Abstract
We explore the principles of pressure tolerance in enzymes of deep‐sea fishes using lactate dehydrogenases (LDH) as a case study. We compared the effects of pressure on the activities of LDH from hadal snailfishes Notoliparis kermadecensis and Pseudoliparis swirei with those from a shallow‐adapted Liparis florae and an abyssal grenadier Coryphaenoides armatus. We then quantified the LDH content in muscle homogenates using mass‐spectrometric determination of the LDH‐specific conserved peptide LNLVQR. Existing theory suggests that adaptation to high pressure requires a decrease in volume changes in enzymatic catalysis. Accordingly, evolved pressure tolerance must be accompanied with an important reduction in the volume change associated with pressure‐promoted alteration of enzymatic activity ( ). Our results suggest an important revision to this paradigm. Here, we describe an opposite effect of pressure adaptation—a substantial increase in the absolute value of in deep‐living species compared to shallow‐water counterparts. With this change, the enzyme activities in abyssal and hadal species do not substantially decrease their activity with pressure increasing up to 1–2 kbar, well beyond full‐ocean depth pressures. In contrast, the activity of the enzyme from the tidepool snailfish, L. florae, decreases nearly linearly from 1 to 2500 bar. The increased tolerance of LDH activity to pressure comes at the expense of decreased catalytic efficiency, which is compensated with increased enzyme contents in high‐pressure‐adapted species. The newly discovered strategy is presumably used when the enzyme mechanism involves the formation of potentially unstable excited transient states associated with substantial changes in enzyme–solvent interactions.
Keywords: high‐pressure adaptation, hydrostatic pressure, lactate dehydrogenase, Liparidae, Macrouridae, pressure‐tolerant enzymes
The studies of lactate dehydrogenase of hadal and abyssal fishes vs. pressure revealed an unexpected aspect of piezophilic adaptation—an increase in specific volume change of pressure‐induced deactivating transition concomitant with an increase in the pressure of half‐inactivation (P 1/2). As a result, the enzymes from deep‐living species remain unaffected by pressures up to 1–2 kbar, in contrast to their shallow‐water counterpart.
Abbreviations
- GPDH
glyceraldehyde‐3‐phosphate dehydrogenase
- LDH
lactate dehydrogenase
- MDH
malate dehydrogenase
- MRM
multiple reaction monitoring
Introduction
Pressures in the deepest parts of the ocean, the hadal zone (6000–11 000 m), reach up to 1100 times higher than atmospheric pressure. The study of how such high hydrostatic pressures affect biological systems has fascinated scientists for well over a century. Naturally, the deep sea is home to a wide array of physiological adaptations to life under high hydrostatic pressures (see Ref. [1, 2, 3] for review). High‐pressure‐adapted species (piezophiles) in the deep sea show profound adaptation of both biochemical and physiological systems [1, 2, 5]. Most investigations into piezophily have focused on pressure‐sensing systems and pressure‐regulated genes or have explored high‐pressure adaptation in biological membranes [6, 7, 8]. However, the adaptations that allow proteins of piezophiles, and piezophilic enzymes in particular, to fold and function at high hydrostatic pressure have received less attention [2, 5, 9].
Increased hydrostatic pressure impacts biological systems by affecting all transitions that involve changes in the system volume [2, 10]. High pressure (P) enhances biochemical processes that are accompanied by a decrease in system volume, according to Le Chatelier’s principle. Conversely, high pressure inhibits processes in which system volume increases. These volume changes during protein transitions are closely linked with solvent interactions [11, 12, 13, 14, 15, 16], including water penetration into enzyme cavities, constriction of water around solvent‐exposed polar groups, and changes in water network around solvent‐exposed hydrophobic patches [10, 16, 17, 18]. The enhancement of protein hydration is known to play a central role in nondenaturing transitions induced by pressure [13, 19, 20, 21].
The dynamics of protein–solvent interactions [12, 13, 22, 23, 24, 25, 26, 27, 28] and conformational flexibility [29, 30] are fundamental to enzymatic mechanisms. Therefore, the changes in protein conformational landscapes caused by enhanced hydration at increased hydrostatic pressure present large challenges for deep‐sea adaptation. According to the concept of conformational selection in enzymatic mechanisms [31, 32], an enzyme population comprises a dynamic ensemble of preexisting conformers, including those involved in substrate binding and catalysis. Under pressure, this conformational landscape changes, resulting in a redistribution of functionally important conformers that can jeopardize enzyme function [5]. Deep‐adapted species must counteract pressure effects by changing the energy landscape of their proteins and redistributing the population of enzyme conformers to optimize function at habitat pressure [5, 33, 34].
Considering enzymes as dynamic ensembles of multiple conformers existing in a pressure‐dependent equilibrium helps interpreting the effects of pressure on enzyme catalysis. Canonically, the effect of pressure on the rate of a chemical reaction is determined by the activation volume (ΔV≠), the difference between the volume of the transition state and the volume of the reactants. In this model, the natural logarithm of the rate constant under pressure (k p) follows a linear relationship [35, 36]:
| (1) |
where k p is the rate constant at pressure P and k 0 is the rate constant extrapolated to zero pressure, ΔV ≠ is the activation volume of the reaction, T is the temperature, and R is the universal gas constant. However, in experimental observations of enzyme reactions, the plots of ln(k p) against pressure are routinely nonlinear [37, 38, 39, 40, 41, 42]. Common explanations for this nonlinear relationship include pressure‐induced conformational transitions or a change in the rate‐limiting step of enzymatic reaction [36]. However, a more general explanation may be possible if we consider enzymes as existing in dynamic pressure‐dependent equilibrium between multiple conformers. In other words, the effect of pressure on the enzyme may be considered as a pressure‐induced reshaping of the protein conformational landscape. This concept not only provides a more realistic representation for a suggested ‘pressure‐induced conformational change’, but also serves as a mechanistic background for a possible change in the rate‐limiting step.
If the pressure‐induced changes in the equilibrium between multiple conformers with different catalytic efficiencies determine an enzyme’s response to pressure, the system must follow the fundamental relationship between the equilibrium constant (K eq) for a reversible transition and the respective changes in standard free energy (ΔG°), internal energy (ΔE°), volume (ΔV°), and entropy (ΔS°):
| (2) |
The mechanisms of high‐pressure adaptation that allow the enzymes of piezophiles to cope with the effects of pressure on the protein conformational landscapes are particularly important for multimeric enzymes. Due to the large positive volume changes inherent to protein–protein interactions [16, 36, 43, 44], pressure commonly induces dissociation of protein oligomers. This dissociation is one of the causes underlying the inhibitory effects of pressure on multimeric enzymes, such as lactate dehydrogenase (LDH), glyceraldehyde‐3‐phosphate dehydrogenase (GPDH), and malate dehydrogenase (MDH). Slow process of pressure‐induced dissociation of LDH, a tetrameric enzyme, has been used to evaluate the dissociation constant of its tetramers and to study the kinetic mechanism of their re‐assembly [45, 46, 47, 48, 49].
However, the pressure effects on LDH are not limited to pressure‐induced dissociation. According to current understanding, the catalytic mechanisms of LDH include multiple conformational rearrangements, and those involved in the rate‐limiting step of closing the active site loop after substrate binding are of particular importance. [50, 51]. This vital step in the enzyme mechanism is inherently associated with large volume changes [52] and thus will be sensitive to changes in hydrostatic pressure. Therefore, by comparing the effects of pressure on LDH from organisms with different degrees of piezophilic adaptation [49, 53, 54, 55], we can elucidate the general mechanisms of structural and functional adaptation in the enzymes of deep‐living species.
The present study extends our recent comparative investigation of high‐pressure adaptation in four metabolic enzymes of teleost fishes [56]. Here, we compare the specific content, catalytic efficiency, and pressure susceptibility of LDH in muscle tissues of four fish species adapted to different habitat pressures to understand physiological adaptation to the deep‐sea environment. We chose the snailfishes (family Liparidae) as the core study group because of their bathymetric range that extends from the intertidal to the hadal zone (0–> 8000 m) and because of their constraining phylogeny [57, 58]. We examined LDHs from the shallow‐adapted Liparis florae Jordan & Starks 1895 and two hadal species from ~ 6000–> 8000 m: Notoliparis kermadecensis Nielsen 1964 from the Kermadec Trench, and the recently discovered Pseudoliparis swirei Gerringer & Linley 2017 from the Mariana Trench, the deepest‐living vertebrate known [59]. These three confamilial species are complemented with an abyssal grenadier (family Macrouridae), Coryphaenoides armatus Hector 1875, which has been the subject of several studies on pressure adaptation [49, 53, 60, 61].
To understand pressure tolerance in deep‐sea enzymes, we analyze the effects of pressure based on a simplified model. Here, we consider pressure inactivation of LDH to be a result of a pressure‐promoted displacement of the equilibrium between two states with different catalytic efficiencies. Of these two states, one corresponds to the ensemble of conformations predominating at the fish’s habitat pressures, while the second one represents the endpoint of the nondenaturing pressure‐promoted displacement of the conformational equilibria. The actual transitions behind this tentative two‐state model may have a very complex physical nature and include a multitude of different conformational rearrangements in the enzyme. Nevertheless, this simplified semi‐empirical model serves as a satisfactory approximation for the pressure dependencies we observe experimentally and provides a robust protocol for the quantitative analysis of pressure tolerance and pressure‐induced inactivation in enzymes.
Our results reveal a new aspect of high‐pressure adaptation in proteins, namely a substantial increase in volume changes associated with the protein transition to a pressure‐promoted conformational state (ΔV PP) that is evolved in deep‐living species. In agreement with previous reports [49, 55, 62], we demonstrate that this increase in the pressure stability of LDH comes at the expense of its catalytic efficiency, which is compensated by increased enzyme content in deep‐sea species. Our results may have important implications beyond understanding the mechanisms of piezophilic adaptation and provide new insights for engineering pressure‐tolerant enzymes for biotechnological applications [35, 63, 64, 65].
