Abstract
The Rab family of small GTPases regulates intracellular membrane trafficking by orchestrating the biogenesis, transport, tethering, and fusion of membrane‐bound organelles and vesicles. Like other small GTPases, Rabs cycle between two states, an active (GTP‐loaded) state and an inactive (GDP‐loaded) state, and their cycling is catalyzed by guanine nucleotide exchange factors (GEFs) and GTPase‐activating proteins (GAPs). Because an active form of each Rab localizes on a specific organelle (or vesicle) and recruits various effector proteins to facilitate each step of membrane trafficking, knowing when and where Rabs are activated and what effectors Rabs recruit is crucial to understand their functions. Since the discovery of Rabs, they have been regarded as one of the central hubs for membrane trafficking, and numerous biochemical and genetic studies have revealed the mechanisms of Rab functions in recent years. The results of these studies have included the identification and characterization of novel GEFs, GAPs, and effectors, as well as post‐translational modifications, for example, phosphorylation, of Rabs. Rab functions beyond the simple effector‐recruiting model are also emerging. Furthermore, the recently developed CRISPR/Cas technology has enabled acceleration of knockout analyses in both animals and cultured cells and revealed previously unknown physiological roles of many Rabs. In this review article, we provide the most up‐to‐date and comprehensive lists of GEFs, GAPs, effectors, and knockout phenotypes of mammalian Rabs and discuss recent findings in regard to their regulation and functions.
Keywords: effector, GAP, GEF, knockout, membrane traffic, organelle, post‐translational modification, Rab small GTPases
The Rab family of small GTPases regulates intracellular membrane trafficking by localizing on specific organelles (or vesicles) and recruiting various effector proteins to facilitate each step of membrane trafficking. In this review article, we provide the most up‐to‐date and comprehensive lists of guanine nucleotide exchange factors, GTPase‐activating proteins, effectors, and knockout phenotypes of mammalian Rabs and discuss recent findings in regard to their regulation and functions.
Abbreviations
- DENN
differentially expressed in normal and neoplastic cells
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ER
endoplasmic reticulum
- EV
extracellular vesicle
- GAP
GTPase‐activating protein
- GDF
GDI displacement factor
- GDI
GDP dissociation inhibitor
- GEF
guanine nucleotide exchange factor
- Hh
Hedgehog
- HPS
Hermansky–Pudlak syndrome
- IMPC
International Mouse Phenotyping Consortium
- KO
knockout
- LCV
Legionella‐containing vacuole
- LRO
lysosome‐related organelle
- LRRK1/2
leucine‐rich repeat kinase 1/2
- MDCK
Madin–Darby canine kidney
- NCBI
National Center for Biotechnology Information
- PD
Parkinson's disease
- PTM
post‐translational modification
- REP
Rab escort protein
- RILPL1/2
Rab interacting lysosomal protein‐like 1/2
- sgRNA
single‐guide RNA
- TBC
Tre‐2/Bub2/Cdc16
- TBK1
TANK‐binding kinase 1
- TGN
trans‐Golgi network
- Vps
vacuolar protein sorting
Introduction
Eukaryotic cells possess a highly organized endomembrane system that compartmentalizes various biochemical reactions and enable the exchange of numerous molecules with the extracellular milieu by means of exocytosis and endocytosis. Many steps of membrane trafficking, including the biogenesis, transport, tethering, and fusion of membrane‐bound organelles and vesicles, are thought to be regulated by the Rab family of small GTPases [1, 2, 3, 4]. Like other small GTPases, Rabs cycle between two states, an active (GTP‐loaded) state and an inactive (GDP‐loaded) state, and the cycling is catalyzed by guanine nucleotide exchange factors (GEFs) and GTPase‐activating proteins (GAPs) (Fig. 1). Switch regions (switch I and II) of Rabs are known to undergo large conformational changes upon binding of GDP or GTP. When activated by a GEF, Rabs localize via their prenylated (or doubly prenylated) C termini to specific membranes of several compartments, such as the endoplasmic reticulum (ER), Golgi apparatus, secretory vesicles, endosomes, or lysosomes, where they recruit effector proteins (i.e., active Rab‐binding partners) that regulate different steps of membrane trafficking. Consequently, knowing when and where Rabs are activated and what kinds of effectors they recruit are crucial to achieving a complete understanding of Rab functions during membrane trafficking. In this review article, we describe recent findings in regard to the regulation and functions of Rabs, and we update the lists of GEFs, GAPs, effectors, and knockout (KO) phenotypes of mammalian Rabs.
