Abstract
Genome stability can be threatened by both endogenous and exogenous agents. Organisms have evolved numerous mechanisms to repair DNA damage, including homologous recombination (HR) and non-homologous end joining (NHEJ). Among the factors associated with DNA repair, the MRE11-RAD50-NBS1 (MRN) complex (MRE11-RAD50-XRS2 in Saccharomyces cerevisiae) plays important roles not only in DNA damage recognition and signaling but also in subsequent HR or NHEJ repair. Upon detecting DNA damage, the MRN complex activates signaling molecules, such as the protein kinase ataxia-telangiectasia mutated (ATM), to trigger a broad DNA damage response, including cell cycle arrest. The nuclease activity of the MRN complex is responsible for DNA end resection, which guides DNA repair to HR in the presence of sister chromatids. The MRN complex is also involved in NHEJ, and has a species-specific role in hairpin repair. This review focuses on the structure of the MRN complex and its function in DNA damage repair.
Keywords: DNA damage repair, MRE11-RAD50-NBS1 (MRN) complex, Homologous recombination, Non-homologous end joining
1 Introduction
The genome is constantly exposed to endogenous (i.e., DNA replication errors and free radicals from cellular metabolism) and exogenous (i.e., ionizing radiation and ultraviolet light) threats (Liu and Huang, 2014; Bian et al., 2019). The most deleterious DNA lesion affecting the DNA backbone is the double-strand break (DSB). If left unrepaired or repaired incorrectly, DSBs will accumulate and lead to genetic instability, cellular senescence, chromosomal aberration, immunodeficiency, and tumorigenesis (Gao et al., 2015; Scully et al., 2019). Therefore, the efficient recognition and repair of DNA DSBs are extremely important. Fortunately, organisms have evolved multiple DNA repair pathways to maintain genome integrity. The main components responsible for DNA repair, including MRE11, are highly conserved from yeasts to mammals, revealing their importance in cell survival (Liu and Huang, 2016).
The major repair pathways include homologous recombination (HR) and canonical non-homologous end joining (C-NHEJ), the latter being a fast, cell-cycle-independent process. In this pathway, DSBs are first recognized by Ku70/80 that recruits the DNA-dependent protein kinase catalytic kinase subunit (DNA-PKcs), which in turn phosphorylates other proteins, like Artemis for end processing (Han and Huang, 2020). Subsequently, X-ray-cross-complementation group 4 (XRCC4) and DNA ligase IV, together with other DNA ligase proteins, ligate the DSB ends directly, regardless of DNA loss or mutation (Han and Huang, 2020). Therefore, C-NHEJ is considered as an error-prone pathway. By contrast, HR is a relative slow process occurring only in the late S and G2 phases, as it requires a sister chromatid as a template following DNA replication. HR starts after the generation of a 3' single-stranded DNA (ssDNA) overhang. The 3' DNA tail is coated by replication protein A (RPA), which is in turn replaced by RAD51 to form a presynaptic filament that is responsible for homology search and subsequent repair (Prakash et al., 2015).
The MRE11-RAD50-NBS1 (MRN) (MRE11-RAD50-XRS2 in Saccharomyces cerevisiae) is an important complex in DNA damage response (DDR). Mutations in either component of the complex lead to severe diseases in humans. For example, a human MRE11 gene mutation results in ataxia telangiectasia-like disorder; mutations in the human NBS1 gene cause Nijmegen breakage syndrome, an autosomal recessive disease characterized by immunodeficiency, intellectual disability, and cancer predisposition (Digweed and Sperling, 2004; Sedghi et al., 2018). The MRN complex can detect altered DNA ends rapidly. It can signal other proteins like ataxia-telangiectasia mutated (ATM) in mammals to trigger other DDRs, including cell cycle arrest (Jin and Oh, 2019). Its nuclease activity contributes to DNA end processing for NHEJ and end resection for HR (Rupnik et al., 2008). HR is inhibited by B-cell lymphoma 2 (Bcl2) in cells through the suppression of MRE11 recruitment to DSB sites, leading to tumorigenesis (Xie et al., 2015). In BRCA1-mutant cells, dynein light chain LC8-type 1 (DYNLL1) also inhibits DNA end resection by interacting with the MRN complex, although the underlying mechanisms are unclear (He et al., 2018). These findings indicate that the MRN complex is essential for DNA repair. Here, we review the structure of the MRN complex and its functions in DNA repair based on recent discoveries, shedding light on the mechanisms of genome maintenance and advancing cancer research.
