Abstract
Proteins are major functional units that are tightly connected to form complex and dynamic networks. These networks enable cells and organisms to operate properly and respond efficiently to environmental cues. Over the past decades, many biochemical methods have been developed to search for protein-binding partners in order to understand how protein networks are constructed and connected. At the same time, rapid development in proteomics and mass spectrometry (MS) techniques makes it possible to identify interacting proteins and build comprehensive protein‒protein interaction networks. The resulting interactomes and networks have proven informative in the investigation of biological functions, such as in the field of DNA damage repair. In recent years, a number of proteins involved in DNA damage response and DNA repair pathways have been uncovered with MS-based protein‒protein interaction studies. As the technologies for enriching associated proteins and MS become more sophisticated, the studies of protein‒protein interactions are entering a new era. In this review, we summarize the strategies and recent developments for exploring protein‒protein interaction. In addition, we discuss the application of these tools in the investigation of protein‒protein interaction networks involved in DNA damage response and DNA repair.
Keywords: Protein‒protein interaction, Interactome, Proteomics, Mass spectrometry, DNA repair, DNA damage response
1 Introduction
Genes and the genome sequences are the carriers of genetic information that are inherited from generation to generation. Their coding products are mostly proteins which carry out structural and enzymatic functions in cells and organisms. The functions of these proteins are regulated and transduced through their communications with other biomolecules, including proteins, DNA, RNA, lipids, and other metabolites, with the majority of these events occurring via protein‒protein interactions. Proteins and their interacting protein partners form a complex network, known as an interactome.
As the key players in most cellular processes, proteins need to act in concert with each other to accomplish various tasks important for the survival and regeneration of cells or organisms. In these processes, communications via protein‒protein interactions are likely the most efficient and direct strategy. Investigation of protein‒protein interaction networks or the protein interactome on a large scale has arrived with the era of omics. Many strategies have been developed to enrich proteins of interest and their binding partners. Enriched proteins with their binding partners are submitted to mass spectrometry (MS) for protein identification. After data filtration with bioinformatics tools, the protein‒protein interaction network or interactome can be established with lists of high-confidence interaction proteins (HCIPs).
Genome instability refers to frequent mutations and other genomic alterations within the genome of a particular cellular lineage. Without the actions of several well-defined DNA damage response and distinct DNA repair pathways, genetic alterations including mutations will accumulate and eventually affect cell survival, expedite the aging process, and result in the development of human diseases, such as cancer or neurodegenerative disease. Therefore, detailed investigation of interactomes involving DNA damage response and DNA repair proteins will facilitate our understanding of the molecular/cellular functions of these proteins, especially how these proteins act together and how the mutation or disruption of just one component in these complex networks may lead to human disease. Furthermore, disease-focused protein‒protein interaction networks can be used for drug development and for identifying the best treatment strategies. In this review, we summarize the current technological developments in MS-based studies of protein‒protein interactions with an emphasis on the investigation of interaction networks involved in DNA damage repair (DDR).
2 Strategies for studying interactomes with mass spectrometry
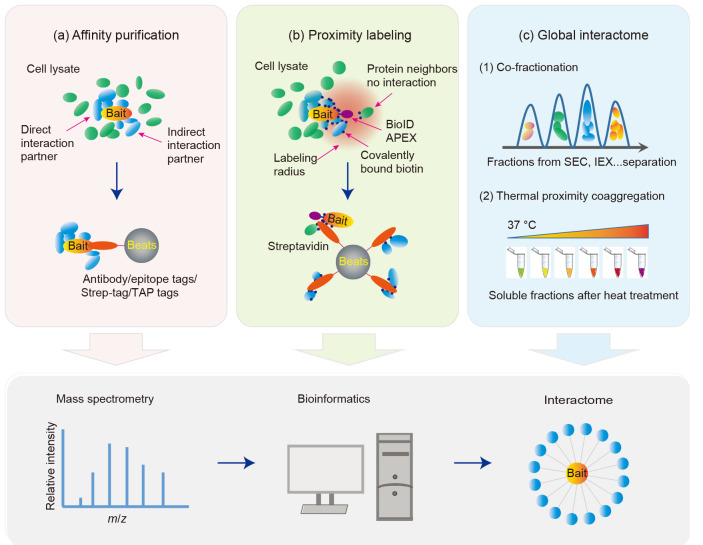
The physical protein‒protein interaction and their interactomes are essential for most if not all biological processes. The fast developing MS techniques enable researchers to quickly identify all proteins in a complex mixture. The combination of bait-prey purification and MS identification workflow facilitates large scale protein‒protein interactome studies. The key in this workflow is obtaining the protein complexes for MS analysis. A few techniques have been developed for the enrichment of proteins (prey) which interact with target proteins of interest (bait).
2.1. Affinity purification
There are different ways to pull down the target protein in a gentle buffer without the disruption of binding partners in order to obtain the interacting proteins for MS analysis (Fig. 1a). Antibody‒antigen interaction relies on the noncovalent binding between the variable region of the antibody and the antigen at one or several sites on the target protein. Using this interaction, researchers developed co-immunoprecipitation (co-IP) to capture the native form of the target protein and its associated protein complex. The components of the complex are then identified by MS. This strategy allows us to purify the protein complex without any manipulation of the target gene and the in vivo environment. This may be important in preserving some specific protein‒protein interactions. For example, a report by Malovannaya et al. (2011) used 1796 primary antibodies which target 1087 unique gene products to analyze the endogenous human coregulatory complexome, which led to the discovery of many unreported protein associations.
Fig. 1. Strategies for studying interactomes with mass spectrometry (MS). (a) Affinity purification using antibody, epitope tags, Strep-tag, or tandem affinity purification (TAP) tags; (b) Proximity labeling process with BioID (biotin identification) or APEX (an engineered peroxidase developed from ascorbate peroxidase); (c) Global interactome analysis using co-fractionation or thermal proximity coaggregation. After enrichment or various processes, the output protein samples are analyzed by MS, bioinformatics tools are used to select for high-confident interacting proteins (HCIPs), and then the protein‒protein interaction network or interactome is generated. IEX: ion-exchange; SEC: size exclusion chromatography.
However, availability of highly specific anti-bodies for the target protein is of the utmost concern for the co-IP method. In many cases, the antibody is either not available or not specific. Cross-reactivity of the primary antibody can severely interfere with the results, since many putative associated proteins turn out to be nonspecific proteins that cross-react with the primary antibody. This issue is particularly challenging since different primary antibodies cross-react with distinct sets of cellular proteins. One way to circumvent this issue is to perform co-IP experiments using two or more validated antibodies that recognize unique epitopes on the target protein. The data from experiments using multiple antibodies recognizing different epitopes may be used to rule out nonspecific interacting proteins (Li et al., 2017). However, as mentioned above, the availability of highly specific antibodies for a given target may be very limited, especially if the target has not been studied extensively. Additionally, antibodies recognizing different epitopes on a target protein may sometimes disrupt selected protein‒protein interactions. These challenges limit the use of co-IP as a general method for the identification of associated proteins.
To circumvent these limitations, epitope tags and specific antibodies recognizing these epitope tags are being used as ways to isolate protein complexes. The commonly used tags or epitope tags include GFP (green fluorescent protein) (Cristea et al., 2005; Hein et al., 2015), HA (hemagglutinin) tag (Sowa et al., 2009), and FLAG tag (Ho et al., 2002). In these methods, the target protein is first tagged with the epitope and the fused protein is expressed in cells. Then, co-IP is conducted with the highly efficient tag-specific antibody incubating with cell lysis to pull down the target protein and its binding partners. These methods are widely used in large-scale interactome studies. For example, Gygi, Harper, and colleagues are continuing the BioPlex interactome project to profile protein interactomes in human cells using the HA tag and anti-HA agarose. BioPlex 1.0 reported interacting partners for 2594 proteins (Huttlin et al., 2015); BioPlex 2.0 increased the bait protein number to 5891 and built the disease networks (Huttlin et al., 2017); BioPlex 3.0 included 10 128 human protein baits and revealed cell-specific remodeling of the human interactome using HEK293T and HCT116 cells (Huttlin et al., 2020).
The antibody for co-IP has also been further improved. Nanobody, which contains one single variable domain (antigen-binding domain), is now being used for interactome studies. This kind of antibody can be easily cloned and has the advantage of being specific, soluble, stable, and having high affinity (Cortez-Retamozo et al., 2004; Muyldermans, 2013; Liu WS et al., 2018). For example, an anti-GFP nanobody has now been applied successfully in co-IP for many interactome studies (Baymaz et al., 2014; Schenstrom et al., 2018).
An alternative approach is the strep-tag system, which tags the target protein with a short peptide (Strep-tag) which binds strongly to streptavidin; then streptavidin beads can be used instead of antibody beads for co-IP experiments. An improved version of streptavidin which is called Strep-tactin has been proven to be highly efficient for the analysis of protein complexes and interactomes ( Schmidt and Skerra, 2007). This technique has been widely used by Krogan and colleagues for several large-scale interactome studies, including virus‒host (Jäger et al., 2012; Davis et al., 2015; Ramage et al., 2015; Batra et al., 2018; Eckhardt et al., 2018; Shah et al., 2018; Diep et al., 2019; Li et al., 2019), bacteria‒host (Penn et al., 2018), and Chlamydia‒host (Mirrashidi et al., 2015) interactions. Their most recent work uses this system to reveal the interactome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins with host proteins and provides interesting leads for repurposing drugs that may prevent the SARS-CoV-2 infection from spreading (Gordon et al., 2020).