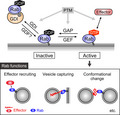
Fig. 1.
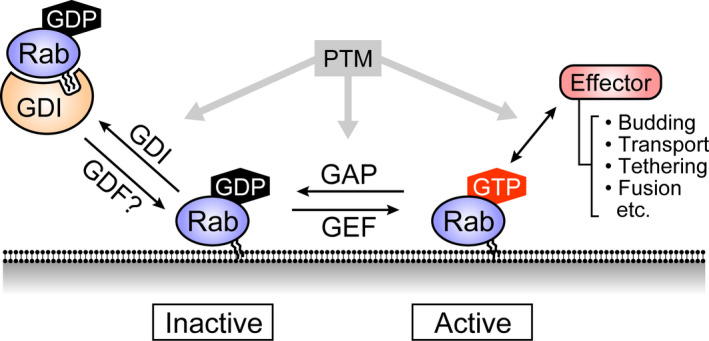
The Rab GTPase cycle. Rab GTPases are activated (GTP‐loaded) by guanine nucleotide exchange factors (GEFs) and inactivated (GDP‐loaded) by GTPase‐activating proteins (GAPs). Inactive Rabs bind to GDP dissociation inhibitor (GDI) and are retained in the cytosol [136]. GDI is thought to be dissociated by GDI displacement factor (GDF) [137], but whether this mechanism is applicable to all Rabs remains unclear. Active Rabs are associated with intracellular membranes and recruit specific effector proteins that regulate various steps of membrane trafficking, including budding, transport, tethering, and fusion of vesicles and organelles. Post‐translational modifications (PTMs), such as phosphorylation, of Rabs are thought to regulate their interaction with GDI, GEFs/GAPs, and effectors [138].
Nomenclature of mammalian Rab genes
Typical Rab genes encode a small GTPase consisting of around 200–250 amino acids, and approximately 60 Rab genes have been identified in mammals to date (Table 1) [5, 6, 7, 8]. The Rab names used in this review are according to the National Center for Biotechnology Information (NCBI) gene symbols; however, the numbering of some Rabs differs among studies (indicated in parentheses in Table 1), and the current Rab names do not always reflect their sequence similarities. A phylogenetic analysis actually suggested that the following Rabs are classified into the same subfamilies (i.e., paralogs) [7, 8]: Rab11 and Rab25; Rab19 and Rab43; Rab22A and Rab31/22B; Rab26 and Rab37; Rab32 and Rab38; Rab34 and Rab36; and Rab39 and Rab42. This classification is well consistent with a classification based on alignments of the switch II region sequences alone (Fig. 2), indicating that the switch II region is the main determinant of not only Rab effector binding specificity [9] but Rab identity as well. The functional relationships between these paralogous Rabs remain unclear and would be worth investigating in the future.
Table 1.
Mouse and human Rab genes. The nomenclature of Rabs in this review is according to the NCBI database. The names of several Rabs in the report by Itoh et al. [6] are different (indicated in parentheses). Representative phenotypes of mutant animals are shown in this table, and all other phenotypes together with their respective references (PubMed ID) are listed in Table S2. The phenotypes of several knockout mice (indicated by asterisks) are available from at the following URL: https://www.mousephenotype.org/. Rab33A (Cat#: RBRC05799) and Rab33B KO mice (Cat#: RBRC05800) (indicated by double asterisks) are also available from the RIKEN BioResource Research Center (https://mus.brc.riken.jp/en/). AJ, adherens junction; AP, autophagosome; C, cilium; E, endosome; EE, early endosome; ERC, endocytic recycling compartment; G, Golgi; KO, knockout; LE, late endosome; LRO, lysosome‐related organelle; LD, lipid droplet; LY, lysosome; MS, melanosome; MT, mitochondrion; N, nucleus; PM, plasma membrane; P, peroxisome; PS, phagosome; RE, recycling endosome; SG, secretory granule; SV, synaptic vesicle; TE, tubular endosome; TGN, trans‐Golgi network; TJ, tight junction.
