Abstract
OBJECTIVE
To identify the core gut microbial features associated with type 2 diabetes risk and potential demographic, adiposity, and dietary factors associated with these features.
RESEARCH DESIGN AND METHODS
We used an interpretable machine learning framework to identify the type 2 diabetes–related gut microbiome features in the cross-sectional analyses of three Chinese cohorts: one discovery cohort (n = 1,832, 270 cases of type 2 diabetes) and two validation cohorts (cohort 1: n = 203, 48 cases; cohort 2: n = 7,009, 608 cases). We constructed a microbiome risk score (MRS) with the identified features. We examined the prospective association of the MRS with glucose increment in 249 participants without type 2 diabetes and assessed the correlation between the MRS and host blood metabolites (n = 1,016). We transferred human fecal samples with different MRS levels to germ-free mice to confirm the MRS–type 2 diabetes relationship. We then examined the prospective association of demographic, adiposity, and dietary factors with the MRS (n = 1,832).
RESULTS
The MRS (including 14 microbial features) consistently associated with type 2 diabetes, with risk ratio for per 1-unit change in MRS 1.28 (95% CI 1.23–1.33), 1.23 (1.13–1.34), and 1.12 (1.06–1.18) across three cohorts. The MRS was positively associated with future glucose increment (P < 0.05) and was correlated with a variety of gut microbiota–derived blood metabolites. Animal study further confirmed the MRS–type 2 diabetes relationship. Body fat distribution was found to be a key factor modulating the gut microbiome–type 2 diabetes relationship.
CONCLUSIONS
Our results reveal a core set of gut microbiome features associated with type 2 diabetes risk and future glucose increment.
Introduction
Type 2 diabetes is a complex disorder influenced by both host genetic and environmental factors (1), and its prevalence is rising rapidly in both developed and developing countries (2). Gut microbiome is considered as a modifiable environmental factor that plays an important role in the development of type 2 diabetes (3). The research interest in identification of gut microbiome–related treatment/prevention targets has recently emerged (4). Although there are a few human studies investigating the association of gut microbiome with type 2 diabetes in the past few years, the results are inconsistent (5). So far, there is sparse human evidence robustly linking specific gut microbiome features to type 2 diabetes.
Machine learning has been widely used in biomedical fields in recent years (6). However, its application in the clinical setting is still limited, as its predictions are usually difficult to interpret. Of note, with the methodology development in the past few years, interpretable algorithms could unlock the traditional “black box” of machine learning results (7). The integration of the new algorithms with large-scale gut microbiome data has the potential to radically unveil the relationship between gut microbiome and type 2 diabetes. Yet, no such investigation has been done.
Therefore, in the current study, we aimed to identify human gut microbiome features associated with type 2 diabetes using a novel interpretable machine learning analytical framework in large-scale human cohorts. We also assessed the correlation between the identified microbes and host blood metabolites to provide insight into the role of type 2 diabetes–related gut microbiota in host metabolism. We further performed a fecal microbiota transfer experiment to confirm the effect of the identified microbes on type 2 diabetes development. As a secondary objective, we aimed to identify potential adiposity, dietary, and lifestyle factors that could modify the type 2 diabetes–related gut microbiome using our longitudinal cohort data.
Research Design and Methods
Study Design
The overview of the study workflow is shown in Supplementary Fig. 1. We included participants from three human cohorts, Guangzhou Nutrition and Health Study (GNHS) (8) as a discovery cohort (n = 1,832, 270 type 2 diabetes cases), the control arm of a case-control study of hip fracture (n = 203, 48 cases) (9), and Guangdong Gut Microbiome Project (GGMP) (n = 7,009, 608 cases) (10) as two validation cohorts.
In the discovery cohort and validation cohort 1, prevalent type 2 diabetes cases were ascertained based on fasting blood glucose ≥7.0 mmol/L or HbA1c ≥6.5% (48 mmol/mol) or current medical treatment for type 2 diabetes at either of the follow-up visits, according to the American Diabetes Association criteria for the diagnosis of diabetes (11). In validation cohort 2, type 2 diabetes was determined by self-report (confirmed with medical history) or fasting blood glucose ≥7.0 mmol/L.
The study design of GNHS has previously been described in detail (8). Stool samples were collected onsite during follow-up visits between 2014 and 2018. Those with measurement of 16S rRNA from stool samples were included in the current study (n = 1,935). Type 2 diabetes cases were ascertained at the same time point or before the stool collection. Study participants were excluded if they had unclear diabetes status (n = 48), chronic renal dysfunction, or self-reported cancers (n = 55). Finally, 1,832 participants (270 case subjects, 117 with the use of medication, 234 with glycemic markers to meet the diagnostic criteria) were included in the present analysis. Among the included participants, there were 249 participants who did not have type 2 diabetes, who were followed up for a median of 3.4 years after the collection of their stool samples (Supplementary Fig. 2). These participants were included in our longitudinal analysis of gut microbiome with glucose increments. All 1,832 participants were included in our longitudinal analysis on the prospective association of baseline factors with gut microbiome (with a median follow-up of 6.2 years).
Detailed information of validation cohort 1 has previously been published (9). Stool samples were collected onsite during follow-up visits between February 2017 and May 2017. Type 2 diabetes cases were ascertained at the same time point or before the stool collection (2015–2017). After adopting the same inclusion and exclusion criteria as used in GNHS, we included 203 participants (48 case subjects, 24 with the use of medication, and 38 glycemic markers to meet the diagnostic criteria) with a measurement of 16S rRNA from stool samples in the present analysis.