Results
Quantification of lactate dehydrogenase in muscle homogenates
We analyzed LDH content of muscle homogenates using mass spectrometry. For this, it was necessary to find a tryptic peptide that would be conserved in LDH proteins of all four species we examined. It should be noted that most vertebrates possess at least three different isoforms of LDH (LDH‐A, LDH‐B, and LDH‐C), of whose only two (LDH‐A and LDH‐B) are usually found in muscle tissues. Although LDH‐A, also termed as M (muscle) isoform, is known to heavily predominate in fish white muscle tissues, it was desirable to find an isoform‐unspecific peptide that allows to determine the total concentration of both LDH‐A and LDH‐B subunits.
Unfortunately, the only known sequence of LDH from these species is a sequence of LDH‐B from C. armatus (accession number CAE75858.1, NCBI Protein Database). Although sequencing of the full genome of P. swirei has been recently reported [66], the only publicly available annotated genome from the family Liparidae remains to be the genome of a shallow‐water snailfish Liparis tanakae. In our search for an LDH‐specific conserved tryptic peptide, we used an alignment of the sequences of LDH‐B from C. armatus with both LDH‐A and LDH‐B from L. tanakae along with the sequences of seven other LDH‐A and LDH‐B proteins from the fishes belonging to the orders Gadiformes and Perciformes (Fig. 1). Despite high overall identity (≥ 85%), our analysis revealed only one completely invariant tryptic peptide longer than two amino acid residues—the peptide LNLVQR located at the position 108 in the sequence of LDH‐B from C. armatus. This peptide corresponds to a part of a functionally important and highly conserved mobile loop over the substrate binding pocket of LDH [50] and is conserved in both LDH‐A and LDH‐B isoforms under comparison. In order to check the uniqueness of this peptide to LDH proteins, we used Motif Search at GenomeNet resource (https://www.genome.jp/tools/motif/MOTIF2.html) to search through the SWISS‐PROT Protein Knowledgebase for a tryptic peptide LNLVQR (LNLVQR peptide that can be singled out by trypsin and thus is preceded by lysine or arginine and not followed by proline). The entries found by the search almost exclusively correspond to LDH enzymes from vertebrates. The two exceptions are the oligomeric Golgi complex subunit 1 from Dictyostelium discoideum (Slime mold) and a protein from the inner membrane of chloroplasts of Pelargonium hortorum (Common geranium). Therefore, the peptide LNLVQR appears to be entirely specific to muscle LDH enzymes under conditions of our experiments. It was therefore used to quantify LDH in the muscle homogenates in our studies.
Fig. 1.
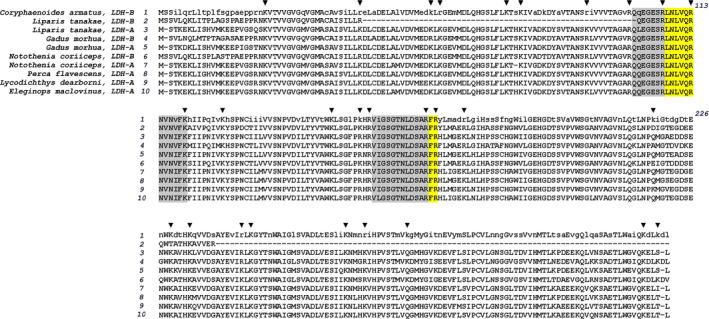
Identification of conserved tryptic peptides in fish LDH. The figure shows an alignment of amino acid sequence of LDH‐B from C. armatus with the sequences of LDH‐A and LDH‐B from the fishes belonging to the orders of Scorpoeniformes (Liparis tanakae),Gadiformes (Gadus morhua), and Perciformes(Notothenia coriiceps,Perca flavescens, L. dearborni and Eleginops maclovinus). Conserved amino acid residues are shown in capital letters in the sequence of LDH‐B from C. armatus. Points of trypsinolysis are marked with black triangles. Fully conserved tryptic peptides are highlighted with yellow shading. Gray shading shows the peptides with only one variable residue. Accession numbers of the sequences shown in the alignment are CAE75858.1, TNN45331.1, TNN87907.1, XP_030209139.1, XP_030222546.1, XP_010787599.1, NP_001290221.1, XP_028440246.1, Q9PW58.3, O93542.3 for the sequences 1–10, respectively.
To perform quantitative multiple reaction monitoring (MRM) measurements, we synthesized the stable isotope‐labeled (Lys 13C6,15N2) peptide analogue and used it as an internal standard. Ratios of the peak areas for labeled and unlabeled peptides were used to calculate the concentration of the target peptide in a sample. As seen from Fig. 2, the selected reaction monitoring profiles of peptides during MRM, the target peptide LNLVQR was found in homogenates from all four fishes. This figure also illustrates a perfect match of the chromatographic traces of the natural peptide and its stable isotope‐labeled standard.
Fig. 2.
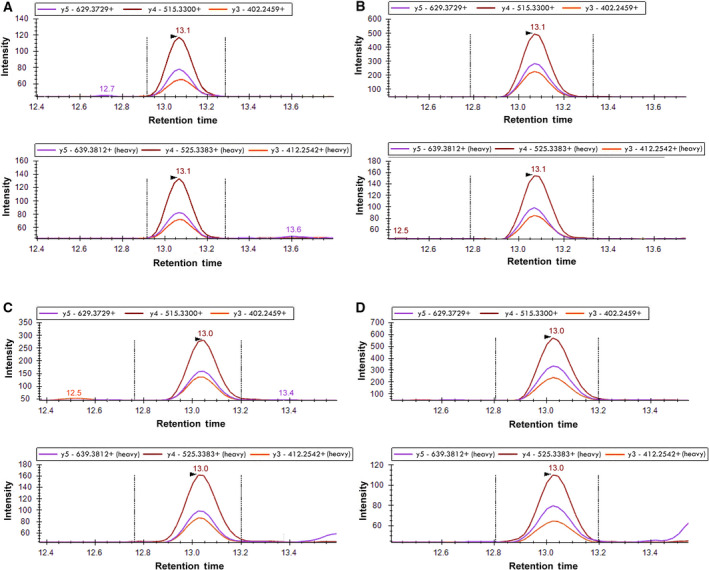
Detection of LDH‐derived peptide LNLVQR in fish muscle homogenates. Examples of MRM profiles of the peptide detected in L. florae (A), C. armatus (B), N. kermadecensis (C), and P. swirei (D). Upper panels in each of A,B,C and D pairs represent the chromatographic traces of the native peptides, while the lower panels show the traces of the stable isotope standard.
The molar concentrations of LDH in the samples estimated from the content of the LNLVQR peptide in tryptic digests are compared in Table 1. The lowest molar content of LDH was found in the shallow‐water L. florae, while the abyssal C. armatus exhibits the highest concentration of LDH (Table 1). Perhaps the most notable observation here is that the three fishes from Liparidae family demonstrate a distinct increase in LDH content with increasing habitat depth.
Table 1.
Content and catalytic efficiency of LDH in muscle homogenates of fish species with different degree of piezophilic adaptation. a
| Species | LDH content, pmol·mg−1 protein b | Protein content in homogenate, mg·mL−1 | LDH concentration in homogenate, nm | Activity of LDH in homogenate, µm·s−1 c | LDH turnover at 1 bar pressure, s‐1 |
|---|---|---|---|---|---|
| L. florae | 16.8 ± 1.8 | 0.75 ± 0.28 | 12.5 ± 2.4 | 47.6 ± 19.4 | 4120 ± 620 |
| C. armatus | 129 ± 28 | 0.72 ± 0.16 | 92.5 ± 15.9 | 87.7 ± 35.6 (0.07) | 987 ± 168 |
| N. kermadecensis | 39.7 ± 8.1 | 0.63 ± 0.25 | 25.0 ± 5.6 | 50.0 ± 26.9 (0.89) | 2240 ± 450 |
| P. swirei | 68.7 ± 8.6 | 0.76 ± 0.18 | 52.0 ± 7.1 | 65.3 ± 25.2 (0.30) | 1640 ± 252 |
The values given in the Table were obtained by averaging results of 3‐6 individual measurements with different samples of fish muscle homogenates and the ‘±’ values show the confidence interval calculated for P = 0.05.
Content of LDH in muscle homogenate determined with mass‐spectroscopic determination of LDH‐specific peptide LNLVQR in tryptic lysates.
Calculated from LDH activity measured at atmospheric pressure (1 bar) at 5 °C and correspond to the µmols of NADH consumed per second per liter of undiluted homogenate. The values in parentheses represent the P‐values of Student’s t‐test for the hypothesis of equality of the respective values of LDH activity to that observed in muscle homogenates from L. florae.
Activity and content of LDH in tissue samples
Protein concentrations, molar content of LDH per milligram of protein, and the LDH activities determined in muscle homogenates allowed us to calculate the catalytic efficiencies (turnover numbers) of the enzymes from four fishes under comparison. The highest catalytic efficiency at atmospheric pressure (1 bar) was found in the enzyme from the shallow‐water L. florae. The LDH from C. armatus had the lowest efficiency (Table 1). Among the three species of liparid, enzyme efficiency at atmospheric pressure gradually decreased with increasing habitat depth. Total activity of LDH in homogenates revealed no statistically significant difference between the three compared snailfishes given this simultaneous increase in LDH concentration in muscle tissue of hadal species.
Effect of pressure on LDH activity
Figure 3 illustrates the pressure dependencies of LDH activity obtained in our experiments. All four fishes demonstrate a pronounced decrease in LDH activity at the highest pressures tested. This pressure‐induced deactivation was partially reversible on decompression. Following a steady, 1–2 min decompression to 1 bar, the reaction rate gradually increased and reached an apparent steady state after ~ 3 min of incubation at atmospheric pressure. The extents of recovery of the enzymatic activity upon decompression from 2.5 to 3 kbar were 49 ± 8% for L. florae, 48 ± 14% for C. armatus, and 76 ± 32% for N. kermadecensis. Recovery of LDH activity after decompression is illustrated in Fig. 4 for the enzyme from N. kermadecensis.
Fig. 3.
