Abstract
Serrated lesions are the precursor lesions of a new model of colorectal carcinogenesis. From a molecular standpoint, the serrated pathway is thought to be responsible for up to 30% of all colorectal cancer cases. The three major processes of this molecular mechanism are alterations in the mitogen-activated protein kinase pathway, production of the CpG island methylation phenotype, and generation of microsatellite instability. Other contributing processes are activation of WNT, alterations in the regulation of tumor suppressor genes, and alterations in microRNAs or in MUC5AC hypomethylation. Although alterations in the serrated pathway also contribute, their precise roles remain obscure because of the various methodologies and definitions used by different research groups. This knowledge gap affects clinical assessment of precursor lesions for their carcinogenic risk. The present review describes the current literature reporting the molecular mechanisms underlying each type of serrated lesion and each phenotype of serrated pathway colorectal cancer, identifying those areas that merit additional research. We also propose a unified serrated carcinogenesis pathway combining molecular alterations and types of serrated lesions, which ends in different serrated pathway colorectal cancer phenotypes depending on the route followed. Finally, we describe some key issues that need to be addressed in order to incorporate the newest technologies in serrated pathway research and to improve overall knowledge for developing specific prevention strategies and new therapeutic targets.
Keywords: Colorectal, Carcinogenesis, Polyps, BRAF, Molecular biology
INTRODUCTION
Colorectal cancer (CRC) is the most prevalent cancer in Western countries and the second cause of cancer-related death, affecting both sexes equally. Furthermore, the number of new CRC cases worldwide is estimated to increase from the 1.4 million reported in 2012 to 2.4 million by the year 2035.1
The vision of a unique carcinogenesis model for CRC has completely changed in the last 25 years. Since the identification of serrated carcinomas by Jass and Smith in 1992,2 the underlying genetic and epigenetic alterations have been described. Although some of these may be shared among the other forms of CRC, specific combinations of the molecular defects will result in the different CRC phenotypes.
Until recently, three molecular pathways of CRC development have been established. The first is the classical adenoma-carcinoma sequence (Fearon and Vogelstein model),3,4 in which carcinomas usually develop chromosomal instability and are microsatellite stable (MSS); this pathway is responsible for 70% to 80% of CRC cases. The second is germline mutations in the mismatch repair (MMR) genes and development of microsatellite instability (MSI); this pathway is responsible for ~3% of CRC cases, and has its paradigm in Lynch syndrome, which is characterized by the inherited mutation in one of the MMR genes. The third is serrated lesions, no longer considered to be lacking malignant potential;5 this pathway is supposed to produce the precursors of the remaining 15% to 30% of all CRC cases through the so-called “serrated pathway.” The serrated pathway colorectal cancers (SPCRCs) are characterized by somatic BRAF mutation and DNA CpG island hypermethylation (CpG island hypermethylator phenotype or CIMP).6,7
In recent years, much research effort has focused on the serrated pathway of carcinogenesis, with the aim of elucidating the molecular evolution from the precursor lesion to CRC. However, those studies on the frequency of mutations in serrated lesions have provided disparate results, making description of the sequence of molecular events somewhat confusing. This situation also makes difficult to stablish a firm correlation between molecular alterations and histology, inducing several grey areas in our knowledge of the serrated pathway. In this review we summarize current knowledge of molecular events present in serrated lesions, describe their prevalence in the different histological subtypes including SPCRC, and identify methodological weaknesses of research. Finally, we propose a conceptual framework to explain the origin of SPCRC, pointing out future areas of research. As a result, many of the published reviews dealing with serrated lesions focus mainly on the endoscopic and histological characteristics of this CRC type, with less attention dedicated to the molecular alterations. In this review, we summarize the most relevant information from the literature on the molecular events underlying the serrated pathway, with the aim of helping to guide future directions in this research. The study protocol was approved by the Hospital La Fe Institutional Review Board (IRB number: 2016/0477).
DEFINING THE LESIONS: HISTOPATHOLOGY AND NOMENCLATURE
The nomenclature and pathological classification of serrated lesions have evolved over the last few years. Longacre and Fenoglio-Preiser8 first used the term “serrated adenoma” in 1990, reporting on its neoplastic nature. Six years later, Torlakovic and Snover9 coined the term sessile serrated adenoma, which is now also used as a synonym for sessile serrated polyp.10 Currently, serrated lesions are pathologically classified into different subtypes using the following World Health Organization (WHO) criteria: sessile serrated adenoma/polyp, with or without cytological dysplasia; traditional serrated adenoma (TSA); and hyperplastic polyp (HP). Additionally, HPs are subdivided into the following three pathological categories: microvesicular (MV)HP; goblet cell (GC)HP; and mucin-poor (MP)HP.11
However, there is lack of uniformity in histopathological terminology, especially for referring to the sessile serrated adenoma and the minimum features required to make a diagnosis. In terms of nomenclature, some authors defend using the word “adenoma” to remark its premalignant nature, but its use generates unnecessary misconceptions because it does not exhibit true cytological dysplasia. Others propose using the word “polyp” instead, but again it would be inaccurate since atypia and polypoid features are not always present. For this reason, in this paper we will use the term “sessile serrated lesion” (SSL) with or without dysplasia in lieu of sessile serrated adenoma/polyp, following the recently published British Society of Gastroenterology position statement.12 This society also recommends using the WHO criteria to diagnose an SSL, which require three crypts or two adjacent crypts showing at least one of the characteristic features: irregular distribution of crypts, dilatation of crypt bases, serration at crypt bases, branched crypts, horizontal extension of crypt bases, dysmaturation of crypts and herniation of crypts through the muscularis mucosa layer.
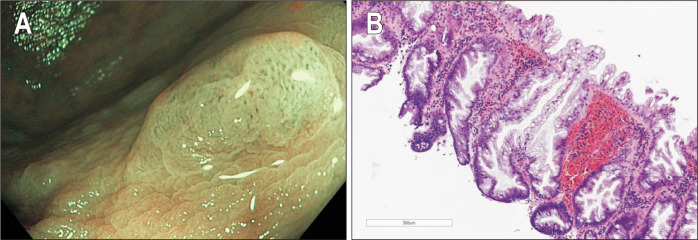
SSLs present distinct endoscopic appearance compared to adenomas or HPs, which includes indistinct borders, cloud-like surface, irregular shape and dark spots inside the crypts. Recognition of these features is key for detection and complete resection of these subtle lesions. Fig. 1 presents typical endoscopic and histologic features of an SSL.