2 Structure of the MRN complex
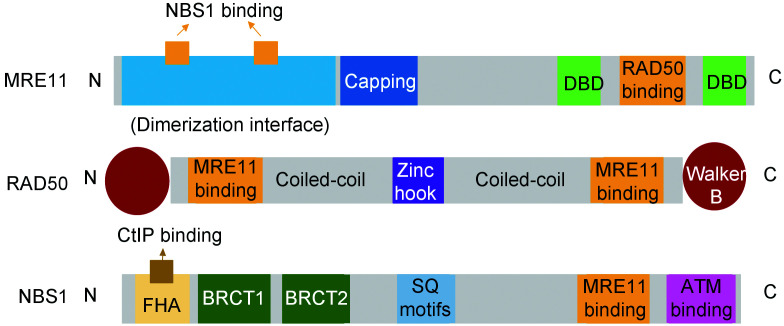
The MRN/X complex is one of the earliest effectors binding to DSB sites. MRE11 core is essential for MRN complex formation, DNA binding, and enzymatic processes (Lamarche et al., 2010). The C-terminal of MRE11 has a RAD50-binding domain between two DNA-binding domains, while the N-terminal contains NBS1-binding sites (Fig. 1)(Lafrance-Vanasse et al., 2015). It can associate with RAD50 and NBS1, which do not interact directly with each other, to form the complex. The MRE11 unit is dimerized through its N-terminal core domains, contributing to the assembly and stabilization of the MRN complex (Williams et al., 2008). Its N-terminal phosphoesterase domain has ssDNA endonuclease and Mn2+-dependent double-stranded DNA (dsDNA) 3'‒5' exonuclease activity, which are important in end resection (Buis et al., 2008). There is a capping domain near the core domain, that can rotate to induce dsDNA unwinding and to orientate the DNA helices for end processing (Rupnik et al., 2008; Lafrance-Vanasse et al., 2015; Gobbini et al., 2018).
Fig. 1. Major domain structure of the MRN complex. The domains involved in MRN complex formation are shown in orange. MRN, MRE11-RAD50-NBS1; DBD, DNA-binding domain; FHA, Forkhead-associated; BRCT, BRCA C-terminal; SQ motifs, Ser-Gln motifs; ATM, ataxia-telangiectasia mutated.
The largest component of the MRN complex is the ATP-binding cassette (ABC)-type ATPase RAD50, which belongs to the structural maintenance of chromosomes (SMC) protein family (Liu and Huang, 2014; Lafrance-Vanasse et al., 2015). It contains Walker A and B nucleotide-binding motifs at its N-terminal and C-terminal ends, respectively (Rojowska et al., 2014). The RAD50 ATPase heads are connected by an anti-parallel coiled-coil domain extending toward the Zn2+-chelating CXXC motif (zinc-hook) (Fig. 1). The zinc-hook of RAD50 is interchangeable with the SMC hinge, indicating its role as a dimerization interface (Tatebe et al., 2020). The dimerization of RAD50 via the coiled-coil domain helps in MRN complex formation and DNA end tethering (Lamarche et al., 2010; Lafrance-Vanasse et al., 2015).
