Abstract
The formation of the biomolecular corona represents a crucial factor in controlling the biological interactions and trafficking of nanomaterials. In this context, the availability of key epitopes exposed on the surface of the corona, and able to engage the biological machinery, is important to define the biological fate of the material. While the full biomolecular corona composition can be investigated by conventional bottom-up proteomics, the assessment of the spatial orientation of proteins in the corona in a high-throughput fashion is still challenging. In this work, we show that labeling corona proteins with isobaric tags in their native conditions and analyzing the MS/MS spectra of tryptic peptides allow an easy and high-throughput assessment of the inner/outer orientation of the corresponding proteins in the original corona. We put our results in the context of what is currently known of the protein corona of graphene-based nanomaterials. Our conclusions are in line with previous data and were confirmed by in silico calculations.
Introduction
The past 20 years have witnessed a steadily increasing research interest in the use of nanomaterials for biological and biomedical applications.1−3 Among nanomaterials, graphene-based nanoflakes have shown interesting potentials for biological applications,4−6 and several works have investigated their properties in terms of interactions with proteins and cells.7−13 In this context, the study of the biological interactions occurring on the nanomaterial surface and their engagement mechanisms with biological systems is of paramount importance to identify successful strategies for the design of effective nanoformulations.14 It is well accepted that the layer of biomolecules, mostly proteins, adsorbing on the surface of nanomaterials, determines the final biological identity of the nanomaterials once in contact with biological fluids in vitro and in vivo (the so-called biomolecular or protein corona).15−20 It has been shown that the biomolecular corona can mask the nanoparticle-designed functionality, strongly affecting the efficacy of the formulation and providing an entirely new set of interacting moieties. The corona composition, generally characterized through mass spectrometry,21−23 is strictly related to several factors such as the core material, the nanomaterial size and shape, the surface chemistry, and the biofluid composition. However, only the biomolecular domains that are exposed at the periphery of the layer participate in the biological engagement with other biomolecules, cell membranes, and cell receptors.21,24−26 Therefore, it is not sufficient to characterize the protein layer in terms of mere composition, but it is of utmost importance to identify how proteins preferentially orient their structure onto the surface of the nanomaterial to identify key epitope domains driving the bionano-interactions. To this aim, several recent works have made use of immunometric mapping techniques based on various types of reporters.27−31 These techniques allow identifying the availability and functionality of specific recognition sites by exploiting the affinity and specificity of antibodies, and it is conceptually possible to multiplex the mapping of antibody signal by using a plethora of different reporters. However, the methodology still presents some practical challenges, and complications arise when considering that the biomolecular corona in situ is not a monolayer, and proteins undergo dynamic exchanges in vivo.(32) Zhang and colleagues31 recently characterized the binding proteins forming the total corona, suggesting that only a minor percentage of proteins is able to bind its target. Based on their findings, they proposed a multilayer corona structure, focusing on the active sites of biological recognitions of the external corona. These approaches, however, do not provide information on the spatial orientation of the whole inner protein structure (in contact with the material surface, the so-called hard corona), an aspect that is commonly investigated in silico.(33,34) In this work, we propose a novel, complementary, high-throughput methodology for the study of protein corona orientation. Graphene-based materials, due to their specific affinity for proteins and large carrier capability, are emerging as a novel class of biomedical nanomaterials for drug delivery, sensing, and in vitro diagnostics.35−37 Given the involvement of our group in the Graphene Flagship, we used two graphene-based materials (with distinct surface chemistries) as models for a proof-of-concept test of our approach. This approach exploits isobaric labeling of proteins,38 followed by conventional bottom-up proteomics, to derive information on the spatial orientation of a given peptide directly from the list of identified proteins. Other methods,39 while similar in the general idea, do not allow easy and high-throughput investigation or multiplexing. The results we obtained for the protein corona of few-layer graphene (FLG, bare graphene surface) and graphene oxide (GO), confirmed by in silico analysis, suggested an intriguing connection between protein preferential adsorption and orientation onto specific graphene-based surface chemistries.
Materials and Methods
Our work relies on a standard approach for bottom-up proteomics investigation of protein corona performed by high-resolution mass spectrometry. In short, the materials were suspended in PBS and incubated with commercial human plasma. Following the labeling at the protein level in native conditions with the first tag, the corona was denatured and labeled with the second tag, then digested with trypsin, and analyzed. The only difference with conventional workflows for corona proteomics is related to the labeling with isobaric tags itself, which is performed following the protocol suggested by the vendor. The full description of our experimental approach is reported in the Supporting Information.
Results and Discussion
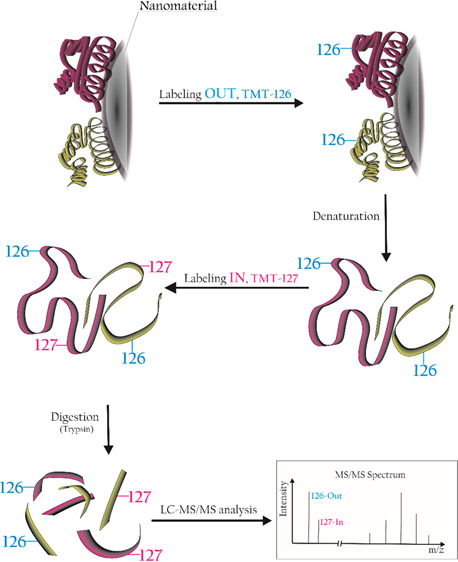
We moved from previous works in the field of protein corona orientation27,30 to figure out a high-throughput way to obtain relevant information directly from conventional bottom-up proteomics experiments used to identify the corona composition. The idea is to employ well-established techniques for protein isobaric labeling, widely used over the past 10 years to quantify protein over- or underexpression in various experimental conditions.40,41 Briefly, this approach uses isotopically labeled tags to covalently label K residues and N-terminal of tryptic peptides from different experimental conditions, say A and B. While each tag is isotopically and specifically labeled with the use of 15N and 13C atoms, the overall mass of the tag is the same for all tryptic peptides deriving from either A or B condition. In the bottom-up proteomics experiments, both peptides elute simultaneously from the chromatographic column and, having the same mass-to-charge ratio (m/z), are selected together for the MS/MS fragmentation. The fragment ions related to the primary sequence of the peptides (allowing one to assign the peptide to a given protein) are the same for both peptides. At the lower end of the MS/MS spectrum, two distinct reporter ions, each at a peculiar m/z ratio, are present. The relative intensities of the reporter ions are directly related to the abundances of the two peptides (and proteins) in the experimental groups. To our purposes, we chose Thermo tandem mass tags (TMT)42 to selectively label the “outer” and “inner” surfaces of the protein corona. This approach holds the advantage to be independent of the complex dynamics happening at the periphery of the layer as it can identify the preferential orientation of a protein onto a certain nanomaterial with given surface chemistry. To maintain the spatial information, we labeled the corona at the protein level rather than at the peptide level: therefore, labeling occurs before digestion with trypsin, opposite to the usual protocol. This procedure results in a generally slightly lower number of positive protein identifications and a lower protein sequence coverage and produces larger peptides as labeled K residues are not recognized by trypsin, resulting in missed cleavage sites. This alternative approach has also been widely used in the past,43,44 including by our group for proteomics on graphene oxide.7 It is important to point out that the information on the orientation of the K residues of each peptide positively assigned to a protein ID is automatically calculated by any proteomics data analysis software and it is retrieved directly from the list of identified proteins. The orientation data thus come automatically, together with all other conventional bottom-up proteomics results, making this methodology suitable for high-throughput screening of nanomaterials. Figure 1 shows a general schematic of the implemented isobaric labeling strategy in the corona orientation workflow.