Fig. 1.
(A) White light high-resolution endoscopic image of a sessile serrated lesion (SSL): dark spots inside the crypts, an irregular shape, indistinct borders and a cloud-like surface. (B) SSL showing typical serration (H&E, ×10). Crypts show pronounced basal dilation and horizontal spreading in an “L” shape.
WHAT ARE THE MAIN MOLECULAR MECHANISMS INVOLVED IN THE SERRATED PATHWAY TO CRC?
The serrated pathway combines three major molecular mechanisms, those being CIMP and MSI, both consequences of epigenetic DNA changes, and alteration of the mitogen-activated protein kinase (MAPK) pathway.
1. Aberrant promoter hypermethylation of CpG islands: CIMP
This concept was first proposed by Toyota and Issa13 in 1999 to describe a subset of CRC that exhibited widespread promoter hypermethylation, an alteration later pointed out as a carcinogenic mechanism within the serrated neoplasia pathway.
CpG islands (cytosine and guanine linked by only one phosphate) are DNA regions where cytosine-guanosine residues appear in a repetitive sequence that is at least 200 bp long and usually longer than 500 bp. In humans, promoters have been found to be embedded in CpG islands in about half of the genome.14 The promoter’s aberrant methylation in CpG islands, known as CIMP, represents one of the most relevant epigenetic mechanisms of tumor suppressor gene inactivation. Cytosines in CpG dinucleotides are transformed into 5-methylcytosine by a methyltransferase inducing a modification in the three-dimensional configuration of DNA, thus producing a defective interaction between transcription factors and the start codon. As a consequence, the involved tumor suppressor gene suffers transcriptional silencing resulting in changes of gene expression.15
In the setting of serrated pathway CRC, CIMP might be the most important oncogenic mechanism. The MLH1 gene and its promoter’s methylation is the most relevant and best characterized in this context, being found in 75% of cases.16 However, methylation can develop in other genes that contribute to the CIMP phenotype as well.
Categorizing the CIMP status is not straightforward. There is some controversy in the literature about the correct methylated loci combination for determining CIMP status in colorectal tumors; moreover, the published studies have used different CIMP panels and data interpretation methods, specifically when grading CIMP according to the number of methylated markers (CIMP-high vs CIMP-low). This makes it difficult to compare outcomes and to get an accurate estimation of the real prevalence of this epigenetic alteration. Therefore, a global standardization of molecular techniques and reporting seems imperative. Table 1 summarizes the most classically used combinations of methylation markers to determine CIMP status.13,17-19
Table 1.
Combinations of Methylation Markers Most Frequently Used to Determine CIMP Status
| Toyota and Issa13,17 | Weisenberger et al.18 | Ogino et al.19 | |
|---|---|---|---|
| Year of publication | 1999 | 2006 | 2007 |
| Remarks/novelties | First publication on this subject | 195 CpG islands screened throughout the human genome | Selection starting from the 195 loci initially explored by Weisenberger et al.18 |
| Type A (age-related): low level of methylation | New cutoff points and new classification | ||
| Type C (cancer-related): high level of methylation → called “CIMP tumors” | |||
| Surrogate markers for epigenetic aberrations | 7 Cancer-associated methylation markers: | Reduced to 5 loci: | Extended to 8: |
| MINT1, MINT2, MINT12, MINT17, MINT25, MINT27, and MINT31 | CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 | CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1, p16CDKN2A, CRABP1, and MLH1 | |
| But a panel with at least RUNX3, CACNA1G, IGF2, and MLH1 can serve as a sensitive and specific marker panel for CIMP-H | |||
| CIMP classification | CIMP-H ≥3 of 7 | CIMP-H ≥3 of 5 | CIMP-0: No methylated markers |
| CIMP-L: 1-5 | |||
| CIMP-H: 6-8 | |||
| Problems/weaknesses | It does not explore MLH1 | Lack of detailed classification of CIMP tumors because it does not explore MLH1 | Sensitive and specific markers for detecting CIMP-H rather than CIMP-L |
| Classic markers MINT1, MINT2 and MINT31 later proved to be nonspecific for BRAF-mutated CIMP tumors19 | Conflicting interpretation for 4-5 positive markers |
CIMP, CpG island methylator phenotype; H, high; L, low.
All of the published panels are invariably subsets of the CpG islands initially explored by Toyota and Issa,13 albeit with some differences, such as for MLH1. Some of the research groups have not routinely studied MLH1 methylation for the following two reasons. First, sporadic (not Lynch-related) cases of MMR deficiency and MSI appear almost exclusively as a consequence of MLH1 methylation in a CIMP-H situation. Second, MLH1 methylation is very rare in MSS tumors, indicating that in this case CIMP is determined by other methylation markers.18,20 On the contrary, other research groups have included MLH1 in order to discriminate CIMP-H among CIMP-positive CRCs.19,21
Most of the research groups have used a commercially available 5- or 8-marker polimerase chain reaction (PCR) assay to determine the presence of promoter hypermethylation. These panels are well designed for classifying CIMP-high (CIMP-H) but not CIMP-low (CIMP-L), and a specific panel to diagnose CIMP-L is not available yet. From our perspective, the 8-marker panel is currently the most useful for research purposes. However, the interpretation of methylation of 4 and 5 markers is still controversial, being better classified into the CIMP-H group than into the CIMP-L according to some researchers.22,23 Current research is focused on expanding the number of markers in an attempt to characterize more accurately the CIMP status of colorectal tumors, but results are not still standardized.24 Genome-wide methylation profiling could be a potential alternative for the extensive exploration of this alteration and to find some similarities with other tumor types.25
2. Microsatellite instability
Microsatellite sequences are short, simple, repetitive tracts of DNA, usually 1 to 6 bp long (although the C and A dinucleotide repeat is the most common in humans). As they are unique and uniform in length in every person, they can be considered as a “DNA fingerprint.” MSI is a hypermutable phenotype caused by the loss of DNA MMR activity, which produces an increased susceptibility to accumulation of microsatellite mutations. Proteins in MMR serve to correct polymerase errors that spontaneously occur during DNA replication (i.e., single base mismatches or short insertions and deletions).26