The NBS1 (XRS2 in S. cerevisiae) subunit is responsible for protein‒protein and protein‒DNA interactions (Zhang et al., 2020). The NBS1 N-terminus has one Forkhead-associated (FHA) domain and two BRCA C-terminal (BRCT) domains, which bind to phosphorylated proteins (Fig. 1). For example, the FHA domain can bind to phosphorylated threonine residues in the Ser-X-Thr motifs of DDR proteins such as Ctp1 (Schiller et al., 2014). Moreover, it can recruit DNA damage checkpoint kinases such as Tel1 and ATM, to DNA damage site in order to pause the cell cycle (Nakada et al., 2003). The NBS1 C-terminus region contains an ATM-binding domain, which helps ATM activation and signaling via the adjacent MRE11-interaction domain (Liu and Huang, 2016). The NBS1 protein can be phosphorylated by ATM in turn through Ser-Gln (SQ) motifs in its central part (Lamarche et al., 2010). It also serves as a regulator in the MRN complex, since it can recruit various protein partners to modulate the complex (Williams et al., 2009).
3 Detection of DSBs and signals to other DNA damage proteins by MRN complex
Following DNA damage, the MRN complex recognizes and binds to DSBs rapidly. It is recruited to DNA damage sites by γ-H2AX and RAD17 (Bian et al., 2019). The former interacts with the FHA/BRCT domain of NBS1 after DNA damage with the help of MDC1 to facilitate NBS1 foci formation (Sharma et al., 2012). Unlike γ-H2AX, RAD17 is independent of MDC1 and is phosphorylated by ATM at T622 sites, which then interacts with NBS1 and helps MRN recruitment (Wang et al., 2014). It also interacts with ATM and forms a positive feedback loop enhancing ATM-related signaling.
Furthermore, the localization of the MRN complex to DNA damage sites requires the MCM8-9 complex. The homozygous depletion of MCM9 makes cells hypersensitive to interstrand-crosslinking agents (Lee et al., 2015). The association of MRE11 with the MCM8-9 complex at damage sites depends on ATP binding and hydrolysis via ATPase motifs. Moreover, depletion of either MCM8-9 or MRE11 had similar negative effects on RPA foci formation, implying that MCM8-9 also participates in end resection (Lee et al., 2015).
Once bound to DSBs, the MRN complex can recruit and activate various DDR proteins, including ATM (Chen et al., 2013). Following its recruitment to DSB sites by NBS1, ATM is activated by monomerization and autophosphorylation (Jin and Oh, 2019). The UFMylation of MRE11 at K282 is needed for MRN complex formation and ATM activation in HR repair (Wang et al., 2019). In turn, the activated ATM phosphorylates targets that include the three subunits of the MRN complex to activate downstream signaling pathways. For example, ATM phosphorylates NBS1 at S278 and S343 to regulate the S-phase checkpoint (Lavin et al., 2015). A recent study found that ATM also phosphorylated Pellino1 to mediate NBS1 ubiquitination (Ha et al., 2019). This positive feedback loop between the MRN complex and ATM is essential for DNA repair.
In addition to ATM, the recruitment of the Bloom syndrome protein (BLM) to DNA damage sites in vivo is also dependent on the MRN complex (Tripathi et al., 2018). In fact, BLM is a tumor suppressor with multiple functions in DNA damage repair. Its helicase activity contributes to recruiting HR- and NHEJ-related proteins in the S and G1 phases, respectively (Bugreev et al., 2007). In the repair phase, however, BLM inhibits HR in the S phase, possibly by counteracting RAD51 localization to DNA damage sites, whereas it inhibits NHEJ in both the S and G1 phases (Patel et al., 2017). The recruitment of BLM in its early phase requires ATM and ssDNA generated by the MRN complex (Tikoo et al., 2013). In the late recruitment phase, the ubiquitylated BLM associates directly with NBS1 and interacts further with MRE11, whose exonuclease activity optimizes BLM recruitment (Tripathi et al., 2018). In conclusion, the MRN complex contributes to DNA repair by sensing DSB and recruiting multiple effectors to DSB sites to perform DDR.