The methods described above use one-step affinity purification, which will identify a relatively long list of proteins with advanced MS instruments that are highly efficient in protein identification. To reduce the amount of non-specific binding proteins, two-step affinity purification, also called tandem affinity purification (TAP), has been developed. In theory, the chance of contaminants being retained in the purified protein mixture after two rounds of successive affinity purifications should be minimal. Different pairs of tags are used in this strategy, and they include calmodulin-binding peptide (CBP)-protein A (Rigaut et al., 1999; Gavin et al., 2002, 2006; Krogan et al., 2006), GFP-S protein (Cheeseman et al., 2004; Cheeseman and Desai, 2005), and Strep-S protein (Kim et al., 2007b; Li et al., 2015; Chen et al., 2016, 2019, 2020; Srivastava et al., 2018). TAP methods have been proven to be efficient at enriching for the target protein and its binding partners, while reducing contaminants.
A common issue in the above-mentioned affinity purification methods is that the interacting proteins may dissociate from the target protein during the experimental procedures. It may be due to the weak or dynamic binding between these proteins. This problem may be a concern when conducting TAP pro-cedures, since further dilution during the second step of purification may lead to dissociation of weak protein complexes. Another issue in affinity purification is the membrane protein complex, which is difficult to extract using moderate detergents/conditions. To address these problems, one solution is to use crosslink reactions which stabilize the protein complexes. Several crosslink reagents are used for the studies of protein‒protein interactions, including formaldehyde (Larance et al., 2016), biotin-aspartate proline-N-hydroxyphthalimide (BDP-NHP) (Schweppe et al., 2017), and disuccinimidyl sulfoxide (DSSO) (Liu F et al., 2018). Crosslinks work efficiently in identifying protein interactomes (Liu and Heck, 2015; Liu F et al., 2015, 2018; Wu et al., 2016); however, the drawback is that it is difficult to know the precise timing and amount of crosslinking agents to use in order to preserve specific interactions but not increase non-specific contaminants. Using crosslinking reagents during affinity purification is also beneficial for structural biology studies. When it is used in conjunction with in silicomodeling of the structures of protein complexes, crosslinking enables the observation of the physical proximity of different parts of the proteins, such as conformational and dynamics changes in a complex (Schweppe et al., 2015; Chavez et al., 2016; Wu et al., 2016). Therefore, crosslink strategy can be utilized for both interactome and structural biology studies. However, the efficiency and specificity of the crosslinking agents, strategy for enrichment of cross-linked peptides, and dedicated data analysis software are required before we can use it routinely and with ease (O'Reilly and Rappsilber, 2018).
2.2. Proximity labeling-based interactome studies
A new strategy has been developed to partially address the problems associated with affinity purification methods. Because enzyme‒substrate interaction can only occur within a short distance, when the target protein is tagged with an enzyme, it should be able to label the neighbor proteins with a small molecule, such as fluorescein or biotin (Rees et al., 2015; Kim and Roux, 2016; Trinkle-Mulcahy, 2019). The proximity labeling strategy is a powerful approach to study the protein interactome (Fig. 1b), since the labeling occurs in vivo in cells before cell lysis and therefore can be used to capture transient and weak interactions that may be easily lost during cell lysis and/or various purification procedures as mentioned above.
Proximity-dependent biotin identification (BioID), developed by Roux and colleagues, uses a promiscuous biotin ligase, Escherichia coli protein BirA* (BirA with R118G mutation). When it is fused to a protein of interest and then expressed in cells, this fusion protein can biotinylate endogenous proteins within an estimated surrounding radius of approximately 10 nm (Roux et al., 2012; Kim et al., 2014). BioID facilitates the identification not only of direct interacting proteins, but also proteins in the vicinity that do not physically interact with the bait protein. BioID has been successfully applied to studies of interactomes of different functional protein groups. Pelletier and colleagues reported the centrosome-cilium interface interactome landscape (Gupta et al., 2015) and later the centriolar satellites interactome using BioID (Gheiratmand et al., 2019). Gingras and colleagues applied BioID to study the interactomes of splicing networks (Gonatopoulos-Pournatzis et al., 2018), Rho-family GTPases (Bagci et al., 2020), messenger RNA (mRNA) biology proteins ( Youn et al. 2018), and phosphatases (St-Denis et al., 2016). Christopher and colleagues reported an extensive BioID-based proximity map of a human cell with 192 subcellular markers (Go et al., 2019). Additionally, the virus‒host global interactome established with the use of BioID uncovered extensive organelles targeting by the Zika virus (Coyaud et al., 2018).
These studies suggest that BioID is a powerful technique for establishing large scale interactomes. However, some limitations exist. A major drawback of BioID is the extremely long reaction time, i.e.,16‒18 h, needed for biotinylation (Roux et al., 2013). Another issue is that the addition of a 35-kDa biotin ligase to a bait protein may impair its localization and function (Kim and Roux, 2016). Several groups have developed new versions of the BioID tag-BioID2 (Kim et al., 2016), BASU (Ramanathan et al., 2018), and TurboID/miniTurboID (Branon et al., 2018). BioID2 replaces the original biotin ligase with a smaller one from Aquifex aeolicus with human codon optimization and a conserved residue mutation (R40G) (Kim et al., 2016). BioID2 requires less biotin supplementation, and enhances the labeling efficiency. Furthermore, this new version uses a smaller size BioA (27 kDa), which may improve the localization and/or function of the target protein (Kim et al., 2016). BASU is a newer mutated BirA* engineered from Bacillus subtilis with >1000-fold faster kinetics and >30-fold increased signal-to-noise ratio over the first generation of BioID (Ramanathan et al., 2018). TurboID and miniTurboID, which were developed in Ting's lab, and used directed evolution of E. coli biotin ligase to generate new promiscuous variants, which resulted in TurboID and miniTurboID that enable proximity labeling reactions in as little as 10 min (Branon et al., 2018).
Another proximity labeling strategy involves the use of engineered ascorbate peroxidase 2 (APEX2). APEX is an engineered peroxidase developed from ascorbate peroxidase (APX), a class I cytosolic plant peroxidase, that has been used for electron microscopy (Martell et al., 2012). APEX2 is a new version of APEX, which is a monomeric APX that catalyzes the oxidation of biotin-phenol to the biotin-phenoxyl radical in the presence of H2O2. This reaction results in biotinylation of neighboring proteins ( Lam et al., 2015). APEX2 was used in several recent studies. As examples, APEX2 was used to profile autophagosome content and revealed a role for microtubule-associated proteins 1A/1B light chain 3C (MAP1LC3C) in maintaining basal mitochondrial homeostasis (le Guerroué et al., 2017). Additionally, APEX2 was employed to elucidate the Weibel-Palade bodies (WPBs, a secretory organelle)-associated proteins (Holthenrich et al., 2019), and capture in vivo protein‒protein interaction at chlamydial inclusion (Olson et al., 2019). The advantage of APEX2 over BioID is its significantly faster reaction time, which is as short as 1 min (Hung et al., 2016; Trinkle-Mulcahy, 2019). However, the APEX2 method uses H2O2 treatment to activate the peroxidase. The concern is that such treatment may affect cellular oxidative status and induce a stress response (Trinkle-Mulcahy, 2019).
More recently, the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) knock-in technique has made it possible to tag the target gene at its endogenous locus. Junctophilin-2 (JPH2) interactome was studied with knock-in of BioID2 (Feng et al., 2020); the endogenous interaction networks of breast cancer type 1 susceptibility protein (BRCA1), tumor suppressor p53-binding protein 1 (TP53BP1), and mediator of DNA damage checkpoint protein 1 (MDC1) were also studied using knock-in of APEX2 (Gupta et al., 2018). The endogenous tagging technique avoids the overexpression of an exogenous fusion protein. The expression level and the regulation of the endogenously tagged protein are the same or similar to its endogenous counterpart. This helps reveal the physiological regulation and function of the target protein.
The application of the proximity labeling method has been expanded in other studies. Spatial proteome research using BioID or APEX has been conducted to study the dynamic protein localizations at subcellular levels (Rhee et al., 2013; Hung et al., 2014; Liu XN et al., 2018; Go et al., 2019). Split-BioID (de Munter et al., 2017; Schopp et al., 2017; Schopp and Béthune, 2018), Split-APEX2 (Xue et al., 2017; Han et al., 2019), or Split-TurboID (Cho et al., 2020) divide the labeling enzymes into N- and C-terminal fragments so that the proximity labeling reaction can only be restored when the two divided fragments are brought into proximity. This method has been employed to validate protein‒protein interaction or explore interactomes under certain conditions. The proximity labeling strategy was also adopted to label RNA-associated protein complexes for the studies of RNA‒protein interactions (Castello et al., 2016; Ramanathan et al., 2018). In addition, BioID or APEX is combined with a deactivated CRISPR-Cas9 strategy for the study of chromatin-associated protein complexes at a target genomic locus (Schmidtmann et al., 2016; Gao et al., 2018; Myers et al., 2018). These innovative approaches further expand the use of proximity labeling techniques to address many biological questions.
2.3. Global interactome profiling
The interactome studies with affinity purification and proximity labeling are scalable from a few to tens of bait genes. However, the amount of work increases dramatically when attempting to study hundreds or thousands of target genes at once. Therefore, we still need methods to investigate interactomes on a global proteome level. A method called co-fractionation MS has been developed to make it possible to construct the whole network of protein‒protein interactions in the cell (Fig. 1c). In brief, protein extracts from cells or tissues are extensively fractionated to separate protein complexes, and then the components of each fraction are identified and quantified by MS. A network of 13 993 high-confidence physical interactions among 3006 stably associated soluble human proteins and 622 putative protein complexes was built using quantitative proteomics analysis of more than one thousand biochemical fractions separated by chromatography (Havugimana et al., 2012). Another group combined quantitative proteomics and size-exclusion chromatography and mapped 291 co-eluting protein complexes (Kristensen et al., 2012). Additionally, software predicting interactomes from co-elution (PrInCE) was developed for predicting interactomes from co-fractionation experiments (Stacey et al., 2017). However, it remains difficult to identify subunits within a multi-protein complex using limited fractions. Further optimization of the fractionation process and data analysis are needed before this method can be adopted as a routine experimental procedure.