The MSI mechanism is not specific for the serrated course27 and consequently it would not be helpful as diagnostic filter. In fact, it was first described in relation to Lynch syndrome, where germline mutations take place in specific MMR genes, such as MLH1, MSH2, MSH6, and PMS2. But Lynch syndrome represents only 3% out of the global 15% of CRC cases that exhibit MSI. The remaining 12% involve the serrated pathway, where the DNA MMR alteration is due to epigenetic silencing by sporadic, acquired hypermethylation of the MLH1 gene promoter.28 In this pathway, the MSI occurs at later stages of the carcinogenetic sequence and represents a key point in it, as demonstrated by the association of MSI with the rapid development of dysplasia and malignancy.29
To determine MSI status, five microsatellite markers are used; this is called the Bethesda panel and consists of 2 mononucleotide (BAT 25 and BAT 26) and 3 dinucleotide (D2S123, D5S346, and D17S250) markers. When 2 or more of the markers show instability, the tumor is classified as MSI-high. When only 1 marker is affected, the tumor is classified as MSI-low. If no instability is found for any of the markers, the tumor is considered MSS.30
3. Alterations in MAPK signaling pathway genes: BRAF and KRAS mutations
MAPK pathways modulate extracellular signals from the cell surface to the nucleus to control growth, proliferation, differentiation, migration, and apoptosis. The RAS and RAF families are among the constituent proteins of this RAS-RAF-MEK-MAPK pathway. Mutations in these two oncogenes are frequent in many types of cancers and their activation is known to lead to increased cell proliferation and reduced apoptosis.31 Moreover, BRAF and KRAS activating mutations have been found to be mutually exclusive in both CRC32 and the precursor lesions.6,33-35
1) BRAF
In mammals, the RAF kinase family consists of three proteins, those being A-RAF, B-RAF, and C-RAF. In 2006, O’Brien et al.7 reported a high frequency of BRAF mutation in SPCRC and in its precursor lesions. However, this mutation was absent in all of the histologic categories of the traditional adenoma-carcinoma sequence, thus constituting a hallmark of the serrated pathway. This specificity allows exclusion of Lynch syndrome when loss of expression of MLH1 is present.6,7 Within the serrated pathway, BRAF mutation is an early instigating event.36 Indeed, Fang et al.37 have shown BRAF to be directly responsible for CIMP development in colorectal cell lines.
Classically, BRAF mutation has been investigated via molecular methods (e.g., PCR assay, Sanger sequencing, pyrosequencing, etc); however, this might change in the near future, according to the recently developed BRAFV600E mutation-specific antibody. Specifically, published studies on the determination of BRAF status that have used immunohistochemistry (IHC) have yielded results that are comparable to those obtained from DNA analysis; the overall sensitivity and specificity rates for this technique are 100% for BRAFV600E detection.38-40 Its presence is, to date, the most useful marker for demonstrating a serrated origin.
2) KRAS
There are at least three RAS genes, those being H-RAS, N-RAS, and K-RAS.41 KRAS mutation has been found in 30% to 40% of all CRC cases, mainly affecting codons 12 and 13.42 KRAS mutation is known to be related to the classical carcinogenetic pathway, along with chromosomal instability;4 it has been found in serrated cancers as well.43 So, unlike BRAF mutation, KRAS mutation is not pathway-specific. In this context, KRAS mutation harbors intermediate or low methylation status, as seen in MSS tumors.44 Pai et al.34 showed that serrated lesions located specifically in the appendix harbored KRAS mutations in codons 12 and 13 in 51% of cases, suggesting that the serrated pathway could be slightly different at this location.
Similar to BRAF, molecular techniques also represent the gold standard for KRAS mutation detection. In this case, IHC has been evaluated as a diagnostic tool for detection of a mutation in KRAS but the available antibodies have not shown good sensitivity or specificity,45 and currently IHC cannot be recommended as an alternative diagnostic method.
4. Other mechanisms involved
Apart from the well-known alterations that have been already addressed, other mechanisms have been described as part of the serrated cascade, although their role is more controversial. Here we summarize the alterations most extensively studied, which have been proposed as having some role in serrated pathway progression or that could serve as biomarkers.
1) WNT activation
This alteration has been demonstrated in 93% of all CRC cases,46 with the IHC-based detection method generally using β-catenin nuclear staining as a surrogate marker. In conventional adenomas, WNT is activated in early phases through APC mutation. On the contrary, nuclear β-catenin reactivity has been observed to be overwhelmingly more prevalent in dysplastic SSL than in nondysplastic SSL, suggesting that in the serrated pathway WNT activation might have some role during progression rather than in initiation. Since APC mutation is less common in the serrated pathway,47 the WNT activation should occur through: mutation of other genes, such as RNF43 or ZNRF3,48 and DNA methylation changes of the upstream WNT antagonists, including the secreted frizzled-related proteins, the mutated in CRC protein, and AXIN2.16,49
RNF43 is a tumor suppressor gene that down-regulates the WNT/β-catenin signaling pathway and has been reported to be mutated in different cancers. This mutation implies loss of function and therefore WNT activation. RNF43 mutation is not found in the traditional adenoma-carcinoma sequence and is less prevalent in Lynch-CRC than in SPCRC.50 Hashimoto et al.29 described RNF43 mutation as the most prevalent WNT gene mutation in SSLs with cytological dysplasia (SSL-CD), having different mutation profiles according to the MLH1 expression status. However, Tsai et al.51 showed a higher prevalence of RNF43 mutation in TSA (28%) and BRAF-mutated MSS CRC (29%) compared to that in SSL (10%). Specifically, in TSA (with or without dysplasia), RNF43 mutation was shown to be tightly associated with mutation in BRAF but inversely with KRAS.