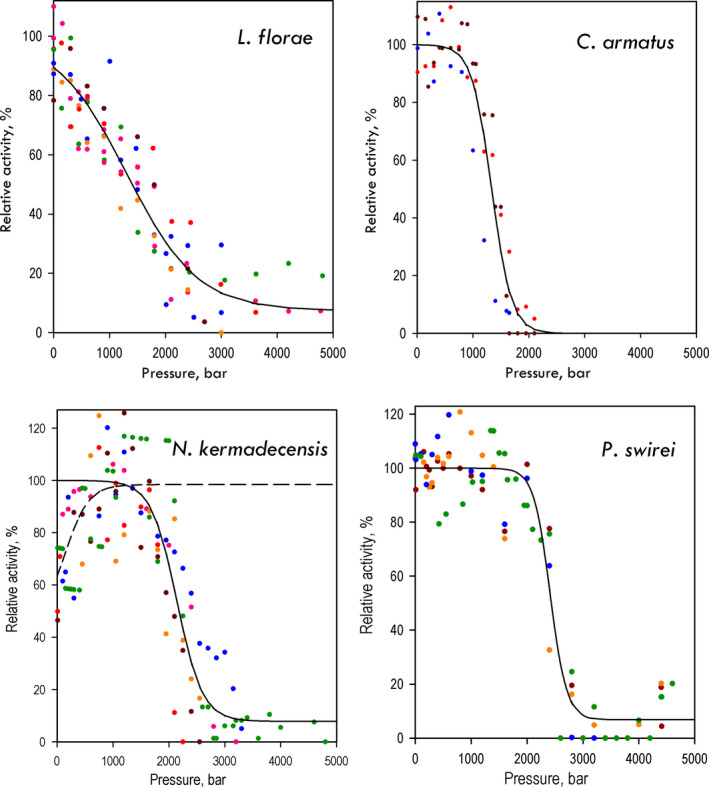
Effects of hydrostatic pressure on LDH activity in muscle homogenates from four fish species with different degrees of piezophilic adaptation. Different colors in the graph correspond to the results of independent experiments. Each individual dataset was scaled to represent the pressure dependence of the percent of the (extrapolated) maximal activity (see Materials and methods). Solid lines represent the results of fitting the combined dataset to Eq. 6. The values of and P ½ and A 0 obtained from these approximations are shown in Table 2. In the case of N. kermadecensis, where the activity of the enzyme exhibits a distinct increase in the interval of 1–1200 bar, the low‐ (1–1500 bar) and high‐pressure (1200–4800 bar) parts of the dataset were fitted separately. Here, the dashed line represents the fitting of the initial part of the dependence, which resulted in the estimates of A 0 (46 ± 3%), A max (52 ± 2%), P ½ (146 ± 97 bar) and (−109 ± 60 mL·mol−1).
Fig. 4.
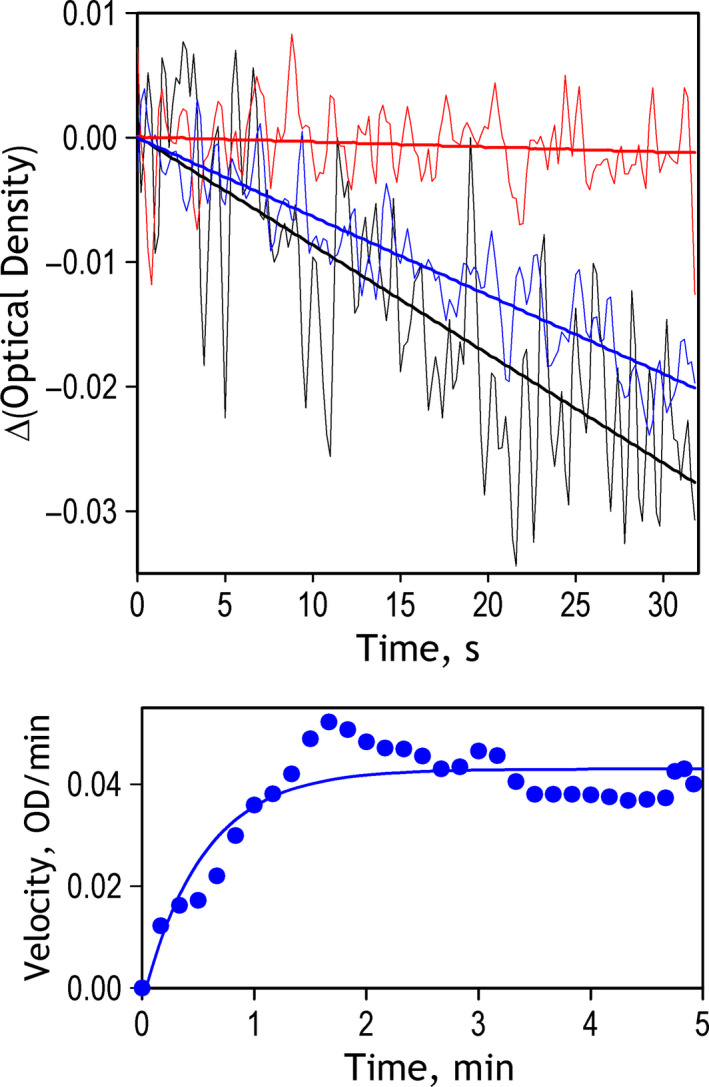
Reversibility of pressure‐induced changes in the rate of catalysis in LDH from N. kermadecensis. Top panel shows the kinetic traces of the decrease in optical density at 350 nm recorded at 1 bar before the experiment (black), at 2.25 kbar (red) and in 200 s after decompression (blue). Bold straight lines show the linear approximations of the recorded traces. Bottom panel shows the time dependence of the reaction rate after decompression. The data were obtained by numerical differentiation of the kinetic curve of the decrease in optical density recorded after decompression. The solid line shows the results of fitting of the dataset to the exponential equation. Characteristic time of the recovery derived from this approximation is equal to 0.54 min.
The four fishes differ considerably in the pressure range of inactivation (Fig. 3). Pressure profoundly inhibited LDH from the shallow‐adapted L. florae. This inhibition begins at modest pressures, with the enzyme exhibiting a nearly linear decrease in activity from 1 to 2000 bar. In contrast, the enzymes from the abyssal and hadal species do not show substantial pressure inhibition up to 1000–2000 bar. Indeed, these enzymes tended to increase their activity with pressure increasing up to 1000–1500 bar. Although in the case of C. armatus and P. swirei this initial activation was marginal and inconsistent, the enzyme from N. kermadecensis exhibited a pronounced, nearly twofold increase in activity when pressure increased from 1 to 1000 bar. This qualitative analysis reveals distinct signs of piezophilic adaptation in LDH of pressure‐adapted species.
Changes in absorbance observed in our experiments were quite subtle and the monitoring of reaction kinetics at each pressure continued for a time interval varying from 30 to 120 s, depending on the rate of reaction and scatter, until a reliable linear kinetic plot was recorded. Consequently, the exact time course of high‐pressure experiments was difficult to reproduce. These factors resulted in a relatively large scatter of data points (Fig. 3). The most probable source of this scatter is variation in the slow pressure‐induced monomerization of LDH. The possibility that the scatter is caused by NADH depletion and/or accumulation of the products (lactate and NAD) is unlikely given our control experiments, where we demonstrated that under these experimental conditions the rate of NADH consumption remains constant for at least 30 min of incubation at ambient pressure (see Materials and methods).
To decrease the effect of this scatter and obtain the most unbiased estimates of and P ½, we used a global fitting of the combined dataset formed from the results of all individual measurements (n = 3–8) performed with each of the four species. Each dataset (single reaction run at varying pressures) was first‐fitted individually to Eq. 6. The obtained estimate of A 0 + A max was then used as a scaling factor to normalize the dataset in relation to the percent of maximum activity observed at each particular pressure. These normalized data were then combined and fitted to Eq. 6 as a single dataset. The results of this fitting are shown in Fig. 3 with solid lines, with the resulting estimates of and P ½ and shown in Table 2.
Table 2.
Parameters of pressure‐induced inactivation of LDH in muscle homogenates of fish species with different degree of piezophilic adaptation. a
| Species | P ½, bar | , mL·mol−1 | A 0, % b | ΔG°, kJ·mol−1 c | K eq° c |
|---|---|---|---|---|---|
| L. florae | 1306 ± 87 | −36.3 ± 4.8 | 7.4 ± 2.2 | −4.7 | 0.148 |
| C. armatus | 1336 ± 53 | −130 ± 34 | 0 | −17.4 | 9.0 × 10−4 |
| N. kermadecensis | 2143 ± 75 | −100 ± 31 | 7.8 ± 3.6 | −21.5 | 1.7 × 10−4 |
| P. swirei | 2413 ± 58 | −157 ± 63 | 6.7 ± 2.4 | −37.8 | 2.4 × 10−7 |
The values given in the Table were obtained by global fitting of data sets composed of the results of 3–8 individual experiments, as shown in Fig. 1. Each of the combined datasets was scaled to have A 0 + A max = 100%. The ‘±’ values show the confidence interval estimated for P = 0.05 from the covariance matrix calculated in the process of nonlinear regression.
The value of A 0 corresponds to the estimate of the percent of maximal activity retained in the pressure‐promoted conformer. In the case of C. armatus, this value was fixed at 0 during optimization in order to prevent the estimated point of extremum from sliding toward the meaningless negative values of A 0.
The values of the change in Gibbs energy (ΔG°) and the respective constant of equilibrium (K eq°) of the apparent transition from the (more active) ‘low‐pressure conformer’ to the pressure‐promoted (low activity) state of the enzyme were calculated from the values of and P ½ using the relationships in Eqs 2 and (4), (5).
Discussion
A newly discovered mode of high‐pressure adaptation in proteins
The most intriguing finding in this study is a prominent increase in the absolute value of of LDH observed with increasing habitat depth of the host species (Table 2). This observation suggests that LDH enzymes from the deep‐sea species undergo more protein hydration during the transition to pressure‐promoted deactivated states than is characteristic of their shallow‐water relatives. Since the molar volume of water is equal to 18 mL, the of −36 mL·mol−1 of the LDH of the shallow‐water L. florae is equivalent to two water molecules penetrating into the protein molecule upon the transition into the ‘pressure‐promoted’ conformational state of the enzyme. In contrast, the volume change in the pressure‐induced transition of LDH from P. swirei (collected from depths of 6898–7996 m) is as high as −156 mL·mol−1. If this volume change is entirely due to water flux into the protein moiety, the number of penetrating water molecules would be as large as eight or nine, fourfold the apparent change in hydration of the shallow‐water enzyme.