GGMP (10) was conducted between 2015 and 2016; 7,009 participants (608 case subjects, 530 fasting blood glucose ≥7.0 mmol/L) with measurement of 16S rRNA from stool samples were included in the current study. Type 2 diabetes ascertainment and stool collection were conducted at the same time point.
Fecal Sample Collection and 16S rRNA Profiling
Microbial DNA extraction, PCR, and amplicon sequencing were performed as previously described (see Supplementary Material). FASTQ files were demultiplexed, merge paired, and quality filtered by QIIME (Quantitative Insights Into Microbial Ecology software (version 1.9.0) (12). Sequences were clustered into operational taxonomic units with 97% similarity and annotated based on the Greengenes Database (version 13.8) (13).
Measurement of Metadata and Metabolome Profiling
Detailed methods for the metadata measurements and shotgun metagenome sequencing are provided in Supplementary Material.
Metadata at the same point in time as the stool sample collection were used as the covariates. Characteristics for each cohort were shown in Table 1.
Table 1.
Characteristics of the participants included in this study
| Discovery cohort | External validation cohort 1 | External validation cohort 2 | |
|---|---|---|---|
| No. of participants | 1,832 | 203 | 7,009 |
| Type 2 diabetes case subjects, n (%) | 270 (14.7) | 48 (23.6) | 608 (8.7) |
| Age (years) | 64.8 (5.9) | 71.7 (6.9) | 52.7 (14.7) |
| Sex, n (%) | |||
| Women | 1,223 (66.9) | 152 (74.9) | 3,848 (54.9) |
| Men | 605 (33.1) | 51 (25.1) | 3,161 (45.1) |
| Marital status, n (%) | |||
| Married | 1,663 (91.0) | 148 (72.9) | 6,322 (90.3) |
| Others | 165 (9.0) | 55 (27.1) | 682 (9.7) |
| Education, n (%) | |||
| Middle school or less | 490 (26.8) | 28 (14.6) | 5,326 (76.0) |
| High school or professional college | 846 (46.3) | 34 (17.7) | 1,398 (19.9) |
| University | 492 (26.9) | 130 (67.7) | 280 (4.0) |
| Unknown | 5 (0.1) | ||
| Income (yuan/month/person), n (%) | |||
| ≤500 | 27 (1.5) | 1 (0.5) | 834 (11.9) |
| 501–1,500 | 388 (21.2) | 3 (1.5) | 2,067 (29.5) |
| 1,501–3,000 | 1,175 (64.3) | 30 (15.1) | 996 (14.2) |
| >3,000 | 238 (13.0) | 165 (82.9) | 481 (6.9) |
| Unknown | 2,631 (37.5) | ||
| Height, cm | 158.4 (10.4) | 154.7 (11.8) | 158.0 (8.5) |
| Weight, kg | 59.4 (10.2) | 58.3 (9.9) | 58.5 (10.9) |
| BMI, kg/m2 | 23.6 (3.4) | 25.5 (15.5) | 23.4 (3.5) |
| Waist circumference, cm | 85.2 (9.3) | 83.5 (9.9) | 80.3 (9.9) |
| Hip circumference, cm | 91.7 (11.6) | 91.3 (6.6) | |
| Neck circumference, cm | 34.0 (3.2) | 33.2 (2.9) | |
| DBP, mmol/L | 74.0 (12.3) | 74.1 (9.5) | 77.7 (11.5) |
| SBP, mmol/L | 120.8 (17.0) | 125.6 (16.3) | 131.7 (21.7) |
| Fasting glucose, mmol/L | 5.5 (1.3) | 5.7 (1.3) | 5.6 (1.7) |
| HDL, mmol/L | 1.5 (0.4) | 1.5 (0.4) | 1.3 (0.5) |
| LDL, mmol/L | 3.6 (1.0) | 3.6 (1.1) | 3.3 (0.9) |
| TC, mmol/L | 5.5 (1.1) | 5.6 (1.3) | 5.3 (0.9) |
| TG, mmol/L | 1.6 (1.1) | 1.7 (1.9) | 1.4 (1.6) |
| Current smoking status, n (%) | 144 (7.9) | 27 (14.1) | 1,815 (26.1) |
| Current tea drinking, n (%) | 1,051 (57.7) | 108 (56.3) | |
| Current alcohol drinking, n (%) | 136 (7.4) | 19 (9.9) | 2,752 (39.3) |
| Physical activity, MET | 40.6 (14.1) | 91.6 (51.1) | |
| Total energy intake, kcal/day | 1,763.1 (568.3) | 1,631.0 (570.5) | |
| Vegetable intake, g/day | 369.4 (176.8) | 427.0 (201.3) | 336.3 (229.2) |
| Fish intake, g/day | 50.5 (51.9) | 43.0 (50.0) | |
| Red and processed meat intake, g/day | 83.6 (62.3) | 72.0 (47.0) | 131.2 (133.8) |
| Fruit intake, g/day | 150.9 (198.5) | 132.1 (84.5) | 79.4 (133.6) |
| Yogurt intake, g/day (dry weight) | 4.7 (15.6) | 3.8 (6.2) |
Data are means (SD) unless otherwise indicated. DBP, diastolic blood pressure, SBP, systolic blood pressure; TC, total cholesterol; TG, triglycerides.