Figure 1.
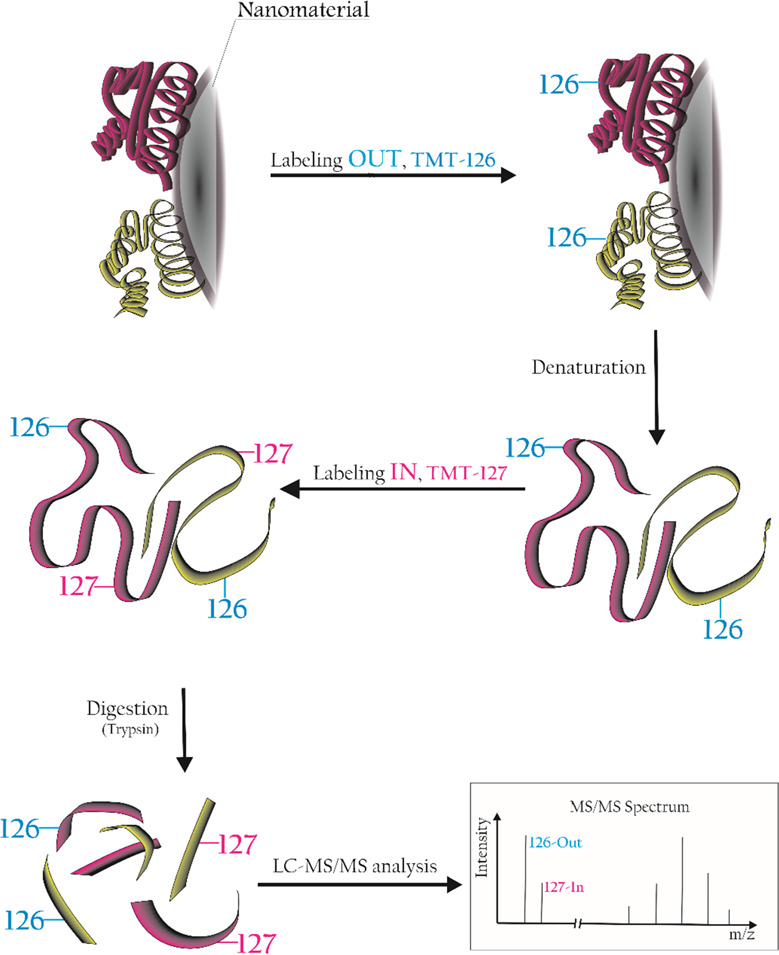
Diagram of the isobaric labeling workflow applied to the investigation of protein orientation.
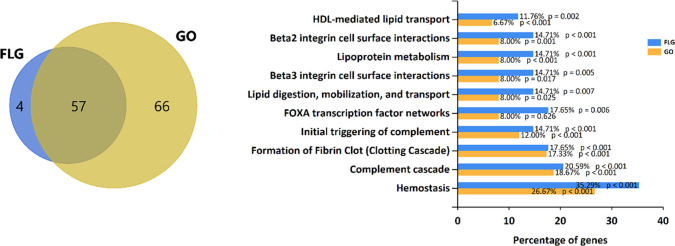
As a proof-of-concept study, two distinct materials, few-layer graphene (FLG) and graphene oxide (GO), were incubated in human plasma (HP) for 48 h to enrich the corona in proteins having the highest affinities.36 The materials characterization is reported in Figure S1. After washing the loosely attached proteins with a series of centrifugal steps in phosphate-buffered saline (PBS; see Materials and Methods), the exposed K residues of the long-lasting corona under its native condition were labeled with the 126 TMT tag (an isobaric tag that produces a 126 m/z fragment ion in the MS/MS spectrum). After removing the unreacted tag, the protein corona was denatured using very harsh conditions (see Materials and Methods). This treatment detached the corona from the material, exposing the “inner” part of the corona that was then labeled with the 127 TMT tag. After precipitation in cold acetone, the labeled proteome was digested with trypsin, and a conventional LC–MS/MS protein identification experiment was performed. As usual, peptides underwent MS/MS experiments to match their backbone fragments against suitable databases for protein identification. Only the peptides carrying labeled K were selected as they were the ones showing the two reporter fragment ions (126 and 127 m/z) in their MS/MS spectrum. The relative intensity of these ions was correlated to the preferential In/Out orientation of the corresponding peptide in the native corona. The samples were analyzed by LC–MS/MS, and all the obtained MS/MS spectra were searched against the Homo sapiens protein database. We first focused on protein identification, retaining those protein-matching all the three replicates of each group. We then compared the protein IDs from the two groups, as reported in Figure 2 (left).
Figure 2.
Left: Venn diagram of the protein identified in the FLG and GO protein corona. Right: Gene enrichment analysis reporting the 10 most significantly altered biological pathways enriched in FLG and GO protein corona.