The idealized curves of pressure‐induced inactivation of LDH of the four fishes contrast drastically in the low‐pressure range (≤ 1 kbar; Fig. 5). There is a three‐ to fivefold difference between the negative value in the LDH of L. florae and the enzymes of hadal and abyssal species. The enzyme from L. florae exhibits a nearly linear decrease in activity from atmospheric pressure to 2.5–3 kbar, where it reaches 5–10% of the maximal activity. In contrast, the enzymes of hadal species exhibit virtually no decline in activity in the pressure range of 0–1 kbar, the entire range of physiologically relevant pressures that deep‐sea organisms experience. Although the upper boundary of the ‘no effect’ range (range in which the pressure‐induced changes are below 1%) of LDH from C. armatus is decreased to 500 bar, it still above the maximum pressure experienced by this abyssal fish (≤ 400 bar). Although the piezophilic enzymes are stable at physiologically relevant pressures, they exhibit an abrupt inactivation in the range of 1.3–2.4 kbar (Table 2). Interestingly, the activity of LDH from the shallow‐water L. florae at very high, nonphysiological pressures (> 3 kbar) is even higher than that of the piezophilic enzymes due to a low value of (Fig. 5).
Fig. 5.
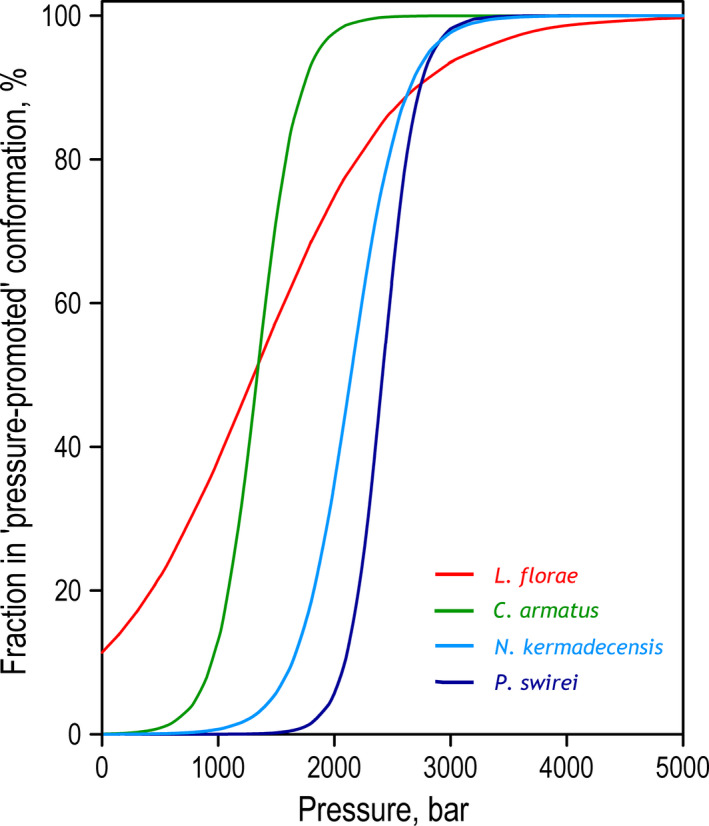
Idealized dependencies of pressure‐induced transition of LDH from L. florae, C. armatus, N. kermadecensis, and P. swirei to pressure‐promoted conformation. The curves were calculated for transitions from 100% ‘optimal activity’ conformer to 100% ‘pressure‐promoted’ state using Eq. 6 and the parameters of pressure‐induced transition shown in Table 2.
It is important to note that the dramatic differences in pressure‐related behavior between these shallow‐ and deep‐adapted LDH enzymes cannot be explained by differences in the effect of pressure on protein oligomerization that may be caused by differences in LDH concentrations in our incubation mixtures. The process of LDH tetramer dissociation into dimers is extremely slow. The first‐order rate constant of this dissociation is as low as 0.002 min−1 in the absence NADH and becomes even slower at saturating concentrations of the cofactor [67]. Therefore, because our experiments were completed within a 20‐min time frame, we expect the effects of pressure‐induced changes in oligomerization equilibrium on these results to be negligible. Even if such a displacement did take place, it would not undermine the observed differences in values. According to the relationship that determines the effects of pressure on equilibrium (Eq. (4), (5)), possible differences in protein concentrations may only change the effective K° (position of equilibrium at standard pressure) and consequently change the values of P ½. In contrast, the values of ΔV° are not dependent on the initial position of equilibrium (and therefore the protein concentration, Eq. (4), (5)). Therefore, our observations of dramatic differences between these four LDH enzymes in values (Table 2) remain valid regardless of the possible dissociation of LDH tetramers during pressure‐induced deactivation and despite certain variation of the enzyme concentrations in the incubation mixtures.
The prevailing hypothesis of piezophilic adaptation in enzymes, as proposed by George Somero and others, suggests that adapting to high pressure requires a decrease in the molar volume changes of all functionally important transitions [2, 55, 68]. This hypothesis was supported by observations with NAD‐dependent dehydrogenases (LDH, GPDH, and MDH) from abyssal fishes that revealed considerably decreased volume changes upon interaction with the cofactor, NADH [53, 55, 62]. Evidently, the term ‘functionally important transitions’ used in this hypothesis must include all transitions of the protein that modify its conformational landscape, change the partitioning of the catalytically competent conformers, and thus affect the enzyme catalytic efficiency. From this point of view, the increase in the volume change of pressure‐promoted protein conformational transitions we observe here in LDH from deep‐living species is surprising. However, this observation aligns with the ‘activity–stability–flexibility’ hypothesis of piezophilic adaptation [5]. Increased conformational flexibility and high compressibility, which this hypothesis considers common themes in protein adaptation to deep‐sea environments, seem to be implicitly associated with increased variability of protein–solvent interactions and increased volume changes in protein transitions.
Increase in volume changes may prevent destabilization of protein structure through increased abundance of excited states
Although a useful model, the above hypothesis of decreased volume changes with high pressure cannot be universally applied. Many functionally important transitions along the conformational trajectory of the enzymatic catalysis are inherently associated with large volume changes and therefore resistant to volume minimization. For example, catalytically important transitions between the ‘open’ and ‘closed’ states of enzymes involve considerable changes in protein interactions with the solvent, which are inherently associated with large volume changes [69]. These transitions result in formation of excited states that are low‐populated in the resting enzyme, but vital for protein functionality as essential intermediates [70, 71, 72].
Conformational rearrangements associated with large volume changes play an exceptional role in catalytic mechanisms of LDH, which became a prototypical object for the studies of conformational landscapes in enzymatic catalysis [50, 73, 74]. According to current understanding, LDH interacts with its substrates through a select‐fit mechanism [50], where the transient opening of the active site controls the rate of formation of the Michaelis complex. Rapid kinetics experiments and molecular dynamics simulations demonstrated that the LDH complex with NADH exists in quite a range of protein conformations with different degree of openness and solvation of the active site, whereby only a minority population of the enzyme is binding competent [50, 51]. Once the enzyme–substrate complex is formed, its further catalytic conversion also proceeds through several different trajectories—the active site conformation of LDH is shown to exhibit important heterogeneity, so that the LDH reaction landscape branches at multiple points creating pathways with varied reactivity [51, 73, 75, 76]. Importantly, the conformational transitions that control the rate of substrate binding and determine the partitioning between different catalytic branches are all associated with substantial changes in protein solvation and, consequently, large volume changes. It makes the catalytic landscape of LDH particularly susceptible to hydrostatic pressure [77] and addition of osmolytes, such as TMAO [52, 75].
When the volume changes associated with catalytic turnover cannot be decreased, as it presumably takes place in the case of LDH, pressure adaptation may occur another way—through the displacement of conformational equilibria toward more catalytically favorable states of the enzyme at higher pressures [33, 78]. In this mode of adaptation, the ‘half‐pressures’ (P ½) of formation of more functionally competent states are increased through a compensatory adjustment of the ΔE − TΔS term (Eq. 2). An example of this displacement occurs in LDH from N. kermadecensis, where we observed a nearly twofold increase in activity from atmospheric pressure to the habitat pressure, which is illustrated in Fig. 3 and discussed in our previous publication [56]. This increase in the activity is presumably caused by pressure‐promoted opening of the active site ‘lid’ and resulting increase in the population of the binding‐competent conformations that takes place at the habitat pressure of N. kermadecensis.
However, this pressure‐induced reshaping of the conformational landscape may also render a destabilizing effect. Higher energy transient conformers promoted by pressure may be prone to further pressure‐induced hydration and conversion into the states with reduced catalytic activity. Therefore, one possible way of evolving pressure‐tolerant enzymes in deep‐sea species is through increasing volume changes associated with the formation of excited transient states of the protein and their further transitions into less active conformers. This mode of piezophilic adaptation is schematically illustrated in Fig. 6. According to our hypothesis, the dramatic increase in observed in pressure‐adapted LDH proteins prevents excessive formation of potentially unstable excited conformations, thus protecting the enzyme transient states from being drawn into further inactivating transitions, at least at the habitat pressure of the host species. Due to increased , the inactivating effect of pressure on LDH of the deep‐sea fishes is negligibly small up to the pressures of 1–2 kbar, in contrast to what is observed with the enzyme from the shallow‐water L. florae, where the inactivation starts at low pressures and continues almost linearly over the all pressure range studied.
Fig. 6.