Interpretable Machine Learning Framework for Data Integration and Explanation
We devised a model based on a gradient boosting framework—Light Gradient Boosting Machine (LightGBM) (14), a gradient boosting decision tree algorithm—to link input features with type 2 diabetes. A total of 297 host features that were potentially related to type 2 diabetes in the literature (metadata, gut microbiota composition, and diversity [see Supplementary Material]) (15) were incorporated into our machine learning model. We also compared our model performance with other widely used methods such as logistic regression and random forest.
We used SHapley Additive exPlanations (SHAP) (7) to unlock the machine learning results. The mean absolute value of the SHAP values for each feature represents their average contribution to the overall model predictions. Thus, features with an average absolute SHAP value >0 were used as selected features. The inflection point of SHAP dependence plots (x-axis represents the feature variable, while y-axis represents the SHAP value for the feature variable) was defined as the optimal threshold for each selected feature.
Classification Analysis
We constructed a classifier based on the identified microbiota features, host genetics, and the traditional type 2 diabetes risk factors separately (see Supplementary Material), and compared their predictive performance. In addition, we excluded patients with prediabetes defined by the criteria from the World Health Organization (16) to reevaluate our microbiome-based classifier performance in the discovery cohort (302 participants with prediabetes) and validation cohort 1 (46 participants with prediabetes). We used DeLong’s test method to test the difference between the classifier’s predictive performance before and after exclusion of the patients with prediabetes. Our predictor is based on code adapted from sklearn 0.15.2 (17) LightGBM class; R package pROC (18) was used for receiver operating characteristic curve analyses, and the method of DeLong was used for area under the curve comparison.
Microbiome Risk Score Construction
We construct a microbiome risk score (MRS) based on the machine learning–selected microbiome features and their SHAP values (for formula, see Supplementary Material). We also constructed another MRS following the conventional method (19) and used a Poisson regression to test the cross-sectional association of the two constructed MRS models with type 2 diabetes risk, respectively.
Gut Microbiota Transplantation
Nine participants were randomly selected as the representative donors according to the level of the MRS (range 0–14). A detailed description of fecal suspension inoculum preparation and transplantation is provided in Supplementary Material.
The study protocols were approved by the local Ethical Review Committee, Guangzhou, China, and all human participants gave written informed consent.
Statistical Analysis
Statistical analysis was performed with Stata 15 (StataCorp, College Station, TX). For the discovery cohort and external validation cohort 1, a multivariable Poisson regression model was used to examine the cross-sectional association with type 2 diabetes for MRS and each machine learning–identified taxa-related feature as a continuous variable or as a binary variable: higher abundance (i.e., at or above the optimal threshold) compared with lower abundance (i.e., below the optimal threshold), with adjustment for age, sex, BMI, waist circumference, household income, marital status, and self-reported educational level, total energy intake, alcohol drinking, and smoking. For external validation cohort 2, all aforementioned covariates except total energy intake (not available) were used in the statistical model. We combined the effect estimates from the three cohorts using random-effects meta-analysis. We repeated the above analysis in the discovery cohort using MRS derived from the same microbial features of metagenomic data to test the robustness of 16S results.
We also used a Poisson regression to estimate the interaction of MRS with age and sex on type 2 diabetes risk and performed subgroup analysis for the MRS–type 2 diabetes relationship stratified by age (<64.3 years vs. ≥64.3 years, with 64.3 years as the median age of this cohort) and sex in the discovery cohort.
We used a linear regression model to explore the association of baseline MRS with glucose increments in the next 3 years, with adjustment for demographic, dietary, and lifestyle factors. Sensitivity analysis was conducted by adding baseline fasting glucose to test the influence of baseline fasting glucose on the performance of the above model.
The association of the MRS with host circulating metabolites was assessed by Spearman correlation. Those MRS-metabolite associations that survived the multiple testing correction (Benjamini-Hochberg method) in the discovery cohort were further chosen for replication in the external validation cohort 1.
In the discovery cohort, linear regression was used to estimate the difference in MRS per-quartile change for continuous dietary factors, per-unit change for adiposity factors, or per-category change for categorical (ordinary) factors in the baseline tested factors, with adjustment for demographic factors and type 2 diabetes medication use and mutual adjustment for the other tested adiposity, dietary, and lifestyle factors. The tested adiposity, dietary, and lifestyle factors included BMI, hip circumference, waist circumference, neck circumference, total energy intake, alcohol drinking, smoking, tea drinking, vegetable intake, fruit intake, fish intake, red and processed meat intake, yogurt intake, and physical activity. The adjusted demographic factors included age, sex, household income, marital status, and educational level.
In both the discovery cohort and the external validation cohort 1, we used a linear regression model to assess the cross-sectional association of MRS with body fat distribution, with adjustment for the demographic, dietary, and lifestyle factors. In both cohorts, Poisson regression was used to estimate interaction of MRS with trunk fat–to–limb fat mass ratio for type 2 diabetes risk, with adjustment for the aforementioned covariates.
For the results of the animal study, ANOVA was used for comparison between multiple groups. The Benjamini-Hochberg method was used to control the false discovery rate. P values <0.05 were considered significant.
Data and Resource Availability
For the discovery and external validation cohort 1, the raw data for 16S rRNA gene sequences are available in CNGB Sequence Archive (CNSA) (https://db.cngb.org/cnsa/) of China National GeneBank (CNGBdb), accession number CNP0000829. For the external validation cohort 2, the raw data for 16S rRNA gene sequences are available from the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/), ac‐cession number PRJEB18535.