4 Initiation of DNA end resection and direction of the repair pathway toward HR by MRN complex
Many factors influence the choice of DSB repair pathway, such as the actual cell cycle phase. The process of NHEJ is available throughout the entire cell cycle, while HR functions only during the S and G2 phases due to the requirement of sister chromatids (Scully et al., 2019). Additionally, the structure of DSBs is also important. Replication-related one-ended DSBs are repaired mainly by HR, since they lack another end for joining (Han and Huang, 2020). A further factor affecting DNA repair choice is DNA end resection, where the MRN complex plays an essential role (Panier and Boulton, 2014). The incision made by MRE11 endonuclease was thought to direct DNA repair to HR. However, failure to extend the initial nick due to the inhibition of both MRE11 and EXO1/BLM exonuclease activity directs the repair toward NHEJ (Shibata et al., 2014). This indicates that the ssDNA region, rather than the initial nick, suppresses NHEJ and commits cells to perform HR (Shibata et al., 2014).
The prerequisite for HR is 3' ssDNA overhangs generated by end resection because an ssDNA overhang can serve as platform for the recruitment of HR repair proteins (i.e., RAD51 recombinase) (Kowalczykowski, 2015). The MRE11 protein is involved in 5' DNA strand degradation (Zhu et al., 2008). Nevertheless, ssDNA tails are generated by 5'‒3' exonuclease, while the exonuclease polarity of MRE11 is the opposite (Stracker and Petrini, 2011). A two-step model has been proposed to explain this phenomenon. In the first step, MRE11 endonuclease and C-terminal binding protein (CtBP)-interacting protein (CtIP) generate an initial resection nick. In the second step, MRE11 exonuclease degrades DNA fragments in a 3'‒5' direction toward DSB ends (Liu and Huang, 2016). However, this model is unable to explain why the 3' ssDNA ends are not degraded by MRE11 exonuclease. A recent study found that MRE11 nuclease activity is regulated by RAD50 through ATP binding and hydrolysis (Cannavo et al., 2019). The RAD50 protein limits MRE11 exonuclease activity in the ATP-bound state to protect 3' ends from degradation. In turn, ATP hydrolysis by RAD50 promotes phosphorylated Sae2 to relieve this restriction and facilitate MRE11 endonuclease activity (Cannavo et al., 2019). In summary, the MRE11 endonuclease is promoted first by Sae2 to generate an ssDNA nick. Next, the 3'‒5' MRE11 exonuclease, regulated by RAD50 and Sae2, digests short DNA segments between endonucleolytic sites. Finally, the long-range nucleases Exo1 and Dna2 are recruited for long resection (Huertas, 2010). The release of MRN and Ku complexes from DNA ends is also dependent on MRE11 nuclease activity and Ctp1 (Langerak et al., 2011). This is important for RPA localization on resected DNA ends, and furthering DSB repair processes.
The MRN complex also influences the DSB repair pathway choice by interacting with DNA-dependent protein kinase (DNA-PK) (Deshpande et al., 2020). The latter is composed of a DNA-PKcs and the DNA end-binding Ku70/80 heterodimer. Due to the high affinity of Ku70/80 to DNA ends, DNA-PK responds rapidly to DSBs and binds to damage sites within seconds. The DNA-PK then stimulates NHEJ, ligating DNA ends by ligase IV together with other proteins. Therefore, DNA-PK seems to compete with the MRN complex over DSB repair choice. However, a recent study showed that DNA-PK promotes DNA end resection by stimulating MRE11 endonuclease activity (Deshpande et al., 2020). In situations where NHEJ failed due to incompatible DNA ends, the stably retained DNA-PK complex on DNA ends works together with phosphorylated CtIP-stimulated MRE11 endonuclease to remove the complex (Deshpande et al., 2020). This reaction triggers further DNA end resection and directs repair toward HR. Once DNA ends have undergone end joining successfully, DNA-PK dissociates and MRN endonuclease activity is not activated (Jette and Lees-Miller, 2015).