| Name (NCBI) | Gene ID | Disease and Mutant animal | Knockout phenotype | Subcellular localization | |
|---|---|---|---|---|---|
| Human | Mouse | ||||
| Rab1A | 5861 | 19324 | EE/ER/G/MS | ||
| Rab1B | 81876 | 76308 | ER/G | ||
| Rab2A | 5862 | 59021 | Preweaning lethality* | G/AP | |
| Rab2B | 84932 | 76338 | G/AP | ||
| Rab3A | 5864 | 19339 | Earlybird [139] | Viable and fertile; perinatal lethality of Rab3A/B/C/D quadruple KO mice; 30% reduction of Ca2+‐triggered synaptic release [140, 141, 142] | Acrosome/MS/SG/SV |
| Rab3B | 5865 | 69908 | Viable and fertile [140] | G/RE/SG/SV/TJ | |
| Rab3C | 115827 | 67295 | Viable and fertile [140] | RE/SG/SV | |
| Rab3D | 9545 | 19340 | Viable and fertile [140] | G/LRO/SG/SV/TGN | |
| Rab4A | 5867 | 19341 | EE/RE | ||
| Rab4B | 53916 | 19342 | Adipocyte hypertrophy and insulin resistance in T cell‐specific KO mice [143] | EE/RE | |
| Rab5A | 5868 | 271457 | EE/early PS | ||
| Rab5B | 5869 | 19344 | EE/PM | ||
| Rab5C | 5878 | 19345 | Preweaning lethality* | EE/early PS | |
| Rab6A | 5870 | 19346 | Embryonic lethal; defects in basement membrane formation in KO embryos [144, 145] | G/P/TGN‐derived vesicle | |
| Rab6B | 51560 | 270192 | G | ||
| Rab6C | 84084 | – | |||
| Rab41 (6D) | 347517 | – | ER/G | ||
| Rab7A (7) | 7879 | 19349 | Charcot–Marie–Tooth type 2B [146] | Embryonic lethal; defects in microautophagy in the visceral endoderm of KO embryos [147] | AP/LE/LY/PS |
| Rab7B (42) | 338382 | 226421 | LE | ||
| Rab8A | 4218 | 17274 | Related to microvillus inclusion disease [148] | Die at postnatal week 4, defects in apical protein localization, and microvillus inclusion bodies [130, 148, 149] | C/RE/SG/TE/TGN |
| Rab8B | 51762 | 235442 | No obvious phenotype; Rab8A/B double KO mice die at postnatal week 3 [130] | AJ/TGN | |
| Rab9A | 9367 | 56382 | LE/TGN | ||
| Rab9B | 51209 | 319642 | LE | ||
| Rab10 | 10890 | 19325 | Embryonic lethal [120] | E/ER/P/RE/TE/TGN | |
| Rab11A | 8766 | 53869 | Embryonic lethal [150, 151, 152] | RE/TGN/TE/TGN‐derived vesicle | |
| Rab11B | 9230 | 19326 | Intellectual disability [153] | RE | |
| Rab12 | 201475 | 19328 | G/SG/RE/LE/LY | ||
| Rab13 | 5872 | 68328 | Viable; reduced lymphocyte numbers and reduced lymophocyte trafficking [121] | RE/TE/TGN/TJ | |
| Rab14 | 51552 | 68365 | EE/G/P/PS/TGN | ||
| Rab15 | 376267 | 104886 | EE/RE | ||
| Rab17 | 64284 | 19329 | RE/MS | ||
| Rab18 | 22931 | 19330 | Warburg Micro syndrome [154] | Viable and fertile; ocular and neurological abnormalities [155] | ER/G/LD/P/SG |
| Rab19 | 401409 | 19331 | G | ||
| Rab20 | 55647 | 19332 | Decreased formation of Mycobacterium tuberculosis‐containing proteolytic phagosomes [119] | ER/G/LE/PS | |