Results
The Identified Combination of Microbes Is Strongly Predictive of Type 2 Diabetes Risk
LightGBM (14), used in the current study, outperformed the logistic regression and random forest in type 2 diabetes prediction (Supplementary Table 1). We identified 21 features that contributed to type 2 diabetes prediction, of which 15 were microbiome features (unweighted_nmds6 and observed_species were indicators of microbial diversity, and others were taxa-related features) (Supplementary Fig. 3). The 21 identified features showed a similar predictive capacity compared with all 297 input features (Supplementary Table 2), and the majority of the selected taxa-related features had a low-to-modest intercorrelation (Supplementary Fig. 4).
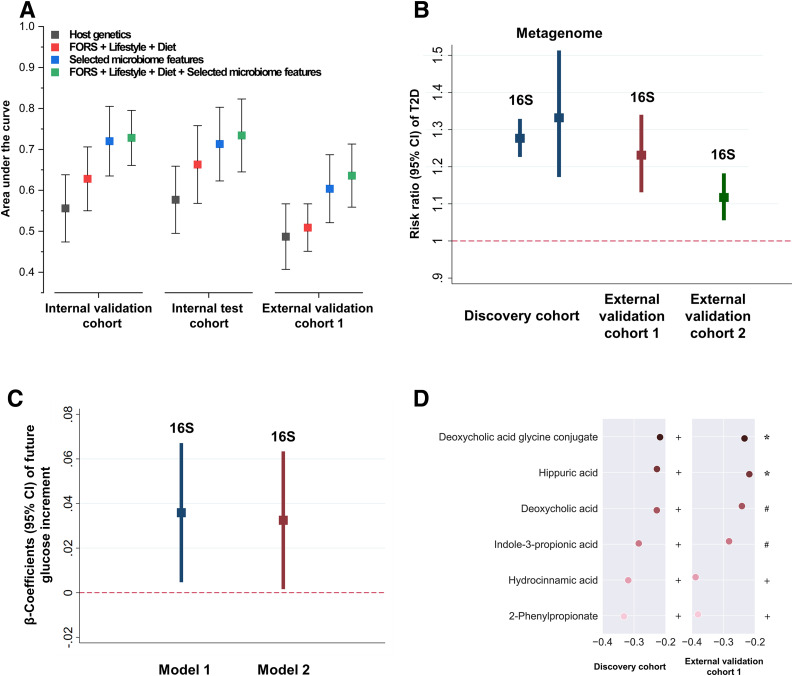
The selected microbiome features showed superior type 2 diabetes prediction accuracy compared with the host genetics and other environmental factors (Framingham-Offspring Risk Score [FORS] + lifestyle + diet) (Fig. 1A). An addition of the selected microbiome features to the model (FORS + lifestyle + diet) increased the area under the curve from 0.63 (95% CI 0.55–0.71) to 0.73 (95% CI 0.66–0.8) in the internal validation cohort (P = 0.0024), 0.66 (95% CI 0.57–0.76) to 0.73 (95% CI 0.65–0.82) in the internal test cohort (P = 0.016), and 0.51 (95% CI 0.45–0.57) to 0.64 (95% CI 0.56–0.71) in the external validation cohort 1 (P = 0.0036), respectively. There was no significant difference in the microbiome-based classifier’s predictive performance before and after exclusion of the patients with prediabetes in the discovery cohort (P = 0.49) and validation cohort 1 (P = 0.14).
Figure 1.
Identified gut microbiota affect type 2 diabetes development and host serum metabolites. A: Algorithm performance in the discovery cohort and external validation cohort 1 based on the selected microbiome features, host genetics, lifestyle and diet, type 2 diabetes traditional risk factors (FORS, including age, sex, parental history of diabetes, BMI, systolic blood pressure, HDL, triglycerides, and waist circumference), and their combination. B: Association of the MRS with type 2 diabetes risk in the discovery cohort, external validation cohort 1, and external validation cohort 2. Poisson regression was used to estimate the RR and 95% CI of type 2 diabetes per 1-unit change in the MRS, with adjustment for demographic, dietary, and lifestyle factors. C: Association between the MRS and prospective glucose increments over 3 years in the discovery cohort. Linear regression was used to estimate the difference in future fasting glucose per unit change in the MRS in a cohort of 249 individuals without type 2 diabetes, with adjustment for demographic, dietary, and lifestyle factors (model 1). Sensitivity analyses were conducted by adding baseline fasting glucose in the above model 1 to test the influence of baseline fasting glucose on the performance of our model (model 2). D: Association of the MRS with host circulating metabolites. Spearman correlation coefficients between the MRS and the host serum metabolites were calculated. The MRS-metabolite associations were further replicated in the external validation cohort 1. *P < 0.05; #P < 0.01; +P < 0.001.
To estimate individual microbiome risk for type 2 diabetes development, we generated an MRS (score range 0–14) based on the 14 identified microbiome features (Table 2). We found that the MRS (per 1-unit change in MRS) consistently showed a positive association with type 2 diabetes risk in the discovery cohort (risk ratio [RR] 1.28, 95% CI 1.23–1.33), external validation cohort 1 (RR 1.23, 95% CI 1.13–1.34), and external validation cohort 2 (RR 1.12, 95% CI 1.06–1.18) (Fig. 1B and Supplementary Table 3). We also found the MRS–type 2 diabetes association could be replicated based on data from 1,068 shotgun metagenomic samples in the discovery cohort (including 159 case subjects). In agreement with the 16S rRNA results, the metagenome-based MRS consistently showed a positive association with type 2 diabetes risk (per-unit change in new MRS: RR 1.33, 95% CI 1.17–1.51) (Fig. 1B). However, results from the conventional method–derived MRS could not be validated in the two external cohorts (Supplementary Fig. 5).