5 Function of the MRN complex in both C-NHEJ and A-NHEJ
In contrast to HR, the function of the MRN complex in NHEJ remains unclear. The process of C-NHEJ requires factors including XRCC4 and the Ku70/80 complex (Davis and Chen, 2013). In the absence of these factors, alternative non-homologous end joining (A-NHEJ) occurs. Short DNA end resection (<100 nucleotides) is the first step of A-NHEJ, followed by microhomology pairing and end ligation (Sallmyr and Tomkinson, 2018). The knockdown of MRE11 in mammalian cells significantly decreased the end-joining efficacy in both the absence and presence of XRCC4, emphasizing the role of MRE11 in both C-NHEJ and A-NHEJ (Rass et al., 2009). One explanation is that the MRE11 nuclease activity contributes to the initial short end resection in A-NHEJ (Lamarche et al., 2010). Alternatively, the MRN/X complex may affect some early processes common to both NHEJ pathways. For example, the MRN/X complex might influence NHEJ via ATM signaling pathways, since the inhibition of ATM reduces NHEJ efficiency (Xie et al., 2009). Based on the observation that end-joining defects in MRE11-deleted S. cerevisiae cells were partially rescued by nuclease-deprived MRE11, MRN/X might have structural functions (Lamarche et al., 2010).
Recent studies revealed that the MRN complex is also involved in the repair of DSBs capped with a hairpin end (Li et al., 2017; Runge and Li, 2018). A DNA hairpin, formed from structures such as palindromes and trinucleotide repeats, can cause chromosomal translocation and genome instability (Deng et al., 2015). The first step in repairing a DSB with a hairpin end is hairpin opening. This requires MRN/X and its associated nuclease Sae2 in S. cerevisiae, but not in Schizosaccharomyces pombe or mammals, in which the exact mechanism is unclear. S. pombe cells lacking the Ku complex, DNA ligase IV, and MRN subunits showed a decreased NHEJ efficiency (Runge and Li, 2018). It is reasonable to think that the enzymatic activity of the MRN complex contributes to NHEJ. However, mutations in MRE11 endonuclease did not cause a significant difference in NHEJ repair compared to a wild-type group (Li et al., 2017). In another study, mutations in the MRN dimerization region impaired NHEJ repair, suggesting that MRN provides structural support in NHEJ (Li et al., 2017). Thus, the MRN complex affected NHEJ by recruiting other nucleases to open the hairpin rather than via its own enzymatic activity in S. pombe. Further research is needed to identify specific nucleases responsible for this pathway.
6 Summary
The MRN complex contributes to the detection and repair of DNA damage, which is essential for genome stability and cell survival. This complex can sense damaged proteins through its NBS1 N-terminal domains and signal other molecules to trigger DDR. An important DDR protein is ATM activated by MRN, which in turn phosphorylates the MRN complex, thereby producing positive feedback. The endonuclease activity of MRE11 helps the initial end resection, whereas its exonuclease activity facilitates the degradation of DNA segments between endonucleolytic sites. The MRN complex, together with other nucleases, generates ssDNA overhangs, directing DNA repair toward HR. The MRN complex is also involved in NHEJ and hairpin repair, although the underlying mechanism of this involvement requires further research.
Acknowledgments
This reasearch was supported by the National Key Research and Development Program of China (No. 2018YFC2000100), the National Natural Science Foundation of China (Nos. 31730021, 31971220, and 31961160725), the Fok Ying Tung Education Foundation, and the China’s Fundamental Research Funds for the Central Universities. We apologize to colleagues whose work could not be cited due to space limitations. We thank all our colleagues in the Huang laboratory for the insightful discussions.
Author contributions
Shan QIU wrote the manuscript. Jun HUANG reviewed and edited the manuscript. Both authors have read and approved the final manuscript and take responsibility for its integrity.