Based on the idea that interaction with other molecules will change the thermal stability of the protein, a new approach, thermal proteome profiling (TPP), was developed. It was first applied in the tracking of cancer drugs in living cells to monitor drug targets and downstream effectors (Savitski et al., 2014). Similarly, as proteins within a complex coaggregate upon heat denaturation, a new strategy of thermal proximity coaggregation (TPCA) has been introduced recently by Tan et al. (2018) (Fig. 1c). Combining with quantitative proteomics, system-wide profiling of protein complex dynamics in cells was conducted (Tan et al., 2018). However, limitations of this method are obvious, as some proteins require extreme temperature conditions. This is not practical when the whole proteome needs to be monitored, and some proteins do not change in their thermal stability with or without interacting with other proteins. In addition, protein‒protein interaction is not the only reason for change in the melting behavior of proteins. For example, small molecules and post-translational modifications also alter this behavior. We need to be careful when drawing conclusions, and independent validation of protein‒protein interaction is required (Mateus et al., 2020). Further work is needed to address these major limitations.
3 Application of interactome determination in the investigation of DNA repair pathways
Genome instability is one of the pervasive characteristics of most human cancers, which may be linked with increased DNA damage and decreased DNA repair capacity in cancers (Negrini et al., 2010; Lord and Ashworth, 2012). The integrity of the genome suffers from insults due to various exogenous (such as ultraviolet (UV) and ionizing radiation) and endogenous (such as reactive oxygen species and replication errors) damages (Friedberg et al., 2004). These DNA lesions require efficient and precise repair processes, the failure of which leads to reduced viability, genome instability, cancer development, senescence or aging (Hoeijmakers, 2009). Major causes of defective DNA repair are due to dysfunctional DNA repair genes and their associated functional networks. One efficient way to understand the molecular mechanisms underlying DNA damage response and DNA repair is to carry out interactome studies. Through the analysis of protein‒protein interaction networks, new functional partners, regulatory components, or signaling pathways can be uncovered. Moreover, the detailed physical and functional interactomes will facilitate the identification of key drug targets and improve current therapies.
Cells encountering endogenous lesions or exogenous genotoxic agents acquire various types of DNA damage. These include base and sugar damages at nucleotide level, single-strand and double-strand breaks, as well as DNA‒protein adducts. From the characteristics of DNA lesions, cells employ one or multiple DNA repair pathways to fix these lesions. Many key DNA repair proteins or protein complexes are revealed by interactome studies, which together elucidate complex DNA repair networks that are connected and coordinated to repair DNA damage and prevent genomic instability. Here, we present a few examples which applied MS to study protein‒protein interaction in the investigation of DNA repair pathways.
3.1. Direct reversal repair
Direct reversal repair is the most energy efficient way to repair DNA damage, but is only applicable to DNA damage with chemical changes in the bases (Yi and He, 2013). Methyl guanine methyl transferase (MGMT) is the key enzyme that reverses base methylation. Affinity purification of MGMT was carried out, which led to the identification of more than 60 MGMT-interacting proteins (Niture et al., 2005). Many of these MGMT-binding partners participate in DNA replication, DNA repair, and cell cycle progression. The association between proliferating cell nuclear antigen (PCNA) and MGMT was further investigated (Mostofa et al., 2018), revealing the relationship between this simple repair pathway and other cellular processes.
3.2. Repair of DNA damage on one strand
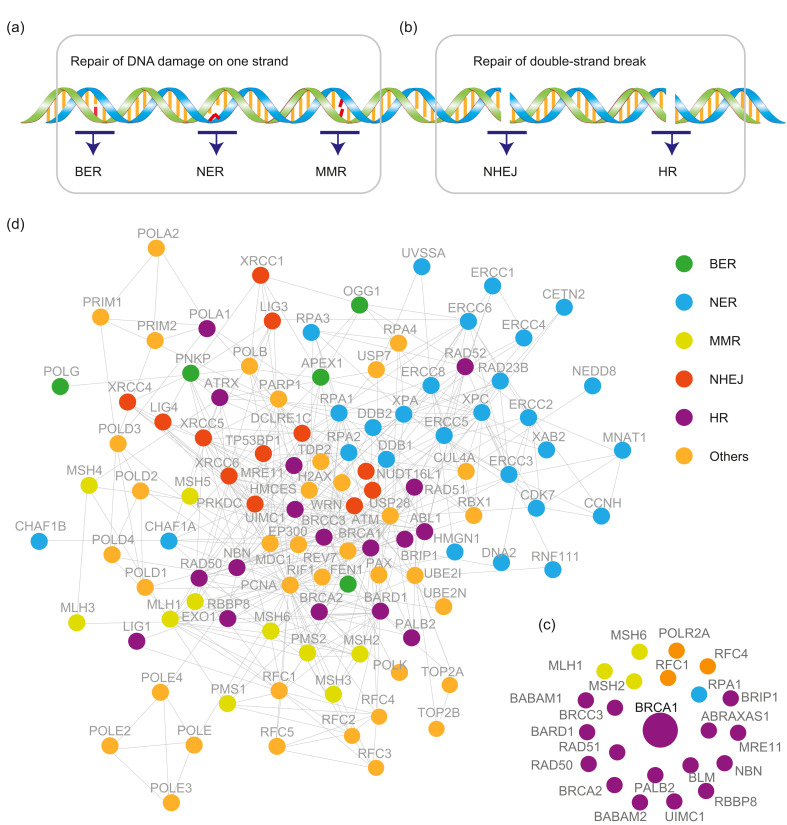
When damage occurs on one strand, the complementary intact DNA strand is used as the template to correct the DNA lesion. To replace the damaged or misincorporated nucleotide, three strategies may be used: base excision repair (BER), nucleotide excision repair (NER), and mismatch repair (MMR), as shown in Fig. 2a (Fleck and Nielsen, 2004).
Fig. 2. Interactome of DNA repair genes. (a, b) The major DNA repair pathways of DNA damage on one strand (a) or double strands (b). (c) The interactome of BRCA1 generated by Cytoscape 3.7.2 using the data from IPA and reference mining. (d) The interactome of DNA damage repair genes. The network was generated by the software IPA with manual check. In the figure, orange dots labeled as “others” indicate that the proteins play a role in multiple DNA repair pathways. BER: base excision repair; BRCA1: breast cancer type 1 susceptibility protein; HR: homologous recombination; IPA: Ingenuity Pathway Analysis; MMR: mismatch repair; NER: nucleotide excision repair; NHEJ: non-homologous end joining.
BER operates when a single base or nucleotide is damaged. BER machinery removes damaged bases, followed by the formation of a nick by apurinic/apyrimidinic (AP) endonucleases and insertion of the correct base. BER is the most used DNA repair pathway to correct base lesions caused by DNA damage, such as oxidation, alkylation, deamination, and depurinatiation/depyrimidination damages (Robertson et al., 2009). Co-IPs using FLAG-antibody were used in studying the interactome of several BER components. For example, DNA polymerase β is the key enzyme in BER, since it uses the complementary strand to fill the single nucleotide gap during the repair. Its interacting proteins were identified using anti-FLAG affinity purification, which led to the discovery of many well-known BER components including poly(ADP-ribose) polymerase 1 (PARP1), X-ray repair cross-complementing protein 1 (XRCC1), DNA ligase III (LIG3), bifunctional polynucleotide phosphatase/kinase (PNKP), and tyrosyl-DNA phosphodiesterase 1 (TDP1) (Prasad et al., 2012, 2015). DNA-(apurinic or apyrimidinic site) lyase (APEX1) functions as the AP endonuclease in the BER pathway. With the same co-IP strategy, an APEX1 protein‒protein interaction network was built, which revealed that the signature of APEX1 interactome is associated with poor prognosis in several types of cancers (Ayyildiz et al., 2020).
NER deals with bulky, helix-distorting damage. It removes an approximately 12‒24 nucleotide-long strand including the DNA lesion and then resynthesizes over the removed DNA region using the other strand as the template (Reardon and Sancar, 2006). DNA repair protein complementing XP-C cells (XPCs) acts as the DNA damage sensor in NER. The XPC protein interactome was first built with yeast two-hybrid screening and 49 novel interactors were identified (Lubin et al., 2014). The XPC interactome was again defined later with the use of immunoprecipitation (IP) coupled with MS analysis, which led to the hypothesis that the XPC‒PARP1 interaction may facilitate the search of XPC for DNA damage on chromatin DNA (Robu et al., 2017). The interactomes of other genes involved in the NER pathway are presented in large-scale protein‒protein interaction studies, such as the BioPlex Network database. This study was conducted with affinity purification (Huttlin et al., 2015, 2017) and the CoFrac database which used co-fractionation MS strategy (Havugimana et al., 2012; Wan et al., 2015). These datasets include a large number of baits, which comprise of some of the NER pathway genes/proteins, such as DNA excision repair protein ERCC-1 (ERCC1) or ERCC4. Given the nature of these large-scale studies, it is likely that more detailed interactome analysis of the NER pathway is needed to further reveal the detailed network associated with this repair pathway.
MMR is primarily responsible for the repair mismatches and insertion/deletion generated during DNA replication and recombination (Li, 2008). It involves at least two major steps before the action of polymerase and ligase. The first step is the detection of the damage, which requires the mutator S (MutS) protein complex (MSH2-MSH6 or MSH2-MSH3 heterodimer). The second step is recruitment of endonuclease to make a nick and then remove the newly synthesized DNA strand close to the damage site via exonuclease 1 (EXO1). The second step requires MutL heterodimeric complexes MutLα (MLH1-PMS2), MutLβ (MLH1-PMS1), or MutLγ (MLH1-MLH3) (Jiricny, 2006). The endonuclease involved remains unclear, but the data indicate that PMS2 may have the endonuclease active site (Kadyrov et al., 2006). The protein‒protein interactions of human MLH1, PMS1, and PMS2 have been reported with TAP-MS strategy, and the majority of them are new interacting partners (Cannavo et al., 2007). We used ten MMR pathway genes as baits, performed TAP-MS analysis, and built an interactome network of MMR with 230 HCIPs that may bind directly or indirectly to one or more components of the MMR pathway (Chen et al., 2016). We further validated a novel MSH2-MSH3/MSH6 complex-binding partner WDHD1 and showed that it is indeed involved in MMR (Chen et al., 2016).
