Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is widely used for targeted genomic and epigenomic modifications and imaging in cells and organisms, and holds tremendous promise in clinical applications. The efficiency and accuracy of the technology are partly determined by the target binding affinity and residence time of Cas9-single-guide RNA (sgRNA) at a given site. However, little attention has been paid to the effect of target binding affinity and residence duration on the repair of Cas9-induced DNA double-strand breaks (DSBs). We propose that the choice of DSB repair pathway may be altered by variation in the binding affinity and residence duration of Cas9-sgRNA at the cleaved target, contributing to significantly heterogeneous mutations in CRISPR/Cas9 genome editing. Here, we discuss the effect of Cas9-sgRNA target binding and residence on the choice of DSB repair pathway in CRISPR/Cas9 genome editing, and the opportunity this presents to optimize Cas9-based technology.
Keywords: CRISPR/Cas9 genome editing, Double-strand break (DSB) repair pathway choice, Target binding affinity, Target residence
1 Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR) was originally discovered in the genomes of bacteria and archaea as a defense system to combat virus infection (Horvath and Barrangou, 2010). Partnering with CRISPR-associated (Cas) nucleases, CRISPR detects and cleaves viral DNA to remove invading viruses from hosts. Among diverse CRISPR systems, both CRISPR/Cas9 and CRISPR/Cas12a (also called Cpf1) are single-subunit effectors initially developed as powerful tools in genome editing due to their ability to target genomic DNA in eukaryotic cells and induce a site-specific DNA double-strand break (DSB) at the target (Hsu et al., 2014; Jiang and Doudna, 2017). Both systems comprise a nuclease and a single-guide RNA (sgRNA). For the widely used Cas9 system from Streptococcus pyogenes, the sgRNA is created by fusing CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). The sgRNA component contains a spacer segment that can be designed to match a DNA sequence of interest and guide the Cas protein to the target, allowing induction of site-specific DSBs by the enzyme. This two-component genome editing technology is easy to use, highly efficient, and reliable in DNA cleavage, and has revolutionized the fields of biology, agriculture, and medicine since its inception (Hsu et al., 2014; Jiang and Doudna, 2017).
CRISPR-mediated genome editing is eventually achieved by cellular repair of site-specific DSBs induced by CRISPR nucleases such as Cas9 and Cas12a, generating the desired DNA editing, including substitution, insertion, deletion, or translocation, among a variety of repair products. In eukaryotes, DSBs are repaired primarily by homology-directed repair (HDR) and non-homologous end joining (NHEJ), two major evolutionarily conserved repair mechanisms (Jasin and Haber, 2016; Gallagher and Haber, 2018). HDR requires a homologous sequence as a template for repair and resolves DSBs primarily in the S and G2 phases when sister chromatids are available as homologous templates, whereas NHEJ operates throughout the cell cycle. NHEJ can be further divided into at least two sub-pathways, classical and alternative (Boboila et al., 2012). Classical NHEJ (c-NHEJ) is the primary NHEJ pathway and requires several core NHEJ factors including DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Ku70/Ku80, X-ray repair cross-complementing protein 4 (XRCC4), and DNA ligase 4 to catalyze ligation of DNA ends. Alternative NHEJ (a-NHEJ) operates independently of either of these core factors and often uses microhomology to mediate rejoining of DNA ends. When limited homologous sequence is available in close vicinity of the break site, the other two pathways, microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA), are engaged, and the choice may be determined by the length of microhomology (Bhargava et al., 2016; Seol et al., 2018).
In CRISPR genome editing, upon site-specific DNA cleavage, different repair pathways compete for repair of the DSB. Cell cycle stage is a primary determinant of the choice of DSB repair pathway (Symington and Gautier, 2011). Chemicals that arrest the cell cycle have been explored to promote HDR in CRISPR genome editing (Lin et al., 2014). Other factors, including DNA end configuration, surrounding chromatin structure, and local DNA metabolism, also participate in the regulation of repair pathway choice (Symington and Gautier, 2011). Adding to this complexity, unique DSB induction by CRISPR nucleases appears to play an important role, part of which is the distinct binding of the Cas-sgRNA complex to its target. As controlling repair pathway choice has become an important strategy to enhance the efficiency and accuracy of CRISPR genome editing (Yeh et al., 2019), we focus here on S. pyogenes Cas9 (SpCas9) to discuss the effect of CRISPR target binding on DSB repair pathway choice and its potential for improving CRISPR genome editing.
2 Unique DSB induction and repair in CRISPR/Cas9 genome editing
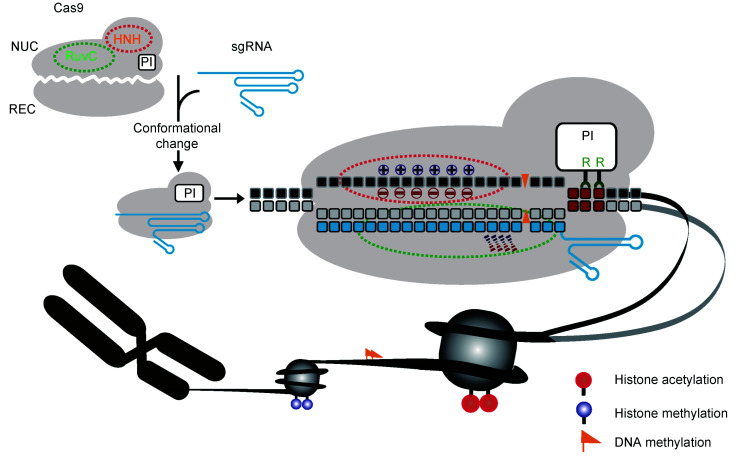
Cas9 contains two distinct lobes, the nuclease (NUC) lobe and the recognition (REC) lobe. The NUC lobe is composed of an HNH nuclease domain, a RuvC-like nuclease domain, and a C-terminal protospacer adjacent motif (PAM)-interacting (PI) domain. To bind its target, the Cas9-sgRNA complex first searches through the entire genome for a PAM via the PI domain of Cas9, and then initiates unwinding of duplexed DNA, starting at the nucleotide most proximal to the PAM and moving toward the PAM-distal nucleotide (Anders et al., 2014; Sternberg et al., 2014; Jiang and Doudna, 2017). Both DNA strands are trapped by the two-lobed Cas9 architecture and accommodated within a positively charged channel between the two lobes (Anders et al., 2014; Nishimasu et al., 2014; Jiang and Doudna, 2017). Along with unwinding of DNA duplex, sgRNA displaces the non-target DNA strand with a 20-nucleotide (nt) spacer sequence that is annealed to the target strand of DNA by base pairing to form an R-loop structure, which stabilizes the Cas9-sgRNA-DNA complex. A perfect or near-perfect match between the seed region of the sgRNA spacer and the target strand leads to cleavage of both the target and non-target DNA strands. The seed region is defined as the PAM-proximal 10‒12 nt located in the 3'-end of the spacer sequence (Anders et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014; Jiang and Doudna, 2017). The HNH and RuvC-like nuclease domains respectively cleave target and non-target DNA strands between the third and fourth nucleotides upstream of the PAM, forming a blunt-ended DSB (Anders et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014, 2015; Jiang and Doudna, 2017). It remains unclear how Cas9-sgRNA is dissociated from cleaved DNA to expose Cas9-induced DSBs for repair. However, in vitroexperiments have shown that the Cas9-sgRNA complex remains tightly bound to target DNA for several hours after DNA cleavage (Sternberg et al., 2014; Richardson et al., 2016).