| Rab21 | 23011 | 216344 | Preweaning lethality* | EE | |
| Rab22A | 57403 | 19334 | EE/RE/PS/TE | ||
| Rab22B (31) | 11031 | 106572 | G/TGN/PS | ||
| Rab23 | 51715 | 19335 | Carpenter syndrome [156], open brain [157] | Embryonic lethality; neural‐tube defects [157] | GAS‐containing AP/E/PM/C |
| Rab24 | 53917 | 19336 | Canine hereditary ataxia [158] | AP/ER/G/LE/N | |
| Rab25 | 57111 | 53868 | Increased tumor formation [159, 160] | RE | |
| Rab26 | 25837 | 328778 | Decreased microvascular barrier function [161] | AP/LY/SG | |
| Rab27A | 5873 | 11891 | Griscelli syndrome type 2 [162], ashen [163] | Hypopigmentation; immunodeficincy [164, 165, 166] | MS/LRO/SG |
| Rab27B | 5874 | 80718 | Secretory defects in various endocrine, exocrine, and immune cells [167, 168, 169] | MS/LRO/SG/SV | |
| Rab28 | 9364 | 100972 | Cone‐rod dystrophy [170] | Retina degeneration [171] | |
| Rab29 (7L1) | 8934 | 226422 | Association with Parkinson's disease [172] | Renal enlargement and discoloration [173] | G/RE/TGN |
| Rab30 | 27314 | 75985 | G | ||
| Rab32 | 10981 | 67844 | Increased susceptibility to Salmonella Typhi [117]; coat and eye pigment dilution, some enlarged lung multilamellar bodies, and prolonged bleeding in Rab32/38 DKO mice [114] | ER/G/LRO/MS/MT | |
| Rab33A | 9363 | 19337 | Viable and fertile** | G/SG/SV precursor | |
| Rab33B | 83452 | 19338 | Dyggve–Melchior–Clausen syndrome [174, 175] | Viable and fertile** | AP/G |
| Rab34 | 83871 | 19376 | Preweaning lethality; polydactyly; cleft‐lip/palate; ciliogenesis defect [124, 125] | G/PS | |
| Rab35 | 11021 | 77407 | Somatic mutations in human tumors [176] | Preweaning lethality* | EE/PM/RE |
| Rab36 | 9609 | 76877 | G/RE | ||
| Rab37 | 326624 | 58222 | SG | ||
| Rab38 | 23682 | 72433 | chocolate [177], Ruby [178] | Hypopigmentation; prolonged bleeding; pulmonary fibrosis [179]; coat and eye pigment dilution, some enlarged lung multilamellar bodies, and prolonged bleeding in Rab32/38 DKO mice [114] | G/LRO/MS |
| Rab39A | 54734 | 270160 | Viable; reduced cross‐presentation by dendritic cells [122] | G/LE/PS | |
| Rab39B | 116442 | 67790 | X‐linked mental retardation/Parkinson's disease [180, 181] | ER/G | |
| Rab40A | 142684 | ‐ | |||
| Rab40B | 10966 | 217371 | TGN‐derived vesicle | ||
| Rab40C | 57799 | 224624 | Subviable* | ERC/LD | |
| Rab40AL | 282808 | – | G | ||
| Rab42 (43) | 115273 | 242681 | G | ||
| Rab43 (41) | 339122 | 69834 | Association with a hereditary liver–colon cancer syndrome [182] | Viable and fertile; reduced cross‐presentation by dendritic cells [183] | G |
Fig. 2.