The above interactome studies of repair pathways not only improve our understanding of these pathways, but also reveal their key modules and connections. As we expand the network of distinct DNA repair pathways, it becomes clear that many of them are interconnected and also join directly or indirectly with DNA replication and/or the use of DNA replication machinery. Studies are needed to further define these connections and reveal mechanistically how DNA replication and various DNA repair pathways intersect and communicate to ensure genome integrity.
3.3. Double-strand break repair
Double-strand breaks occur on both strands of the DNA double helix, and are particularly hazardous to cells. Several double-strand break repair mechanisms to repair the damage and maintain genome integrity have been discovered. Non-homologous end joining (NHEJ) and homologous recombination (HR) are the two most studied (Fig. 2b). Genetic mutation or disruption of any one of these pathways leads to genomic instability, which may result in cancer, developmental disorders, or aging (Scully et al., 2019).
NHEJ is believed to be the predominant double-strand break repair pathway in humans. It joins the two ends directly by recruiting DNA break-binding heterodimeric protein complex lupus Ku autoantigen protein p70 (KU70)/KU80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and the ligation complex that consists of DNA ligase IV, XRCC4, and XRCC4-like factor (XLF) (Brandsma and van Gent, 2012). Systematic study of 19 FLAG-tagged NHEJ proteins using co-IP strategy was conducted by Xing and colleagues. This led to the discovery of a new NHEJ factor XRCC4-like small protein (PAXX) as a KU complex-binding partner (Xing et al., 2015). The analysis of the PAXX interactome using GFP-tag and anti-GFP nanobody identified several known NHEJ factors (Tadi et al., 2016). More recently, the interactomes of PAXX, XLF, XRCC4, and DNA-PKcs were investigated and Pol λ was identified as a novel downstream effector of PAXX function (Craxton et al., 2018). The interactome of the KU complex was analyzed in depth with a combination of BioID and affinity purification methods, which revealed new protein complexes involved in the KU complex function (Abbasi and Schild-Poulter, 2019). The known NHEJ component Artemis forms a complex with DNA-PKcs and possesses exonuclease and endonuclease activity. PAX-interacting protein 1 (PAXIP1 or PTIP) was found to interact with Artemis via a TAP-MS approach, which trims DNA ends and promotes NHEJ (Wang et al., 2014). In the study of an AMP-activated protein kinase (AMPK) interactome, Artemis was also identified as an AMPK-binding protein, which implies a new function of AMPK in NHEJ (Chen et al., 2020). The interactome studies of the NHEJ pathway start to come together to establish an NHEJ interaction network, which will help in systematical analysis of this pathway in future.
HR is a highly accurate strategy for double-strand break repair. It requires the presence of an identical or nearly identical donor sequence for the repair of a DNA break, and this often involves the intact sister chromatid. HR plays critical roles not only in DNA repair, but also in the rescue of a stalled DNA replication fork, meiotic chromosome segregation, and telomere maintenance (Sung and Klein, 2006). BRCA1 and BRCA2 are well known tumor suppressors involved in the maintenance of genome stability, specifically for their roles in HR repair of DNA double-stand breaks. There are numerous interaction studies focusing on BRCA1, BRCA2, and other HR factors. We select only a few studies as examples here. Several well-known BRCA complex-binding proteins were first identified using yeast two-hybrid screening, co-IP-western, or other biochemical processes, such as the BRCA1‒BARD1 (BRCA1-associated RING domain protein 1) interaction (Wu et al., 1996), BRCA1‒RAD51 (DNA repair protein RAD51 homolog 1) (Scully et al., 1997), BRCA2‒RAD51 (Sharan et al., 1997), BRCA1‒BRCA2 (Chen et al., 1998), BRCA1‒RBBP8 (DNA endonuclease RBBP8, CtIP) (Wong et al., 1998; Yu et al., 1998; Li et al., 1999), BRCA1‒ATM (Serine-protein kinase ATM) (Cortez et al., 1999), and BRCA1‒MRN complex (MRE11-RAD50-NBS1) (Zhong et al., 1999; Wu et al., 2000). The arrival of MS technology greatly accelerated the identification of BRCA1/2 interaction proteins. At the beginning of MS, only single or several gel bands separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel could be analyzed due to the throughput of the MS technology. Analyzing a single band at about 130 kDa in the SDS-PAGE gel by MS, the Fanconi anemia group J protein (FANCJ, BRIP1, or BACH1) was identified as the directly interacting protein of BRCA1. This contributes to a key BRCA1 activity (Cantor et al., 2001). With the improvement and increased throughput of MS, it became possible to study the whole interactome of BRCA complexes. Wang et al. (2000) purified the BRCA1 complex through antibody affinity purification, which led to the identification of the BRCA1-associated genome surveillance complex (BASC). Many components identified in BASC play roles in the recognition of abnormal or damaged DNA, which suggests that BRCA1 and BASC may function in the maintenance of genomic integrity. BRCA1/BRCA2-containing complex (BRCC) was identified using FLAG-tagged BARD1, which is a multiprotein complex containing BRCA1, BARD1, BRCA2, and RAD51. This interactome study also led to the discovery of two novel components of this complex, BRCC3 and BRCC4 (Dong et al., 2003). The partner and localizer of BRCA2 (PALB2) was first identified as a BRCA2-interacting protein using antibody co-IP and MS (Xia et al., 2006). Our lab applied TAP and MS to further uncover the interactome of PALB2 and elucidated that not only BRCA2, but also BRCA1 interacted with PALB2 (Sy et al., 2009). The BRCA1‒PALB2 interaction was also discovered using affinity purification and MS (Zhang et al., 2009). The results from these two studies revealed that PALB2 is an integral component of the BRCA complex, which is important for the HR repair. Our group and others discovered BRCA1-A complex subunit Abraxas 1 (ABRAXAS1 or CCDC98) and BRCA1-A complex subunit RAP80 (UIMC1 or RAP80) as partners of BRCA1 using affinity purification or TAP methods. This suggests that a ubiquitination-dependent signaling pathway is involved in DNA damage response (Kim et al., 2007a, 2007b; Liu et al., 2007; Sobhian et al., 2007; Wang et al., 2007). The interactome of BRCA1 is summarized in Fig. 2c based on the data from the Ingenuity Pathway Analysis (IPA) software and reference mining. Besides the BRCA complex interactome, the interactomes of other HR repair-related genes have also been studied. The interactome of histone H2AX (H2AX) was explored with the TAP method and revealed many well-known DDR proteins as the top H2AX-interacting proteins (Leung et al., 2017). A new H2AX-binding protein zinc finger MYM-type protein 3 (ZMYM3) was further studied and shown to promote HR-mediated repair by regulating BRCA1 localization (Leung et al., 2017). The MRN complexes are key proteins involved in the recognition of double-strand breaks (DSBs) and the initiation of HR repair. The interactomes of MRE11, NBS1, and MDC1 were investigated using FLAG-tagged baits and IP-MS strategy, and identified USP7 as a binding partner of MRN and MDC1 and a participant in HR repair (Su et al., 2018).
Altogether, the interactomes of many proteins involved in DNA double-strand break repair provide new insights into these key DNA repair pathways that are important for survival, genome maintenance, and suppression of tumorigenesis.
3.4. Other key proteins in DNA repair
PCNA plays a key role in DDR by being conveniently positioned at replication forks to coordinate DNA replication with DNA repair and DNA damage tolerance pathways. The PCNA interactome was first constructed with affinity chromatography and revealed nearly 20 PCNA-binding proteins, including replication protein A 1 (RPA1), replication factor C subunits (RFC1/2/3/4/5), MSH3/6, POLD1/3, POLE, KU70, LIG1, and CHTF18 (Ohta et al., 2002). TAP-MS was applied later to identify novel PCNA-interacting proteins. For example, SprT-like domain-containing protein Spartan (SPRTN or C1orf124) was found binding to mono-ubiquitinated PCNA and participating in polymerase switch for cell survival following UV damage (Ghosal et al., 2012); SIVA1 was identified to interact with both RAD18 and PCNA and regulate RAD18-dependent PCNA monoubiquitination (Han et al., 2014); and TRAIP was shown to interact with PCNA to regulate replication fork recovery and progression (Feng et al., 2016). Combining BioID and TAP strategies, PCNA interactomes under normal and stressed conditions were also explored, which led to the discovery of many players involved in replication and replication stress response. Furthermore, two candidates, protein Wiz (WIZ) and Sal-like protein 1 (SALL1), were validated (Srivastava et al., 2018). In addition, a previously unknown protein abasic site processing protein HMCES (HMCES or C3orf37) was also identified as a PCNA-interacting protein in this study; this protein, HMCES, was later proven to be a critical component of the replication stress response which facilitates resolution of oxidative base damage (Srivastava et al., 2018, 2020). The binding between PCNA and HMCES was also confirmed by others using FLAG-tagged HMCES and co-IP study (Mohni et al., 2019).