Structural and biochemical studies have demonstrated that DSB induction by Cas9-sgRNA is distinct from DSB induction by ionizing radiation (IR), radiomimetic chemicals, and meganucleases such as I-SceI, the HO homing endonuclease, zinc finger nuclease (ZFN), and transcription activator-like effector nuclease (TALEN) (Anders et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014, 2015; Richardson et al., 2016; Jiang and Doudna, 2017; Gallagher and Haber, 2018; Yeh et al., 2019). Thus, the cellular DNA damage response (DDR) and repair machineries have to respond to unique features in DSB induction by Cas9. One of these unique features is the variability of the target binding affinity of the Cas9-sgRNA complex, which is dependent upon sgRNA-DNA base pairing and the interactions of Cas9 with sgRNA and target DNA (Jiang and Doudna, 2017; Kim et al., 2019). In contrast, meganucleases bind to target DNA solely via protein‒DNA interaction (Jasin and Haber, 2016; Gallagher and Haber, 2018). Consequently, variation in the target binding affinity of Cas9-sgRNA could have a significant influence on repair pathway choice, contributing to the heterogeneity of mutation profiles in CRISPR genome editing. Although the target binding of Cas9-sgRNA can tolerate some mismatches between the target strand and the complementary sgRNA spacer, target binding affinity is highly sensitive to the number and position of the mismatches (Doench et al., 2016; Boyle et al., 2017; Kim et al., 2019). Differences in mismatch number and position could lead to different repair choices between on-target and off-target sites in CRISPR/Cas9 genome editing.
Additionally, unlike meganucleases, cleavage of DNA strands by CRISPR/Cas9 is asymmetric because the HNH domain cleaves the target strand of the RNA-DNA hybrid and the RuvC-like domain cleaves the non-target strand of DNA within the R-loop of the Cas9-sgRNA-DNA complex (Garneau et al., 2010; Jinek et al., 2012; Sternberg et al., 2014; Szczelkun et al., 2014). While HNH cuts the DNA strand of the RNA-DNA hybrid stringently between the third and fourth nucleotides upstream of the PAM, the RuvC-like domain is more flexible and cleaves DNA at different positions 3 nt or more upstream of the PAM, leading to a high level of predictable templated insertions in repair products (Garneau et al., 2010; Jinek et al., 2012; Guo et al., 2018; Lemos et al., 2018; Shou et al., 2018; Allen et al., 2019; Chakrabarti et al., 2019). Nevertheless, Cas9 generates mostly blunt-ended DSBs, whereas Cas12a induces DSBs with 5'-overhanging ends (Zetsche et al., 2015). Each type of end configuration appears to require different accommodation for end recognition and repair. However, our current knowledge about repair of site-specific DSBs is based mainly on studies of DSBs with 4-nt 3'-overhanging ends induced by meganucleases such as I-SceI or the HO homing endonuclease (Chang et al., 2016; Gallagher and Haber, 2018). Therefore, we expect that our current understanding of site-specific DSB repair may not be completely applicable to those caused by the CRISPR/Cas systems.
Moreover, the Cas9-sgRNA complex is not only bound to the target DNA for several hours, but also remains tightly bound to the cleaved DNA products for a period of time (Kim et al., 2014; Sternberg et al., 2014; Ma et al., 2016; Richardson et al., 2016). Cas9-sgRNA residing at the cleaved DNA conceals the DSBs from access by the DDR and repair machineries, which can be fully activated only after Cas9-sgRNA leaves the DSBs. A long residence duration of Cas9-sgRNA at the cleaved DNA may increase the probability of collision with DNA replication and transcription, stalling the replication fork or generating a transcription-associated R-loop. This may help explain why Fanconi anemia complementation group D2 (FANCD2) is localized to the target sites in the presence of Cas9-sgRNA, although it is unclear whether FANCD2 enrichment is dependent upon the nuclease activity of Cas9 (Richardson et al., 2018). Of note, like Cas9-sgRNA, the target binding of nuclease-dead Cas9 (dCas9)-sgRNA also generates the R-loop structure, which itself is a source for DSBs and a cause of genome instability (Jiang and Doudna, 2017; Hegazy et al., 2020). Before complete dissociation of Cas9-sgRNA from the DSB, the 3'-end of the cleaved non-target strand is released from the Cas9-sgRNA-DSB complex immediately upon DNA nicking (Richardson et al., 2016, 2018; Jiang and Doudna, 2017). This free single-stranded DNA (ssDNA) end can be annealed by a complementary ssDNA template to enhance HDR (Richardson et al., 2016). This HDR-mediated incorporation of single-stranded donor oligodeoxynucleotide (ssODN) is surprisingly independent of Rad51, but requires the Fanconi anemia (FA) pathway, which is normally implicated in resolving interstrand cross-links (ICLs) (Ceccaldi et al., 2016; Richardson et al., 2018). Unlike SpCas9, both the Cas12a-sgRNA complex and the Staphylococcus aureus Cas9 (SaCas9)-sgRNA complex may release the PAM-distal double-stranded DNA (dsDNA) ends, but remain bound to the PAM-proximal end after cleavage of the target DNA (Singh et al., 2018; Strohkendl et al., 2018; Zhang SQ et al., 2020). This asymmetric release of two DNA ends of a DSB is expected to have differential impacts on Cas12a-and SaCas9-induced DSB repair and genome editing.
3 Determinants of target binding and target residence of Cas9-sgRNA
Structural comparison between apo-Cas9 unbound with sgRNA and Cas9 in complex with sgRNA reveals the autoinhibited state of apo-Cas9 and the shift to an active Cas9 conformation competent for target search (Sternberg et al., 2014). The sequence and structure of sgRNA for the Cas9 partner are conserved, except for the spacer segment, and predetermine the binding between Cas9 and sgRNA. After the formation of a stable Cas9-sgRNA effector complex, Cas9-sgRNA searches through the genome for the PAM, but it is the spacer of sgRNA that identifies the target for Cas9 and promotes the formation of a stable Cas9-sgRNA-DNA complex (Anders et al., 2014; Jiang and Doudna, 2017). Within this complex, target binding of Cas9-sgRNA is intrinsically supported by the interaction between the PI domain and the PAM, the base pairing between the spacer of the sgRNA and the target strand of DNA, and non-specific interactions between Cas9 and both the target and non-target DNA strands. The efficiency and accuracy of CRISPR/Cas9 genome editing are influenced by these interactions, which can be engineered to generate novel Cas9 variants with improved target specificity and expanded PAM compatibility (Kleinstiver et al., 2015a, 2015b; Hu et al., 2018).