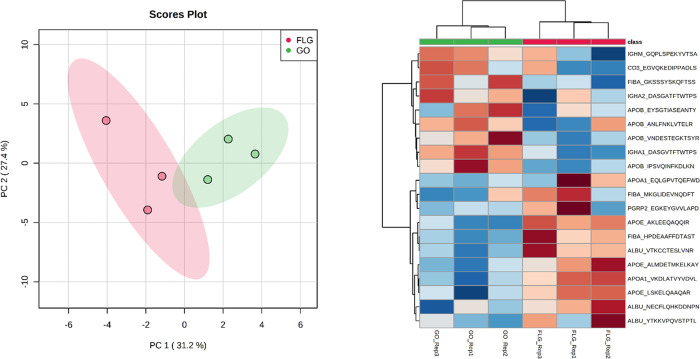
A total of 57 proteins were identified in both groups, while 4 and 66 were exclusively identified in the FLG and GO groups, respectively (see Supporting File 1 for the complete list of proteins). With gene enrichment analysis (Figure 2, right), we highlighted the top 10 most significant biological pathways differently represented by the proteins forming corona over the two materials. Figures S2 and S3 show the corresponding most enriched biological processes and molecular functions, respectively. We then focused our attention on the 57 proteins shared by the two materials, visually scrutinizing the peptides assigned to each ID to determine the influence of the surface chemistry on the protein preferential orientation. The full peptide dataset is reported in Supporting File 2. We identified 46 peptides, assigned to 22 of the common proteins, observed in both samples (FLG and GO) in all the three replicates. All these peptides have K in their sequence and are thus carrying information about their In/Out orientation. From a visual inspection (Supporting File 2), it is challenging to spot precise trends in protein orientation: the same peptide almost always shows opposite In/Out ratios in the three replicates, indicating no preferential orientation of the corresponding protein on the graphene material surface. Indeed, only 4 out of 46 peptides (8%) showed concordant In/Out values in all the six samples. With the aim to have a general overview of the whole dataset, we then used principal component analysis (PCA) using the observed In/Out ratios of the 46 peptides as variables. We then performed t-test statistics for the two groups and a heatmap analysis to highlight differential orientation trends. Figure 3 summarizes our findings.
Figure 3.
Left: PCA analysis of the 46 peptide dataset. The two groups appear to be separated by unsupervised data analysis. Right: Heatmap analysis of the In/Out orientation for the top 20 most significant peptides from a t-test comparison of FLG and GO corona. The In/Out ratios are <1 for “cold” samples (blue) and >1 for “hot” samples (red).
The PCA shows that despite the highly complicated appearance of the quantitative dataset, it is indeed possible to observe patterns in the In/Out orientation of the 46 peptides, as also depicted in the heatmap analysis, which shows orthogonality between the orientations on the two surfaces for some of the proteins. For example, two of the three mapped albumin peptides (NECFLQHKDDNPNLPR and VTKCCTESLVNR) appear to be oriented “In” when in FLG corona (red values) and “Out” when in GO corona (blue values). The third peptide (YTKKVPQVSTPTLVEVSR) generated apparently random In/Out ratios, with blue and red values mixed in the replicates. In this case, the covalent labeling is capturing a nonspecific orientation of this part of the sequence. This happens because that part of the sequence is flexible enough to be dynamically moving on the corona structure. Similar conclusions can be drawn for the APOE peptide LSKELQ AAQAR and for the HPDEAAFFDTASTGKTFPGFFSPMLGEFVSETESR peptide of fibrinogen alpha chain (FIBA). An opposite trend (“Out” for FLG and “In” for GO) is observed for the DASGVTFTW TPSSGKSAVQGPPER peptide of immunoglobulin heavy constant alpha 1. Our method is, in this case, capturing kinetic processes ongoing for parts of the sequences of corona proteins. The observed trends in the In/Out ratios might thus be associated with preferential orientations, or most likely configurations, that these sequences assume in the corona. Based on this assumption, we then hypothesized that proteins showing high specificity for a given material might have a specific affinity for its surface, therefore assuming more univocally oriented conformation in the corresponding corona. We then looked into the 4 and 66 proteins exclusively observed in FLG or GO, respectively. We used the Venn analysis of the peptides (Figure S4) to select those peptides unique to each of the two materials. In the end, a list of 42 peptides was present in all three replicates of each group (the details of the analysis are reported in Supporting File 2). The proteins corresponding to these peptides present a more specific orientation onto the surface of the two graphene materials. Indeed, 19 out of 42 peptides (45%) show concordant In/Out ratios in all three replicate samples. This suggests a link between the protein affinity for given surface chemistries and preferential orientation of the protein onto that surface. Quite interestingly, all the concordant peptides were observed in the GO group only, perhaps suggesting that this material can induce a more defined orientation of the specific proteins in its corona. Table 1 reports the summary of these 17 concordant peptides (14 corresponding proteins).
Table 1. List of 17 Peptides Unique for GO, Having Concordant In/Out Ratios in All Three Replicatesa.
| In/Out
ratio |
||||
|---|---|---|---|---|
| protein | peptide | Rep1 | Rep2 | Rep3 |
| APOA4 | LTPYADEFKVKIDQTVEELR | 0.51 | 0.55 | 0.66 |
| LAKDSEKLKEEIGKELEELR | 0.42 | 0.74 | 0.51 | |
| GNTEGLQKSLAELGGHLDQQVEEFRR | 0.97 | 0.63 | 0.81 | |
| RVEPYGENFNKALVQQMEQLR | 0.43 | 0.61 | 0.76 | |
| APOC2 | DLYSKSTAAMSTYTGIFTDQVLSVLKGEE | 0.16 | 0.41 | 0.35 |
| CBPN | IHILPSMNPDGYEVAAAQGPNKPGYLVGR | 0.34 | 0.45 | 0.67 |
| CO4A | NVNFQKAINEKLGQYASPTAKR | 0.62 | 0.72 | 0.93 |
| CO5 | GEQIQLKGTVYNYR | 1.67 | 2.75 | 4.66 |
| CXCL7 | NLAKGKEESLDSDLYAELR | 0.41 | 0.60 | 0.63 |
| HBA | VLSPADKTNVKAAWGKVGAHAGEYGAEALER | 0.44 | 0.61 | 0.84 |
| HV323 | DNSKNTLYLQMNSLR | 0.61 | 0.56 | 0.95 |
| IBP3 | EMEDTLNHLKFLNVLSPR | 0.46 | 0.49 | 0.41 |
| ITIH1 | VQSWKGSLVQASEANLQAAQDFVR | 1.58 | 1.29 | 1.84 |
| KNG1 | VQVVAGKKYFIDFVAR | 0.28 | 0.26 | 0.49 |
| LBP | ITDKGLQYAAQEGLLALQSELLR | 1.02 | 1.07 | 1.77 |
| RL40 | LIFAGKQLEDGR | 0.41 | 0.91 | 0.90 |
| TRFE | NTYEKYLGEEYVKAVGNLR | 0.36 | 0.60 | 0.67 |
To confirm our findings, we modeled the interaction of some of these proteins with the GO surface by performing docking simulations for proteins showing an In/Out peptide ratio smaller than 1 and a known experimental structure. In general, we observed a good agreement between the measured In/Out ratio of the peptides and the predicted docked orientation. Figure 4 reports the highest-ranking docked poses of five proteins: carboxypeptidase N1 (CBPN), hemoglobin a (HBA), immunoglobulin heavy variable 3-23 (HV323), serotransferrin (TRFE), and ubiquitin 60S (RL40) on GO, along with the average measured In/Out ratio of the corresponding observed peptides (represented as yellow spheres).