RPA is a heterotrimeric complex composed of three subunits, RPA1 (RPA70), RPA2 (RPA32), and RPA3 (RPA14). RPA is the major protein complex that binds to single-stranded DNA (ssDNA). It is involved in NER, BER, and MMR repair pathways that repair lesions on one strand of DNA, as well as in HR repair pathway of DNA double-strand breaks (Maréchal and Zou, 2015). The interactome of RPA has been studied extensively with proteomics tools and many new binding proteins and their functions were reported. For example, with the TAP purific-ation approach, E3 ubiquitin-protein ligase RFWD3 (RFWD3) was identified as an RPA-binding protein, which mediates checkpoint activation and repair at stalled replication forks (Gong and Chen, 2011). The RPA interactome also led to the discovery of a DNA-directed primase/polymerase protein (PRIMPOL or CCDC111), which possesses primase and DNA polymerase activity, and binds to RPA in response to DNA replication stress (Wan et al., 2013). Additionally, a proteomic screen using biotinylated ssDNA coated with RPA proteins to capture RPA-binding partners uncovered pre-mRNA-processing factor 19 (PRPF19) as an ATR activator, which senses RPA-ssDNA during DDR (Maréchal et al., 2014). Affinity purification of FLAG-tagged RPA1 identified the RPA-binding protein Ewing's tumor-associated antigen 1 (ETAA1) which functions in parallel to the TOPBP1/RAD9/HUS1/RAD1 pathway to regulate ATR and maintains genome integrity (Bass et al., 2016). The interaction between RPA and ETAA1 was also discovered by TAP purification (Lee et al., 2016). Schlafen family member 11 (SLFN11) was identified as an RPA-interacting protein in a TAP-MS analysis and was shown to inhibit HR repair (Mu et al., 2016). Using an antibody of E3 ubiquitin-protein ligase HERC2 (HERC2) to perform co-IP and MS analysis, the RPA complex was identified with strong binding to HERC2, and this interaction plays a role in regulation of the G-quadruplex (Wu et al., 2018).
The Fanconi anemia (FA) pathway is a biochemical network that assists DNA interstrand crosslink repair in the genome (Rodríguez and D'Andrea, 2017). The FA pathway network has been shown to involve at least 22 FA or FA-like genes. These genes were named starting with the letters "FANC" except for some which have familiar recommended names, such as BRCA2 (FANCD1), BRIP1 (FANCJ), PALB2 (FANCN), RAD51C (FANCO), SLX4 (FANCP), XPF(FANCQ), BRCA1 (FANCS), UBE2T (FANCT), XRCC2 (FANCU), REV7 (FANCV), and RFWD3 (FANCW). The interactome study of FA genes helped elucidate their roles in DDR. Meetei et al. (2003) indicated that five core complex FA proteins (FANCA, FANCC, FANCE, FANCF, and FANCG) were co-IP in a complex together with Bloom syndrome protein (BLM), topoisomerase IIIα (TOPIIIα), and RPA, suggesting functional connections between FA and BLM/TOPIII. Understanding of key compositions of the FA core complex was revealed by IP of the whole complex through the pull-down of FANCA and protein identification by MS (Thomashevski et al., 2004). Similar approaches were used for the discovery of several FA-associated proteins such as Fanconi anemia core complex-associated protein 20 (FAAP20) (Ali et al., 2012; Leung et al., 2012; Yan et al., 2012) and FAAP100 (Ling et al., 2007). FANCM-MHF association and function in genome maintenance were revealed with co-IP and proteomics analysis (Yan et al., 2010). Using TAP purification or GFP-trap beads, Fanconi-associated nuclease 1 (FAN1) was identified as a FANCD2-binding protein that acts as a DNA repair nuclease participating in interstrand crosslink repair (Liu et al., 2010; Mackay et al., 2010). Affinity purification of FLAG-FANCD2 together with MS analysis uncovered additional FANCD2-associated proteins, including MCM proteins (Lossaint et al., 2013) and CtIP (Unno et al., 2014) that link the functions of FANCD2 to replisome and DNA interstrand crosslink repair, respectively. The interactome of SLX4 was studied by several groups, which clearly demonstrated its role as the key mediator that associates with several structural specific nucleases involved in distinct repair pathways (Andersen et al., 2009; Fekairi et al., 2009; Muñoz et al., 2009; Svendsen et al., 2009). More recently, TAP strategy was used to define SLX4 and SLX4IP interactomes, which further support the functional connection between SLX4/SLX4IP and XPF-ERCC1 complexes (Zhang et al., 2019).
TP53BP1 plays a key role in DNA double-strand break repair in response to DNA damage by promoting NHEJ-mediated repair and specifically counteracting HR repair in the absence of BRCA1 (Chapman et al., 2012). TP53BP1 was first found to co-localize with phosphorylated H2AX and the two biochemically interact with each other after radiation, suggesting that TP53BP1 is involved early in DDR (Rappold et al., 2001). Zhang et al. (2006) identified ubiquitin carboxyl-terminal hydrolase 28 (USP28) as a new TP53BP1-binding protein in responding to DNA damage. Additionally, the Tudor-interacting repair regulator protein (TIRR or NUDT16L1) was discovered when the TP53BP1 complex was purified using TAP (Zhang et al., 2017) or double co-IP purification methods (using FLAG- and HA-epitope tags to do co-IP) (Drané et al., 2017). NUDT16L1 turns out to be a new factor that influences double-strand break repair by masking the binding of TP53BP1 to methylated histone (Drané et al., 2017; Botuyan et al., 2018). High resolution interaction neighborhood maps of TP53BP1, BRCA1, and MDC1 were delineated by APEX2-tagged endogenously expressed proteins (Gupta et al., 2018), which led to the discovery of a novel protein complex shieldin (REV7 plus RINN1, RINN2, and RINN3) associated with TP53BP1. The shieldin complex has been validated by several groups to function as a downstream effector of TP53BP1-RIF1 in DNA repair (Ghezraoui et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018).
PARP1 and PARP2 are the poly-ADP-ribosyltransferases that mediate the poly-ADP-ribosylation of themselves as well as their substrates, and play key roles in DNA repair. PARG is the poly-ADP-ribose glycohydrolase that reverses the action of PARP1/PARP2. The interactomes of these three proteins were conducted by co-IP using anti-PARP1 antibody or anti-FLAG antibody for the other two genes, which revealed 179 interactions, 139 of which are novel interactions (Isabelle et al., 2010). The interactome of PARG was also established and a novel non-canonical PCNA-interacting protein motif (PIP)-box-mediated interaction between PARG and PCNA was unearthed (Kaufmann et al., 2017).
Taken together, the interactome studies focusing on DNA repair have been conducted extensively using single genes or small groups of genes. These studies are excellent examples to guide future comprehensive investigation of DDR interactomes. It is anticipated that advanced technologies and large-scale interactome analysis on DDR will help build an all-inclusive network of DDR. The establishment of DDR networks should benefit research and treatment for human diseases associated with defects in DNA damage response and DNA repair. In Fig. 2d, we organize the data from IPA analysis, including BER, NER, MMR, NHEJ, and HR DNA repair pathway genes. Using these data together with reference mining, we built an interactome network of many DNA repair genes.
4 Future perspectives
Interactome studies armed with advanced proteomics tools launch a new era of discovery-driven research and have dramatically accelerated the pace of discovering new knowledge in the area. The strategies for enrichment of interacting proteins and tools for proteomics studies are developing rapidly and becoming more powerful; however, the main challenge remains in data processing and analysis. All protein‒protein interaction experiments are presented with many non-specific contaminants in the raw identification lists. Efficient bioinformatics analysis and algorithms are needed to recognize the useful information in the sea of specific and non-specific signals. The continual expansion of datasets with small- to large-scale interaction studies will facilitate the detection of true binding proteins and build reliable interaction networks. Furthermore, the integration of interactome data with other omics data, such as proteome, transcriptome, and genome-wide screening with CRISPR, will offer us multiple ways to analyze these data and make new discoveries. For example, combined analysis of interactome and transcriptome highlighted genes that are differentially expressed in lung cancer tissue, which may provide therapeutic targets in cancer treatment (Wachi et al., 2005). Another example is integrated interactome, transcriptome, proteome, ubiquitinome, and phosphoproteome to study the influence of SARS-CoV-2 on an infected host. The analysis of these multiple-omics data revealed relationships between virus and host, and discovered new targets that can be tested with existing clinically approved drugs (Stukalov et al., 2020).
Although more and more players involved in DDR and genome maintenance have been uncovered due to the fast progress of interactome research, it is still interesting to discover new DDR-binding partners, which may show relatively weaker binding ability. Another future direction is the dynamics of DNA repair. The dynamic changes of protein‒protein interactions are known to be critical for checkpoint activation and DNA repair following DNA damage. The interactome dynamics will reveal precisely how these interactions are controlled and promote distinct cellular functions following DNA damage. Major challenges in dynamics interactome research are how to capture these binding partners and quantify their changed levels. The rapidly evolving proximity labeling techniques, such as APEX2 and TurboID, help uncover relatively weak, transient-associated proteins as well as adjacent proteins. These techniques can label the surrounding proteins in a short time period, and help to capture fast-changing protein‒protein interactions after DNA damage. At the same time, quantitative proteomics and MS are becoming faster, more precise, and more accurate. It is now possible to correctly quantify the low abundance proteins. The development of these techniques helps research on dynamic interactomes in response to any stimuli, including but not limited to DNA damage.
The majority of the interactome studies were carried out in well-established cell lines, such as the HEK293T cells. This is informative but always comes with the concern that the identified protein‒protein interaction may not happen in vivo. Performing protein‒protein studies with more relevant cell lines or in vivo may obtain additional information for building the interaction network. Model organisms or clinical samples may be used to address this concern, but sample- handing techniques require further development for interactome research. We need to find efficient ways to extract proteins from fresh tissue or Formalin-fixed samples, since in vivo tissues, especially the clinical samples, are always limited. Along with fast developing ultra-sensitive MS techniques, highly sensitive protein‒protein interaction assays need to be further improved. As targeting DDR has rapidly expanded to disease treatment (Pilié et al., 2019) and protein‒protein interactions are increasingly targeted by drug discovery groups (Scott et al., 2016), it is time to expand interactome studies in vivoand/or in more relevant tissues/samples to fulfil their promise.
Cells can be viewed as a complex web with constant and regulated macromolecular interactions. With these controlled and dynamic interactions, cellular processes can be carried out quickly and precisely according to endogenous and environmental cues. Disturbing these interactions and their regulations will likely lead to disorders or diseases. Moreover, precise understanding of the interactomes involved in DNA repair genes may be used in drug development and the design of new treatments for patients. Indeed, a recent study reported an SARS-CoV-2 virus-host protein‒protein interaction map, which may help in the screening of drugs for coronavirus disease 2019 (COVID-19) treatment (Gordon et al., 2020). This kind of interactome study concentrated on a group of genes that are implicated in the same disease will help us build disease-specific protein‒protein interaction networks. Such networks will be useful in uncovering the molecular mechanisms underlying disease development and in selecting key targets for disease treatment.