Table 2.
List of components included in the MRS construction
| Microbiome | Taxa annotation |
|---|---|
| f__lactobacillaceae | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__lactobacillaceae |
| c__alphaproteobacteria | p__Proteobacteria; c__alphaproteobacteria |
| f__mogibacteriaceae | p__Firmicutes; c__Clostridia; o__Clostridiales; f__mogibacteriaceae |
| g__clostridiaceae spp | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiaceae; g__ |
| c__deltaproteobacteria | p__Proteobacteria; c__deltaproteobacteria |
| g__butyrivibrio | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__butyrivibrio |
| o__lactobacillales | p__Firmicutes; c__Bacilli; o__lactobacillales |
| f__comamonadaceae | p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__comamonadaceae |
| g__roseburia | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__roseburia |
| g__megamonas | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Veillonellaceae; g__megamonas |
| g__mogibacteriaceae spp | p__Firmicutes; c__Clostridia; o__Clostridiales; f__mogibacteriaceae; g__ |
| g__dorea | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__dorea |
| s__dispar | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Veillonellaceae; g__Veillonella; s__dispar |
| Observed species (an indicator of the gut microbial diversity) |
The MRS is generated based on 14 microbiome features, including 13 taxa and 1 microbial alpha-diversity index (i.e., observed species).
There was no significant interaction between MRS and age (Pinteraction = 0.8) or MRS and sex (Pinteraction = 0.3) for type 2 diabetes risk in the discovery cohort. Subgroup analysis for the MRS–type 2 diabetes relationship stratified by age and sex produced similar results (Supplementary Table 4).
Factors Underlying Type 2 Diabetes Prediction
Our results indicated that individuals age >66.7 years or with waist circumference >84.6 cm were at high risk of type 2 diabetes (Supplementary Fig. 6). This is consistent with the standards of medical care for type 2 diabetes in China (20,21), which suggests that individuals >65 years old or with waist circumference >85 cm (male) or 80 cm (female) are at high risk of type 2 diabetes.
We identified the optimal threshold of the identified 13 taxa-related features according to their SHAP dependence plots (Supplementary Table 5). Eight of 13 taxa-related features showed statistically significant associations with type 2 diabetes when they were treated as binary variables—high abundance (i.e., at or above the optimal threshold) compared with low abundance (i.e., below the optimal threshold) (Supplementary Fig. 7A)—while only 3 taxa-related features showed significant association with type 2 diabetes if the abundance of the selected microbiome was treated as a continuous variable (Supplementary Fig. 7B).
The Identified Combination of Microbes Is Longitudinally Associated With Glucose Increments
We conducted a prospective investigation among 249 participants with normal fasting glucose (fasting glucose <7 mmol/L) at baseline. The mean (SD) initial and end blood glucose level of this group was 5.2 (0.5) mmol/L and 5.4 (0.6) mmol/L, respectively. Our results showed that MRS was significantly positively associated (P < 0.05) with future glucose increments in two statistical models (Fig. 1C).
Correlation of the Identified Combination of Microbes With Host Blood Metabolome
We performed targeted metabolomics profiling of serum samples from the discovery cohort (n = 903) and external validation 1 (n = 113) and assessed the correlation of the type 2 diabetes–related combination of microbes (i.e., MRS) with 199 serum metabolites. Participants with a history of type 2 diabetes medication use were excluded from this analysis. The serum samples were collected at the same point in time as the stool samples. We found that the MRS was consistently correlated with six metabolites in the discovery cohort and external validation cohort 1 (Fig. 1D).
The MRS was negatively correlated with 2-phenylpropionate, hydrocinnamic acid, and indole-3-propionic acid, which were all associated with gut microbiome metabolism (22–24). Deoxycholic acid and deoxycholic acid glycine conjugate are secondary bile acids produced by the action of enzymes existing in the microbial flora of the colonic environment (25). Recent studies have revealed that alteration of gut microbiota not only could affect the bile acid pool but also could influence the bile acid receptor signaling (i.e., FXR and TGR5). The FXR has been reported to be involved in glucose homeostasis, energy expenditure, and lipid metabolism (26). These observations provide insight into the potential function and mechanism of our identified microbial features, represented by the MRS, in host metabolism.
The Identified Combination of Microbes Affects Type 2 Diabetes Development in Germ-Free Mice
There was no significant difference in basal fasting glucose among the four experimental groups (P = 0.11). Mice transplanted with the gut microbiota from high-MRS individuals, of either non–type 2 diabetes or type 2 diabetes status, showed a significant increase in fasting glucose levels compared with levels of the low-MRS individuals or germ-free control mice (Supplementary Fig. 8A–C). There was no significant difference in fasting glucose between the germ-free control group and the low-MRS group.