2) Altered regulation of the tumor suppressor gene p16(ink4a) and TP53
Following activation of MAPK (BRAF and KRAS), proliferation at the aberrant crypt foci is induced, but then a mechanism driven by the p16 tumor suppressor gene (CDKN2A) halts cell proliferation and induces cell senescence.52 According to the results presented by Kriegl et al.,53 p16 appears to be up-regulated in the first phases of the serrated cascade, leading to an increased secretion of insulin-like growth factor binding protein 7 (IGFBP7) in the aberrant crypt foci. However, in an investigation of the CDKN2A promoter, as the serrated lesion progressed towards malignancy, a CIMP-induced aberrant methylation was found to facilitate overcoming the p16 senescence barrier. This alteration has also been demonstrated (by IHC) to be present in BRAF-mutated SSL-CD or carcinoma, showing a loss of expression of the p16 protein. p16 silencing has also been demonstrated in BRAF mutant TSA (but not in those presenting KRAS mutation).54 Furthermore, IGFBP7 methylation has been reported as strongly associated with BRAF mutation and the presence of CIMP.55
Mutation of the TP53 tumor suppressor gene can be evaluated by IHC detection of its product, the p53 protein. It is classically described in the classic adenoma-carcinoma model but it can also be mutated in CIMP tumors or its function can be lost due to epigenetic silencing (methylation of IGFBP7). BRAF-mutated MSS-CRC has been reported to have a significantly higher rate of TP53 mutation then the MSI-CRC (40.6% vs 17.0%).56 This finding was later confirmed by an Australian group, who suggested a possible association of this mutation with the poor prognosis of these cancers.16
3) MicroRNAs
These small molecules are known to be involved in the progression towards malignancy, although their specific role in the different stages of the serrated pathway is still to be elucidated. They might play an important role in the development of SSL, because miR-31 expression has been shown to be associated with CIMP status in serrated lesions with BRAF mutation.57 To date several studies have found a unique miRNA signature in different serrated precursors. Some authors have found that HPs and SSLs profile of miRNAs appears to be down-regulated from normal colonic mucosa.58 Another group assessed miRNA patterns in 109 colon screening biopsies and identified five different histopathologic groups (including normal mucosa, SSL or adenomas). The expression of specific miRNAs allowed to discriminate serrated polyp histology from others. Also, some of them were identified as potential biomarkers of the serrated pathway development (i.e., miR-335, miR-222, and miR-214).59 More recently, Kanth et al.60 performed small RNA sequencing in 108 different colon samples to characterize miRNA profiles and found specific miRNAs differentially expressed in serrated lesions compared to normal colon and HPs.
4) MUC5AC hypomethylation
MUC5AC hypomethylation has been described as an early event in this pathway. Recently, Renaud et al.61 showed that its presence, as detected by IHC, might represent a novel marker for detecting serrated precursor lesions with BRAF mutation, CIMP-H, or MSI. The positive predictive value was 100% and the negative predictive value was 81.4% for the identification of serrated lesions with malignant potential. In this sense, it might also be useful in discrimination of goblet cell hyperplastic polyp (GCHP) from microvesicular hyperplastic polyp (MVHP) and SSL.
WHAT IS THE DISTRIBUTION OF THE MAIN MOLECULAR ALTERATIONS IN THE DIFFERENT TYPES OF PRECURSOR SERRATED LESIONS?
There is a great heterogeneity among the published studies on the prevalence of all these molecular alterations in serrated lesions. Indeed, it is usual to find contradictory information or even large gaps in the knowledge. Both of these features ultimately complicate interpretation of the available data.
1. Prevalence of molecular alterations in serrated precursor lesions
HPs represent 80% to 90% of all serrated lesions, and among them MVHP is the most frequent subtype. It is generally agreed that MVHPs have a high frequency of BRAF mutation, while KRAS mutation alone is much rarer.7,35,62,63 However, the described prevalence of the CIMP-H phenotype varies substantially (Table 2).7,11,35,62-65 Some authors have reported on the existence of the CIMP phenotype only, without differentiating CIMP-L/H; this represents a major obstacle against standardizing results. Regarding GCHP, KRAS mutation is the most important genetic marker, but again, a great variability in prevalence has been reported (8% to 73%).7,35,62,64 A significant proportion of BRAF mutation has been described in these polyps as well, usually around 20%, but in some studies it is even higher than that of KRAS mutation.62,65 Reports on the prevalence of CIMP indicate a much lower rate than that described for MVHP, accounting for 0% to 18%.35,62,66
Table 2.
Frequencies of Molecular Alterations Depending on Lesion Subtype
| MVHP | GCHP | MPHP | SSL | SSL-CD | TSA | SPCRC | |
|---|---|---|---|---|---|---|---|
| Prevalence, % | 45–94* | 6–55* | 0–9* | 10–25† | Rare | 1–5† | 15–30‡ |
| Location | Variable | Distal | Distal | Proximal | Proximal | Distal | Distal |
| BRAF mutant, % | 67–88 | 10–83 | 25 | 61–83 | 93 | 67 | 82 |
| KRAS mutant, % | 6–22 | 8–73 | 25 | 0–25 | 1 | 22 | 0–45 |
| CIMP-positive, % | 10–73 | 0–18 | 75 (CIMP-H) | 44–77 | 93 | 43–80 | 90 (CIMP-H) |
| MSI-H, % | 0 | 0 | 0 | - | - | - | 82 |
MVHP, microvesicular hyperplastic polyp; GCHP, goblet cell hyperplastic polyp; MPHP, mucin-poor hyperplastic polyp; SSL, sessile serrated lesion; SSL-CD, SSL with cytological dysplasia; TSA, traditional serrated adenoma; SPCRC, serrated pathway colorectal cancer; CIMP, CpG island methylator phenotype; H, high; MSI, microsatellite instability.
Only one study to date has evaluated the prevalence of all these molecular alterations in MPHP, itself the least prevalent subtype of HP. Those researchers found an identical percentage of BRAF and KRAS mutations (25%), and expression of the CIMP-H in 75% of cases. However, their results must be interpreted with caution, since only four polyps in total were evaluated.35
Regarding SSL, BRAF mutation and the presence of very high levels of CIMP are the hallmarks of these lesions.7,35,62-65 Conversely, KRAS mutation is unusual and some authors have reported data suggesting that KRAS-mutated SSL frequently corresponds to a misclassified large GCHP. Others think that expression of higher levels in SSL and frequency of CIMP could be features of these lesions that distinguish them from HPs, their potential precursors. In this sense, Burnett-Hartman et al.66 described CIMP-H in approximately 50% of SSLs, but in only 10% of MVHP and none in GCHP.
The genetic alterations that have been described for the TSAs are very heterogeneous, and its ability (and mechanism) to progress to cancer remains to be clarified. The prevalence of CIMP and BRAF and KRAS mutations has varied widely among the published series. The largest one included 200 lesions and showed a high BRAF mutation prevalence, of around 67%, mainly involving proximally located lesions; meanwhile, mutated KRAS was present in only 22% of the lesions, with a distal preponderance. In that same paper, the investigators reported 16% of KRAS-mutant TSAs to be CIMP-H and 44% to be CIMP-L.54 However, other authors have shown a prevalence of CIMP in 43%62 and 80%17 of cases.
MSI-H status is almost invariably not identified in precursor serrated lesions,7,62,67 suggesting that this is a late event in the molecular evolution of serrated lesions.