3.1. Interaction between the PI domain and the PAM
Cas9-sgRNA randomly slides through the genome to search for PAM sites (Sternberg et al., 2014). SpCas9 recognizes a canonical 5'-NGG-3' PAM sequence by two conserved arginine residues (R1333 and R1335) in the PI domain forming major groove interaction with the GG dinucleotide in the non-target strand (Fig. 1) (Anders et al., 2014). Additional Cas9 interactions with the minor groove of the PAM duplex are also induced to help unwind the DNA target. PAM recognition by the PI domain of Cas9 allows the unwinding and interrogation of flanking DNA to match the target strand with the spacer of the sgRNA and, if perfectly or near-perfectly matched, form a stable R-loop structure (Anders et al., 2014; Jiang and Doudna, 2017). This model of PAM search is also applicable to other CRISPR/Cas systems (Jeon et al., 2018; Singh et al., 2018; Strohkendl et al., 2018). Because PAM recognition is a critical prerequisite for Cas9 target binding and Cas9-mediated DNA cleavage, expanding PAM compatibility is necessary to broaden the targeting range for the application of CRISPR/Cas9 genome editing. The PAM spectrum has been expanded by computational engineering of key residues that directly contact or help contact the GG dinucleotide (Kleinstiver et al., 2015a, 2015b), or by artificially accelerated protein evolution (Hu et al., 2018). These modifications may however attenuate the nuclease activity and binding affinity of Cas9 variants (Bolukbasi et al., 2015; Kleinstiver et al., 2015a, 2015b; Hu et al., 2018).
Fig. 1. Targeted binding of Cas9 to genomic DNA in the context of chromatin. Cas9 undergoes a conformational change in complex with sgRNA and binds a genomic target in the context of chromatin via several interactions, including the interaction of the PI domain of Cas9 with the PAM sequence of the target, the 20-nt Watson-Crick base pairing between the sgRNA spacer and the target strand, and non-specific interactions between Cas9 and target DNA. Two arginine residues (R1333 and R1335) in the PI domain directly contact the conserved GG dinucleotide in the PAM sequence. Epigenetic modifications, such as histone acetylation, histone methylation, and DNA methylation, affect Cas9-sgRNA targeting to DNA. Cas9: clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9; NUC: the nuclease lobe; REC: the recognition lobe; PAM: protospacer adjacent motif; PI: PAM-interacting; sgRNA: single-guicle RNA.
3.2. Base pairing between the spacer of sgRNA and the target strand
Upon recognition of a PAM sequence, directional unwinding of the adjacent protospacer allows the concomitant hybridization of the 20-nt sgRNA spacer to the target strand and the displacement of the non-target strand, forming an R-loop structure (Szczelkun et al., 2014). The 10 ‒12 bp PAM-proximal seed region of the protospacer is the earliest to unwind and determines the target binding specificity (Cong et al., 2013; Sternberg et al., 2014; Jiang and Doudna, 2017). The R-loop structure, with full complementarity of the seed sequences between the sgRNA spacer and the target strand of the protospacer, leads to conformational change in Cas9 that triggers concerted cleavage of the target and non-target strands (Sternberg et al., 2015; Zhu et al., 2019). Mismatches between the seed sequence of the sgRNA spacer and that of the target strand may inhibit progression of full R-loop formation with the entire spacer sequence annealed to the target, thus dramatically reducing or even abolishing the target binding affinity and cleavage activity of Cas9-sgRNA (Hsu et al., 2014; Doench et al., 2016; Boyle et al., 2017; Kim et al., 2019). Mismatches are more tolerated outside the seed region (Hsu et al., 2014; Doench et al., 2016; Boyle et al., 2017; Kim et al., 2019). However, while the PAM-proximal seed region determines the formation of the Cas9-sgRNA-DNA complex, the dissociation of the complex may be controlled primarily by the PAM-distal region (Boyle et al., 2017; Zhang Q et al., 2019).
Considering the contribution of the base pairing between the spacer of the sgRNA and the target strand to the stability of the Cas9-sgRNA-DNA complex, we expect that a shorter spacer would reduce the target binding affinity of Cas9-sgRNA, thereby lowering the specificity of Cas9 to its target. Paradoxically, compared to a 20-nt spacer, extension of the spacer sequence does not improve Cas9 target specificity, but truncated sgRNA, with a shorter spacer of 17‒19 nt, minimizes off-target effects without greatly affecting the on-target activity of Cas9-sgRNA (Fu et al., 2014). This suggests that a 20-nt spacer may already provide excess affinity for target binding of Cas9-sgRNA and the activation of the Cas9 endonuclease.
3.3. Non-specific interactions between Cas9 and target DNA
The contacts between Cas9 and target DNA are non-specific, but add an additional layer of regulation of the stability and activity of the Cas9-sgRNA-DNA complex. For Cas9 to perform different functions such as target recognition, R-loop formation, catalytic reactions in DNA cleavage and target dissociation, the Cas9-sgRNA-DNA complex adopts several conformational states, each requiring different Cas9-DNA contacts for their respective functions (Sternberg et al., 2015; Jiang and Doudna, 2017; Zhu et al., 2019). Target binding of Cas9-sgRNA leads to an early pre-catalytic conformational change with opening of the central channel in Cas9. The central channel accommodates the newly formed R-loop structure with the interactions between the REC3 domain of the REC lobe and the PAM-distal end of the target strand (Anders et al., 2014). In another pre-catalytic state of conformation, the negatively charged non-target strand is stabilized in a positively charged groove formed between the HNH, RuvC-like and PI domains of Cas9 via extensive hydrogen bonds and electrostatic attractions (Fig. 1) (Nishimasu et al., 2014; Zhu et al., 2019). Neutralization of the positively charged residues or disruption of hydrogen bonds reduces non-specific DNA binding energy, generating Cas9 variants that retain efficient nuclease activity, but with weaker off-target effects (Slaymaker et al., 2016; Chen et al., 2017; Casini et al., 2018). In addition, structural studies have revealed direct hydrogen bonds and hydrophobic interactions between Cas9 and the backbone of the target strand (Anders et al., 2014; Nishimasu et al., 2014). These contacts have been modified to reduce the off-target activity of Cas9-sgRNA (Kleinstiver et al., 2016). Upon DNA cleavage, the HNH domain of Cas9 is shifted into a catalytically competent conformation in which the side chains of several key residues such as D839, H840, and N863 can form hydrogen bonds with the target strand (Zhu et al., 2019). Even in a post-catalytic conformational state, several interactions, including those between REC3 and the RNA-DNA hybrid, persist from the preceding conformation. The interactions between REC3 and the RNA-DNA hybrid assist HNH nuclease activation and can also be exploited to improve Cas9 specificity (Chen et al., 2017). Strong interactions exhibited within the post-catalytic conformation suggest tight binding of Cas9-sgRNA to cleaved DNA and a long residence time of Cas9 at the DSB even after DNA scission (Zhu et al., 2019). As dCas9 does not cleave target DNA due to inactivation of its nuclease activity, dCas9 may not adopt a post-catalytic conformational change mimicking that of Cas9, and its dissociation from target DNA may not be the same as that of Cas9.