Baseline Adiposity and Dietary Factors Can Modulate the Type 2 Diabetes–Related Microbiome
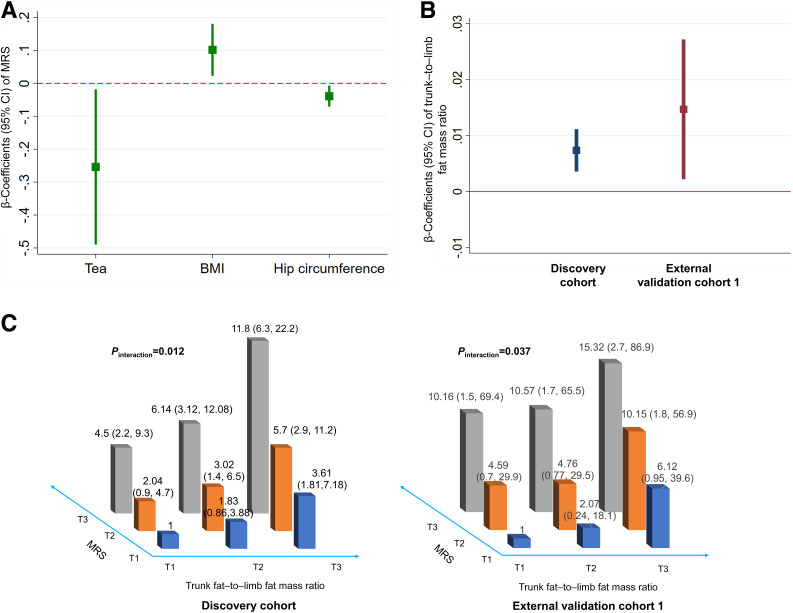
In the longitudinal analysis of the discovery cohort, baseline BMI was positively associated with the MRS, while hip circumference and tea drinking were inversely associated (Fig. 2A and Supplementary Table 6).
Figure 2.
Adiposity and dietary factors modulate the association between gut microbiome and type 2 diabetes. A: Association of baseline adiposity and dietary factors with the microbiome risk score (MRS). Linear regression was used to estimate the difference in MRS per quartile (for continuous dietary factors), per unit (for adiposity factors), or per category (for ordinary factors) change in the baseline tested factors, with adjustment for demographic factors and type 2 diabetes medication use and mutual adjustment for the other tested adiposity, dietary, and lifestyle factors. We only present those adipose, dietary, or lifestyle factors showing significant association with the MRS in the figure. B: Association between MRS and trunk fat–to–limb fat mass ratio in the discovery cohort and external validation cohort 1. Linear regression was used to estimate the difference in trunk fat–to–limb fat mass ratio per unit change in the MRS, with adjustment for demographic, dietary, and lifestyle factors. C: Interaction between MRS and trunk fat–to–limb fat mass ratio for type 2 diabetes risk. Poisson regression was used to estimate the interaction of MRS and trunk fat–to–limb fat mass ratio for type 2 diabetes risk, with adjustment for demographic, dietary, and lifestyle factors. RR (95% CI) of type 2 diabetes by MRS stratified by trunk fat–to–limb fat mass ratio tertile (T) or RR (95% CI) of type 2 diabetes by trunk fat–to–limb fat mass ratio stratified by MRS tertile is presented.
Body Shape Is Associated With Gut Microbiome, Modulating the Association of Gut Microbiome With Type 2 Diabetes
Obesity is the most important risk factor for type 2 diabetes (27). As BMI and hip circumference are closely correlated with MRS in our study, we hypothesized that the relationship of gut microbiome with type 2 diabetes might be modulated by adiposity status. MRS (per 1-unit change in MRS) was positively associated (P < 0.05) with the distribution of trunk–to–limb fat ratio in the discovery cohort (β 0.007, 95% CI 0.0037–0.011) and external validation cohort 1 (β 0.015, 95% CI 0.0023–0.03) (Fig. 2B and Supplementary Tables 7 and 8). We found a significant interaction between MRS and trunk–to–limb fat mass ratio for type 2 diabetes risk in the discovery cohort (Pinteraction = 0.012) and external validation cohort 1 (Pinteraction = 0.037), with adjustment for potential confounders (Fig. 2C). In the discovery cohort, adjusted RR (95% CI) of type 2 diabetes was 1 (reference), 1.83 (0.86–3.88), and 3.61 (1.81–7.18) for tertiles 1, 2, and 3 of trunk-to-limb fat mass ratio among individuals in the lowest MRS levels (MRS tertile 1). It was 4.5 (2.21–9.17), 6.14 (3.12–12.08), and 11.79 (6.28–22.16) among individuals in the highest MRS tertile. Similar interaction results were found in the external validation cohort 1 (Fig. 2C).
Conclusions
In the current study, we identify a robust combination of gut microbes associated with type 2 diabetes by integrating a cutting-edge interpretable machine learning framework with large-scale human cohort studies. The combination of gut microbes shows superior type 2 diabetes prediction accuracy compared with host genetics or traditional risk factors. Additionally, we construct a novel risk score for the gut microbiome and successfully replicate the MRS–type 2 diabetes association in another two independent cohorts. We then reveal that the MRS is correlated with a few gut microbiota–derived blood metabolites. The fecal microbiota transfer experiment further confirms the effect of the identified combination of microbes on type 2 diabetes development. Finally, we identify potential modulating factors that could modulate the type 2 diabetes–related microbiome features and demonstrate that the relationship between MRS and type 2 diabetes could be modified by body fat distribution.
Microbiome data are highly dimensional, underdetermined, and overdispersed. These features challenge standard statistical tools, making results from both traditional parametric and nonparametric models unsatisfactory (28). On the other hand, multiple host anthropometric, dietary, and lifestyle factors play important roles in shaping the microbiome composition (29), while large human cohorts that take into account these confounders are necessary but are thus far sparse. Here, we used a machine learning algorithm (LightGBM) (14) to link multiple features with type 2 diabetes. We also interpret the results of the “black box” machine learning models with a recently developed novel tool: SHAP (7). Compared with other interpreting methods such as gain, split count, and permutation method, SHAP has been theoretically verified as the only consistent and locally accurate method to interpret machine learning results (30). We demonstrated that our new analytic framework could effectively integrate data from different dimensions and help unlock the machine learning results. This analytic framework could be used for other multiomics research as well—beyond gut microbiome.