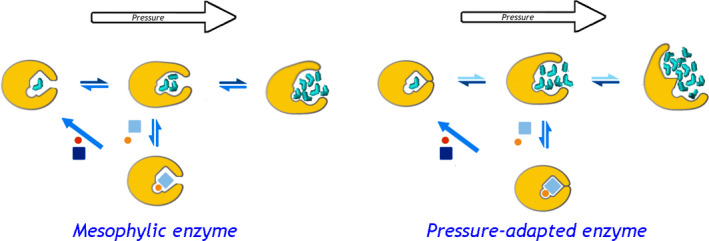
A schematic representation of the proposed mode of piezophilic adaptation in LDH through increasing volume changes in its conformational transitions. The top row shows the enzyme monomer (yellow) in three different conformations: closed (‘ground state’, left), open substrate‐free (‘excited state’, middle) and deactivated (‘pressure‐promoted’, top right). The Michaelis complex (bottom) formed through binding the substrate (pyruvate, orange circle) and the cofactor (NADH, light‐blue rectangle) proceeds through the catalytic step (diagonal arrow) followed by release of the product (lactate) and the oxidized cofactor (red circle and dark‐blue rectangle). Bidirectional arrows indicate reversible conformational transitions, where the color depth symbolizes the position of equilibrium at atmospheric pressure. Water molecules filling the enzyme cavities are shown in cyan. The left diagram shows the position of equilibria and degree of protein hydration in the mesophilic enzyme, while situation observed in the pressure‐adapted enzyme is illustrated on the left.
As a side effect of this adaptation, such an increase in volume changes may reduce catalytic efficiency due to decreased steady‐state concentrations of the functionally essential excited intermediates. This increase in volume changes also results in an abrupt inactivation at pressures commensurate with P ½ (1300–2400 bar, see Table 2). However, this adaptation ensures pressure tolerance at the pressures below this level, including full habitat range pressures (< 1 kbar).
It is important to note that the intrinsic mode of high‐pressure adaptation of LDH that occurs through increase in , and a displacement of P ½ of pressure‐promoted deactivation to higher pressures appears to be complemented with the extrinsic adaptation of biochemical systems of piezophiles through increasing cellular concentrations of natural cosolvents (such as trimethylamine N‐oxide, TMAO) [3]. Increase in osmotic pressure via addition of cosolvents is known to have a profound effect on the activity and stability of LDH. Thus, addition of TMAO was shown to increase the thermal stability of LDH from the mackerel icefish (Champsocephalus gunnari Lönnberg 1905) muscle, while simultaneously reducing its catalytic activity [75]. A study with LDH from rabbit muscle showed a displacement of P ½ of its inactivation to higher pressure by an addition of TMAO. A study with LDH from porcine heart demonstrated that the addition of TMAO displaces the conformational equilibrium in the enzyme toward the closed conformation of the active site loop and thus strongly decreases the pyruvate binding rate [52]. Thus, the effect of increasing concentrations of osmolytes appears to be complementary to the effects of intrinsic piezophilic adaptation in LDH. Both ways of adaptation render their effects on the enzyme by displacing its conformational equilibria in LDH toward more stable conformations, while simultaneously decreasing the enzyme activity.
Pressure‐induced formation of destabilized states of LDH may eventually result in its irreversible inactivation through dissociation of its oligomers
Why, though, would the pressure‐induced conversion of LDH to its pressure‐promoted and less active conformers at the level of only 40% (which LDH from L. florae would experience being exposed to hadal pressures) be so unfavorable? This modest decrease could be compensated through an increase in LDH cellular concentration. Why would the hadal fishes have evolved to prevent this by increasing instead?
The most established and widely discussed mechanism of inactivation of LDH at high pressures is through the quasi‐irreversible inactivation of monomers after pressure‐promoted dissociation of the tetrameric enzyme [44, 46, 47, 48, 79]. However, due to an extremely slow rate of dissociation [67] and very low K D (dissociation constant) values of tetramers (< 10−9 m, [67]), this inactivation proceeds very slowly. Thus, 50% inactivation of porcine LDH at enzyme concentration of 290 nm requires 1 h of incubation at 1 kbar [48]. Most of the previous experiments with pressure‐induced inactivation of LDH were performed through measuring the residual activity of the enzyme at ambient pressure after prolonged exposure to pressure [48, 49, 53, 80].
In contrast, our experiments were designed to minimize high‐pressure exposure time to focus on pressure‐induced changes in protein conformational landscapes that precedes pressure‐induced dissociation. Therefore, the majority of our observed pressure effects result from a rapid displacement of conformational equilibria between multiple enzyme conformers with different catalytic efficiencies [51, 81, 82, 83].
However, the two components of the pressure effects—rapid effects on protein conformation and slow inactivation through dissociation of its tetramers—are likely interconnected. The excessive formation of the pressure‐promoted hydrated state of tetrameric enzyme may represent an initial stage of pressure‐induced dissociation, which eventually results in irreversible inactivation. Therefore, we hypothesize that a 40% increase in the population of the pressure‐promoted conformer at 1000 bar observed in our experiments with L. florae will result in an important increase in the rate constant of dissociation. This increased dissociation would promote complete inactivation of the enzyme at prolonged exposure to deep‐sea pressures. In contrast, in high‐pressure‐adapted hadal and abyssal species, this potential instability at elevated pressures is prevented by increasing volume changes in the transitions that precede dissociation.
The proposed mechanism of high‐pressure adaptation in enzymes through increasing volume changes during potentially destabilizing (but functionally important) transitions may complement the mechanism based on a decrease in volume changes in critical steps of the catalytic cycle proposed by George Somero and others [2, 55, 68]. Although these two opposite tacks in high‐pressure adaptation may seem contradictory, they are closely interrelated.
Higher cellular concentrations of LDH compensate for reduced catalytic efficiency in deep‐sea species
Another important insight in the mechanisms of high‐pressure adaptation may be derived from our measurements of the molar content of LDH in muscle homogenates and the estimated catalytic efficiencies of these enzymes. To our knowledge, this study provides the first example of determination of the content of LDH in muscle tissue of deep‐sea fishes with mass‐spectroscopic techniques. This determination allowed us to compare turnover numbers specific to the enzymes from four fish species differing in the degree of piezophilic adaptation without purification of these proteins.
Catalytic efficiency of LDH significantly decreased with the fish’s increasing habitat depth within the family Liparidae (Table 1). The observed decrease corroborates previous observations of LDH from bathyal and abyssal fishes in other families [49, 55, 62]. In agreement with previous studies, we documented a substantial increase in cellular concentration of LDH in deep‐living snailfishes, which compensates the above‐mentioned decrease of its catalytic efficiency. As a result, the activities of LDH in muscle homogenates are kept at comparable levels in all four fishes (Table 1).
Certainly, the comparison of the levels of LDH activities in different species is complicated by possible differences in their lifestyle and consequent variations in their burst locomotor activities. LDH activities in the macrourids, including C. armatus, are known to be much higher than enzyme activities in the liparids, due to intraspecific differences in metabolic rates [84]. However, three of the four species under comparison belong to one and the same family (Liparidae) and share similar lifestyles. Swimming speeds of the shallow and deep‐water snailfishes were found to be comparable when standardized for temperature [56]. There were also no significant differences in LDH activity at atmospheric pressure with habitat depth in six snailfishes ranging from the intertidal to hadal zone [56], suggesting that the comparisons within the family are appropriate. Further, according to the visual interactions hypothesis, the metabolic rates of deep‐sea species are expected to be even lower than those of their shallow‐water counterparts [85, 86], so that the need for a compensatory increase due to this pressure adaptation may be only compromised (but not increased) from shallow‐water to deep‐sea species. From this perspective, a multifold increase in the LDH concentration in the deep‐sea species observed in our studies might be positively interpreted as a compensatory adaptation to decreased catalytic activity of pressure‐tolerant LDH variants.
Concluding remarks
Our study reveals an unexpected mode of piezophilic adaptation in abyssal and hadal fishes. In contrast to previous understanding of pressure tolerance, this adaptation involves a substantial increase in volume changes in potentially destabilizing conformational transitions during enzyme function. The volume increase makes the enzyme conformational landscape virtually pressure‐insensitive at physiological pressures of habitation of the host species. In return for this pressure tolerance in lower pressure ranges (< 1 kbar), this adaptation causes an abrupt displacement of equilibrium toward the pressure‐promoted conformer(s) on approaching pressures commensurate with P ½.of the deactivating transition. In deep‐adapted fishes, the half‐pressure of conversion, P ½, is displaced to pressures significantly higher than those found at the fishes’ habitat depths. This mode of piezophilic adaptation comes at the expense of decreased catalytic efficiency of pressure‐adapted enzymes, which in turn compensated by the increased enzyme content in deep‐sea species demonstrated in our study.
This mode of high‐pressure adaptation complements the one based on a decrease in reaction volume changes that has been put forward as a universal strategy in early studies [2, 68]. It is apparently used in situations when substantial changes in the enzyme interactions with solvent are innately associated with the enzyme mechanism and cannot be eliminated.
It should be noted, however, that despite the compelling demonstration of a dramatic increase in volume change in protein transitions evolved in pressure‐adapted LDH variants, our study does not reveal the specific nature of the respective adaptations in the protein structure. In particular, the effect of this adaptation on pressure‐induced monomerization of the protein and its subsequent unfolding remains largely unclear. These structural and mechanistic aspects could not be addressed in the present study made with muscle homogenates due to methodological limitations. Further exploration of pressure dependencies of LDH activity, structure, and oligomeric state using purified recombinant proteins of deep‐sea species would provide insights into the structural interpretation of the observed mode of adaptation.
Materials and methods
Reagents
Tris(hydroxymethyl)aminomethane hydrochloride (TRIS), ethylenediaminetetraacetic acid (EDTA), sodium pyruvate and DTT were sourced from Sigma‐Aldrich (St. Louis, MO, USA). 4‐(2‐Hydroxyethyl)‐1‐piperazine ethanesulfonic acid (HEPES) was obtained from Indofine Chemical Company (Hillsborough, NJ, USA).
Sample collection
Specimens of the abyssal fish C. armatus (collection depths 3569–4149 m) and hadal snailfishes—N. kermadecensis (6456–7554 m) and the recently discovered P. swirei (6898–7996 m) from the Mariana Trench [57, 59]—were collected near and from the Kermadec (April–May 2014) and Mariana (November–December 2014) trenches using a free‐vehicle trap baited with mackerel. Sample collection in this study followed the University of Washington Institutional Animal Care and Use Committee (IACUC) guidelines on animal welfare in research. Tissue collection was preapproved by IACUC under the IACUC #4238‐03 protocol. Once on the ship, whole fish were kept on ice or in a cold room (4 °C) and processed as quickly as possible. As a shallow‐adapted comparison from the same family, specimens of L. florae, the tide pool snailfish, were collected from Puget Sound (Cornell University Museum of Vertebrates Fish: 98020) by trawl and hand net (July 2014). Each individual was weighed and measured fresh. White muscle tissue was taken from the anterior portion of the epaxial muscle. Care was taken to ensure no red muscle or gelatinous tissue [87] was included in the sample. Abyssal and hadal tissues were flash‐frozen in liquid nitrogen and stored at −80 °C shipboard and in the laboratory prior to analysis. L. florae was placed in a −80 °C freezer directly.