3.4. External effectors
In eukaryotes, DNA targets for CRISPR genome editing are assembled into the chromatin structure, in which genomic DNA wraps around "core" histone octamers to form nucleosomes and is further packaged into a more compact unit (Fig. 1). This poses a serious obstacle for Cas9 target search and binding (Verkuijl and Rots, 2019). In fact, the Cas9 activity is dramatically inhibited when a DNA target site is positioned within the nucleosome core, but not affected if the binding site is located in a linker DNA region (Hinz et al., 2015). However, once the Cas9-sgRNA-DNA complex is formed, it is unclear whether the local chromatin context, including nucleosome position, histone modifications, and chromatin structure, influences the stability of the Cas9-sgRNA-DNA complex and thus the activity of Cas9. As chromatin is inherently dynamic and highly malleable, the influence of the local chromatin context, if present, would not be static (Isaac et al., 2016; Verkuijl and Rots, 2019).
Cas9 bound to target DNA persists for an extended time and, therefore, creates a window in which it can be challenged by local DNA metabolism, including DNA replication, transcription, and recombination, in addition to local chromatin activity. On the one hand, molecular motor proteins in DNA metabolism and chromatin activity are capable of generating mechanical forces on target DNA and altering DNA topology (Bustamante et al., 2003). Recent studies have shown that mechanical perturbations, such as DNA torsion and DNA stretching, affect R-loop stability and Cas9 cleavage specificity (Szczelkun et al., 2014; Newton et al., 2019; Ivanov et al., 2020). An in vitro study has shown that a Bloom syndrome helicase (BLM) downstream of a PAM displaces dCas9 from its target more easily than a BLM from the upstream side of a PAM (Zhang Q et al., 2019). Therefore, by applying mechanical forces on target DNA, local DNA metabolism and chromatin activity may alter base pairing within the RNA-DNA hybrid and interactions of Cas9 with the PAM and target DNA, destabilizing the Cas9-sgRNA-DNA complex and even dislodging Cas9 from its target. On the other hand, in addition to generating DNA torsion or stretching, replication fork or transcription machinery may directly collide with Cas9-sgRNA (Qi et al., 2013; Jones et al., 2017; Clarke et al., 2018). Coordination between hexameric helicases and DNA polymerases in eukaryotic DNA replication may lead to an extremely powerful collision with Cas9-sgRNA from both directions (Patel et al., 2011). However, the collision force alone may not be sufficient to dissociate Cas9 from its target, as the bacteriophage Phi29 DNA polymerase (DNAP) and T7 RNA polymerase (RNAP) dislodge dCas9 from its target in a strand-biased manner in vitro (Clarke et al., 2018; Zhang Q et al., 2019). It is possible that the unwinding of the RNA-DNA hybrid by RNAP or DNAP after collision may be required to assist in the process of dislodging Cas9 or dCas9 from the DNA target.
4 Effect of CRISPR/Cas9 target binding and residence on genome editing
CRISPR offers an opportunity to modify genomes both genetically and epigenetically in a manner more precise and more efficient than ever before. While a CRISPR/Cas9-induced site-specific DSB at a target leads to the desired genetic edit in the genome among repair products, dCas9-based platforms have been developed for genomic imaging, transcription regulation, epigenetic modification, and base editing (Wang et al., 2016; Rees and Liu, 2018). To advance the applications of CRISPR systems, the precision and efficiency of these Cas9-based tools have also been improved by modifying either the Cas9 protein or the sgRNA scaffold (Kim et al., 2019). In fact, Cas9 and sgRNA have been optimized by various methods to increase the efficiency of genome editing or minimize off-target effects while retaining robust on-target activity (Kim et al., 2019). As the target binding affinity of Cas9-sgRNA is thought to be excessive for activating cleavage of target DNA, one key idea is to weaken the binding affinity of Cas9-sgRNA to reduce off-target effects (Fu et al., 2014; Kim et al., 2019). Given that tight binding and a long residence time at a target appear to affect the functionality of Cas9-sgRNA, we can also broaden the applications of CRISPR by enhancing the target binding of Cas9-sgRNA.
4.1. dCas9-based applications
By altering the sequence of the spacer, sgRNA can easily target the Cas9 nuclease to new sites for genome editing in a site-specific manner. This leads to the idea of using dCas9-sgRNA to recruit additional effectors to genomic sites for sequence-specific DNA or chromatin imaging or modification (Wang et al., 2016; Rees and Liu, 2018). Both dCas9 and sgRNA can be modified and turned into a scaffold to recruit transcriptional activators and repressors for site-specific transcriptional regulation (Gilbert et al., 2013; Perez-Pinera et al., 2013). Similarly, by fusing to dCas9, epigenetic modifiers can also be recruited to a given site to edit chromatin and reshape the epigenome at the relevant locus (Hilton et al., 2015; Kearns et al., 2015). In addition, the ability to visualize sequence-specific genomic segments in living cells is important for tracking the dynamics of the genome and identifying the functions of specific genomic architecture within cells. Thus, after dCas9-based imaging using dCas9-green fluorescent protein (GFP) fusion proteins was developed to label endogenous genomic loci (Chen et al., 2013), this strategy was further modified and improved to expand the applications, such as for studying the Cas9 target search in living cells in real time and tracking the dynamics of multiple genomic loci simultaneously (Knight et al., 2015; Ma et al., 2015). Another important dCas9-based genome editing platform is base editors that, by fusing dCas9 or Cas9 D10A nickase (nCas9) with a nucleobase deaminase, such as the cytidine deaminase APOBEC3 and the adenosine deaminase TadA, can directly convert one base into another, inducing point mutations in both dividing and non-dividing cells without DSB induction (Komor et al., 2016; Gaudelli et al., 2017; Rees and Liu, 2018).