Figure 4.
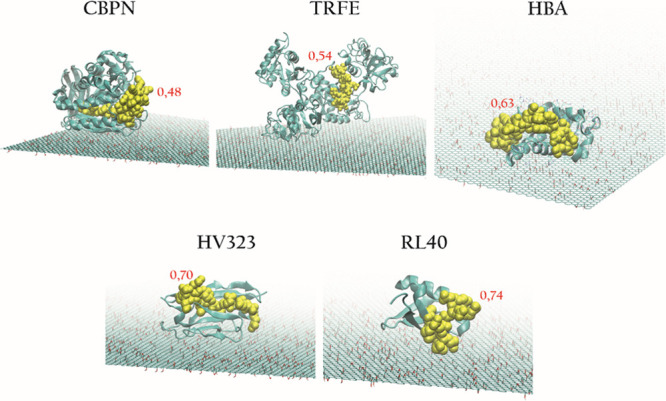
Docking of five proteins on GO. The corresponding peptide is represented as yellow spheres, and the average In/Out ratio measured by untargeted proteomics is indicated in red.
As expected from the experimental proteomics data, the peptides with In/Out < 1 are mostly located on the periphery of the protein, thus indicating that the preferential orientation of specific proteins is efficiently “captured” by this labeling strategy and proteomics workflow. When comparing the results obtained with published proteomics data for FLG and GO, some difficulties are expected, given the use of different graphene-based materials (GRMs), experimental setups, and protein identification databases. We compared our results to those of two recent publications on protein corona for bare graphene nanoflakes in the human serum11 and GO in human plasma.35 The extensive comparison of protein IDs of our study with those reported in these two papers is reported in Supporting File 3. We first noticed that despite the already discussed limitations of protein-level labeling, the total numbers of identified proteins in these studies (100 for the work of Castagnola et al.11 and 153 for the work of Di Santo et al.35) are only slightly higher compared to those found in this study (61 for FLG and 123 for GO). While comparing our GO data with the work of Di Santo et al. (Figure S5A), roughly 50% of the IDs match, indicating that our GO corona composition is comparable. As far as FLG is concerned, the comparison with the human serum protein corona from Castagnola et al.11 (Figure S5B) indicates that 35% of the corona proteins are shared by all three materials. GRM materials are not standard nanomaterials used for proteomics studies as they do not present a regular geometrical nanoshape, and they are mostly a collection of “flakes” with different sizes and shapes. Moreover, GRMs present diverse dispersion stability in relation to their surface chemistry and hydrophobicity level. It is interesting to note that despite the intrinsic complexity in handling these materials, a trend in the preferential protein affinity for different GRM surfaces can be extrapolated from data coming both from the present study and other reports, even though the GRMs and biofluids employed came from different sources (see Figure S6). Thirty-one proteins are reported in Table 2, which are recurrent in the protein corona of these four GRMs.
Table 2. Details of the 31 Proteins Observed in the Protein Corona of Four GRMs (Two from Our Study and Two from the Literature).
| entry name | protein name | gene name | length |
|---|---|---|---|
| ALBU_HUMAN | albumin | ALB | 609 |
| APOA1_HUMAN | apolipoprotein A-I (Apo-AI) (ApoA-I) (apolipoprotein A1) | APOA1 | 267 |
| APOE_HUMAN | apolipoprotein E (Apo-E) | APOE | 317 |
| VTNC_HUMAN | vitronectin (VN) (S-protein) (serum-spreading factor) | VTN | 478 |
| APOA4_HUMAN | apolipoprotein A-IV (Apo-AIV) (ApoA-IV) (apolipoprotein A4) | APOA4 | 396 |
| CO3_HUMAN | complement C3 C3 and PZP-like alpha-2-macroglobulin domain-containing protein 1 | C3 CPAMD1 | 1663 |
| HRG_HUMAN | histidine-rich glycoprotein (histidine-proline-rich glycoprotein) (HPRG) | HRG | 525 |
| CLUS_HUMAN | clusterin (aging-associated gene 4 protein) (apolipoprotein J) (Apo-J) | CLU APOJ CLI | 449 |
| IGJ_HUMAN | immunoglobulin J chain (joining chain of multimeric IgA and IgM) | JCHAIN IGCJ | 159 |
| GELS_HUMAN | gelsolin (AGEL) (actin-depolymerizing factor) (ADF) (brevin) | GSN | 782 |
| ITIH2_HUMAN | inter-alpha-trypsin inhibitor heavy chain H2 (ITI heavy chain H2) (ITI-HC2) | ITIH2 IGHEP2 | 946 |
| SAA4_HUMAN | serum amyloid A-4 protein (constitutively expressed serum amyloid A protein) | SAA4 CSAA | 130 |
| A2MG_HUMAN | alpha-2-macroglobulin (alpha-2-M) | A2M CPAMD5 | 1474 |
| KNG1_HUMAN | kininogen-1 (alpha-2-thiol proteinase inhibitor) (Fitzgerald factor) | KNG1 BDK | 644 |
| ITIH4_HUMAN | inter-alpha-trypsin inhibitor heavy chain H4 (ITI heavy chain H4) (ITI-HC4) | ITIH4 IHRP | 930 |
| APOC4_HUMAN | apolipoprotein C-IV (Apo-CIV) (ApoC-IV) (apolipoprotein C4) | APOC4 | 127 |
| ITIH1_HUMAN | inter-alpha-trypsin inhibitor heavy chain H1 (ITI heavy chain H1) (ITI-HC1) | ITIH1 IGHEP1 | 911 |
| FINC_HUMAN | fibronectin (FN) (cold-insoluble globulin) (CIG) | FN1 FN | 2477 |
| FETUA_HUMAN | alpha-2-HS-glycoprotein (alpha-2-Z-globulin) (Ba-alpha-2-glycoprotein) (fetuin-A) | AHSG FETUA | 367 |
| APOB_HUMAN | apolipoprotein B-100 (Apo B-100) [cleaved into: apolipoprotein B-48 (Apo B-48)] | APOB | 4563 |
| CFAB_HUMAN | complement factor B (EC 3.4.21.47) (C3/C5 convertase) | CFB BF BFD | 764 |
| PLMN_HUMAN | plasminogen (EC 3.4.21.7) [cleaved into: plasmin heavy chain A] | PLG | 810 |
| TTHY_HUMAN | transthyretin (ATTR) (prealbumin) (TBPA) | TTR PALB | 147 |
| APOL1_HUMAN | apolipoprotein L1 (apolipoprotein L) (Apo-L) (ApoL) (apolipoprotein L-I) (ApoL-I) | APOL1 APOL | 398 |
| CO9_HUMAN | complement component C9 | C9 | 559 |