Preparation of muscle homogenates
White muscle samples (0.1 g) were homogenized using a Potter–Elvehjem glass pestle homogenizer in 1 mL of 50 mm Tris–HCl buffer, pH 7.5, containing 1 mm EDTA, and 1 mm DTT. Homogenates were centrifuged for 10 min at 2000 g at 4 °C. The protein concentration in homogenates was determined with microbiuret assay [88] with bovine serum albumin as a standard.
Mass‐spectrometric determination of the content of LDH in muscle homogenates
Quantification of the LDH content was performed with a triple quadrupole mass spectrometer using the MRM method. LDH quantity was determined based on content of the peptide LNLVQR. This peptide was chosen because it is the only invariant tryptic peptide available in the alignment of sequences of LDHs A and B from nine fish species belonging to the orders Gadiformes and Perciformes (see Results).
Peptides from digested fish muscle homogenates were separated using the UPLC Agilent 1290 system, which included a pump and an autosampler. The sample was loaded into the analytical column Eclipse Plus SBC‐18 (2.1 × 100 mm, 1.8 µm, 100 A). Peptides were eluted with a mixture of solvents A and B. Solvent A was HPLC grade water with 0.1% (v/v) formic acid, and solvent B was 80% (v/v) HPLC grade acetonitrile/water with 0.1% (v/v) formic acid. The separations were performed by applying a linear gradient from 3% to 32% solvent B over 50 min, then from 32% to 53% solvent B over 3 min at 300 μL·min−1 followed by a washing step (5 min at 90% solvent B) and an equilibration step (5 min at 3% solvent B). Ten microliters of each sample was applied on a chromatographic column. The quantitative analysis was performed using an Agilent 6495 Triple Quadrupole (Agilent, Santa Clara, CA USA) equipped with a Jet Stream ionization source. The following parameters were used for the Agilent Jet Stream ionization source: a temperature of the drying gas of 280 °C, 18 psi pressure in the nebulizer, 14 L·min−1 flow rate of the drying gas, and 3000 V voltage on the capillary.
The standard sample of the target peptide was obtained using the solid‐phase peptide synthesis on the Overture (Protein Technologies, Tucson, AZ, USA) according to the published method [89]. The isotopically labeled lysine (13C6,15N2) was used for isotopically labeled peptide synthesis instead of the unlabeled lysine. Concentration of the synthesized peptide was quantified through acidic hydrolysis followed by analysis of derived amino acids with fluorometric detection [90].
LDH activity
Activity of LDH in muscle homogenates at ambient pressure was determined by measuring the changes in NADH fluorescence with the use of Cary Eclipse fluorometer (Agilent Technologies) equipped with a Peltier thermostated multicell holder. Excitation and emission wavelengths were set at 340 and 465 nm with the bandwidths of 5 and 20 nm, respectively. Each measurement was performed in 500 µL of 100 mm Na‐HEPES buffer, pH 7.5 at 2.25 mm pyruvate, and 200 mm NADH placed in a 5 × 5 mm quartz cell controlled to a temperature of 5 °C. Kinetics of decrease in NADH fluorescence was monitored for 3–4 min after adding 1 µL of muscle homogenate. The reaction rate was determined from the slope of the recorded linear trace.
The effect of pressure on LDH activity
Pressure‐induced changes in the activity of LDH were studied with a rapid scanning MC2000‐2 spectrophotometer (Ocean Optics, Inc., Dunedin, FL, USA) using a pulsing PX‐2 Xe arc lamp (Ocean Optics Inc) as a light source. A flexible optic guide connected the instrument to a custom‐made high‐pressure cell with sapphire windows [91]. The pressure was generated with a manual pressure generator (high‐pressure equipment CO, Erie, PA, USA) capable of generating a pressure up to 6000 bar. This setup used a cylindrical quartz cell with the path length of 5 mm and internal volume of 300 µL, covered with a Teflon® membrane. Pressure was transmitted through the liquid n‐Pentane. Data were collected and fitted with the use of our spectralab software [92] (.http://cyp3a4.chem.wsu.edu/spectralab.html).
The high‐pressure experiments were carried out at 4 °C in 80 mm Tris–HCl buffer (pH 7.5 at 5 °C) at 4 mm pyruvate. In the experiments with C. armatus, N. kermadecensis, and P. swirei, the initial concentration of NADH was equal to 600 µm and reactions were monitored through the difference in optical densities at 350 and 400 nm (ε350–400 = 4.9 mm −1cm−1). Displacement of the monitoring wavelength out of the maximum of NADH absorbance was necessary to avoid extremely high noise of signal that would be observed with monitoring at 340 nm, where the transmittance of 600 µm solution of NADH in 5 mm optical cell is as low as 1.4%. Shifting the wavelength to 350 nm increases transmittance to 3.4% and makes the intensity of light high enough to be steadily monitored. In the experiments with L. florae, the concentration of NADH was 250 µm and the difference of optical densities at 340 and 400 nm (ε350–400 = 6.1 mm −1·cm−1) was used for reaction monitoring. The amount of tissue homogenate added to the cell (0.5–2.5 µL) was adjusted to ensure an NADH consumption rate of 5–15 µm·min−1. Control experiments demonstrated that at this reaction rate and the NADH concentration used in our assays, NADH was consumed linearly at ambient pressure (1 bar) for at least 30 min. The reaction was initiated by the addition of protein homogenate. After initiation, the pressure cell was closed rapidly. Reaction monitoring began 30–45 s after the addition of the homogenate. Pressure was increased in steps of 150–300 bar, with 12–18 data points covering the pressure ranges from 1–1800 to 1–5400 bar collected in each experiment. Each pressure was maintained for ~ 0.5–2 min, with the whole experiment taking 15–20 min. In all cases, the residual concentration of NADPH at the end of experiment was not < 100 µm.
Interpretation of the effect of pressure on protein equilibra
The interpretation implemented in this article is based on the assumption that the pressure dependence of the enzymatic activity reflects the pressure‐induced displacement of the equilibrium between enzyme conformers with different catalytic efficiencies. This effect is therefore expected to obey the general equation for the pressure dependence of the equilibrium ([93], Eq. 1):
| (3) |
or in integral form ([94], p. 212, eq. 9:
| (4) |
Here, K eq is the equilibrium constant of the reaction at pressure P, P ½ is the pressure at which K eq = 1 (‘half‐pressure’ of the conversion), ΔV° is the standard molar reaction volume, and is the equilibrium constant extrapolated to zero pressure, .
For the equilibrium A⇌B and = [B]/[A], Eq. (4), (5) may be transformed into the following relationship:
| (5) |
where C 0 = [A] + [B]. To determine the ΔV° and P ½ parameters from the experimental datasets describing pressure‐induced changes in enzymatic activity (A p), this equation was complemented with the offset (A 0) and scaling factor (A max) parameters and used in the following form:
| (6) |
Fitting of the experimental data to this equation with a combination of Neilder–Mead and Marquardt nonlinear regression algorithms resulted in a set of A 0, A max, ΔV° and P ½ parameters.
All data treatment and fitting, including data acquisition in pressure–perturbation experiments, were performed using our spectralab software [92, 95]. The software package is available for download at http://cyp3a4.chem.wsu.edu/spectralab.html. It may be also obtained from the author (DRD) upon request.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
DRD, MEG, PHY, and VGZ designed the study. MEG, DRD, PHY, NEV, and OVT conducted the experiments. DRD, MEG, and VGZ analyzed the data. DRD and MEG wrote the manuscript with the participation of VGZ and PHY. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Doug Bartlett (UCSD) and Jeff Jones (WSU) for opening their laboratories to this work and Jeffrey Drazen (University of Hawaiʻi) and Logan Peoples (UCSD) for research support. We are grateful for Thomas Linley (Newcastle University), Alan Jamieson (Newcastle University), Matteo Ichino (University of Southampton), Chloe Weinstock (Whitman College), and the science parties of the National Science Foundation (NSF) and Schmidt Ocean Institute’s (SOI) HADES Program cruises, who assisted with collection and processing of fish samples at sea. Thanks to Matt Tietbohl (KAUST), Stacy Farina (Howard University), and Adam Summers (Friday Harbor Labs) for their contributions to L. florae collection and dissection. The authors thank the captains and crews of the R/Vs Thompson, R/V Centennial, and R/V Falkor. Authors are also grateful to the ‘Human Proteome’ Core Facility, Institute of Biomedical Chemistry (IBMC), which is supported by the Ministry of Education and Science of the Russian Federation (agreement 14.621.21.0017, unique project ID RFMEFI62117X0017).
This research was supported by the grants from National Science Foundation (NSF‐OCE 1130494 to P. Yancey and 1130712 and 1508760 to J. Drazen covering M. Gerringer’s work) and Schmidt Ocean Institute (FK141109 to J. Drazen and P. Yancey). M. Gerringer is grateful for the support of the National Science Foundation’s Graduate Research Fellowships Program, the Seaver Institute, and the State University of New York at Geneseo. Mass‐spectrometric part of this work was done within the framework of the Russian State Academy of Sciences Fundamental Scientific Research Program for 2013–2020. The funding sources had no involvement in the study design, collection, analysis, and interpretation of data, as well as in the decision to submit the article for publication.