In principle, the persistent binding of dCas9-sgRNA to its target determines not only the target specificity of the effectors recruited by dCas9-sgRNA, but also the duration of their action at the target. Therefore, strategies reducing excess contacts between Cas9-sgRNA and DNA, although increasing the specificity, should not be applied to dCas9-based platforms. Instead, to increase the efficiency of dCas9-based tools, dCas9-sgRNA should be modified to strengthen the binding and retention of dCas9 at its target, improving the efficiency of dCas9-based applications. It has been shown that dCas9-sgRNA can be used to physically block RNAP-promoter binding and RNAP translocation during transcription, mainly by tight target binding and robust retention at a given site, thus inhibiting gene expression (Qi et al., 2013; Clarke et al., 2018). However, it appears that translocating RNAP can remove sgRNA annealed to the transcription template strand more easily than sgRNA annealed to the transcription non-template strand (Qi et al., 2013; Clarke et al., 2018). As a result, transcription blockage by dCas9-sgRNA exhibits a strand-biased effect. In addition, sgRNA has been reconstructed to improve the stability of a dCas9-sgRNA complex conjugated with fluorescent proteins and to provide brighter and sharper fluorescence signals for site-specific genomic imaging in living cells (Chen et al., 2013). It is possible that this optimized dCas9-sgRNA binds its target better than the original. Recently, the ssDNA-binding domain of Rad51 was fused to various base editors to improve the binding affinity to the target DNA, especially the non-target strand, and possibly prolong the residence of base editors at their targets (Zhang XH et al., 2020). These re-engineered base editors exhibit enhanced efficiency and a widened range for base editing. The scope of dCas9-based applications could be expanded by further efforts to engineer or evolve a Cas9 variant with a stronger binding affinity, or with longer residence at its target.
4.2. Choice of repair pathway
Normally, Ku70/Ku80, Mre11/Rad50/NBS1, or RPA is recruited to DNA ends upon a DSB and activates three major phosphatidylinositol 3-kinase-related kinases (PI3KKs) including DNA-PKcs, ataxia telangiectasia- mutated (ATM), and ATM and Rad3-related (ATR) (Ciccia and Elledge, 2010; Blackford and Jackson, 2017). Activation of these three kinases initiates the DDR kinase signaling cascades to coordinate with cell cycle checkpoints and cell death to ensure proper repair of DSBs, maintaining genome integrity (Ciccia and Elledge, 2010; Blackford and Jackson, 2017). However, Cas9-sgRNA remains tightly bound to the cleaved target for a period of time after DNA cleavage (Sternberg et al., 2014; Richardson et al., 2016). Structural studies have revealed persistent base pairing between sgRNA and the target strand of DNA, and persistent interactions between Cas9 and target DNA within the post-catalytic conformational state of Cas9-sgRNA bound with the cleaved DNA (Sternberg et al., 2014; Jiang and Doudna, 2017; Zhu et al., 2019). It has been shown that chromatin perturbations are sufficient to induce ATM kinase activation and ATM-dependent cell cycle checkpoints in the absence of DSBs (Bakkenist and Kastan, 2015). If a post-catalytic conformational change perturbs local chromatin, ATM might be activated, preparing for exposure of DSBs from the Cas9-sgRNA-DNA complex. In addition, the non-target strand within the post-catalytic Cas9-sgRNA-DNA complex appears freed from the Cas9-sgRNA complex upon nicking by the RuvC-like domain of Cas9. It is yet to be determined whether this non-target strand can recruit RPA and ATR and initiate some degree of DDR (Sternberg et al., 2014; Richardson et al., 2016). Nevertheless, without clear DSB exposure, it is conceivable that the DDR machinery would not recognize or start to repair DSBs buried within the Cas9-sgRNA-DNA complex, and the DNA damage checkpoint response, including the p53-mediated checkpoint, would not be triggered (Zhang YX et al., 2019). Indeed, time-course analyses of genome editing via direct Cas9-sgRNA ribonucleoprotein (RNP) delivery demonstrated that it takes about 20 h to complete repair of Cas9-induced DSBs in mammalian cells (Kim et al., 2014).
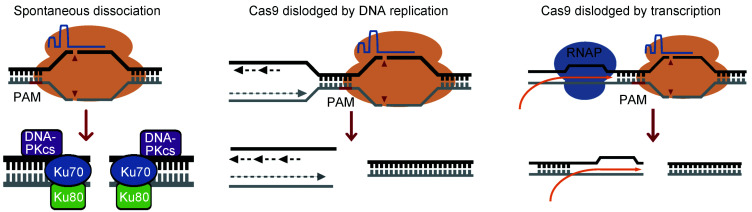
After Cas9-induced DNA cleavage, a gradual conformation change is observed by structural studies, but the mechanism underlying dissociation of Cas9-sgRNA from cleaved DNA has not been elucidated (Sternberg et al., 2014; Jiang and Doudna, 2017; Zhu et al., 2019). However, due to the difference in target binding affinity and residence duration as well as in the molecular activity near the Cas9-sgRNA-DNA complex, Cas9-sgRNA bound to its target could be released from the cleaved DNA product in at least two different ways: spontaneous dissociation, in which Cas9-sgRNA is freely dissociated from its target, or passive dissociation, in which Cas9-sgRNA is released from its target by forces imposed by DNA or chromatin metabolism, including DNA replication, transcription, and chromatin remodeling (Fig. 2). These two forms of dissociation generate different end configurations and can be influenced by the cell cycle stage, which is a primary determinant of DSB repair pathway choice. It is likely that the choice of repair pathway differs for Cas9-induced DSBs exposed by different dissociations. If Cas9-sgRNA is released from the cleaved DNA spontaneously, the exposed DSBs present two clean ends, which are recognized and bound easily by Ku70/Ku80 or Mre11/Rad50/NBS1 (Blackford and Jackson, 2017; Ciccia and Elledge, 2010). Ku70/Ku80 promotes c-NHEJ and Mre11/Rad50/NBS1 may facilitate the processing of the ends for either HDR or a-NHEJ (Lieber, 2010; Jasin and Rothstein, 2013). However, if Cas9 is passively dissociated, the ends may be unfavorable for binding of core NHEJ factors such as Ku70/Ku80 and XRCC4/DNA ligase 4.
Fig. 2. Distinct end configurations generated by different forms of Cas9-sgRNA dissociation from cleaved DNA. Spontaneous release of Cas9-sgRNA from a cleaved target generates a conventional two-ended DSB that can be directly recognized by core NHEJ factors. Persistent Cas9-sgRNA target binding increases the probability of encountering local DNA replication or transcription. Collision with a replication fork may generate a DSB with three ends: a blunt end at the leading strand, a 3’-overhanging end at the lagging strand, and a blunt end away from the replication fork. Transcription may also dislodge Cas9-sgRNA from the cleaved target site, forming a potentially unique end configuration. Cas9: clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9; DNA-PKcs: DNA-dependent protein kinase catalytic subunit; DSB: double-strand break; NHEJ: non-homologous end joining; PAM: protospacer adjacent motif; RNAP: RNA polymerase; sgRNA: single-guide RNA.