| THRB_HUMAN | prothrombin (EC 3.4.21.5) (coagulation factor II) | F2 | 622 |
| ANGT_HUMAN | angiotensinogen (Serpin A8) | AGT SERPINA8 | 485 |
| CERU_HUMAN | ceruloplasmin (EC 1.16.3.1) (ferroxidase) | CP | 1065 |
| A1BG_HUMAN | alpha-1B-glycoprotein (alpha-1-B glycoprotein) | A1BG | 495 |
| C1S_HUMAN | complement C1s subcomponent (EC 3.4.21.42) (C1 esterase) | C1S | 688 |
| AMBP_HUMAN | protein AMBP [cleaved into: alpha-1-microglobulin (protein HC)] | AMBP HCP | 352 |
This comparison suggests that our orientation data obtained with TMT experiments were generated on a protein corona that is representative of what is currently known for other GRMs. We also focused on the corona orientation data reported by Castagnola et al.11 Based on a positive recognition of the corona proteins by an antibody targeting the 113–243 sequence of ApoA1, the authors show that this (large) part of the protein (54% of the sequence) is exposed on the outer surface of the corona. It is difficult to compare results from such different techniques: the broad part of a protein recognized by an antibody might be globally oriented outward, while individual K residues present in its sequence might still be facing the material. We nevertheless searched our orientation data on ApoA1 for the corona of FLG, the closest material to the graphene nanoflakes used in this paper. The sequence (155–173) QKLHELQEKLSPLGEEMR shows an apparently random orientation in our data, with an average In/Out ratio of 1.1. The preceding peptide 108–140 QEMSKDLEEVKAKVQPYLDDFQKKWQEEMELYR, on the contrary, was not immediately present in our proteomics dataset. This type of very large peptide (this one has a mass of 5325 Da with the TMT tags) is not usually taken into consideration in bottom-up proteomics experiments. We then manually searched our RAW data, looking for MS and MS/MS data of this individual peptide. Indeed, we found evidence of this peptide (Figure S7A) as demonstrated by the presence of the extracted ion current of its charge state 6+ ion at 887.98 m/z. The mass spectrum (Figure S7B) shows that this peptide is detectable as charge states 6+, 5+, and 4+. It is normally difficult to obtain good MS/MS data from such large peptides, but by dedicated experiments, we managed to confirm the sequence of this molecule (Figure S8). Despite the presence of five K residues in its sequence, each in principle having a different orientation, we retrieved the data on its global In/Out ratio that, in our experiments, was constantly <1 for the three FLG replicates (Figure S9). This result supports the global “outer” orientation of this part of ApoA1, and it is in agreement with previous data.
Conclusions
This work represents a proof-of-concept study highlighting an innovative strategy to simultaneously investigate the composition and the orientation of the proteins forming the biomolecular corona onto nanomaterial surfaces. Our corona composition data are in line with the current knowledge of the protein corona of widely used GRMs (FLG and GO), and the orientation data we obtained with the TMT tags match the poses calculated by molecular docking. Our method allows us to obtain data on protein orientation directly from large-scale proteomics datasets, thus in a high-throughput way. In addition to conventional bottom-up proteomics experiments for protein corona, this approach is (1) fast (a few additional hours of treatment are required), (2) easy to perform, (3) relatively cheap, and (4) minimally invasive toward post-translational modification analysis (several tens of phosphorylation sites were identified in our data). Moreover, our method can probe individual K residues, and it thus provides a higher resolution compared to conventional immunometric mapping, which normally relies on the antibody recognition of large sequences of corona proteins. For a better understanding, Table 3 gives a general overview and a comparison of the advantages of our method compared to others currently available.
Table 3. Comparison of Different Experimental Methods to Assess Protein Corona Orientation (*, Poor; **, Acceptable; ***, Good)a.
| method | cost-effective | easy to operate | easy analysis | high-throughput | MS proteomics coupled | full serum analysis | in situb | single-point determination of individual epitopes |
|---|---|---|---|---|---|---|---|---|
| isobaric labeling | *** | *** | *** | *** | YES | YES | * | partiallyc |
| immunoAu27,28 | * | * | ** | * | NO | YES | * | YES |
| immunoQD29 | * | * | * | * | NO | YES | *** | YES |
| QCM30,45,46 | * | ** | ** | ** | NO | YES | ** | YES |
| FCS47,48 | ** | * | * | * | NO | NO | *** | NO |
| SANS49 | * | * | * | * | NO | NO | *** | NO |
| NMR50,51 | *** | ** | ** | * | NO | NO | *** | NO |
QCM, quartz crystal microbalance; FCS, fluorescence correlation spectroscopy; SANS, small-angle neutron scattering; NMR, nuclear magnetic resonance spectroscopy.
Compared to conventional MS-based proteomics; allowing one to capture soft interactions.
Only if K residues are in the epitope.