Compliance with ethics guidelines
Shan QIU and Jun HUANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by either of the authors.
References
- Bian L, Meng YL, Zhang MC, et al. , 2019. MRE11-RAD50-NBS1 complex alterations and DNA damage response: implications for cancer treatment. Mol Cancer, 18: 169 10.1186/s12943-019-1100-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, et al. , 2007. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev, 21(23): 3085-3094. 10.1101/gad.1609007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng YB, et al. , 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell, 135(1): 85-96. 10.1016/j.cell.2008.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Reginato G, Cejka P, 2019. Stepwise 5' DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proc Natl Acad Sci USA, 116(12): 5505-5513. 10.1073/pnas.1820157116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang GL, Huang NJ, et al. , 2013. Suppression of DNA-damage checkpoint signaling by Rsk-mediated phosphorylation of Mre11. Proc Natl Acad Sci USA, 110(51): 20605-20610. 10.1073/pnas.1306328110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Chen DJ, 2013. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res, 2(3): 130-143. 10.3978/j.issn.2218-676X.2013.04.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng SK, Yin Y, Petes TD, et al. , 2015. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell, 60(3): 500-508. 10.1016/j.molcel.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Myler LR, Soniat MM, et al. , 2020. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv, 6(2): eaay0922 10.1126/sciadv.aay0922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M, Sperling K, 2004. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair, 3(8-9): 1207-1217. 10.1016/j.dnarep.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Gao R, Singh R, Kaul Z, et al. , 2015. Targeting of DNA damage signaling pathway induced senescence and reduced migration of cancer cells. J Gerontol: Ser A, 70(6): 701-713. 10.1093/gerona/glu019 [DOI] [PubMed] [Google Scholar]
- Gobbini E, Cassani C, Vertemara J, et al. , 2018. The MRX complex regulates Exo1 resection activity by altering DNA end structure. EMBO J, 37(16): e98588 10.15252/embj.201798588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha GH, Ji JH, Chae S, et al. , 2019. Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat Commun, 10: 1577 10.1038/s41467-019-09641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Huang J, 2020. DNA double-strand break repair pathway choice: the fork in the road. Genome Instab Dis, 1(1): 10-19. 10.1007/s42764-019-00002-w [DOI] [Google Scholar]
- He YJ, Meghani K, Caron MC, et al. , 2018. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature, 563(7732): 522-526. 10.1038/s41586-018-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, 2010. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol, 17(1): 11-16. 10.1038/nsmb.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP, 2015. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol, 117(2-3): 194-205. 10.1016/j.pbiomolbio.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MH, Oh DY, 2019. ATM in DNA repair in cancer. Pharmacol Ther, 203: 107391 10.1016/j.pharmthera.2019.07.002 [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol, 7(11): a016410 10.1101/cshperspect.a016410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrance-Vanasse J, Williams GJ, Tainer JA, 2015. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol, 117(2-3): 182-193. 10.1016/j.pbiomolbio.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, Weitzman MD, 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett, 584(17): 3682-3695. 10.1016/j.febslet.2010.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, et al. , 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet, 7(9): e1002271 10.1371/journal.pgen.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Kozlov S, Gatei M, et al. , 2015. ATM-dependent phosphorylation of all three members of the MRN complex: from sensor to adaptor. Biomolecules, 5(4): 2877-2902. 10.3390/biom5042877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Im JS, Shibata E, et al. , 2015. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat Commun, 6: 7744 10.1038/ncomms8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Wang JY, Zhou G, et al. , 2017. Nonhomologous end-joining with minimal sequence loss is promoted by the Mre11-Rad50-Nbs1-Ctp1 complex in Schizosaccharomyces pombe . Genetics, 206(1): 481-496. 10.1534/genetics.117.200972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Huang J, 2014. Quality control of homologous recombination. Cell Mol Life Sci, 71(19): 3779-3797. 10.1007/s00018-014-1649-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Huang J, 2016. DNA end resection: facts and mechanisms. Genomics Proteomics Bioinform, 14(3): 126-130. 