Acknowledgments
Junjie CHEN received support from the Pamela and Wayne Garrison Distinguished Chair in Cancer Research, the Cancer Prevention & Research Institute of Texas (Nos. RP160667 and RP180813) and the National Institutes of Health (NIH) (Nos. CA210929, CA216911, and CA216437). We apologize to many colleagues whose outstanding work was not cited because of the page limitation. We thank all the members of the CHEN laboratory for their help and constructive discussion. We thank Mrinal SRIVASTAVA and Samuel K. SWISHER for critically reading the manuscript and insightful discussions.
Author contributions
Zhen CHEN and Junjie CHEN wrote and edited the manuscript and created the figures, and have read and approved the final version.
Compliance with ethics guidelines
Zhen CHEN and Junjie CHEN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by either of the authors.
References
- Abbasi S, Schild-Poulter C, 2019. Mapping the Ku interactome using proximity-dependent biotin identification in human cells. J Proteome Res, 18(3): 1064-1077. 10.1021/acs.jproteome.8b00771 [DOI] [PubMed] [Google Scholar]
- Ali AM, Pradhan A, Singh TR, et al. , 2012. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood, 119(14): 3285-3294. 10.1182/blood-2011-10-385963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, et al. , 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell, 35(1): 128-135. 10.1016/j.molcel.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyildiz D, Antoniali G, D'Ambrosio C, et al. , 2020. Architecture of the human APE1 interactome defines novel cancers signatures. Sci Rep, 10: 28 10.1038/s41598-019-56981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagci H, Sriskandarajah N, Robert A, et al. , 2020. Mapping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mechanisms. Nat Cell Biol, 22(1): 120-134. 10.1038/s41556-019-0438-7 [DOI] [PubMed] [Google Scholar]
- Bass TE, Luzwick JW, Kavanaugh G, et al. , 2016. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol, 18(11): 1185-1195. 10.1038/ncb3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra J, Hultquist JF, Liu DD, et al. , 2018. Protein interaction mapping identifies RBBP6 as a negative regulator of Ebola virus replication. Cell, 175(7): 1917-1930. e13 10.1016/j.cell.2018.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymaz HI, Spruijt CG, Vermeulen M, 2014. Identifying nuclear protein‒protein interactions using GFP affinity purification and SILAC-based quantitative mass spectrometry. In: Warscheid B (Ed.), Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC): Methods and Protocols. Humana Press, New York, p.207-226. 10.1007/978-1-4939-1142-4_15 [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Cui GF, Drané P, et al. , 2018. Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein. Nat Struct Mol Biol, 25(7): 591-600. 10.1038/s41594-018-0083-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma I, van Gent DC, 2012. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr, 3: 9 10.1186/2041-9414-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, et al. , 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol, 36(9): 880-887. 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Gerrits B, Marra G, et al. , 2007. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem, 282(5): 2976-2986. 10.1074/jbc.M609989200 [DOI] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, et al. , 2001. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell, 105(1): 149-160. 10.1016/s0092-8674(01)00304-x [DOI] [PubMed] [Google Scholar]
- Castello A, Horos R, Strein C, et al. , 2016. Comprehensive identification of RNA-binding proteins by RNA interactome capture. In: Dassi E (Ed.), Post-Transcriptional Gene Regulation. Humana Press, New York, p.131-139. 10.1007/978-1-4939-3067-8_8 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, et al. , 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci, 125: 3529- 3534. 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez JD, Schweppe DK, Eng JK, et al. , 2016. In vivo conformational dynamics of Hsp90 and its interactors. Cell Chem Biol, 23(6): 716-726. 10.1016/j.chembiol.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A, 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE, 2005( 266): pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, et al. , 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev, 18(18): 2255-2268. 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Silver DP, Walpita D, et al. , 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell, 2(3): 317-328. 10.1016/s1097-2765(00)80276-2 [DOI] [PubMed] [Google Scholar]
- Chen Z, Tran M, Tang MF, et al. , 2016. Proteomic analysis reveals a novel mutator S (MutS) partner involved in mismatch repair pathway. Mol Cell Proteomics, 15(4): 1299-1308. 10.1074/mcp.M115.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Lei CQ, Wang C, et al. , 2019. Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat Commun, 10: 104 10.1038/s41467-018-08004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang C, Jain A, et al. , 2020. AMPK interactome reveals new function in non-homologous end joining DNA repair. Mol Cell Proteomics, 19(3): 467-477. 10.1074/mcp.RA119.001794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KF, Branon TC, Rajeev S, et al. , 2020. Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci USA, 117( 22): 12143-12154. 10.1073/pnas.1919528117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, et al. , 1999. Requirement of ATM-dependent phosphorylation of BRCA1 in the DNA damage response to double-strand breaks. Science, 286(5442): 1162-1166. 10.1126/science.286.5442.1162 [DOI] [PubMed] [Google Scholar]
- Cortez-Retamozo V, Backmann N, Senter PD, et al. , 2004. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res, 64(8): 2853-2857. 10.1158/0008-5472.can-03-3935 [DOI] [PubMed] [Google Scholar]
- Coyaud E, Ranadheera C, Cheng D, et al. , 2018. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics, 17(11): 2242-2255. 10.1074/mcp.TIR118.000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A, Munnur D, Jukes-Jones R, et al. , 2018. PAXX and its paralogs synergistically direct DNA polymerase λ activity in DNA repair. Nat Commun, 9: 3877 10.1038/s41467-018-06127-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, et al. , 2005. Fluorescent proteins as proteomic probes. Mol Cell Proteomics, 4(12): 1933-1941. 10.1074/mcp.M500227-MCP200 [DOI] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, et al. , 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell, 57(2): 349-360. 10.1016/j.molcel.2014.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munter S, Görnemann J, Derua R, et al. , 2017. Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett, 591(2): 415-424. 10.1002/1873-3468.12548 [DOI] [PubMed] [Google Scholar]
- Diep J, Ooi YS, Wilkinson AW, et al. , 2019. Enterovirus pathogenesis requires the host methyltransferase SETD3. Nat Microbiol, 4(12): 2523-2537. 10.1038/s41564-019-0551-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YS, Hakimi MA, Chen XW, et al. , 2003. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell, 12(5): 1087-1099. 10.1016/s1097-2765(03)00424-6 [DOI] [PubMed] [Google Scholar]
- Drané P, Brault ME, Cui GF, et al. , 2017. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature, 543(7644): 211-216. 10.1038/nature21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M, Zhang W, Gross AM, et al. , 2018. Multiple routes to oncogenesis are promoted by the human papillomavirus-host protein network. Cancer Discov, 8(11): 1474-1489. 10.1158/2159-8290.CD-17-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, et al. , 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell, 138(1): 78-89. 10.1016/j.cell.2009.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Liu CZ, Spinozzi S, et al. , 2020. Identifying the cardiac dyad proteome in vivo by a BioID2 knock-in strategy. Circulation, 141(11): 940-942. 10.1161/circulationaha.119.043434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng WJ, Guo YY, Huang J, et al. , 2016. TRAIP regulates replication fork recovery and progression via PCNA. Cell Discov, 2: 16016 10.1038/celldisc.2016.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck O, Nielsen O, 2004. DNA repair. J Cell Sci, 117(4): 515-517. 10.1242/jcs.00952 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, McDaniel LD, Schultz RA, 2004. The role of endogenous and exogenous DNA damage and mutagenesis. Curr Opin Genet Dev, 14(1): 5-10. 10.1016/j.gde.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Gao XD, Tu LC, Mir A, et al. , 2018. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat Methods, 15(6): 433-436. 10.1038/s41592-018-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, et al. , 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415(6868): 141-147. 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, et al. , 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature, 440(7084): 631-636. 10.1038/nature04532 [DOI] [PubMed] [Google Scholar]
- Gheiratmand L, Coyaud E, Gupta GD, et al. , 2019. Spatial and proteomic profiling reveals centrosome-independent features of centriolar satellites. EMBO J, 38(14): e101109 10.15252/embj.2018101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Oliveira C, Becker JR, et al. , 2018. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature, 560(7716): 122-127. 10.1038/s41586-018-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Leung JWC, Nair BC, et al. , 2012. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J Biol Chem, 287(41): 34225-34233. 10.1074/jbc.M112.400135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go CD, Knight JDR, Rajasekharan A, et al. , 2019. A proximity biotinylation map of a human cell. bioRxiv, preprint. 10.1101/796391 [DOI] [Google Scholar]
- Gonatopoulos-Pournatzis T, Wu MK, Braunschweig U, et al. , 2018. Genome-wide CRISPR-Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Mol Cell, 72(3): 510-524. e12 10.1016/j.molcel.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Gong ZH, Chen JJ, 2011. E3 ligase RFWD3 participates in replication checkpoint control. J Biol Chem, 286(25): 22308-22313. 10.1074/jbc.M111.222869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, et al. , 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816): 459-468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, É Coyaud, Gonçalves J, et al. , 2015. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell, 163(6): 1484-1499. 10.1016/j.cell.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Somyajit K, Narita T, et al. , 2018. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell, 173(4): 972-988. e23 10.1016/j.cell.2018.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Liu T, Huen MS, et al. , 2014. SIVA1 directs the E3 ubiquitin ligase RAD18 for PCNA monoubiquitination. J Cell Biol, 205(6): 811-827. 10.1083/jcb.201311007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YS, Branon TC, Martell JD, et al. , 2019. Directed evolution of split APEX2 peroxidase. ACS Chem Biol, 14(4): 619-635. 10.1021/acschembio.8b00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, et al. , 2012. A census of human soluble protein complexes. Cell, 150(5): 1068-1081. 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, et al. , 2015. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell, 163(3): 712-723. 10.1016/j.cell.2015.09.053 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, et al. , 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415(6868): 180-183. 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH, 2009. DNA damage, aging, and cancer. N Engl J Med, 361(15): 1475-1485. 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Holthenrich A, Drexler HCA, Chehab T, et al. , 2019. Proximity proteomics of endothelial Weibel-Palade bodies identifies novel regulator of von Willebrand factor secretion. Blood, 134(12): 979-982. 10.1182/blood.2019000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee HW, et al. , 2014. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell, 55(2): 332-341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Udeshi ND, Lam SS, et al. , 2016. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc, 11(3): 456-475. 10.1038/nprot.2016.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, et al. , 2015. The BioPlex network: a systematic exploration of the human interactome. Cell, 162(2): 425-440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, et al. , 2017. Architecture of the human interactome defines protein communities and disease networks. Nature, 545(7655): 505-509. 10.1038/nature22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Navarrete-Perea J, et al. , 2020. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. bioRxiv, preprint. 10.1101/2020.01.19.905109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M, Moreel X, Gagné JP, et al. , 2010. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci, 8: 22 10.1186/1477-5956-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, et al. , 2012. Global landscape of HIV-human protein complexes. Nature, 481(7381): 365-370. 10.1038/nature10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J, 2006. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol, 7(5): 335-346. 10.1038/nrm1907 [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, et al. , 2006. Endonucleolytic function of MutLα in human mismatch repair. Cell, 126(2): 297-308. 10.1016/j.cell.2006.05.039 [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Grishkovskaya I, Polyansky AA, et al. , 2017. A novel non-canonical PIP-box mediates PARG interaction with PCNA. Nucleic Acids Res, 45(16): 9741-9759. 10.1093/nar/gkx604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Roux KJ, 2016. Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol, 26(11): 804-817. 10.1016/j.tcb.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu WH, et al. , 2014. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA, 111(24): E2453-E2461. 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, et al. , 2016. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell, 27(8): 1188-1196. 10.1091/mbc.E15-12-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Huang J, Chen JJ, 2007a. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol, 14(8): 710-715. 10.1038/nsmb1277 [DOI] [PubMed] [Google Scholar]
- Kim H, Chen JJ, Yu XC, 2007b. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science, 316(5828): 1202-1205. 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Gsponer J, Foster LJ, 2012. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods, 9(9): 907-909. 10.1038/nmeth.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu HY, et al. , 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature, 440(7084): 637-643. 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, et al. , 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods, 12(1): 51-54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M, Kirkwood KJ, Tinti M, et al. , 2016. Global membrane protein interactome analysis using in vivo crosslinking and mass spectrometry-based protein correlation profiling. Mol Cell Proteomics, 15(7): 2476-2490. 10.1074/mcp.O115.055467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Guerroué F, Eck F, Jung J, et al. , 2017. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol Cell, 68(4): 786-796. e6 10.1016/j.molcel.2017.10.029 [DOI] [PubMed] [Google Scholar]
- Lee YC, Zhou Q, Chen JJ, et al. , 2016. RPA-binding protein ETAA1 is an ATR activator involved in DNA replication stress response. Curr Biol, 26(24): 3257-3268. 10.1016/j.cub.2016.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JWC, Wang YC, Fong KW, et al. , 2012. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci USA, 109(12): 4491-4496. 10.1073/pnas.1118720109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JWC, Makharashvili N, Agarwal P, et al. , 2017. ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes Dev, 31(3): 260-274. 10.1101/gad.292516.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, 2008. Mechanisms and functions of DNA mismatch repair. Cell Res, 18(1): 85-98. 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- Li MH, Johnson JR, Truong B, et al. , 2019. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol, 4(6): 985-995. 10.1038/s41564-019-0375-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen PL, Subramanian T, et al. , 1999. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J Biol Chem, 274(16): 11334-11338. 10.1074/jbc.274.16.11334 [DOI] [PubMed] [Google Scholar]
- Li X, Wang WQ, Wang JD, et al. , 2015. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol, 11(1): 775 10.15252/msb.20145504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao M, Choi JM, et al. , 2017. Clustered, regularly interspaced short palindromic repeats (CRISPR)/Cas9-coupled affinity purification/mass spectrometry analysis revealed a novel role of neurofibromin in mTOR signaling. Mol Cell Proteomics, 16(4): 594-607. 10.1074/mcp.M116.064543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Ishiai M, Ali AM, et al. , 2007. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J, 26(8): 2104-2114. 10.1038/sj.emboj.7601666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Heck AJR, 2015. Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr Opin Struct Biol, 35: 100-108. 10.1016/j.sbi.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Liu F, Rijkers DTS, Post H, et al. , 2015. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods, 12(12): 1179-1184. 10.1038/nmeth.3603 [DOI] [PubMed] [Google Scholar]
- Liu F, Lössl P, Rabbitts BM, et al. , 2018. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol Cell Proteomics, 17(2): 216-232. 10.1074/mcp.RA117.000470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan JS, et al. , 2010. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science, 329(5992): 693-696. 10.1126/science.1192656 [DOI] [PubMed] [Google Scholar]
- Liu WS, Song HP, Chen Q, et al. , 2018. Recent advances in the selection and identification of antigen-specific nanobodies. Mol Immunol, 96: 37-47. 10.1016/j.molimm.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Liu XN, Salokas K, Tamene F, et al. , 2018. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat Commun, 9: 1188 10.1038/s41467-018-03523-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZX, Wu JX, Yu XC, 2007. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol, 14(8): 716-720. 10.1038/nsmb1279 [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A, 2012. The DNA damage response and cancer therapy. Nature, 481(7381): 287-294. 10.1038/nature10760 [DOI] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, et al. , 2013. FANCD2 binds MCM proteins and controls replisome function upon activation of S phase checkpoint signaling. Mol Cell, 51(5): 678-690. 10.1016/j.molcel.2013.07.023 [DOI] [PubMed] [Google Scholar]
- Lubin A, Zhang L, Chen H, et al. , 2014. A human XPC protein interactome—a resource. Int J Mol Sci, 15(1): 141-158. 10.3390/ijms15010141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay C, Déclais AC, Lundin C, et al. , 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell, 142(1): 65-76. 10.1016/j.cell.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, et al. , 2011. Analysis of the human endogenous coregulator complexome. Cell, 145(5): 787-799. 10.1016/j.cell.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Zou L, 2015. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res, 25(1): 9-23. 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Li JM, Ji XY, et al. , 2014. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell, 53(2): 235-246. 10.1016/j.molcel.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, et al. , 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol, 30(11): 1143-1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus A, Kurzawa N, Becher I, et al. , 2020. Thermal proteome profiling for interrogating protein interactions. Mol Syst Biol, 16(3): e9232 10.15252/msb.20199232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, et al. , 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet, 35(2): 165-170. 10.1038/ng1241 [DOI] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, et al. , 2018. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature, 560(7716): 112-116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrashidi KM, Elwell CA, Verschueren E, et al. , 2015. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe, 18(1): 109-121. 10.1016/j.chom.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohni KN, Wessel SR, Zhao RX, et al. , 2019. HMCES maintains genome integrity by shielding abasic sites in single-strand DNA. Cell, 176(1-2): 144-153. e13 10.1016/j.cell.2018.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostofa AGM, Punganuru SR, Madala HR, et al. , 2018. S-phase specific downregulation of human O 6-methylguanine DNA methyltransferase (MGMT) and its serendipitous interactions with PCNA and p21cip1 proteins in glioma cells. Neoplasia, 20(4): 305-323. 10.1016/j.neo.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu YH, Lou JM, Srivastava M, et al. , 2016. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep, 17(1): 94-109. 10.15252/embr.201540964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais AC, et al. , 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell, 35(1): 116-127. 10.1016/j.molcel.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Muyldermans S, 2013. Nanobodies: natural single-domain antibodies. Annu Rev Biochem, 82: 775-797. 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- Myers SA, Wright J, Peckner R, et al. , 2018. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat Methods, 15(6): 437-439. 10.1038/s41592-018-0007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD, 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol, 11(3): 220-228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Niture SK, Doneanu CE, Velu CS, et al. , 2005. Proteomic analysis of human O 6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem Biophys Res Commun, 337(4): 1176-1184. 10.1016/j.bbrc.2005.09.177 [DOI] [PubMed] [Google Scholar]
- Noordermeer SM, Adam S, Setiaputra D, et al. , 2018. The shieldin complex mediates 53BP1-dependent DNA repair. Nature, 560(7716): 117-121. 10.1038/s41586-018-0340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Shiomi Y, Sugimoto K, et al. , 2002. A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs 2. ‒5 complex as a novel PCNA-binding protein. J Biol Chem, 277(43): 40362-40367. 10.1074/jbc.M206194200 [DOI] [PubMed] [Google Scholar]
- Olson MG, Widner RE, Jorgenson LM, et al. , 2019. Proximity labeling to map host-pathogen interactions at the membrane of a bacterium-containing vacuole in Chlamydia trachomatis-infected human cells. Infect Immun, 87(11): e0053719 10.1128/IAI.00537-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly FJ, Rappsilber J, 2018. Cross-linking mass spectrometry: methods and applications in structural, molecular and systems biology. Nat Struct Mol Biol, 25(11): 1000-1008. 10.1038/s41594-018-0147-0 [DOI] [PubMed] [Google Scholar]
- Penn BH, Netter Z, Johnson JR, et al. , 2018. An Mtb-human protein‒protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol Cell, 71(4): 637-648. e5 10.1016/j.molcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilié PG, Tang C, Mills GB, et al. , 2019. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol, 16(2): 81-104. 10.1038/s41571-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Williams JG, Hou EW, et al. , 2012. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res, 40(22): 11571-11582. 10.1093/nar/gks898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Dyrkheeva N, Williams J, et al. , 2015. Mammalian base excision repair: functional partnership between PARP-1 and APE1 in AP-site repair. PLoS ONE, 10(5): e0124269. 10.1371/journal.pone.0124269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Kumar GR, Verschueren E, et al. , 2015. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell, 57(2): 329-340. 10.1016/j.molcel.2014.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M, Majzoub K, Rao DS, et al. , 2018. RNA‒protein interaction detection in living cells. Nat Methods, 15(3): 207-212. 10.1038/nmeth.4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, et al. , 2001. Tumor suppressor p53 binding protein 1 (53bp1) is involved in DNA damage‒signaling pathways. J Cell Biol, 153(3): 613-620. 10.1083/jcb.153.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Sancar A, 2006. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol, 408: 189-213. 10.1016/S0076-6879(06)08012-8 [DOI] [PubMed] [Google Scholar]