One limitation of many of the studies on the prevalence of genetic alterations is that information about size of the included lesions is not provided. Size could be an important confounding factor, as large serrated lesions are more prone to show higher prevalence of all these molecular alterations and to develop malignancy.66,68
2. Prevalence of molecular alterations in SSL-CD
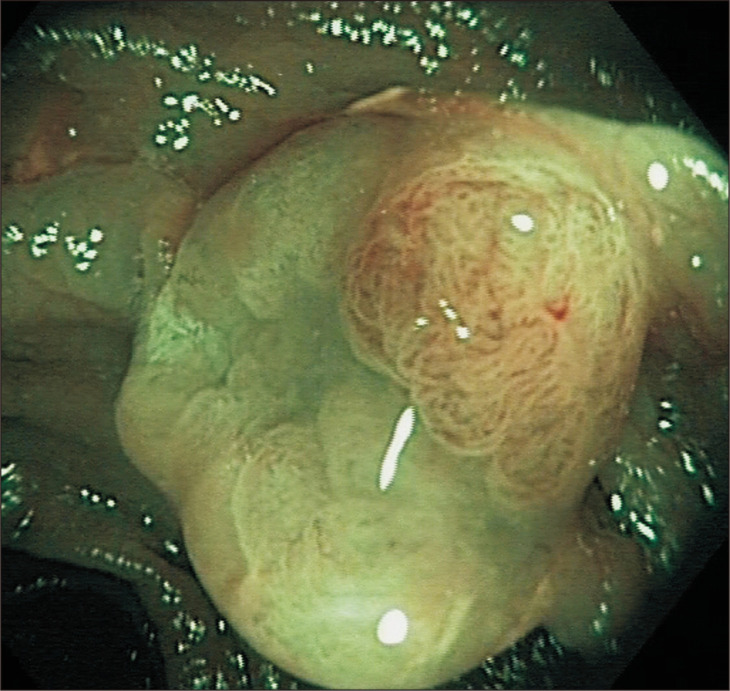
Because of its low prevalence, the molecular characterization of SSL-CD has been less explored. Bettington et al.16 conducted the first large prospective study relating clinicopathological and molecular features of SSLs with a focus of dysplasia or carcinoma. Of 137 cases, 93% had the BRAFV600E mutation and also 93% expressed CIMP-H. On the other hand, KRAS mutation appeared only in 1% of cases. The investigators had found 74.5% of SSLs to have MLH1 methylation as they developed dysplasia; the remaining quarter of SSLs was supposed to pass through other possible molecular pathways in their progression towards malignancy. In this context, SSL-CDs were observed in patients who were older by 17 years (mean age) than those with SSL without dysplasia, and there was no substantial difference in polyp size between the two groups. In fact, the SSL-CDs were generally smaller than 10 mm. Similar observations have been reported by Hashimoto et al.,29 suggesting that the evolution from HP or SSL to SSL-CD may take a long time, but once the lesion has acquired MLH1 methylation and dysplasia is present, the development of MSI and progression towards SPCRC occurs very rapidly. This accelerated progression to malignancy makes a reliable detection, characterization and complete resection of these lesions a main objective in the prevention of CRC. In the endoscopic examination, the dysplastic area may become more apparent than the rest of the lesion (Fig. 2) increasing the chances of incompletely removing and leaving behind a part of the SSL-CD, which could be directly responsible for an unplanned interval cancer.
Fig. 2.
Endoscopic appearance of a sessile serrated lesion with cytological dysplasia. The nodular element (containing dysplasia) is more apparent than the rest of the lesion, increasing the likelihood of incomplete resection of the whole lesion.
HOW ARE THESE MOLECULAR ALTERATIONS EXPRESSED IN THE SPCRC?
The main problem regarding SPCRCs is that, although up to 30% of all CRCs are thought to come from serrated precursors, only about 10% of them have conserved a true serrated morphology at diagnosis.27,69 As a consequence, the proportion of all CRCs that originate through the serrated pathway is still unknown. The term “serrated carcinoma” is actually a catch-all term used in the literature to refer to two different situations: (1) tumors arising via the serrated pathway; (2) tumors that show a serrated morphology on pathological diagnosis-although it is not always known if they actually originated in a serrated precursor. Rhee et al.70 suggested that a CRC could be considered as an SPCRC in circumstances in which “cancers are in direct contiguity with serrated adenomas, cancers are CIMP-H, or cancers are serrated adenocarcinomas.”
This uncertainty in definitions shows that sole reliance on morphological criteria is not sufficient to identify and classify SPCRCs. An accurate SPCRC classification can only be obtained through a comprehensive description of the correlation between molecular markers and clinical phenotype. In this sense, different molecular tracks within the serrated pathway have been postulated, which would ultimately result in different subsets of SPCRCs (Table 3).70,71
Table 3.
Subsets, Molecular Features and Characteristics of Serrated Pathway CRC
| Contribution to CRC70,71 | MAPK mutation | CIMP status | Microsatellites | Pathway-phenotype | Proposed precursor |
|---|---|---|---|---|---|
| 9%–12% | BRAF mutation | High | Instable | Serrated pathway | SSL |
| MLH1 methylation | Proximal location | ||||
| Female sex | |||||
| Proximal location | |||||
| 6%–8% | BRAF mutation | High | Stable | Serrated pathway | SSL |
| No MLH1 methylation | Older age | ||||
| Female sex | |||||
| Proximal location | |||||
| p16 and p53 silencing | |||||
| WNT pathway | |||||
| 15%–20% | KRAS mutation | Low | Stable | Challenging group: different pathways | TSA/adenomas |
| MGMT methylation | CIN | ||||
| Other TSG silenced | Male sex | ||||
| Distal location |
CRC, colorectal cancer; MAPK, mitogen-activated protein kinase; CIMP, CpG island methylator phenotype; SSL, sessile serrated lesion; TSG, tumor suppressor gene; TSA, traditional serrated adenoma; CIN, chromosomal instability.