10.1016/j.gpb.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K, 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev, 17(16): 1957-1962. 10.1101/gad.1099003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ, 2014. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol, 15(1): 7-18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Patel DS, Misenko SM, Her J, et al. , 2017. BLM helicase regulates DNA repair by counteracting RAD51 loading at DNA double-strand break sites. J Cell Biol, 216(11): 3521-3534. 10.1083/jcb.201703144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng WR, et al. , 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol, 7(4): a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, et al. , 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol, 16(8): 819-824. 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Rojowska A, Lammens K, Seifert FU, et al. , 2014. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J, 33(23): 2847-2859. 10.15252/embj.201488889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge KW, Li YH, 2018. A curious new role for MRN in Schizosaccharomyces pombe non-homologous end-joining. Curr Genet, 64(2): 359-364. 10.1007/s00294-017-0760-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik A, Grenon M, Lowndes N, 2008. The MRN complex. Curr Biol, 18(11): R455-R457. 10.1016/j.cub.2008.03.040 [DOI] [PubMed] [Google Scholar]
- Sallmyr A, Tomkinson AE, 2018. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem, 293(27): 10536-10546. 10.1074/jbc.TM117.000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Seifert FU, Linke-Winnebeck C, et al. , 2014. Structural studies of DNA end detection and resection in homologous recombination. Cold Spring Harb Perspect Biol, 6(10): a017962 10.1101/cshperspect.a017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, et al. , 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 20(11): 698-714. 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedghi M, Salari M, Moslemi AR, et al. , 2018. Ataxia-telangiectasia-like disorder in a family deficient for MRE11A, caused by a MRE11 variant. Neurol Genet, 4(6): e295 10.1212/NXG.0000000000000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Singh K, Almasan A, 2012. Histone H2AX phosphorylation: a marker for DNA damage. In: Bjergbæk L (Ed.), DNA Repair Protocols. Methods in Molecular Biology. Humana Press, Totowa, NJ, p.613-626. 10.1007/978-1-61779-998-3_40 [DOI] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, et al. , 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell, 53(1): 7-18. 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JHJ, 2011. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol, 12(2): 90-103. 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Lim CT, Konno H, et al. , 2020. Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge. Nat Commun, 11: 370 10.1038/s41467-019-14025-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo S, Madhavan V, Hussain M, et al. , 2013. Ubiquitin-dependent recruitment of the Bloom Syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J, 32(12): 1778-1792. 10.1038/emboj.2013.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Agarwal H, Priya S, et al. , 2018. MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways. Nat Commun, 9: 1016 10.1038/s41467-018-03393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QH, Goldstein M, Alexander P, et al. , 2014. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J, 33(8): 862-877. 10.1002/embj.201386064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Gong YM, Peng B, et al. , 2019. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res, 47(8): 4124-4135. 10.1093/nar/gkz110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, et al. , 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell, 135(1): 97-109. 10.1016/j.cell.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, et al. , 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell, 139(1): 87-99. 10.1016/j.cell.2009.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie AY, Kwok A, Scully R, 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol, 16(8): 814-818. 10.1038/nsmb.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie MH, Park D, You S, et al. , 2015. Bcl2 inhibits recruitment of Mre11 complex to DNA double-strand breaks in response to high-linear energy transfer radiation. Nucleic Acids Res, 43(2): 960-972. 10.1093/nar/gku1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Tang ZH, Li LJ, et al. , 2020. NBS1 is required for SPO11-linked DNA double-strand break repair in male meiosis. Cell Death Differ, 27(7): 2176-2190. 10.1038/s41418-020-0493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, et al. , 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell, 134(6): 981-94. 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]