- Rees JS, Li XW, Perrett S, et al. , 2015. Protein neighbors and proximity proteomics. Mol Cell Proteomics, 14(11): 2848-2856. 10.1074/mcp.R115.052902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, et al. , 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science, 339(6125): 1328-1331. 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, et al. , 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol, 17(10): 1030-1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, et al. , 2009. DNA repair in mammalian cells. Cell Mol Life Sci, 66(6): 981-993. 10.1007/s00018-009-8736-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M, Shah RG, Purohit NK, et al. , 2017. Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc Natl Acad Sci USA, 114(33): E6847-E6856. 10.1073/pnas.1706981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A, D'Andrea A, 2017. Fanconi anemia pathway. Curr Biol, 27(18): R986-R988. 10.1016/j.cub.2017.07.043 [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, et al. , 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol, 196(6): 801-810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Burke B, 2013. BioID: a screen for protein‒protein interactions. Curr Protoc Protein Sci, 74(1): 19.23.1-19.23.14. 10.1002/0471140864.ps1923s74 [DOI] [PubMed] [Google Scholar]
- Savitski MM, Reinhard FBM, Franken H, et al. , 2014. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science, 346(6205): 1255784 10.1126/science.1255784 [DOI] [PubMed] [Google Scholar]
- Schenstrom SM, Rebula CA, Tatham MH, et al. , 2018. Expanded interactome of the intrinsically disordered protein Dss1. Cell Rep, 25(4): 862-870. 10.1016/j.celrep.2018.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TGM, Skerra A, 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protocols, 2(6): 1528-1535. 10.1038/nprot.2007.209 [DOI] [PubMed] [Google Scholar]
- Schmidtmann E, Anton T, Rombaut P, et al. , 2016. Determination of local chromatin composition by CasID. Nucleus, 7(5): 476-484. 10.1080/19491034.2016.1239000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp IM, Béthune J, 2018. Split-BioID—proteomic analysis of context-specific protein complexes in their native cellular environment. J Vis Exp, (134): e57479 10.3791/57479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp IM, Amaya Ramirez CC, Debeljak J, et al. , 2017. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat Commun, 8: 15690 10.1038/ncomms15690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Harding C, Chavez JD, et al. , 2015. Host-microbe protein interactions during bacterial infection. Chem Biol, 22(11): 1521-1530. 10.1016/j.chembiol.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Chavez JD, Lee CF, et al. , 2017. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci USA, 114(7): 1732-1737. 10.1073/pnas.1617220114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DE, Bayly AR, Abell C, et al. , 2016. Small molecules, big targets: drug discovery faces the protein‒protein interaction challenge. Nat Rev Drug Discov, 15(8): 533-550. 10.1038/nrd.2016.29 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen JJ, Plug A, et al. , 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88(2): 265-275. 10.1016/s0092-8674(00)81847-4 [DOI] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, et al. , 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 20(11): 698-714. 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PS, Link N, Jang GM, et al. , 2018. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell, 175(7): 1931-1945. e18 10.1016/j.cell.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, et al. , 1997. Embryonic lethality and radiation hypersensitivity mediated by RAD51 in mice lacking BRCA2 . Nature, 386(6627): 804-810. 10.1038/386804a0 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao GZ, Lilli DR, et al. , 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science, 316(5828): 1198-1202. 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, et al. , 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell, 138(2): 389-403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Chen Z, Zhang HM, et al. , 2018. Replisome dynamics and their functional relevance upon DNA damage through the PCNA interactome. Cell Rep, 25(13): 3869-3883. e4 10.1016/j.celrep.2018.11.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Su D, Zhang HM, et al. , 2020. HMCES safeguards replication from oxidative stress and ensures error-free repair. EMBO Rep, 21(6): e49123 10.15252/embr.201949123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey RG, Skinnider MA, Scott NE, et al. , 2017. A rapid and accurate approach for prediction of interactomes from co-elution data (PrInCE). BMC Bioinformatics, 18: 457 10.1186/s12859-017-1865-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis N, Gupta GD, Lin ZY, et al. , 2016. Phenotypic and interaction profiling of the human phosphatases identifies diverse mitotic regulators. Cell Rep, 17(9): 2488-2501. 10.1016/j.celrep.2016.10.078 [DOI] [PubMed] [Google Scholar]
- Stukalov A, Girault V, Grass V, et al. , 2020. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv, preprint. 10.1101/2020.06.17.156455 [DOI] [Google Scholar]
- Su DX, Ma S, Shan L, et al. , 2018. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J Clin Invest, 128(10): 4280-4296. 10.1172/JCI120518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Klein H, 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol, 7(10): 739-750. 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, et al. , 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell, 138(1): 63-77. 10.1016/j.cell.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SMH, Huen MSY, Chen JJ, 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA, 106(17): 7155-7160. 10.1073/pnas.0811159106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadi SK, Tellier-Lebègue C, Nemoz C, et al. , 2016. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep, 17(2): 541-555. 10.1016/j.celrep.2016.09.026 [DOI] [PubMed] [Google Scholar]
- Tan CSH, Go KD, Bisteau X, et al. , 2018. Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science, 359(6380): 1170-1177. 10.1126/science.aan0346 [DOI] [PubMed] [Google Scholar]
- Thomashevski A, High AA, Drozd M, et al. , 2004. The Fanconi anemia core complex forms four complexes of different sizes in different subcellular compartments. J Biol Chem, 279(25): 26201-26209. 10.1074/jbc.M400091200 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, 2019. Recent advances in proximity-based labeling methods for interactome mapping. F1000Res, 8: 135 10.12688/f1000research.16903.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno J, Itaya A, Taoka M, et al. , 2014. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep, 7(4): 1039-1047. 10.1016/j.celrep.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Wachi S, Yoneda K, Wu RE, 2005. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics, 21(23): 4205-4208. 10.1093/bioinformatics/bti688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CH, Borgeson B, Phanse S, et al. , 2015. Panorama of ancient metazoan macromolecular complexes. Nature, 525(7569): 339-344. 10.1038/nature14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Lou JM, Xia YS, et al. , 2013. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep, 14(12): 1104-1112. 10.1038/embor.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, et al. , 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science, 316(5828): 1194-1198. 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Aroumougame A, Lobrich M, et al. , 2014. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev, 28(24): 2693-2698. 10.1101/gad.252478.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, et al. , 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev, 14(8): 927-939. 10.1101/gad.14.8.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AKC, Ormonde PA, Pero R, et al. , 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene, 17(18): 2279-2285. 10.1038/sj.onc.1202150 [DOI] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, et al. , 1996. Identification of a ring protein that can interact in vivo with the BRCA1 gene product. Nat Genet, 14(4): 430-440. 10.1038/ng1296-430 [DOI] [PubMed] [Google Scholar]
- Wu WW, Rokutanda N, Takeuchi J, et al. , 2018. HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-quadruplex DNA. Cancer Res, 78(22): 6371-6385. 10.1158/0008-5472.CAN-18-1877 [DOI] [PubMed] [Google Scholar]
- Wu X, Petrini JH, Heine WF, et al. , 2000. Independence of R/M/N focus formation and the presence of intact BRCA1. Science, 289(5476): 11 10.1126/science.289.5476.11a [DOI] [PubMed] [Google Scholar]
- Wu X, Chavez JD, Schweppe DK, et al. , 2016. In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat Commun, 7: 13414 10.1038/ncomms13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, et al. , 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell, 22(6): 719-729. 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Xing MT, Yang MR, Huo W, et al. , 2015. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat Commun, 6: 6233 10.1038/ncomms7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue MM, Hou JJ, Wang LL, et al. , 2017. Optimizing the fragment complementation of APEX2 for detection of specific protein‒protein interactions in live cells. Sci Rep, 7: 12039 10.1038/s41598-017-12365-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan ZJ, Delannoy M, Ling C, et al. , 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell, 37(6): 865-878. 10.1016/j.molcel.2010.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan ZJ, Guo R, Paramasivam M, et al. , 2012. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell, 47(1): 61-75. 10.1016/j.molcel.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CQ, He C, 2013. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol, 5: a012575 10.1101/cshperspect.a012575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, et al. , 2018. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell, 69(3): 517-532. e11 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, et al. , 1998. The C-terminal (BRCT) domains of brca1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem, 273(39): 25388--25392. 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- Zhang AL, Peng B, Huang P, et al. , 2017. The p53-binding protein 1-Tudor-interacting repair regulator complex participates in the DNA damage response. J Biol Chem, 292(16): 6461-6467. 10.1074/jbc.M117.777474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, et al. , 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell, 126(3): 529-542. 10.1016/j.cell.2006.06.039 [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma JL, Wu JX, et al. , 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol, 19(6): 524-529. 10.1016/j.cub.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Chen Z, Ye Y, et al. , 2019. SLX4IP acts with SLX4 and XPF-ERCC1 to promote interstrand crosslink repair. Nucleic Acids Res, 47(19): 10181-10201. 10.1093/nar/gkz769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, et al. , 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science, 285(5428): 747-750. 10.1126/science.285.5428.747 [DOI] [PubMed] [Google Scholar]