The workflow we developed could be routinely implemented into protein corona proteomics studies, and it would represent a valuable tool in line with the great efforts for nanomaterial biointeraction screening and classification promoted by several EU nanosafety projects (Nanosolutions: https://nanosolutionsfp7.com/, NanoREG I and II: http://nanoreg2.eu/, Nanoclassifier: https://cordis.europa.eu/project/id/324519/it, etc.). These efforts would greatly benefit from easy-to-obtain information on the epitopes of corona proteins that are exposed to biological interactions. Furthermore, our approach is multiplexable: TMT technology (Thermo) allows us to label 2, 6, or 10 different conditions, and ITRAQ (Sciex) allows 4 or 8 multiplexed experiments. In this study, we used the TMT-6plex with the 126 and 127 tags. All proteomics data analysis software allow us to easily handle the plexing multiplicity, customize the reporting in many ways, and retrieve different In/Out ratios directly from the protein lists. This protocol opens up possibilities for analysis of increasing complexity to get closer to the realistic in vivo scenario. For example, different “layers” of corona proteins can be analyzed in the same experiment by detaching treatments of increasing strength followed by selective labeling with the corresponding multiplexed tags. This might give an opportunity to investigate proteins that are more loosely bound to the material surface and prone to dynamic exchange equilibria with the solvent and the biological environment (so-called soft corona).52 This investigation is currently performed with in situ spectroscopic techniques,29,53−55 given the difficulty in tuning the incubation/removal conditions to discriminate and isolate the different corona layers, i.e., the outer layers of the corona, loosely bound to the material. While our method can generate orientation data with small additional effort compared to routine proteomics experiments on protein corona, the graphical representation of protein orientation will require dedicated software solutions. Finally, to achieve the highest sequence coverage (thus orientation data), overcoming the limitations of missed cleavages at K, multidimensional chromatographic separations, both online and offline,56 could also be implemented.
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement nos. 785219 Graphene Flagship, Core2, and 881603 Graphene Flagship, Core3. M.B. acknowledges the European Union’s Horizon 2020 under the Marie Skłodowska-Curie Action-COFUND Athenea3i (grant agreement no. 754446). Ester Vázquez Fernández-Pacheco (Universidad de Castilla-La Mancha) and her research group are gratefully acknowledged for providing FLG and helpful discussion. Michele Dipalo, member of the Plasmon Nanotechnology line at the Fondazione Istituto Italiano di Tecnologia (IIT, Genova, Italy), is gratefully acknowledged for the use of Raman spectroscopy. The authors acknowledge the electron microscopy facility members of the Nanophysics Department at IIT Genova for the assistance with electron imaging.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c03134.
Chemicals, materials, and solvents; materials characterization; protein corona formation, labeling with tandem mass tags (TMT), and digestion with trypsin; data acquisition; data analysis; docking calculations (PDF)
Raman spectra and representative TEM micrographs for FLG and GO in water; gene enrichment analyses reporting the 10 most significantly altered biological processes and 10 most significantly altered molecular functions; Venn diagram for the assigned peptides; comparison of the protein corona composition data generated by our work with data from two other papers published; extracted ion current of charge state 6+, full scan mass spectrum and overlapped isotopic distribution of peptide; MS/MS spectrum of charge state 6+ of peptide; In/Out ratio of the MS/MS spectrum of charge state 6+ of peptide (PDF)
Complete list of proteins (XLSX)
Full peptide dataset (XLSX)
Extensive comparison of protein IDs of the present study with those reported in the literature (XLSX)
Author Contributions
N.L. performed all wet lab activities. L.M. performed all docking experiments. V.C. performed materials characterization and critically revised the interpretation of data. M.B. and F.B. critically revised the work and manuscript. A.A. wrote the paper and supervised the project.
The authors declare no competing financial interest.
Notes
All the RAW data files related to this project are freely accessible through the ProteomeXchange Consortium (via the PRIDE partner repository) with the dataset identifier PXD019524.
Supplementary Material
References
- Moghimi S. M.; Hunter A. C.; Murray J. C. Nanomedicine: current status and future prospects. FASEB J. 2005, 19, 311–330. 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- Stark W. J. Nanoparticles in biological systems. Angew. Chem. Int. Ed. Engl. 2011, 50, 1242–1258. 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]
- Wagner V.; Dullaart A.; Bock A. K.; Zweck A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006, 24, 1211–1217. 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- Chakrabarti M.; Kiseleva R.; Vertegel A.; Ray S. K. Carbon Nanomaterials for Drug Delivery and Cancer Therapy. J. Nanosci. Nanotechnol. 2015, 15, 5501–5511. 10.1166/jnn.2015.10614. [DOI] [PubMed] [Google Scholar]
- Chung C.; Kim Y. K.; Shin D.; Ryoo S. R.; Hong B. H.; Min D. H. Biomedical applications of graphene and graphene oxide. Acc. Chem. Res. 2013, 46, 2211–2224. 10.1021/ar300159f. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Li Z.; Wang J.; Li J.; Lin Y. Graphene and graphene oxide: biofunctionalization and applications in biotechnology. Trends Biotechnol. 2011, 29, 205–212. 10.1016/j.tibtech.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramini M.; Sacchetti S.; Armirotti A.; Rocchi A.; Vázquez E.; León Castellanos V.; Bandiera T.; Cesca F.; Benfenati F. Graphene Oxide Nanosheets Disrupt Lipid Composition, Ca2+ Homeostasis, and Synaptic Transmission in Primary Cortical Neurons. ACS Nano 2016, 10, 7154–7171. 10.1021/acsnano.6b03438. [DOI] [PubMed] [Google Scholar]