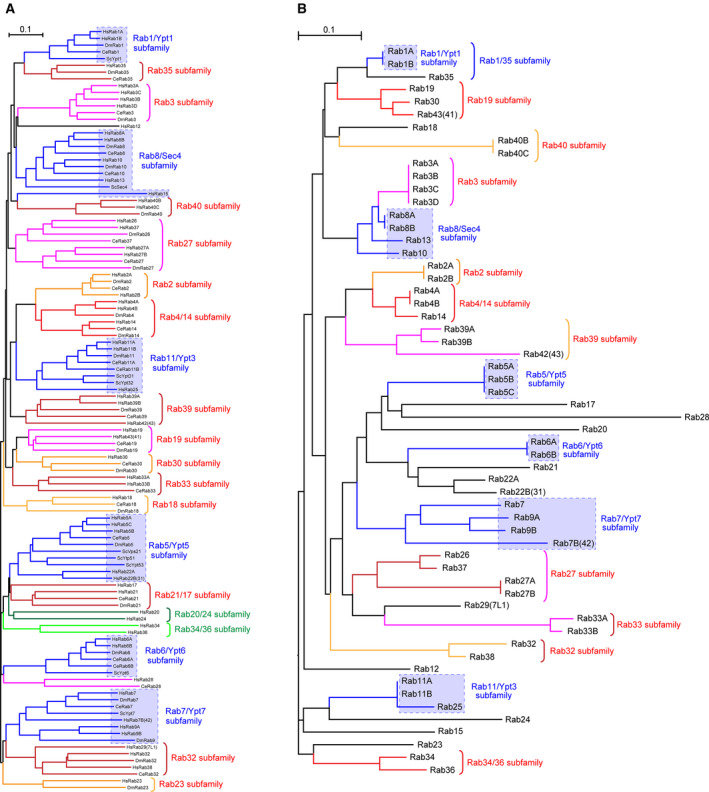
Phylogenetic analysis of Rabs. Amino acid sequences of the full length (A) or the switch II region (B) of human Rabs have been aligned using the clustalw software program (version 2.1; available at http://clustalw.ddbj.nig.ac.jp/top‐e.html) set at the default parameters and their phylogenetic tree was drawn by the neighbor‐joining method. All Rab sequences used for the phylogenetic analysis were obtained from the NCBI database (see also Figs S1 and S2). Note that the classifications of subfamilies are similar to each other and to the classification based on the full‐length sequences of more than 7600 Rabs from various species [8]. The Rab subfamily members that have been conserved from budding yeasts to humans are enclosed in boxes composed of dashed blue lines. (A) includes budding yeast (Sc) Ypts, Caenorhabditis elegans (Ce) Rabs, and Drosophila melanogaster (Dm) Rabs that are also conserved in humans. Human‐ or primate‐specific Rabs, that is, Rab6C, Rab41/6D, Rab40A, and Rab40AL, have been excluded from this figure.
In addition to the above typical Rabs, there are several Rab‐related proteins (nevertheless referred to as Rabs by the NCBI) that fall outside the classical Rab category. Rab44, Rab45, and CRACR2A/Rab46 have long N‐terminal regions containing EF‐hand and coiled‐coil domains in addition to the C‐terminal Rab‐like GTPase domains (Fig. 3). The functions of these ‘large Rab GTPases’ have only recently begun to be investigated [10, 11, 12], and they are not discussed in this review. There are also six ‘Rab‐like’ proteins (Rabl2A, Rabl2B, Rabl3, Rabl4/Ift27, Rabl5/Ift22, and Rabl6), which contain only a Rab‐like GTPase domain, but they are not expected to regulate membrane trafficking, because they lack C‐terminal prenylation, which is required for membrane insertion. Actually, several Rab‐like proteins, such as Rabl4 and Rabl5, have been shown to regulate intraflagellar transport in cilia (i.e., the bidirectional transport of protein complexes required for cilium formation along axonemal microtubules) [13].
Fig. 3.