References
- 1. Bartlett DH (2002) Pressure effects on in vivo microbial processes. Biochim Biophys Acta 1595, 367–381. [DOI] [PubMed] [Google Scholar]
- 2. Somero GN (1992) Adaptations to high hydrostatic pressure. Ann Rev Physiol 54, 557–577. [DOI] [PubMed] [Google Scholar]
- 3. Yancey PH & Siebenaller JF (2015) Co‐evolution of proteins and solutions: protein adaptation versus cytoprotective micromolecules and their roles in marine organisms. J Exp Biol 218, 1880–1896. [DOI] [PubMed] [Google Scholar]
- 4. Somero GN (2003) Protein adaptations to temperature and pressure: complementary roles of adaptive changes in amino acid sequence and internal milieu. Comp Biochem and Physiol 136, 577–591. [DOI] [PubMed] [Google Scholar]
- 5. Ichiye T (2018) Enzymes from piezophiles. Semin Cell Dev Biol 84, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prieur D, Jebbar M, Bartlett D, Kato C & Oger P (2009) Piezophilic prokaryotes In Comparative High Pressure Biology(Sebert P, ed), pp. 281–318. CRC Science Publishers, Enfield, New Hampshire. [Google Scholar]
- 7. Kato C (2012) Microbiology of Piezophiles in Deep‐sea Environments In Extermphiles Microbiology and Biotechnology (Anitori RP, ed). Caiser Academic Press, Norfolk, UK. [Google Scholar]
- 8. Kato C & Bartlett DH (1997) The molecular biology of barophilic bacteria. Extremophiles 1, 111–116. [DOI] [PubMed] [Google Scholar]
- 9. Somero GN (1992) Biochemical ecology of deep‐sea animals. Experientia 48, 537–543. [DOI] [PubMed] [Google Scholar]
- 10. Mozhaev VV, Heremans K, Frank J, Masson P & Balny C (1996) High pressure effects on protein structure and function. Proteins 24, 81–91. [DOI] [PubMed] [Google Scholar]
- 11. Masson P & Reybaud J (1988) Hydrophobic interaction electrophoresis under high hydrostatic pressure: study of the effects of pressure upon the interaction of serum albumin with a long‐chain aliphatic ligand. Electrophoresis 9, 157–161. [DOI] [PubMed] [Google Scholar]
- 12. Low PS & Somero GN (1975) Protein hydration changes during catalysis ‐ new mechanism of enzymic rate‐enhancement and ion activation inhibition of catalysis. Proc Natl Acad Sci USA 72, 3305–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Low PS & Somero GN (1975) Activation volumes in enzymic catalysis ‐ their sources and modification by low‐molecular‐weight solutes. Proc Natl Acad Sci USA 72, 3014–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weber G & Drickamer HG (1983) The effect of high pressure upon proteins and other biomolecules. Q Rev Biophys 16, 89–112. [DOI] [PubMed] [Google Scholar]
- 15. Shimizu S (2004) Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc Natl Acad Sci USA 101, 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boonyaratanakornkit BB, Park CB & Clark DS (2002) Pressure effects on intra‐ and intermolecular interactions within proteins. Biochim Biophys Acta 1595, 235–249. [DOI] [PubMed] [Google Scholar]
- 17. Imai T & Hirata F (2005) Hydrophobic effects on partial molar volume. J Chem Phys 122, 094509. [DOI] [PubMed] [Google Scholar]
- 18. Chalikian TV & Macgregor RB Jr (2009) Origins of pressure‐induced protein transitions. J Mol Biol 394, 834–842. [DOI] [PubMed] [Google Scholar]
- 19. Mentre P & Hoa GHB (2001) Effects of high hydrostatic pressures on living cells: a consequence of the properties of macromolecules and macromolecule‐associated water. Int Rev Cytol 201, 1–84. [DOI] [PubMed] [Google Scholar]
- 20. Kornblatt JA & Kornblatt MJ (2002) The effects of osmotic and hydrostatic pressures on macromolecular systems. Biochim Biophys Acta 1595, 30–47. [DOI] [PubMed] [Google Scholar]
- 21. Helms V (2007) Protein dynamics tightly connected to the dynamics of surrounding and internal water molecules. ChemPhysChem 8, 23–33. [DOI] [PubMed] [Google Scholar]
- 22. Loftfield RB, Eigner EA, Pastuszyn A, Lovgren TNE & Jakubowski H (1980) Conformational‐changes during enzyme catalysis ‐ role of water in the transition‐state. Proc Natl Acad Sci USA 77, 3374–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pavlic MR (1987) The role of hydration in an enzyme reaction. Arch Biochem Biophys 253, 446–452. [DOI] [PubMed] [Google Scholar]
- 24. Rupley JA & Careri G (1991) Protein hydration and function. Adv Protein Chem 41, 37–172. [DOI] [PubMed] [Google Scholar]
- 25. Pocker Y (2000) Water in enzyme reactions: biophysical aspects of hydration‐dehydration processes. Cell Mol Life Sci 57, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kornblatt JA & Kornblatt MJ (2002) Water as it applies to the function of enzymes. Int Rev Cytol 215, 49–73. [DOI] [PubMed] [Google Scholar]
- 27. Rand RP (2004) Probing the role of water in protein conformation and function. Philos Trans R Soc Lond B Biol Sci 359, 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imai T (2009) Roles of water in protein structure and function studied by molecular liquid theory. Front Biosci 14, 1387–1402. [DOI] [PubMed] [Google Scholar]
- 29. Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf‐Watz M, Bosco DA, Skalicky JJ, Kay LE & Kern D (2005) Intrinsic dynamics of an enzyme underlies catalysis. Nature 438, 117–121. [DOI] [PubMed] [Google Scholar]
- 30. Henzler‐Wildman K & Kern D (2007) Dynamic personalities of proteins. Nature 450, 964–972. [DOI] [PubMed] [Google Scholar]
- 31. Weikl TR & Paul F (2014) Conformational selection in protein binding and function. Prot Sci 23, 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammes GG, Chang YC & Oas TG (2009) Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci USA 106, 13737–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davydov DR (2012) Merging thermodynamics and evolution: how the studies of high‐pressure adaptation may help to understand enzymatic mechanisms. J Thermodynam Cat 3, 1000e110. [Google Scholar]
- 34. Ohmae E, Gekko K & Kato C (2015) Environmental adaptation of dihydrofolate reductase from deep‐sea bacteria In High Pressure Bioscience: Basic Concepts, Applications and Frontiers (Akasaka K. & Matsuki H, eds), pp. 423–442. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- 35. Eisenmenger MJ & Reyes‐de‐Corcuera JI (2009) High pressure enhancement of enzymes: a review. Enz Microb Technol 45, 331–347. [Google Scholar]
- 36. Masson P & Balny C (2005) Linear and non‐linear pressure dependence of enzyme catalytic parameters. Biochim Biophys Acta 1724, 440–450. [DOI] [PubMed] [Google Scholar]
- 37. Dallet S & Legoy MD (1996) Hydrostatic pressure induces conformational and catalytic changes on two alcohol dehydrogenases but no oligomeric dissociation. Biochim Biophys Acta 1294, 15–24. [DOI] [PubMed] [Google Scholar]
- 38. Gross M, Auerbach G & Jaenicke R (1993) The catalytic activities of monomeric enzymes show complex pressure‐dependence. FEBS Lett 321, 256–260. [DOI] [PubMed] [Google Scholar]
- 39. Schmid G, Luedemann HD & Jaenicke R (1975) High pressure effects on the activity of glycolytic enzymes. Biophys Chem 3, 90–98. [DOI] [PubMed] [Google Scholar]
- 40. Weingand‐Ziade A, Ribes F, Renault F & Masson P (2001) Pressure‐ and heat‐induced inactivation of butyrylcholinesterase: evidence for multiple intermediates and the remnant inactivation process. Biochem J 356, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohmae E, Tatsuta M, Abe F, Kato C, Tanaka N, Kunugi S & Gekko K (2008) Effects of pressure on enzyme function of Escherichia coli dihydrofolate reductase. Biochim Biophys Acta 1784, 1115–1121. [DOI] [PubMed] [Google Scholar]
- 42. Hamajima Y, Nagae T, Watanabe N, Ohmae E, Kato‐Yamada Y & Kato C (2016) Pressure adaptation of 3‐isopropylmalate dehydrogenase from an extremely piezophilic bacterium is attributed to a single amino acid substitution. Extremophiles 20, 177–186. [DOI] [PubMed] [Google Scholar]
- 43. Ingr M, Kutalkova E, Hrncirik J & Lange R (2016) Equilibria of oligomeric proteins under high pressure ‐ A theoretical description. J Theor Biol 411, 16–26. [DOI] [PubMed] [Google Scholar]
- 44. Gross M & Jaenicke R (1994) Proteins under pressure. The influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur J Biochem 221, 617–630. [DOI] [PubMed] [Google Scholar]
- 45. Muller K, Ludemann HD & Jaenicke R (1981) Reconstitution of lactic‐dehydrogenase from pig‐heart after reversible high‐pressure dissociation. Biochemistry 20, 5411–5416. [DOI] [PubMed] [Google Scholar]
- 46. Muller K, Ludemann HD & Jaenicke R (1982) Thermodynamics and mechanism of high‐pressure deactivation and dissociation of porcine lactic‐dehydrogenase. Biophys Chem 16, 1–7. [DOI] [PubMed] [Google Scholar]
- 47. Schade BC, Ludemann HD, Rudolph R & Jaenicke R (1980) Kinetics of reconstitution of porcine muscle lactic‐dehydrogenase after reversible high‐pressure dissociation. Biophys Chem 11, 257–263. [DOI] [PubMed] [Google Scholar]
- 48. Schade BC, Rudolph R, Ludemann HD & Jaenicke R (1980) Reversible high‐pressure dissociation of lactic‐dehydrogenase from pig muscle. Biochemistry 19, 1121–1126. [DOI] [PubMed] [Google Scholar]
- 49. Hennessey JPJ & Siebenaller JF (1985) Pressure inactivation of tetrameric lactate dehydrogenase homologues of confamilial deep‐living fishes. J Comp Physiol B 155, 647–652. [DOI] [PubMed] [Google Scholar]
