledgements
Not applicable.
Abbreviations
- qRT-PCR
Quantitative real-time polymerase chain reaction
- DDX21
DEAD cassette helicase 21
- MMP7
matrix metalloproteinase-7
- MMP13
matrix metalloproteinase-13
Authors’ contributions
KW, XY, BT and JQ involved in performing the experiments and analyzed the data. KW and XY prepared the manuscript. The final manuscript has been read and approved by all the authors.
Funding
Not applicable.
Availability of data and materials
The data and materials are available when the request is reasonable.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kehao Wang and Xiaoyu Yu are co-first authors.
References
- Cao J, Wu N, Han Y, Hou Q, Zhao Y, Pan Y, Xie X, Chen F. DDX21 promotes gastric cancer proliferation by regulating cell cycle. Biochem Biophys Res Commun. 2018;505:1189–1194. doi: 10.1016/j.bbrc.2018.10.060. [DOI] [PubMed] [Google Scholar]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lei T, Chen X, Gu J, Huang J, Lu B, Wang Z. Long non-coding RNA in lung cancer. Clin Chim Acta. 2020;504:190–200. doi: 10.1016/j.cca.2019.11.031. [DOI] [PubMed] [Google Scholar]
- Chi J, Liu T, Shi C, Luo H, Wu Z, Xiong B, Liu S, Zeng Y. Long non-coding RNA LUCAT1 promotes proliferation and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis. Biomed Pharmacother. 2019;118:109201. doi: 10.1016/j.biopha.2019.109201. [DOI] [PubMed] [Google Scholar]
- Eichelberger L, Murphy G, Etemadi A, Abnet CC, Islami F, Shakeri R, Malekzadeh R, Dawsey SM. Risk of gastric cancer by water source: evidence from the Golestan case-control study. PLoS ONE. 2015;10:e0128491. doi: 10.1371/journal.pone.0128491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Kitago M, Aiura K, Shinoda M, Yagi H, Abe Y, Oshima G, Hori S, Nakano Y, Itano O, Fukada J, Masugi Y, Kitagawa Y. Efficacy and safety of preoperative 5-fluorouracil, cisplatin, and mitomycin C in combination with radiotherapy in patients with resectable and borderline resectable pancreatic cancer: a long-term follow-up study. World J Surg Oncol. 2019;17:145. doi: 10.1186/s12957-019-1687-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattahi S, Kosari-Monfared M, Golpour M, Emami Z, Ghasemiyan M, Nouri M, Akhavan-Niaki H. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: A novel approach to personalized medicine. J Cell Physiol. 2020;235:3189–3206. doi: 10.1002/jcp.29260. [DOI] [PubMed] [Google Scholar]
- Gao Y, Nan X, Shi X, Mu X, Liu B, Zhu H, Yao B, Liu X, Yang T, Hu Y, Liu S. SREBP1 promotes the invasion of colorectal cancer accompanied upregulation of MMP7 expression and NF-κB pathway activation. BMC Cancer. 2019;19:685. doi: 10.1186/s12885-019-5904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Goyal A, Myacheva K, Groß M, Klingenberg M, Duran Arqué B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45:e12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Xia J, Deng K. Long non-coding RNAs: emerging players in gastric cancer. Tumour Biol. 2014;35:10591–10600. doi: 10.1007/s13277-014-2548-y. [DOI] [PubMed] [Google Scholar]
- Hu J, Ni G, Mao L, Xue X, Zhang J, Wu W, Zhang S, Zhao H, Ding L, Wang L. LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis. Onco Targets Ther. 2019;12:7865–7875. doi: 10.2147/OTT.S189471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, Mo YY, Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, Seo J, Kim B, Jung Y, Kim WK, Chun HK, Lee WY, Kim J. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- Li XM, Cao LL. Identification of GOLM1 as a positively regulator of tumor metastasis by regulating MMP13 in gastric carcinoma. Cancer Biomark. 2019;26:421–430. doi: 10.3233/CBM-190301. [DOI] [PubMed] [Google Scholar]
- Lin X, Yang F, Qi X, Li Q, Wang D, Yi T, Yin R, Zhao X, Zhong X, Bian C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. 2019;58:2286–2296. doi: 10.1002/mc.23117. [DOI] [PubMed] [Google Scholar]
- Liu S, Huang M, Chen Z, Chen J, Chao Q, Yin X, Quan M. FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp Cell Res. 2020;389:111894. doi: 10.1016/j.yexcr.2020.111894. [DOI] [PubMed] [Google Scholar]
- Luzón-Toro B, Fernández RM, Martos-Martínez JM, Rubio-Manzanares-Dorado M, Antiñolo G, Borrego S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci Rep. 2019;9:14374. doi: 10.1038/s41598-019-50913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Li X, Xie L, Li S, Liu J, Jia L, Dong X, Ren X, Xiao J, Yang C, Zhou Y, Chen Z. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 2016;44:6423–6433. doi: 10.1093/nar/gkw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wang X, Hua Y. LncRNA GACAT1 Promotes Gastric Cancer Cell Growth, Invasion And Migration By Regulating MiR-149-mediated Of ZBTB2 And SP1. J Cancer. 2018;9:3715–3722. doi: 10.7150/jca.27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang P, Yu J, Zhang F, Dai W, Yi S. Matrix metalloproteinase 7 promoted Schwann cell migration and myelination after rat sciatic nerve injury. Mol Brain. 2019;12:101. doi: 10.1186/s13041-019-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol. 2019;11:1758835919874651. doi: 10.1177/1758835919874651. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wei X, Gu X, Ma M, Lou C. Long noncoding RNA HCP5 suppresses skin cutaneous melanoma development by regulating RARRES3 gene expression via sponging miR-12. Onco Targets Ther. 2019;12:6323–6335. doi: 10.2147/OTT.S195796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TS, Wang XF, Fairweather M, Sun YH, Mamon HJ, Wang JP. The survival benefit from the addition of radiation to chemotherapy in gastric cancer patients following surgical resection. Clin Oncol (R Coll Radiol) 2020;32:110–120. doi: 10.1016/j.clon.2019.09.047. [DOI] [PubMed] [Google Scholar]
- Yu S, Cai L, Lin F, Wu X, Zhang C, Liu X, Li W. Durable response after combination of concurrent chemoradiotherapy and anti-PD-1 therapy in HER2-negative advanced gastric adenocarcinoma: a case report. Onco Targets Ther. 2019;12:7691–7698. doi: 10.2147/OTT.S221436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Shen HM, Fang DM, Meng QJ, Xin YH. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22:4812–4819. doi: 10.26355/eurrev_201808_15616. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Baysac KC, Yee LF, Saporita AJ, Weber JD. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014;16:449. doi: 10.1186/s13058-014-0449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang Y, Chen C, Zhu X, Zhang C, Xia Y, Zhao Y, Andrisani OM, Kong L. A double-negative feedback loop between DEAD-box protein DDX21 and Snail regulates epithelial-mesenchymal transition and metastasis in breast cancer. Cancer Lett. 2018;437:67–78. doi: 10.1016/j.canlet.2018.08.021. [DOI] [PubMed] [Google Scholar]
- Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Ren Q, Zhao Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett. 2019;17:5335–5342. doi: 10.3892/ol.2019.10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials are available when the request is reasonable.