- Chiacchiaretta M.; Bramini M.; Rocchi A.; Armirotti A.; Giordano E.; Vázquez E.; Bandiera T.; Ferroni S.; Cesca F.; Benfenati F. Graphene Oxide Upregulates the Homeostatic Functions of Primary Astrocytes and Modulates Astrocyte-to-Neuron Communication. Nano Lett. 2018, 18, 5827–5838. 10.1021/acs.nanolett.8b02487. [DOI] [PubMed] [Google Scholar]
- Guarnieri D.; Sánchez-Moreno P.; Del Rio Castillo A. E.; Bonaccorso F.; Gatto F.; Bardi G.; Martín C.; Vázquez E.; Catelani T.; Sabella S.; Pompa P. P. Biotransformation and Biological Interaction of Graphene and Graphene Oxide during Simulated Oral Ingestion. Small 2018, 14, e1800227 10.1002/smll.201800227. [DOI] [PubMed] [Google Scholar]
- Papi M.; Palmieri V.; Digiacomo L.; Giulimondi F.; Palchetti S.; Ciasca G.; Perini G.; Caputo D.; Cartillone M. C.; Cascone C.; Coppola R.; Capriotti A. L.; Laganà A.; Pozzi D.; Caracciolo G. Converting the personalized biomolecular corona of graphene oxide nanoflakes into a high-throughput diagnostic test for early cancer detection. Nanoscale 2019, 11, 15339–15346. 10.1039/C9NR01413F. [DOI] [PubMed] [Google Scholar]
- Castagnola V.; Zhao W.; Boselli L.; Lo Giudice M. C.; Meder F.; Polo E.; Paton K. R.; Backes C.; Coleman J. N.; Dawson K. A. Biological recognition of graphene nanoflakes. Nat. Commun. 2018, 9, 1577. 10.1038/s41467-018-04009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.; Peng C.; Lv M.; Li X.; Zhang Y.; Chen N.; Fan C.; Huang Q. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano 2011, 5, 3693–3700. 10.1021/nn200021j. [DOI] [PubMed] [Google Scholar]
- Alnasser F.; Castagnola V.; Boselli L.; Esquivel-Gaon M.; Efeoglu E.; McIntyre J.; Byrne H. J.; Dawson K. A. Graphene Nanoflake Uptake Mediated by Scavenger Receptors. Nano Lett. 2019, 19, 1260–1268. 10.1021/acs.nanolett.8b04820. [DOI] [PubMed] [Google Scholar]
- Petros R. A.; DeSimone J. M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Ke P. C.; Lin S.; Parak W. J.; Davis T. P.; Caruso F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Monopoli M. P.; Åberg C.; Salvati A.; Dawson K. A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Monopoli M. P.; Walczyk D.; Campbell A.; Elia G.; Lynch I.; Bombelli F. B.; Dawson K. A. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- Casals E.; Puntes V. F. Inorganic nanoparticle biomolecular corona: formation, evolution and biological impact. Nanomedicine (Lond) 2012, 7, 1917–1930. 10.2217/nnm.12.169. [DOI] [PubMed] [Google Scholar]
- Tenzer S.; Docter D.; Kuharev J.; Musyanovych A.; Fetz V.; Hecht R.; Schlenk F.; Fischer D.; Kiouptsi K.; Reinhardt C.; Landfester K.; Schild H.; Maskos M.; Knauer S. K.; Stauber R. H. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Farokhzad O. C.; Mahmoudi M. Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona. Trends Biotechnol. 2017, 35, 257–264. 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Patel P. C.; Giljohann D. A.; Daniel W. L.; Zheng D.; Prigodich A. E.; Mirkin C. A. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem. 2010, 21, 2250–2256. 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Åberg C.; Mahon E.; Dawson K. A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Lundqvist M.; Stigler J.; Elia G.; Lynch I.; Cedervall T.; Dawson K. A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 14265–14270. 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara S.; Alnasser F.; Polo E.; Garry D.; Lo Giudice M. C.; Hristov D. R.; Rocks L.; Salvati A.; Yan Y.; Dawson K. A. Identification of Receptor Binding to the Biomolecular Corona of Nanoparticles. ACS Nano 2017, 11, 1884–1893. 10.1021/acsnano.6b07933. [DOI] [PubMed] [Google Scholar]
- Shannahan J. H.; Podila R.; Aldossari A. A.; Emerson H.; Powell B. A.; Ke P. C.; Rao A. M.; Brown J. M. Formation of a protein corona on silver nanoparticles mediates cellular toxicity via scavenger receptors. Toxicol. Sci. 2015, 143, 136–146. 10.1093/toxsci/kfu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiacomo L.; Cardarelli F.; Pozzi D.; Palchetti S.; Digman M. A.; Gratton E.; Capriotti A. L.; Mahmoudi M.; Caracciolo G. An apolipoprotein-enriched biomolecular corona switches the cellular uptake mechanism and trafficking pathway of lipid nanoparticles. Nanoscale 2017, 9, 17254–17262. 10.1039/C7NR06437C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P. M.; Åberg C.; Polo E.; O’Connell A.; Cookman J.; Fallon J.; Krpetić Z.; Dawson K. A. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat. Nanotechnol. 2015, 10, 472–479. 10.1038/nnano.2015.47. [DOI] [PubMed] [Google Scholar]
- Herda L. M.; Hristov D. R.; Lo Giudice M. C.; Polo E.; Dawson K. A. Mapping of Molecular Structure of the Nanoscale Surface in Bionanoparticles. J. Am. Chem. Soc. 2016, 139, 111–114. 10.1021/jacs.6b12297. [DOI] [PubMed] [Google Scholar]
- Lo Giudice M. C.; Herda L. M.; Polo E.; Dawson K. A. In situ characterization of nanoparticle biomolecular interactions in complex biological media by flow cytometry. Nat. Commun. 2016, 7, 13475. 10.1038/ncomms13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianneli M.; Polo E.; Lopez H.; Castagnola V.; Aastrup T.; Dawson K. A. Label-free in-flow detection of receptor recognition motifs on the biomolecular corona of nanoparticles. Nanoscale 2018, 10, 5474–5481. 10.1039/C7NR07887K. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Wu J. L. Y.; Lazarovits J.; Chan W. C. W. An Analysis of the Binding Function and Structural Organization of the Protein Corona. J. Am. Chem. Soc. 2020, 142, 8827–8836. 10.1021/jacs.0c01853. [DOI] [PubMed] [Google Scholar]
- Chen F.; Wang G.; Griffin J. I.; Brenneman B.; Banda N. K.; Holers V. M.; Backos D. S.; Wu L.; Moghimi S. M.; Simberg D. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 2017, 12, 387–393. 10.1038/nnano.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.; Zhou J. Understanding the curvature effect of silica nanoparticles on lysozyme adsorption orientation and conformation: a mesoscopic coarse-grained simulation study. Phys. Chem. Chem. Phys. 2016, 18, 23500–23507. 10.1039/C6CP01478J. [DOI] [PubMed] [Google Scholar]