Replication-coupled dissociation of Cas9-sgRNA from cleaved DNA is restricted to the S phase of the cell cycle in which HDR is a preferred pathway for DSB repair. Once a replication fork collides with Cas9-sgRNA bound to DNA, the collision may release the Cas9-sgRNA from the cleaved target, collapsing the fork and resulting in three-ended DSBs: a blunt end of a sister chromatid at the leading strand, a staggered end of the other sister chromatid with a long 3'-ssDNA overhang at the lagging strand, and a blunt end away from the colliding replication fork (Fig. 2). The three-ended DSBs provide an opportunity for the DNA ends of two sister chromatids to rejoin, creating a giant palindromic chromosome (Tanaka and Yao, 2009). Due to the special configuration of the three-ended DSB structure and the cell cycle stage in which it occurs, the choices between HDR and NHEJ, and even between different DSB repair sub-pathways, are dictated not only by repair factors recognizing different end configurations, but also by a cell cycle-dependent decision (Scully et al., 2019). For example, it is unlikely that the staggered end with a long 3'-ssDNA overhang can readily engage c-NHEJ factors such as Ku70/Ku80 for c-NHEJ repair, whereas HDR may be preferentially promoted by availability of sister chromatids. In vitro study has indicated that Cas9-sgRNA bound with the cleaved target prevents access of Ku70/Ku80 to DNA ends (Clarke et al., 2018).
Translocating RNAP in transcription can dislodge Cas9-sgRNA from the cleaved DNA upon collision, but may form a DNA end containing nascent messenger RNA (mRNA)-DNA heteroduplex and a non-template single DNA strand (Fig. 2). The RNA-DNA hybrid itself can function as a platform to recruit repair factors, such as excision repair cross-complementing gene 6 (ERCC6), breast cancer type 1 susceptibility protein (BRCA1) and Rad52 (Ouyang et al., 2017; Puget et al., 2019). Otherwise, nascent mRNA at the end is either released spontaneously or removed by RNase H1 to allow recognition by DDR factors and repair pathway choice. As transcription-active regions may prefer HDR for DSB repair, it is important to discover how Cas9-induced DSBs exposed by transcription are repaired. Unlike DNA replication and transcription, chromatin remodeling releases Cas9-sgRNA from cleaved DNA possibly by applying forces through DNA torsion and stretching to destabilize the Cas9-sgRNA-DNA complex context (Verkuijl and Rots, 2019). This generates relatively clean DNA ends of Cas9-induced DSBs for repair (Feng et al., 2017). Without the effect of end configuration, chromatin remodeling may influence DSB repair pathway choice by modifying the local chromatin context (Feng et al., 2016, 2017; Verkuijl and Rots, 2019).
Cas9 residence at targeted DNA affects Cas9-sgRNA dissociation from a cleaved target and influences the choice of repair pathway for Cas9-induced DSBs at a given site. Changes in target binding affinity and residence duration could thus alter the choice of repair pathway at a specific target or between different targets, introducing significant variation into Cas9-mediated genome editing and generating highly heterogeneous mutational profiles, which are difficult to predict, even with big data analysis or deep machine learning (Abadi et al., 2017; Chuai et al., 2018). Therefore, it is important to identify the factors involved in repair of Cas9-induced replication- or transcription-coupled DSBs and elucidate the underlying repair mechanisms. This will help identify and control deleterious repair events in applications of CRISPR/Cas9 genome editing.
4.3. Off-target effects
Prominent off-target effects are a serious problem in CRISPR genome editing and have greatly limited clinical use of this technology (Kim et al., 2019). Off-target cuts occur because Cas9 is recruited to sites that are not perfectly matched with the spacer segment of sgRNA, but which can still be annealed by sgRNA to generate the R-loop structure and activate DNA cleavage mediated by the complexed Cas9 nuclease (Jiang and Doudna, 2017; Rees and Liu, 2018; Kim et al., 2019). However, at off-target sites, a single mismatch or multiple mismatches alter the base pairing between the sgRNA and off-target DNA, often reducing the binding affinity and residence duration of Cas9-sgRNA at the off-target sites (Bisaria et al., 2017; Kim et al., 2019). Thus, Cas9-sgRNA at off-target sites is more easily dissociated from the cleaved DNA and is less likely to encounter local DNA replication, transcription, or chromatin remodeling. As a result, compared to on-target sites, spontaneous dissociation of Cas9-sgRNA from cleaved DNA at off-target sites is more frequent and generates more ends suitable for engaging c-NHEJ, which is mostly accurate for repair of Cas9-induced DSBs. Combined with the possibility that DNA re-cleavage is rare at off-target sites, inhibition of c-NHEJ, which is widely used for improving CRISPR/Cas9-mediated HDR-based knock-in and gene correction (Chu et al., 2015; Maruyama et al., 2015; Yeh et al., 2019), could generate more mutagenic NHEJ events at off-target sites, causing stronger off-target effects. Therefore, when a chemical or genetic approach is used to increase the efficiency of genome editing at a given target in cells or organisms, off-target effects may be exacerbated and should be taken into account for evaluating the efficacy of the improvement approach.
Tight target binding of Cas9-sgRNA is generally required for efficient DNA cleavage, which is later transformed into efficient genome editing, but at the same time yields a high risk of off-target effects (Bisaria et al., 2017; Kim et al., 2019). At some sites, this tight binding is excessive, providing an opportunity for minimizing off-target effects while retaining robust on-target activity. Strategies include truncating the 20-nt spacer of sgRNA to 17‒18 nt and mutating the Cas9 residues that are important for non-specific interactions of Cas9 with the non-target strand of DNA and the RNA-DNA hybrid, and have been successfully tested to remove the excessive binding and improve the specificity of the modified Cas9-sgRNA (Fu et al., 2014). In addition, paired Cas9 nickases have been used as a strategy to minimize off-target effects in CRISPR/Cas9 genome editing because the probability of off-target double-nicking is almost zero and DNA nicks at off-target sites can be readily and precisely repaired (Ran et al., 2013). Once Cas9-sgRNA variants are generated with a stronger target binding affinity or longer target residence to open up new applications for dCas9- or nCas9-based platforms, these strategies that minimize off-target effects have to be reassessed for altered repair pathway choices or different off-target genetic and epigenetic editing consequences. This may yield a new opportunity for creating Cas9-sgRNA variants that not only bind their targets more tightly and for a longer time, but also have minimal off-target effects.