As we previously mentioned, BRAF mutation is a specific marker for a serrated pathway, originating from a hyperplastic polyp (MVHP) and ending up in a CIMP-H, MSI carcinoma.7 The BRAFV600E mutation would be related to a CIMP-H status, and when the MLH1 promoter becomes methylated, an MSI carcinoma would be generated. However, if methylation affects other genes, the tumor will be an MSS.6,7,20,32 Therefore, most sporadic MSI CRCs will exhibit a CIMP phenotype, but on the contrary not all CIMP tumors will express MLH1 methylation. The CIMP-H phenotype has shown strong association with female sex, proximal location, and older age20,23 and its development has been specifically linked to the serrated pathway.72
Approximately 70% of CRCs containing activated KRAS express the CIMP phenotype, and therefore many tumor suppressor genes, such as p14(ARF), p15(INK4B) and p16(INK4A), are silenced through methylation.17,73 For instance, MGMT silencing through promoter hypermethylation strongly correlates with KRAS mutation. However, the arising tumors are CIMP-L74,75 and predominantly MSS.71 The attempts to relate CIMP-L tumors with patient characteristics have been less successful. However, a possible association of CIMP-L tumors with KRAS mutation and male sex has been described.74
CAN WE BUILD A MOLECULAR FRAMEWORK TO EXPLAIN THE EVOLUTION FROM PRECURSOR LESIONS TO CRC THROUGH THE SERRATED PATHWAY?
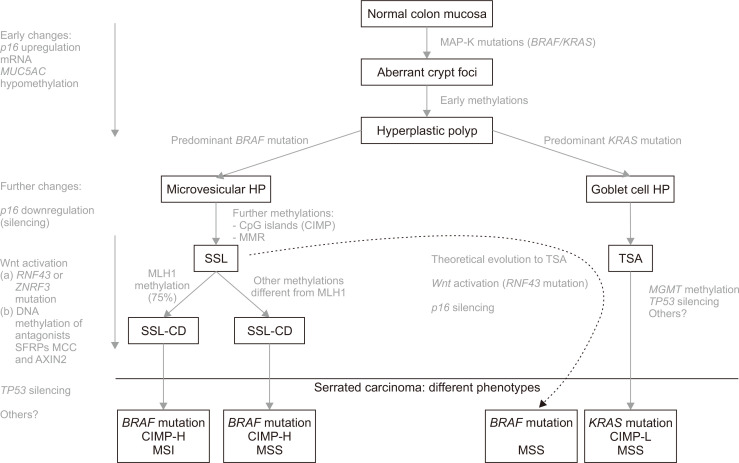
Given the diverse rate of reported prevalence for the described molecular alterations and the distinct results obtained by different research groups, providing a conceptual framework could at best be considered speculative at the moment (Fig. 3). The initiating event in the neoplastic cascade seems to be expression of either mutated BRAF or KRAS in the normal mucosa of the colon, leading to an excessive cell proliferation through altered MAPK signaling. This change can induce the emergence of aberrant hyperplastic crypt foci as the first histological lesion76 and, along with some early methylations, the formation of an HP. Depending on which MAPK mutation appears to be predominant in HPs, it has been postulated that they could be precursor lesions of the other two subtypes of serrated lesions through two different molecular tracks. Although these are normally treated as separate models, they can sometimes overlap.
Fig. 3.
Schematic view of serrated pathway progression.
mRNA, microRNA; HP, hyperplastic polyp; CIMP, CpG island hypermethylator phenotype; MMR, mismatch repair system; SSL, sessile serrated lesion; SSL-CD, SSL with cytological dysplasia; CIMP-H, CIMP-high; MSI, microsatellite instability; MSS, microsatellite stability; TSA, traditional serrated adenoma; CIMP-L, CIMP-low.
1. BRAF mutation predominant
In this context, an MVHP could be originated and then evolve into SSL after acquisition of the CIMP phenotype and further accumulation of aberrantly methylated, and thus silenced, genes. This theory is strengthened by the fact that similar frequencies of BRAF mutation have been reported for MVHPs, SSLs and CRCs, suggesting that this mutation is an early event and remains throughout the carcinogenetic pathway. The concept of BRAF mutation as an early event is also supported by a recent report from Bettington et al.72 Those investigators found that, although around 10% of all CRCs harbored BRAF mutation,6,77 the proportion of BRAF-mutated CRCs differed according to age, with less than 40% occurring before the age of 50 years but 40% occurring in patients of age greater than 80 years. Because the precursor lesion (SSL) was present in similar frequencies among all age groups, the authors concluded that SSL likely develops at a young age but may remain in an early phase for years before the accumulation of other genetic alterations, followed by development of dysplasia and cancer. The CIMP phenotype was also reported as determinative and highly correlated with BRAF mutation, both alterations being genuine markers of this pathway. In general, SSL is also believed to be able to arise de novo.
Later on, the key step towards evolution of SSL to SSL-CD (the aberrant hypermethylation of MLH1) occurs and, through this, the accumulation of mutations in microsatellites ultimately results in MSI. Evidence has shown that, after a prolonged stable phase as an SSL and the appearance of dysplasia, an MSI-SPCRC can arise very rapidly.16 The fact that MSI is absent in precursor lesions supports the theory that this alteration may be a final step in the progression of the serrated pathway. But, in some other cases, methylation in SSL-CDs may occur in genes other than MLH1 (itself accounting for 25%), and the arising SPCRC will be an MSS instead.
2. KRAS mutation predominant
In the context, the GCHP may evolve into a TSA, the latter of which is thought to be a precursor of MSS SPCRC with evolution through MGMT methylation. But, it has also been proposed that a non-MLH1 mutating SSL could progress to a TSA and ultimately develop into a BRAF-mutated MSS tumor (Fig. 3, dashed line).16,54 The question as to whether TSAs with KRAS mutation should be considered similar in terms of CRC risk to those TSAs with BRAF mutation needs further investigation.
CONCLUSION AND FUTURE CHALLENGES
Serrated lesions constitute a heterogeneous group of lesions that can potentially be the origin of a broad spectrum of CRC phenotypes. The various molecular profiles must be interpreted as a combination of genetic and epigenetic alterations. Many years have passed since the identification and description of the serrated pathway and, despite of a huge research effort, grey areas and gaps in the pathway knowledge remain. The main consequence of this situation is that the real malignant potential of each serrated lesions is largely unknown, hampering the strength of recommendations of surveillance guidelines which are mainly based in expert opinions.
Identifying and classifying SPCRCs is key and also understanding the route the serrated lesions follow towards malignancy. But to this purpose histological analysis has proven to be insufficient, and its combination with molecular markers seems to be the best approach. However, as we have shown current research methods do not seem to be accurate enough to fulfill these objectives. In the near future, traditional Sanger sequencing and panels of specific markers will probably be abandoned, with the field moving to a genome-wide approach with next-generation sequencing. This would allow for the identification of new biomarkers and combinations that could boost our knowledge of the serrated pathway. Nowadays, we can easily obtain sequence data on the whole genome, specific exomes, the methylome, and the transcriptome. The ability to have such vast genomic information, obtained at a very fast pace and in a cost-effective manner, opens a wide range of new possibilities for this field.