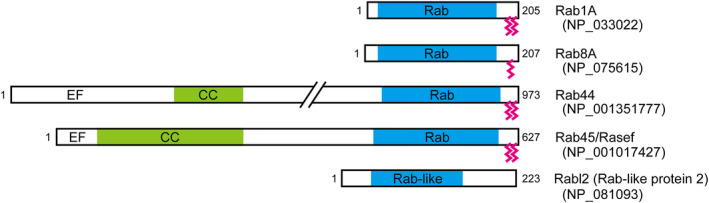
Typical Rabs and Rab‐related proteins. A comparison between typical Rabs (mouse Rab1A and Rab8A) and Rab‐related proteins (mouse Rab44, Rab45, and Rabl2) and their domain architecture are shown. The numbers in parentheses are the protein ID numbers of the respective proteins in the NCBI. Rab44 and Rab45 have long N‐terminal regions containing EF‐hand and coiled‐coil (CC) domains, in addition to their C‐terminal Rab‐like GTPase domains. Rabl2, an example of ‘Rab‐like’ proteins, has a Rab‐like GTPase domain but lacks a C‐terminal prenylation site (indicated by the pink lines), which is required for membrane insertion.
Rab regulation
This section summarizes recent studies on newly discovered Rab regulation mechanisms and the life cycle of Rab proteins.
mRNA compartmentalization
One member of the Rab family, Rab13, undergoes unique mRNA regulation. A genome‐wide microarray analysis revealed that Rab13 mRNA is enriched in cell protrusions induced by migratory stimuli [14]. The 3′‐untranslated region of Rab13 mRNA has been shown to be necessary and sufficient for transport to and accumulation at the tip of the protrusions in a microtubule‐dependent manner, which presumably facilitates local translation of Rab13. Moreover, Rab13 mRNA has been found to be highly enriched (> 200‐fold) in extracellular vesicles (EVs) in comparison with the cell bodies of KRAS‐mutated colorectal cancer cells [15]. These EVs can be transferred to recipient cells and enable Rab13 translation within them. Because Rab13 has been implicated in cancer progression (reviewed in Ref. [16]), the relation between such mRNA regulation and cancer biology would be worth investigating in the future.
Prenylation and activation
After Rab proteins are translated, they first bind to Rab escort protein (REP) and undergo geranylgeranyl transferase‐mediated prenylation at their C terminus, which enables them to be inserted into hydrophobic lipid bilayers (reviewed in Ref. [17]). Because inactive (GDP‐loaded) Rabs are extracted from membranes by GDP dissociation inhibitor (GDI), which masks their prenylated C terminus, Rabs need to be activated (GTP‐loaded) by GEFs to associate with specific membranes after release from GDI (Fig. 1). Consequently, Rab‐GEFs are thought to be major determinants of the specific Rab localizations to various organelles. For this reason, it is necessary to identify Rab‐GEFs in order to understand the spatiotemporal regulation of Rabs. To date, at least Drr1 (Rab1‐GEF), Rabex‐5 (Rab5‐GEF), Rabin8 (Rab8‐GEF), Rab3GAP1/2 (Rab18‐GEF), DENN/Rab3GEP (Rab27‐GEF), and HPS1/4 (Rab32/38‐GEF) have been shown to be necessary and/or sufficient for the proper localization of the corresponding Rabs [18, 19, 20, 21]. In addition, some Rab‐GEFs have been shown to also act as effectors of other Rabs (e.g., Rab8‐GEFs Rabin8 and GRAB act as Rab11 effectors). Such Rab‐GEFs mediate the transition of membrane (or organelle) identity from a ‘Rab A’‐positive membrane compartment to a ‘Rab B’‐positive membrane compartment by activating Rab B via a Rab A effector, which also functions as a Rab B‐GEF. These sequential activation mechanisms are called ‘Rab cascades’ (see reviews [22, 23] for details). The C‐terminal tail of yeast Ypt1 (mammalian Rab1 homolog) and of yeast Ypt31/32 (mammalian Rab11 homolog) has recently been shown to be critical for differential activation by the TRAPPIII complex and TRAPPII complex, respectively [24]. Remarkably, although these multisubunit complexes share the same catalytic subunits, the TRAPPII‐specific bulky subunits seem to keep the catalytic site of TRAPPII farther from the membrane surface than those of TRAPPIII. Thus, only Ypt31/32, which has a longer C‐terminal tail than Ypt1 does, is able to reach the catalytic site of TRAPPII, thereby ensuring specific activation of Ypt31/32, but not Ypt1, on the trans‐Golgi network (TGN). Mammalian TRAPPII/III complexes may also function as specific Rab‐GEFs, but their GEF activity and specificity remain to be determined.