5 Concluding remarks
CRISPR/Cas9 genome editing technology is powerful and revolutionary but still needs more efficiency, less off-target activity, and broader application to fulfill its tremendous promise in precision medicine. As Cas9-sgRNA binds DNA tightly and remains bound to its target for a period of time even after DNA cleavage, one key issue that is often ignored in CRISPR/Cas9 development is the possible effects of target binding and residence time on the efficiency and specificity of CRISPR/Cas9 genome editing. In particular, the binding affinity and the post-cleavage residence of Cas9-sgRNA vary among different sites, including on-target and off-target sites, or even at a given site due to local DNA and chromatin activity. This may thereby influence repair pathway choice for Cas9-induced DSBs, resulting in significant variation in mutational profiles of CRISPR/Cas9 genome editing. Due to variable target binding and residence, Cas9-sgRNA is released from the cleaved DNA either spontaneously or by forces imposed by DNA replication, transcription, and chromatin remodeling. We propose that different forms of Cas9-sgRNA dissociation from the cleaved DNA may expose Cas9-induced DSBs with different end configurations, thus modulating repair pathway choice for Cas9-induced DSBs at a given site or among different sites. On the other hand, the efficiency of Cas9-based platforms relies partly on tight and persistent binding of dCas9-sgRNA to its target. To improve the efficiency of dCas9-based applications including genomic imaging, transcription regulation, epigenetic modification, and base editing, efforts should be made to generate Cas9 or sgRNA variants with a stronger target binding affinity or longer target residence. However, a stronger binding affinity and longer residence of Cas9-sgRNA at a target have potential to exacerbate off-target effects, which limit therapeutic applications of CRISPR/Cas9 for human health. Therefore, the effect of Cas9-sgRNA residence on off-target sites should also be considered in genome editing assessments. Optimization of CRISPR/Cas9 is thus a balancing act between improving target binding and residence of Cas9-sgRNA for higher efficiency and minimizing the off-target activity of Cas9-sgRNA.
Acknowledgments
The researchAcknowledgement: is supported by the National Natural Science Foundation of China (Nos. 31671385 and 31870806), the Zhejiang Provincial Natural Science Foundation of China (Nos. LY18C050001 and LQ20C050004), and the Fundamental Research Funds for the Central Universities in China (No. 2019QNA7031). We thank members of the XIE’s lab for their contributions and helpful discussions. We sincerely apologize to authors who are not quoted in references due to space limitations.
Author contributions
Anyong XIE, Yili FENG, and Sicheng LIU wrote the manuscript. Yili FENG drew the figures. Anyong XIE, Yili FENG, Sicheng LIU, and Ruodan CHEN edited the manuscript. All authors have read and approved the final manuscript.
Compliance with ethics guidelines
Yili FENG, Sicheng LIU, Ruodan CHEN, and Anyong XIE declare that they have no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Abadi S, Yan WX, Amar D, et al. , 2017. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol, 13(10): e1005807 10.1371/journal.pcbi.1005807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Crepaldi L, Alsinet C, et al. , 2019. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol, 37(1): 64-72. 10.1038/nbt.4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, et al. , 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature, 513(7519): 569-573. 10.1038/nature13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB, 2015. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair, 36: 8-12. 10.1016/j.dnarep.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, Stark JM, 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet, 32(9): 566-575. 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaria N, Jarmoskaite I, Herschlag D, 2017. Lessons from enzyme kinetics reveal specificity principles for RNA-guided nucleases in RNA interference and CRISPR-based genome editing. Cell Syst, 4(1): 21-29. 10.1016/j.cels.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP, 2017. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell, 66(6): 801-817. 10.1016/j.molcel.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Boboila C, Alt FW, Schwer B, 2012. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol, 116: 1-49. 10.1016/B978-0-12-394300-2.00001-6 [DOI] [PubMed] [Google Scholar]
- Bolukbasi MF, Gupta A, Oikemus S, et al. , 2015. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods, 12(12): 1150-1156. 10.1038/nmeth.3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Andreasson JOL, Chircus LM, et al. , 2017. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci USA, 114(21): 5461-5466. 10.1073/pnas.1700557114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Bryant Z, Smith SB, 2003. Ten years of tension: single-molecule DNA mechanics. Nature, 421(6921): 423-427. 10.1038/nature01405 [DOI] [PubMed] [Google Scholar]
- Casini A, Olivieri M, Petris G, et al. , 2018. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol, 36(3): 265-271. 10.1038/nbt.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D'Andrea AD, 2016. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol, 17(6): 337-349. 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- Chakrabarti AM, Henser-Brownhill T, Monserrat J, et al. , 2019. Target-specific precision of CRISPR-mediated genome editing. Mol Cell, 73(4): 699-713. e6 10.1016/j.molcel.2018.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Watanabe G, Gerodimos CA, et al. , 2016. Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J Biol Chem, 291(47): 24377-24389. 10.1074/jbc.M116.752329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Gilbert LA, Cimini BA, et al. , 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155(7): 1479-1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, et al. , 2017. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature, 550(7676): 407-410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, et al. , 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol, 33(5): 543-548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Chuai GH, Ma HH, Yan JF, et al. , 2018. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol, 19: 80 10.1186/s13059-018-1459-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ, 2010. The DNA damage response: making it safe to play with knives. Mol Cell, 40(2): 179-204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Heler R, MacDougall MS, et al. , 2018. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol Cell, 71(1): 42-55. e8 10.1016/j.molcel.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121): 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, et al. , 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 34(2): 184-191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YL, Xiang JF, Kong N, et al. , 2016. Buried territories: heterochromatic response to DNA double-strand breaks. Acta Biochim Biophys Sin (Shanghai), 48(7): 594-602. 10.1093/abbs/gmw033 [DOI] [PubMed] [Google Scholar]
- Feng YL, Xiang JF, Liu SC, et al. , 2017. H2AX facilitates classical non-homologous end joining at the expense of limited nucleotide loss at repair junctions. Nucleic Acids Res, 45(18): 10614-10633. 10.1093/nar/gkx715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YF, Sander JD, Reyon D, et al. , 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol, 32(3): 279-284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher DN, Haber JE, 2018. Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem Biol, 13(2): 397-405. 10.1021/acschembio.7b00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, et al. , 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature, 468(7320): 67-71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, et al. , 2017. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551(7681): 464-471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154(2): 442-451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Feng YL, Xiao JJ, et al. , 2018. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol, 19: 170 10.1186/s13059-018-1518-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy YA, Fernando CM, Tran EJ, 2020. The balancing act of R-loop biology: the good, the bad, and the ugly. J Biol Chem, 295(4): 905-913. 10.1074/jbc.REV119.011353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, et al. , 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol, 33(5): 510-517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Laughery MF, Wyrick JJ, 2015. Nucleosomes inhibit Cas9 endonuclease activity in vitro . Biochemistry, 54(48): 7063-7066. 10.1021/acs.biochem.5b01108 [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R, 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science, 327(5962): 167-170. 