Recently, Lannagan et al.78 went a step further and generated novel preclinical models of SPCRC by combining next-generation sequencing, organoid culture, gene editing and orthotopic transplantation techniques. They first cultured organoids from colonic cells that were then transplanted into mice. Then, using CRISPR/cas9 genome engineering, they modelled DNA alterations that have been found in SPCRC. The resultant tumor behaved like an SPCRC (clinically, histologically, and genetically). This preclinical model is expected to contribute to the overall understanding of SPCRC biology and open new avenues for testing of potential treatments.
However, before we adopt these novelties with enthusiasm, a deep reflection is needed. As has been shown, interpretation of results from the current molecular techniques is challenging, and this situation could be worsened by the incorporation of more potent technologies. To avoid the previous mistakes, the molecular methods and their results should be reported in a standardized manner in order to be comparable across studies and groups.
An accurate identification and classification of serrated lesions and SPCRCs would also allow for the incorporation of epidemiological and endoscopic features to define risk profiles that will be applicable in clinical practice. In this sense, the combination of epidemiological risk factors, number and histological type of serrated lesions and specific molecular profiles could allow to individualize therapeutic and preventive strategies, constituting one of the best examples of precision medicine. Specific prevention strategies and potential individualized therapeutic targets would emerge as the necessary second step, giving rise to a completely new scenario of individualized treatments and precision medicine.
ACKNOWLEDGEMENTS
This study was supported by a grant from the Spanish Society of Gastrointestinal Endoscopy.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jass JR, Smith M. Sialic acid and epithelial differentiation in colorectal polyps and cancer: a morphological, mucin and lectin histochemical study. Pathology. 1992;24:233–242. doi: 10.3109/00313029209068874. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–150. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 6.Kambara T, Simms LA, Whitehall VL, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 8.Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas: a distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14:524–537. doi: 10.1097/00000478-199006000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Torlakovic E, Snover DC. Serrated adenomatous polyposis in humans. Gastroenterology. 1996;110:748–755. doi: 10.1053/gast.1996.v110.pm8608884. [DOI] [PubMed] [Google Scholar]
- 10.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. Volume 3. 4th ed. International Agency for Research on Cancer; Lyon: 2010. p. 417. [Google Scholar]
- 12.East JE, Atkin WS, Bateman AC, et al. British Society of Gastroenterology position statement on serrated polyps in the colon and rectum. Gut. 2017;66:1181–1196. doi: 10.1136/gutjnl-2017-314005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 16.Bettington M, Walker N, Rosty C, et al. Clinicopathological and molecular features of sessile serrated adenomas with dysplasia or carcinoma. Gut. 2017;66:97–106. doi: 10.1136/gutjnl-2015-310456. [DOI] [PubMed] [Google Scholar]
- 17.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Bae JM, Kim JH, Kwak Y, et al. Distinct clinical outcomes of two CIMP-positive colorectal cancer subtypes based on a revised CIMP classification system. Br J Cancer. 2017;116:1012–1020. doi: 10.1038/bjc.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae JM, Rhee YY, Kim KJ, et al. Are clinicopathological features of colorectal cancers with methylation in half of CpG island methylator phenotype panel markers different from those of CpG island methylator phenotype-high colorectal cancers? Hum Pathol. 2016;47:85–94. doi: 10.1016/j.humpath.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka N, Huttenhower C, Nosho K, et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. Am J Pathol. 2010;177:2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez-Vega F, Gotea V, Chen YC, Elnitski L. CpG island methylator phenotype in adenocarcinomas from the digestive tract: methods, conclusions, and controversies. World J Gastrointest Oncol. 2017;9:105–120. doi: 10.4251/wjgo.v9.i3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 27.Tuppurainen K, Mäkinen JM, Junttila O, et al. Morphology and microsatellite instability in sporadic serrated and non-serrated colorectal cancer. J Pathol. 2005;207:285–294. doi: 10.1002/path.1850. [DOI] [PubMed] [Google Scholar]
- 28.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto T, Yamashita S, Yoshida H, et al. WNT pathway gene mutations are associated with the presence of dysplasia in colorectal sessile serrated adenoma/polyps. Am J Surg Pathol. 2017;41:1188–1197. doi: 10.1097/PAS.0000000000000877. [DOI] [PubMed] [Google Scholar]
- 30.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 33.Sakai E, Fukuyo M, Ohata K, et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int J Cancer. 2016;138:1634–1644. doi: 10.1002/ijc.29903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai RK, Hartman DJ, Gonzalo DH, et al. Serrated lesions of the appendix frequently harbor KRAS mutations and not BRAF mutations indicating a distinctly different serrated neoplastic pathway in the appendix. Hum Pathol. 2014;45:227–235. doi: 10.1016/j.humpath.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 35.Kim KM, Lee EJ, Ha S, et al. Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol. 2011;35:1274–1286. doi: 10.1097/PAS.0b013e318224cd2e. [DOI] [PubMed] [Google Scholar]
- 36.Beach R, Chan AO, Wu TT, et al. BRAF mutations in aberrant crypt foci and hyperplastic polyposis. Am J Pathol. 2005;166:1069–1075. doi: 10.1016/S0002-9440(10)62327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 39.Dinges HC, Capper D, Ritz O, et al. Validation of a manual protocol for BRAF V600E mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol. 2015;23:382–388. doi: 10.1097/PAI.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 40.Affolter K, Samowitz W, Tripp S, Bronner MP. BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes Chromosomes Cancer. 2013;52:748–752. doi: 10.1002/gcc.22070. [DOI] [PubMed] [Google Scholar]