The largest group of mammalian Rab‐GEFs is differentially expressed in normal and neoplastic cells (DENN) domain‐containing proteins [25], whose involvement in Rab regulation was first revealed by a genetic study using C. elegans [26]. Other Rab‐GEFs include vacuolar protein sorting 9 (Vps9) domain‐containing proteins, Sec2 domain‐containing proteins, and some heterodimeric or multimeric complexes (see reviews [27, 28] for details). Furthermore, the Fuzzy/Inturned complex and Plekhg5 have recently been added to the Rab‐GEF list as Rab23‐GEF and Rab26‐GEF, respectively [29, 30] (see Table S1).
Phosphorylation
Phosphorylation is one of the most common post‐translational modifications (PTMs) of proteins. Previous studies on Rab phosphorylation are summarized in Table 2. The first report on Rab phosphorylation dates back to 1991, and it states that Rab1 and Rab4 are phosphorylated by a mitotic kinase CDK1, but that Rab2 or Rab6 is not [31]. In particular, almost the entire pool of Rab4 becomes phosphorylated during mitosis, thereby preventing it from associating with endosomal membranes [32]. These findings are consistent with the temporary cessation of membrane trafficking that occurs during mitosis. Rab7A has recently been shown to be phosphorylated under the following cellular conditions. Upon strong epidermal growth factor (EGF) stimulation, the activated epidermal growth factor receptors (EGFRs) are rapidly endocytosed and degraded in lysosomes. Phosphorylation of Rab7A at Y183 is induced during this event and is required for EGFRs to be efficiently delivered to lysosomes, rather than being recycled back to the plasma membrane [33]. Although the kinase responsible has yet to be identified, another report has stated that Src is capable of phosphorylating the Y183 of Rab7A [34]. On the other hand, Rab7A is phosphorylated at S72 by TANK‐binding kinase 1 (TBK1), a critical kinase for PINK1‐PARKIN‐mediated mitophagy [35]. Mitochondrial depolarization induces TBK1‐dependent phosphorylation of Rab7A, which leads to increased binding to its interactor folliculin [36], which is also required for mitophagy. The S72 of Rab7A is also phosphorylated by leucine‐rich repeat kinase 1 (LRRK1) [37], and both phosphorylated residues of Rab7A (S72 and Y183) have been shown to be dephosphorylated by PTEN [38]. Thus, the function of Rab7A is regulated by context‐dependent phosphorylation and dephosphorylation.
Table 2.
Rab phosphorylation. Only studies that clarified the effect of phosphorylation on Rab function are shown. Because of space limitations, only the first two reports of Rab phosphorylation by LRRK2 (see text for details) [39, 40] are listed in this table.
| Kinase | Phosphorylated Rab (residue) |
Effect (−), inhibition; (+), promotion |
Condition | Reference |
|---|---|---|---|---|
| CDK1 | 1A, 4A (S196) | (−) membrane association | During mitosis | [31, 32] |
| LRRK1 | 7A (S72) | (+) effector (RILP) binding | Expressing a hyperactive mutant of LRRK1 (Y944F) | [37] |
| LRRK2 | 3, 8, 10, 12, 29, 35, 43, etc. (T73 of Rab10 and the equivalent S/Ts of other Rabs) |
(−) GDI binding (−) GEF (Rabin8) binding |
Expressing a pathogenic mutant of LRRK2 (G2019S) | [39, 40] |
| PKC (α/β/γ) | 6 | (−) membrane association | Thrombin‐stimulated platelets | [184] |
| PKCα | 37 (T172) | (−) GTP loading | Lung cancer cells | [185] |
| PKCα, PKCβII | 11 (S177) | (−) TfR recycling | PMA treatment | [186] |
| PKCε | 5A (T7) | (+) endosome trafficking for cell migration | LFA‐1/chemokine‐stimulated T cells | [187] |
| TAK1 | 1A (T75) |
(−) GDI binding (+) Golgi localization |
– | [188] |
| TBK1 | 7A (S72) |
(−) GDI binding (+) effector (folliculin) binding |
Upon mitochondrial depolarization | [35] |
| Src | 7A (Y183) | (−) effector (RILP) binding | EGF treatment | [34] |