- 50. Pineda J, Callender R & Schwartz SD (2007) Ligand binding and protein dynamics in lactate dehydrogenase. Biophys J 93, 1474–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu LL, Gulotta M & Callender R (2007) Lactate dehydrogenase undergoes a substantial structural change to bind its substrate. Biophys J 93, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhadin N & Callender R (2011) Effect of osmolytes on protein dynamics in the lactate dehydrogenase‐catalyzed reaction. Biochemistry 50, 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brindley AA, Pickersgill RW, Partridge JC, Dunstan DJ, Hunt DM & Warren MJ (2008) Enzyme sequence and its relationship to hyperbaric stability of artificial and natural fish lactate dehydrogenases. PLoS ONE, 3, e2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishiguchi Y, Miwa T & Abe F (2008) Pressure‐adaptive differences in lactate dehydrogenases of three hagfishes: Eptatretus burgeri, Paramyxine atami and Eptatretus okinoseanus . Extremophiles 12, 477–480. [DOI] [PubMed] [Google Scholar]
- 55. Somero GN & Siebenaller JF (1979) Inefficient lactate dehydrogenases EC‐1.1.1.27 of deep sea fishes. Nature 282, 100–102. [DOI] [PubMed] [Google Scholar]
- 56. Gerringer ME, Drazen JC & Yancey PH (2017) Metabolic enzyme activities of abyssal and hadal fishes: pressure effects and a re‐evaluation of depth‐related changes. Deep Sea Res Part I Oceanogr Res Pap 125, 135–146. [Google Scholar]
- 57. Linley TD, Gerringer ME, Yancey PH, Drazen JC, Weinstock CL & Jamieson AJ (2016) Fishes of the hadal zone including new species, in situ observations and depth records of Liparidae. Deep Sea Res Part I Oceanogr Res Pap 114, 99–110. [Google Scholar]
- 58. Burke V (1930) Revision of fishes of the family Liparidae. Bullet US Natl Museum 150, 1–204. [Google Scholar]
- 59. Gerringer M, Linley TD, Jameson AJ, Goetze E & Drazen JC (2017) Pseudoliparis swirei sp. nov.: a newly‐discovered hadal snailfish (Scorpaeniformes: Liparidae) from the Mariana Trench. Zootaxa 4358, 161–177. [DOI] [PubMed] [Google Scholar]
- 60. Siebenaller JF, Somero GN & Haedrich RL (1982) Biochemical characteristics of macrourid fishes differing in their depths of distributions. Biol Bullet. 163, 240–249. [Google Scholar]
- 61. Wilson RRJ, Siebenaller JF & Davis BJ (1991) Phylogenetic analysis of species of three subgenera of coryphaenoides teleostei macrouridae by peptide mapping of homologs of LDH‐a‐4. Biochem Syst Ecol 19, 277–288. [Google Scholar]
- 62. Siebenaller JF (1984) Pressure‐adaptive differences in nad‐dependent dehydrogenases of congeneric marine fishes living at different depths. J Comp Physiol 154, 443–448. [Google Scholar]
- 63. Abe F & Horikoshi K (2001) The biotechnological potential of piezophiles. Trends Biotechnol 19, 102–108. [DOI] [PubMed] [Google Scholar]
- 64. Aertsen A, Meersman F, Hendrickx MEG, Vogel RF & Michiels CW (2009) Biotechnology under high pressure: applications and implications. Trends Biotechnol 27, 434–441. [DOI] [PubMed] [Google Scholar]
- 65. Mozhaev VV, Heremans K, Frank J, Masson P & Balny C (1994) Exploiting the effects of high hydrostatic‐pressure in biotechnological applications. Trends Biotechnol 12, 493–501. [Google Scholar]
- 66. Wang K, Shen Y, Yang Y, Gan X, Liu G, Hu K, Li Y, Gao Z, Zhu L, Yan G et al (2019) Morphology and genome of a snailfish from the Mariana Trench provide insights into deep‐sea adaptation. Nat Ecol Evol 3, 823–833. [DOI] [PubMed] [Google Scholar]
- 67. Bartholmes P, Durchschlag H & Jaenicke R (1973) Molecular properties of lactic dehydrogenase ec‐1.1.1.27 under the conditions of the enzymatic test sedimentation analysis and gel filtration in the microgram and nanogram range. Eur J Biochem 39, 101–108. [DOI] [PubMed] [Google Scholar]
- 68. Somero GN (1990) Life at low‐volume change ‐ hydrostatic‐pressure as a selective factor in the aquatic environment. Am Zool 30, 123–135. [Google Scholar]
- 69. Akasaka K (2006) Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev 106, 1814–1835. [DOI] [PubMed] [Google Scholar]
- 70. Agarwal PK, Doucet N, Chennubhotla C, Ramanathan A & Narayanan C (2016) Conformational sub‐states and populations in enzyme catalysis In Computational Approaches for Studying Enzyme Mechanism, Pt B (Voth GA, ed), pp. 273–297. Elsevier, Amsterdam. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Petrovic D, Risso VA, Kamerlin SCL & Sanchez‐Ruiz JM (2018) Conformational dynamics and enzyme evolution. J R Soc Interface 15(144): 20180330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramanathan A, Savol A, Burger V, Chennubhotla CS & Agarwal PK (2014) Protein conformational populations and functionally relevant substates. Acc Chem Res 47, 149–156. [DOI] [PubMed] [Google Scholar]
- 73. Świderek K, Tuñón I, Martí S & Moliner V (2015) Protein conformational landscapes and catalysis. Influence of active site conformations in the reaction catalyzed by L‐Lactate dehydrogenase. ACS Catalusis 5, 1172–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yin H, Li H, Grofe A & Gao J (2019) Active‐site heterogeneity of lactate dehydrogenase. ACS Catalusis 9, 4236–4246. [Google Scholar]
- 75. Khrapunov S, Chang E & Callender RH (2017) Thermodynamic and structural adaptation differences between the mesophilic and psychrophilic lactate dehydrogenases. Biochemistry 56, 3587–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reddish MJ, Callender R & Dyer RB (2017) Resolution of submillisecond kinetics of multiple reaction pathways for lactate dehydrogenase. Biophys J 112, 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Al‐Ayoubi SR, Schummel PH, Cisse A, Seydel T, Peters J & Winter R (2019) Osmolytes modify protein dynamics and function of tetrameric lactate dehydrogenase upon pressurization. Phys Chem Chem Phys 21, 12806–12817. [DOI] [PubMed] [Google Scholar]
- 78. Huang Q, Tran KN, Rodgers JM, Bartlett DH, Hemley RJ & Ichiye T. (2016) A molecular perspective on the limits of life: enzymes under pressure. Condensed Matter Phys 19(2): 22801. [Google Scholar]
- 79. Muller K, Seifert T & Jaenicke R (1984) High‐pressure dissociation of lactate‐dehydrogenase from bacillus‐stearothermophilus and reconstitution of the enzyme after denaturation in 6m guanidine‐hydrochloride. Eur Biophys J Biophy 11, 87–94. [DOI] [PubMed] [Google Scholar]
- 80. Muller K, Ludemann HD & Jaenicke R (1981) Pressure‐induced structural‐changes of pig‐heart lactic‐dehydrogenase. Biophys Chem 14, 101–110. [DOI] [PubMed] [Google Scholar]
- 81. Pan XL & Schwartz SD (2016) Conformational heterogeneity in the michaelis complex of lactate dehydrogenase: an analysis of vibrational spectroscopy using Markov and hidden Markov models. J Phys Chem B 120, 6612–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peng HL, Deng H, Dyer RB & Callender R (2014) Energy landscape of the Michaelis complex of lactate dehydrogenase: relationship to catalytic mechanism. Biochemistry 53, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Peng HL, Egawa T, Chang E, Deng H & Callender R (2015) Mechanism of thermal adaptation in the lactate dehydrogenases. J Phys Chem B 119, 15256–15262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Drazen JC, Friedman JR, Condon NE, Aus EJ, Gerringer ME, Keller AA & Elizabeth Clarke M (2015) Enzyme activities of demersal fishes from the shelf to the abyssal plain. Deep Sea Res Part I Oceanogr Res Pap 100, 117–126. [Google Scholar]
- 85. Drazen JC & Seibel BA (2007) Depth‐related trends in metabolism of benthic and benthopelagic deep‐sea fishes. Limnol Oceanogr 52, 2306–2316. [Google Scholar]
- 86. Childress JJ, Cowles DL, Favuzzi JA & Mickel TJ (1990) Metabolic rates of benthic deep‐sea decapod crustaceans decline with increasing depth primarily due to the decline in temperature. Deep Sea Res Part A Oceanogr Res Pap 37, 929–949. [Google Scholar]
- 87. Gerringer ME, Drazen JC, Linley TD, Summers AP, Jamieson AJ & Yancey PH (2017) Distribution, composition, and functions of gelatinous tissues. Roy Soc Open Sci 4, 171063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Itzhaki R & Gill D (1964) A micro‐biuret method for estimating proteins. Anal Biochem 9, 401–410. [DOI] [PubMed] [Google Scholar]
- 89. Hood CA, Fuentes G, Patel H, Page K, Menakuru M & Park JH (2008) Fast conventional Fmoc solid‐phase peptide synthesis with HCTU. J Pept Sci 14, 97–101. [DOI] [PubMed] [Google Scholar]
- 90. Fekkes D (1996) State‐of‐the‐art of high‐performance liquid chromatographic analysis of amino acids in physiological samples. J Chromatogr B‐Biomed Appl 682, 3–22. [DOI] [PubMed] [Google Scholar]
- 91. Hoa Hui Bon G, Douzou P, Dahan N & Balny C (1982) High pressure spectrometry at subzero temperatures. Anal Biochem 120, 125–135. [DOI] [PubMed] [Google Scholar]
- 92. Davydov DR, Deprez E, Hoa Hui Bon G, Knyushko TV, Kuznetsova GP, Koen YM & Archakov AI (1995) High‐pressure‐induced transitions in microsomal cytochrome P450 2B4 in solution: evidence for conformational inhomogeneity in the oligomers. Arch Biochem Biophys 320, 330–344. [DOI] [PubMed] [Google Scholar]
- 93. Hoa Hui Bon G, McLean MA & Sligar SG (2002) High pressure, a tool for exploring heme protein active sites. Biochim Biophys Acta 1595, 297–308. [DOI] [PubMed] [Google Scholar]
- 94. Weber G (1991) Protein Interactions. Chapman Hall; , New York. [Google Scholar]
- 95. Davydov DR, Yang ZY, Davydova N, Halpert JR & Hubbell WL (2016) Conformational mobility in cytochrome P450 3A4 explored by pressure‐perturbation EPR spectroscopy. Biophys J 110, 1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]