- 41.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter "RASCAL" study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 42.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 43.Stefanius K, Ylitalo L, Tuomisto A, et al. Frequent mutations of KRAS in addition to BRAF in colorectal serrated adenocarcinoma. Histopathology. 2011;58:679–692. doi: 10.1111/j.1365-2559.2011.03821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugai T, Eizuka M, Fujita Y, et al. Molecular profiling based on KRAS/BRAF mutation, methylation, and microsatellite statuses in serrated lesions. Dig Dis Sci. 2018;63:2626–2638. doi: 10.1007/s10620-018-5167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piton N, Borrini F, Bolognese A, Lamy A, Sabourin JC. KRAS and BRAF mutation detection: is immunohistochemistry a possible alternative to molecular biology in colorectal cancer? Gastroenterol Res Pract. 2015;2015:753903. doi: 10.1155/2015/753903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Network, author. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borowsky J, Dumenil T, Bettington M, et al. The role of APC in WNT pathway activation in serrated neoplasia. Mod Pathol. 2018;31:495–504. doi: 10.1038/modpathol.2017.150. [DOI] [PubMed] [Google Scholar]
- 48.Bond CE, McKeone DM, Kalimutho M, et al. RNF43 and ZNRF3 are commonly altered in serrated pathway colorectal tumorigenesis. Oncotarget. 2016;7:70589–70600. doi: 10.18632/oncotarget.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami T, Mitomi H, Saito T, et al. Distinct WNT/β-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol. 2015;28:146–158. doi: 10.1038/modpathol.2014.41. [DOI] [PubMed] [Google Scholar]
- 50.Fennell LJ, Clendenning M, McKeone DM, et al. RNF43 is mutated less frequently in Lynch syndrome compared with sporadic microsatellite unstable colorectal cancers. Fam Cancer. 2018;17:63–69. doi: 10.1007/s10689-017-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai JH, Liau JY, Yuan CT, et al. RNF43 is an early and specific mutated gene in the serrated pathway, with increased frequency in traditional serrated adenoma and its associated malignancy. Am J Surg Pathol. 2016;40:1352–1359. doi: 10.1097/PAS.0000000000000664. [DOI] [PubMed] [Google Scholar]
- 52.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 53.Kriegl L, Neumann J, Vieth M, et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol. 2011;24:1015–1022. doi: 10.1038/modpathol.2011.43. [DOI] [PubMed] [Google Scholar]
- 54.Bettington ML, Walker NI, Rosty C, et al. A clinicopathological and molecular analysis of 200 traditional serrated adenomas. Mod Pathol. 2015;28:414–427. doi: 10.1038/modpathol.2014.122. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki H, Igarashi S, Nojima M, et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- 56.Bond CE, Umapathy A, Ramsnes I, et al. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int J Cancer. 2012;130:1567–1576. doi: 10.1002/ijc.26175. [DOI] [PubMed] [Google Scholar]
- 57.Ito M, Mitsuhashi K, Igarashi H, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 58.Slattery ML, Herrick JS, Wolff RK, Mullany LE, Stevens JR, Samowitz W. The miRNA landscape of colorectal polyps. Genes Chromosomes Cancer. 2017;56:347–353. doi: 10.1002/gcc.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsikitis VL, Potter A, Mori M, et al. MicroRNA signatures of colonic polyps on screening and histology. Cancer Prev Res (Phila) 2016;9:942–949. doi: 10.1158/1940-6207.CAPR-16-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanth P, Hazel MW, Boucher KM, et al. Small RNA sequencing of sessile serrated polyps identifies microRNA profile associated with colon cancer. Genes Chromosomes Cancer. 2019;58:23–33. doi: 10.1002/gcc.22686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Renaud F, Mariette C, Vincent A, et al. The serrated neoplasia pathway of colorectal tumors: identification of MUC5AC hypomethylation as an early marker of polyps with malignant potential. Int J Cancer. 2016;138:1472–1481. doi: 10.1002/ijc.29891. [DOI] [PubMed] [Google Scholar]
- 62.Kim YH, Kakar S, Cun L, Deng G, Kim YS. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123:2587–2593. doi: 10.1002/ijc.23840. [DOI] [PubMed] [Google Scholar]
- 63.Spring KJ, Zhao ZZ, Karamatic R, et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400–1407. doi: 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 64.Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452–1459. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 65.Sandmeier D, Benhattar J, Martin P, Bouzourene H. Serrated polyps of the large intestine: a molecular study comparing sessile serrated adenomas and hyperplastic polyps. Histopathology. 2009;55:206–213. doi: 10.1111/j.1365-2559.2009.03356.x. [DOI] [PubMed] [Google Scholar]
- 66.Burnett-Hartman AN, Newcomb PA, Potter JD, et al. Genomic aberrations occurring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73:2863–2872. doi: 10.1158/0008-5472.CAN-12-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamane LS, Scapulatempo-Neto C, Alvarenga L, et al. KRAS and BRAF mutations and MSI status in precursor lesions of colorectal cancer detected by colonoscopy. Oncol Rep. 2014;32:1419–1426. doi: 10.3892/or.2014.3338. [DOI] [PubMed] [Google Scholar]
- 68.Schreiner MA, Weiss DG, Lieberman DA. Proximal and large hyperplastic and nondysplastic serrated polyps detected by colonoscopy are associated with neoplasia. Gastroenterology. 2010;139:1497–1502. doi: 10.1053/j.gastro.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 69.García-Solano J, Pérez-Guillermo M, Conesa-Zamora P, et al. Clinicopathologic study of 85 colorectal serrated adenocarcinomas: further insights into the full recognition of a new subset of colorectal carcinoma. Hum Pathol. 2010;41:1359–1368. doi: 10.1016/j.humpath.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 70.Rhee YY, Kim KJ, Kang GH. CpG island methylator phenotype-high colorectal cancers and their prognostic implications and relationships with the serrated neoplasia pathway. Gut Liver. 2017;11:38–46. doi: 10.5009/gnl15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogino S, Kawasaki T, Kirkner GJ, Suemoto Y, Meyerhardt JA, Fuchs CS. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56:1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bettington M, Brown I, Rosty C, et al. Sessile serrated adenomas in young patients may have limited risk of malignant progression. J Clin Gastroenterol. 2019;53:e113–e116. doi: 10.1097/MCG.0000000000001014. [DOI] [PubMed] [Google Scholar]
- 73.Yagi K, Akagi K, Hayashi H, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 74.Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosenberg DW, Yang S, Pleau DC, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res. 2007;67:3551–3554. doi: 10.1158/0008-5472.CAN-07-0343. [DOI] [PubMed] [Google Scholar]
- 77.Chen D, Huang JF, Liu K, et al. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e90607. doi: 10.1371/journal.pone.0090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lannagan TRM, Lee YK, Wang T, et al. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut. 2019;68:684–692. doi: 10.1136/gutjnl-2017-315920. [DOI] [PMC free article] [PubMed] [Google Scholar]