- Glukhova O. E.; Prytkova T. R.; Savostyanov G. V. Simulation of High Density Lipoprotein Behavior on a Few Layer Graphene Undergoing Non-Uniform Mechanical Load. J Phys Chem B 2016, 120, 3593–3600. 10.1021/acs.jpcb.5b12648. [DOI] [PubMed] [Google Scholar]
- Di Santo R.; Digiacomo L.; Quagliarini E.; Capriotti A. L.; Laganà A.; Zenezini Chiozzi R.; Caputo D.; Cascone C.; Coppola R.; Pozzi D.; Caracciolo G. Personalized Graphene Oxide-Protein Corona in the Human Plasma of Pancreatic Cancer Patients. Front. Bioeng. Biotechnol. 2020, 8, 491. 10.3389/fbioe.2020.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey C. D.; Chan W. C. W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. 10.1039/C1CS15233E. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Robinson J. T.; Tabakman S. M.; Yang K.; Dai H. Carbon materials for drug delivery & cancer therapy. Mater. Today 2011, 14, 316–323. 10.1016/S1369-7021(11)70161-4. [DOI] [Google Scholar]
- Romero R.; Kusanovic J. P.; Gotsch F.; Erez O.; Vaisbuch E.; Mazaki-Tovi S.; Moser A.; Tam S.; Leszyk J.; Master S. R.; Juhasz P.; Pacora P.; Ogge G.; Gomez R.; Yoon B. H.; Yeo L.; Hassan S. S.; Rogers W. T. Isobaric labeling and tandem mass spectrometry: a novel approach for profiling and quantifying proteins differentially expressed in amniotic fluid in preterm labor with and without intra-amniotic infection/inflammation. J. Matern. Fetal Neonatal. Med. 2010, 23, 261–280. 10.3109/14767050903067386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S.; Nuffer J. H.; Siegel R. W.; Dordick J. S. Position-specific chemical modification and quantitative proteomics disclose protein orientation adsorbed on silica nanoparticles. Nano Lett. 2012, 12, 1583–1587. 10.1021/nl2044524. [DOI] [PubMed] [Google Scholar]
- Gupta D.; Mohammed M.; Mekala L. P.; Chintalapati S.; Chintalapati V. R. iTRAQ-based quantitative proteomics reveals insights into metabolic and molecular responses of glucose-grown cells of Rubrivivax benzoatilyticus JA2. J. Proteomics 2019, 194, 49–59. 10.1016/j.jprot.2018.12.027. [DOI] [PubMed] [Google Scholar]
- Moulder R.; Bhosale S. D.; Goodlett D. R.; Lahesmaa R. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom. Rev. 2018, 37, 583–606. 10.1002/mas.21550. [DOI] [PubMed] [Google Scholar]
- Zecha J.; Satpathy S.; Kanashova T.; Avanessian S. C.; Kane M. H.; Clauser K. R.; Mertins P.; Carr S. A.; Kuster B. TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Mol. Cell. Proteomics 2019, 18, 1468–1478. 10.1074/mcp.TIR119.001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engmann O.; Campbell J.; Ward M.; Giese K. P.; Thompson A. J. Comparison of a protein-level and peptide-level labeling strategy for quantitative proteomics of synaptosomes using isobaric tags. J. Proteome Res. 2010, 9, 2725–2733. 10.1021/pr900627e. [DOI] [PubMed] [Google Scholar]
- Nie S.; Lo A.; Zhu J.; Wu J.; Ruffin M. T.; Lubman D. M. Isobaric protein-level labeling strategy for serum glycoprotein quantification analysis by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2013, 85, 5353–5357. 10.1021/ac400838s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramesh M.; Shimoni O.; Ostrikov K.; Prawer S.; Cervenka J. Surface charge effects in protein adsorption on nanodiamonds. Nanoscale 2015, 7, 5726–5736. 10.1039/C5NR00250H. [DOI] [PubMed] [Google Scholar]
- Wang X.; Wang M.; Lei R.; Zhu S. F.; Zhao Y.; Chen C. Chiral Surface of Nanoparticles Determines the Orientation of Adsorbed Transferrin and Its Interaction with Receptors. ACS Nano 2017, 11, 4606–4616. 10.1021/acsnano.7b00200. [DOI] [PubMed] [Google Scholar]
- Shang L.; Nienhaus G. U. In Situ Characterization of Protein Adsorption onto Nanoparticles by Fluorescence Correlation Spectroscopy. Acc. Chem. Res. 2017, 50, 387–395. 10.1021/acs.accounts.6b00579. [DOI] [PubMed] [Google Scholar]
- Maffre P.; Brandholt S.; Nienhaus K.; Shang L.; Parak W. J.; Nienhaus G. U. Effects of surface functionalization on the adsorption of human serum albumin onto nanoparticles - a fluorescence correlation spectroscopy study. Beilstein J. Nanotechnol. 2014, 5, 2036–2047. 10.3762/bjnano.5.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner J.; Wu Y.; Jestin J.; Shelton W. A.; Findenegg G. H.; Bharti B. pH-Induced reorientation of cytochrome c on silica nanoparticles. Soft Matter 2019, 15, 350–354. 10.1039/C8SM01909F. [DOI] [PubMed] [Google Scholar]
- Assfalg M.; Ragona L.; Pagano K.; D’Onofrio M.; Zanzoni S.; Tomaselli S.; Molinari H. The study of transient protein-nanoparticle interactions by solution NMR spectroscopy. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 102–114. 10.1016/j.bbapap.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Lin W.; Insley T.; Tuttle M. D.; Zhu L.; Berthold D. A.; Král P.; Rienstra C. M.; Murphy C. J. Control of protein orientation on gold nanoparticles. J. Phys. Chem. C. Nanomater. Interfaces 2015, 119, 21035–21043. 10.1021/acs.jpcc.5b07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani S.; Bombelli F. B.; Pitek A. S.; Dawson K. A.; Rädler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano 2012, 6, 2532–2541. 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- Vilanova O.; Mittag J. J.; Kelly P. M.; Milani S.; Dawson K. A.; Rädler J. O.; Franzese G. Understanding the Kinetics of Protein-Nanoparticle Corona Formation. ACS Nano 2016, 10, 10842–10850. 10.1021/acsnano.6b04858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclăuş T.; Bochenkov V. E.; Ogaki R.; Howard K. A.; Sutherland D. S. Spatial mapping and quantification of soft and hard protein coronas at silver nanocubes. Nano Lett. 2014, 14, 2086–2093. 10.1021/nl500277c. [DOI] [PubMed] [Google Scholar]
- Srivastava I.; Khan M. S.; Dighe K.; Alafeef M.; Wang Z.; Banerjee T.; Ghonge T.; Grove L. M.; Bashir R.; Pan D. On-Chip Electrical Monitoring of Real-Time “Soft” and “Hard” Protein Corona Formation on Carbon Nanoparticles. Small Methods 2020, 4, 2000099. 10.1002/smtd.202000099. [DOI] [Google Scholar]
- Delahunty C. M.; Yates J. R. III MudPIT: multidimensional protein identification technology. BioTechniques 2007, 43, 563–565. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.