We discovered 11 microbiota taxa as novel predictive factors for type 2 diabetes risk, including c_alphaproteobacteria, c_deltaproteobacteria, o_lactobacillales, f_comamonadaceae, and seven microbiota taxa from the order Clostridiales (f_mogibacteriaceae, g_clostridiaceae spp, g_butyrivibrio, g_megamonas, g_mogibacteriaceae spp, g_dorea, and s_dispar). These seven taxa from the order Clostridiales were enriched in the healthy control subjects rather than type 2 diabetes case subjects in our present study. We also confirmed several taxa (g_roseburia and f_lactobacillacea) that have previously been reported (31–36). The roseburia, which was decreased in our patients with type 2 diabetes, is a butyrate-producing genus and has been shown to causally improve glucose tolerance (31,32). In line with the previous literature indicating that genus lactobacillus might contribute to chronic inflammation in diabetes development (33,34), we also found that the family lactobacillaceae was enriched in the participants with type 2 diabetes and had a strong predictive power for type 2 diabetes. Although based on the different microbiome analysis methods, the two shotgun metagenomics–based studies (35,36) consistently showed a decrease in roseburia species and an increase in lactobacillus species in type 2 diabetes case subjects compared with control subjects. Specially, lactobacillus species had the highest score for the identification of patients with type 2 diabetes in a European study (36). Due to the translational nature of the present project, we did not further investigate the functionality of each identified gut microbial taxa; rather, we were more interested in the role of the overall microbiome combination and pattern.
Some of the microbiota taxa that differ between individuals with and without type 2 diabetes were reported to be related to obesity (37,38). For example, obesity was characterized by an increased abundance of lactobacillaceae and decreased abundance of several groups within class Clostridia in a prior human cohort (37). Animal studies demonstrated that reduction in the diversity and function of the Clostridia might contribute to obesity development potentially via downregulated genes that control lipid absorption (38). Together, the available evidence suggests that the progression from obesity to type 2 diabetes may be, in part, mediated by the gut microbiome.
The prospective investigation of the gut microbiome–glucose association has rarely been conducted in the previous cohort studies, which exclusively investigated a cross-sectional association of gut microbiome with type 2 diabetes or related traits (5,34,35). A recent cross-sectional study demonstrated that the link between the overall gut microbiota composition and fasting glucose was relatively weak (39). In our study, we identified a set of core discriminative gut microbiota between participants with and without type 2 diabetes, which was prospectively associated with fasting glucose in humans, and then the results were confirmed in an animal study. Therefore, our study design was more comprehensive than the previous cross-sectional study (39). Furthermore, our results highlighted that the core gut microbiota, rather than overall gut microbiota composition, was related to fasting glucose and played an important role in type 2 diabetes development. Integration of MRS–blood metabolome analysis revealed the potential mechanism of the MRS–type 2 diabetes association, involving a variety of gut microbiota–derived metabolites, although the detailed mechanism is yet to be discovered.
We further demonstrated that higher BMI or lower hip circumference is positively associated with future MRS levels, which indicates the potential role of adiposity in gut microbiome. The evidence was clearer when we found an interaction between the MRS and trunk–to–limb fat mass ratio, suggesting that adiposity may be an effect modifier for gut microbiome and type 2 diabetes development. Taken together, our results highlight that a healthy body shape may play an important role in maintaining gut health.
This study has several strengths. Firstly, we constructed a robust MRS based on the identified microbiome–type 2 diabetes relationship by applying an interpretable machine learning framework in a large-scale human cohort. In addition, we validated the identified MRS–type 2 diabetes relationship in two independent human cohorts and germ-free mice, which has rarely been achieved in prior studies (5,34,35,38). A major limitation of the current study is that our main results are based on the cross-sectional association between the gut microbiome and type 2 diabetes. Nevertheless, we demonstrated the effect of the identified gut microbiome features on type 2 diabetes development using fecal transplantation with germ-free mice and confirmed the longitudinal association of the microbiome features with glucose increment using prospective data. Another limitation is that in the human cohorts or the animal study, we did not perform the glucose tolerance test or insulin tolerance test, which may provide more information on the MRS–type 2 diabetes relationship. Finally, all participants included in the current study are middle-aged and elderly Chinese, and therefore caution should be taken in extrapolating our findings to other age-groups or ethnic groups.
In summary, we successfully integrated the cutting-edge interpretable machine learning framework and large-scale human cohort studies, identifying a core set of gut microbiome features and their thresholds robustly associated with type 2 diabetes. The newly discovered combination of microbes can potentially be used as type 2 diabetes diagnostic, therapeutic, or preventive targets through diet and lifestyle intervention.
Article Information
Acknowledgments. The authors thank all the participants of the cohorts for contributing stool samples and phenotypes. The authors also thank the Westlake University Supercomputer Center for computational resources and related assistance.
Funding. This study was funded by National Natural Science Foundation of China (82073529, 81903316, 81773416), Zhejiang Province Ten-thousand Talents Program (2019R52039), and the 5010 Program for Clinical Research (2007032) of Sun Yat-sen University (Guangzhou, China).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.-S.Z., W.G., and Y.-m.C. contributed to study conceptualization. J.-S.Z. and W.G. contributed to development of methodology. W.G. and Z.J. contributed to formal analysis. C.-w.L., Y.H., J.-s.L., T.-y.S., and H.-l.Z. contributed to data collection. C.-w.L., Y.H., F.X., and Z.M. contributed to data curation. W.G. and J.-S.Z. contributed to writing the manuscript. J.-S.Z., W.G., Y.F., H.Z., Y.-m.C., Y.H., Z.J, C.-w.L., F.X., Z.M., T.-y.S., J.-s.L., and H.-l.Z. contributed to writing, reviewing, and editing the manuscript. J.-S.Z., Y.-m.C., and H.Z. contributed to funding acquisition. J.-S.Z. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.13148804.