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F, 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157(6): 1262-1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, et al. , 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature, 556(7699): 57-63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac RS, Jiang FG, Doudna JA, et al. , 2016. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife, 5: e13450 10.7554/eLife.13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IE, Wright AV, Cofsky JC, et al. , 2020. Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci USA, 117(11): 5853-5860. 10.1073/pnas.1913445117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R, 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol, 5(11): a012740 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Haber JE, 2016. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair, 44: 6-16. 10.1016/j.dnarep.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Choi YH, Jang YS, et al. , 2018. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat Commun, 9: 2777 10.1038/s41467-018-05245-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang FG, Doudna JA, 2017. CRISPR-Cas9 structures and mechanisms. Ann Rev Biophys, 46: 505-529. 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096): 816-821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Leroy P, Unoson C, et al. , 2017. Kinetics of dCas9 target search in Escherichia coli . Science, 357(6358): 1420-1424. 10.1126/science.aah7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, et al. , 2015. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods, 12(5): 401-403. 10.1038/nmeth.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Luk K, Wolfe SA, et al. , 2019. Evaluating and enhancing target specificity of gene-editing nucleases and deaminases. Annu Rev Biochem, 88: 191-220. 10.1146/annurev-biochem-013118-111730 [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, et al. , 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res, 24(6): 1012-1019. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, et al. , 2015a. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol, 33(12): 1293-1298. 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, et al. , 2015b. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature, 523(7561): 481-485. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, et al. , 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587): 490-495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Xie LQ, Deng WL, et al. , 2015. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science, 350(6262): 823-826. 10.1126/science.aac6572 [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, et al. , 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533(7603): 420-424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos BR, Kaplan AC, Bae JE, et al. , 2018. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci USA, 115(9): E2040-E2047. 10.1073/pnas.1716855115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem, 79: 181-211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, et al. , 2014. Enhanced homology- directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife, 3: e04766 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HH, Naseri A, Reyes-Gutierrez P, et al. , 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA, 112(10): 3002-3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HH, Tu LC, Naseri A, et al. , 2016. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J Cell Biol, 214(5): 529-537. 10.1083/jcb.201604115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang LH, Esvelt KM, et al. , 2013. RNA-guided human genome engineering via Cas9. Science, 339(6121): 823-826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, et al. , 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol, 33(5): 538-542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton MD, Taylor BJ, Driessen RPC, et al. , 2019. DNA stretching induces Cas9 off-target activity. Nat Struct Mol Biol, 26(3): 185-192. 10.1038/s41594-019-0188-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, et al. , 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156(5): 935-949. 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Lan L, Zou L, 2017. Regulation of DNA break repair by transcription and RNA. Sci China Life Sci, 60(10): 1081-1086. 10.1007/s11427-017-9164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Pandey M, Nandakumar D, 2011. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr Opin Chem Biol, 15(5): 595-605. 10.1016/j.cbpa.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, et al. , 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods, 10(10): 973-976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puget N, Miller KM, Legube G, 2019. Non-canonical DNA/RNA structures during transcription-coupled double-strand break repair: roadblocks or Bona fide repair intermediates? DNA Repair, 81: 102661 10.1016/j.dnarep.2019.102661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence- specific control of gene expression. Cell, 152(5): 1173-1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, et al. , 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154(6): 1380-1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Liu DR, 2018. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet, 19(12): 770-788. 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, et al. , 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol, 34(3): 339-344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Richardson CD, Kazane KR, Feng SJ, et al. , 2018. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet, 50(8): 1132-1139. 10.1038/s41588-018-0174-0 [DOI] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, et al. , 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 20(11): 698-714. 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shim EY, Lee SE, 2018. Microhomology-mediated end joining: good, bad and ugly. Mutat Res, 809: 81-87. 10.1016/j.mrfmmm.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou J, Li JH, Liu YB, et al. , 2018. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 71(4): 498-509. e4 10.1016/j.molcel.2018.06.021 [DOI] [PubMed] [Google Scholar]
- Singh D, Mallon J, Poddar A, et al. , 2018. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc Natl Acad Sci USA, 115(21): 5444-5449. 10.1073/pnas.1718686115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao LY, Zetsche B, et al. , 2016. Rationally engineered Cas9 nucleases with improved specificity. Science, 351(6268): 84-88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, et al. , 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature, 507(7490): 62-67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, et al. , 2015. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature, 527(7576): 110-113. 10.1038/nature15544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohkendl I, Saifuddin FA, Rybarski JR, et al. , 2018. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol Cell, 71(5): 816-824. e3 10.1016/j.molcel.2018.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J, 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet, 45: 247-271. 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. , 2014. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA, 111(27): 9798-9803. 10.1073/pnas.1402597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yao MC, 2009. Palindromic gene amplification—an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer, 9(3): 216-224. 10.1038/nrc2591 [DOI] [PubMed] [Google Scholar]
- Verkuijl SAN, Rots MG, 2019. The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr Opin Biotechnol, 55: 68-73. 10.1016/j.copbio.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Wang HF, la Russa M, Qi LS, 2016. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem, 85: 227-264. 10.1146/annurev-biochem-060815-014607 [DOI] [PubMed] [Google Scholar]
- Yeh CD, Richardson CD, Corn JE, 2019. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol, 21(12): 1468-1478. 10.1038/s41556-019-0425-z [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. , 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3): 759-771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wen FC, Zhang SQ, et al. , 2019. The post-PAM interaction of RNA-guided spCas9 with DNA dictates its target binding and dissociation. Sci Adv, 5(11): eaaw9807 10.1126/sciadv.aaw9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SQ, Zhang Q, Hou XM, et al. , 2020. Dynamics of Staphylococcus aureus Cas9 in DNA target association and dissociation. EMBO Rep, 21: e50184 10.15252/embr.202050184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Chen L, Zhu BY, et al. , 2020. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat Cell Biol, 22(6): 740-750. 10.1038/s41556-020-0518-8 [DOI] [PubMed] [Google Scholar]
- Zhang YX, Pan WY, Chen J, 2019. p53 and its isoforms in DNA double-stranded break repair. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 20(6): 457-466. 10.1631/jzus.B1900167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Clarke R, Puppala AK, et al. , 2019. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol, 26(8): 679-685. 10.1038/s41594-019-0258-2 [DOI] [PMC free article] [PubMed] [Google Scholar]