W.G., C.-w.L., Y.H., and Z.J. contributed equally to this work.
H.Z., Y.-m.C., and J.-S.Z. contributed equally to this work.
References
- 1.Franks PW, McCarthy MI. Exposing the exposures responsible for type 2 diabetes and obesity. Science 2016;354:69–73 [DOI] [PubMed] [Google Scholar]
- 2.Zhou B, Lu Y, Kaveh Hajifathalian JB; NCD Risk Factor Collaboration (NCD-RisC) . Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016;387:1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut 2014;63:1513–1521 [DOI] [PubMed] [Google Scholar]
- 4.Petrosino JF The microbiome in precision medicine: the way forward. Genome Med 2018;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurung M, Li Z, You H, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA 2018;319:1317–1318 [DOI] [PubMed] [Google Scholar]
- 7.Lundberg S, Lee SI A unified approach to interpreting model predictions. Advances in Neural Information Processing Systems 30 (NIPS 2017). Accessed 23 November 2020. Available from https://papers.nips.cc/paper/7062-a-unifiedapproach-to-interpretingmodel-predictions [Google Scholar]
- 8.Zhang ZQ, He LP, Liu YH, Liu J, Su YX, Chen YM. Association between dietary intake of flavonoid and bone mineral density in middle aged and elderly Chinese women and men. Osteoporos Int 2014;25:2417–2425 [DOI] [PubMed] [Google Scholar]
- 9.Fan F, Xue WQ, Wu BH, et al. Higher fish intake is associated with a lower risk of hip fractures in Chinese men and women: a matched case-control study. PLoS One 2013;8:e56849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Wu W, Zheng HM, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med 2018;24:1532–1535 [DOI] [PubMed] [Google Scholar]
- 11.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care 2014;37(Suppl. 1):S81–S90 [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke G, Meng Q, Finley T, et al. LightGBM: a highly efficient gradient boosting decision tree. Advances in Neural Information Processing Systems 30 (NIPS 2017). Accessed 23 November 2020. Available from https://papers.nips.cc/paper/6907-lightgbma-highly-efficient-gradient-boosting-decisiontree
- 15.Noble D, Mathur R, Dent T, Meads C, Greenhalgh T. Risk models and scores for type 2 diabetes: systematic review. BMJ 2011;343:d7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization Definition and Diagnosis of Diabetes Mellitus and Intermediate Hyperglycemia: Report of a WHO/IDF Consultation. Geneva,World Health Org., 2006 [Google Scholar]
- 17.Pedregosa F, Weiss R, Brucher M. Scikit-learn machine learning in Python. J Mach Learn Res 2011;12:2825–2830 [Google Scholar]
- 18.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan LM, Massaro JM, D’Agostino RBS Sr. Presentation of multivariate data for clinical use: the Framingham Study risk score functions. Stat Med 2004;23:1631–1660 [DOI] [PubMed] [Google Scholar]
- 20.Jia W, Weng J, Zhu D, et al.; Chinese Diabetes Society . Standards of medical care for type 2 diabetes in China 2019. Diabetes Metab Res Rev 2019;35:e3158. [DOI] [PubMed] [Google Scholar]
- 21.Society CD China guideline for type 2 diabetes (2017 edition). China J Diabetes Mellitus 2018;10:34–86 [Google Scholar]
- 22.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al.; MetaHIT Consortium . Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–381 [DOI] [PubMed] [Google Scholar]
- 23.Aura A Microbial metabolism of dietary phenolic compounds in the colon. Phytochem Rev 2008;7:407–429 [Google Scholar]
- 24.Velagapudi VR, Hezaveh R, Reigstad CS, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res 2010;51:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 2013;17:225–235 [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, Raka F, Adeli K. The role of the gut microbiota in lipid and lipoprotein metabolism. J Clin Med 2019;8:2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006;444:840–846 [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Sun J. Hypothesis testing and statistical analysis of microbiome. Genes Dis 2017;4:138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falony G, Joossens M, Vieira-silva S, et al. Population-level analysis of gut microbiome variation. Science 2016;352:560–564 [DOI] [PubMed] [Google Scholar]
- 30.Lundberg SM, Erion GG, Lee S. Consistent individualized feature attribution for tree ensembles. 2017 [preprint]. arXiv:1802.03888
- 31.Ryan KK, Tremaroli V, Clemmensen C, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanna S, van Zuydam NR, Mahajan A, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 2019;51:600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeuthen LH, Christensen HR, Frøkiaer H. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin Vaccine Immunol 2006;13:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen N, Vogensen FK, van den Berg FWJ, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60 [DOI] [PubMed] [Google Scholar]
- 36.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103 [DOI] [PubMed] [Google Scholar]
- 37.Peters BA, Shapiro JA, Church TR, et al. A taxonomic signature of obesity in a large study of American adults. Sci Rep 2018;8:9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen C, Bell R, Klag KA, et al. T cell-mediated regulation of the microbiota protects against obesity. Science 2019;365:9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, Tremaroli V, Schmidt C, et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab 2020;32:379–390.e3 [DOI] [PubMed] [Google Scholar]