| Src | 34 (Y247) | (+) β3‐integrin recycling for cell migration | During cell adhesion | [189] |
| – | 7A (Y183) | (+) EGFR degradation | EGF treatment | [33] |
| – | 8 (S111) | (−) GEF (Rabin8) binding | Upon mitochondrial depolarization | [50] |
Rab phosphorylation has recently been receiving increasing attention, because LRRK2, a Parkinson's disease (PD)‐associated kinase, has been found to phosphorylate a subset of Rabs. A phosphoproteomic screen designed to identify physiological LRRK2 substrates identified Rab10 as the most promising candidate [39]. Its phosphorylation site (T73) is located within the conserved switch II region, and the equivalent Ser/Thr residue of other Rabs, including Rab3A/B/C/D, Rab8A/B, Rab12, Rab29, and Rab35, can also be phosphorylated by LRRK2 [40], while the phosphorylation sites of Rab8, Rab10, and Rab35 are dephosphorylated by PPM1H phosphatase [41]. Whereas the phosphorylation attenuates the interactions between these Rabs and GDI and several other binding partners, phosphorylated Rab8 and Rab10 strongly bind to Rab interacting lysosomal protein‐like 1 (RILPL1) and RILPL2. In particular, the phospho‐Rab10–RILPL1 interaction negatively regulates primary ciliogenesis, which may contribute to PD pathologies [42]. Rab phosphorylation by LRRK2 is also involved in lysosomal and centrosomal homeostasis and in Rab35‐mediated α‐synuclein propagation [43, 44, 45]. Another remarkable link between Rab and LRRK2 is that Rab29 (also known as Rab7L1), which is encoded within the PD‐linked locus (PARK16), directly binds to LRRK2 and recruits it to the Golgi apparatus, thereby greatly increasing its kinase activity [46, 47, 48, 49]. Moreover, another PD‐associated kinase PINK1 has been shown to induce Rab8 phosphorylation at S111 upon mitochondrial depolarization, which inhibits the interaction between Rab8 and its GEF, Rabin8 [50]. Taken together, the evidence increasingly indicates that phosphorylation is a crucial modulatory mechanism of Rab function in response to a variety of cellular events and that its dysregulation results in diseases such as PD.
Other PTMs
Other known PTMs of Rab proteins include serotonylation, AMPylation, phosphocholination, palmitoylation, and ubiquitination. Non‐neuronal serotonin has been shown to be covalently bound to Rab4 (in platelets) and Rab3/27 (in β‐cells), which presumably contributes to the secretion of α‐granules and insulin, respectively [51, 52]. During Legionella pneumophila infection, Rab1 is AMPylated and phosphocholinated by bacterial proteins DrrA and AnkX, respectively [53, 54, 55, 56]. These modifications occur at the switch II region and affect the interaction of Rab1 with host proteins such as TBC1D20 (Rab1‐GAP), MICAL‐3 (Rab1 effector), and GDI. Rab35 is also phosphocholinated by AnkX and becomes incapable of being activated by its GEF DENND1A [54]. In addition, the palmitoylation of Rab7A at C83/C84 has been shown to be required for efficient interaction with the retromer complex and modulation of endosome‐to‐TGN trafficking of lysosomal sorting receptors [57].
While ubiquitination of transmembrane protein cargos is known to be the sorting signal for endocytic degradation [58], components of the membrane trafficking machinery themselves, including Rabs, are also regulated by ubiquitination. The first report on Rab ubiquitination stated that an E3 ligase, HACE1, ubiquitinates Rab11A at K145, which promotes activation of Rab11A and recycling of a β2‐adrenergic receptor [59]. Rab5A and Rab7A have also subsequently been shown to be ubiquitinated. Structural and biochemical analyses have revealed that ubiquitination of Rab5A impairs binding to its effector, EEA1, but the corresponding E3 ligase is unknown [60]. The K38 of Rab7A has been shown to be ubiquitinated by PARKIN and deubiquitinated by USP32 [61, 62]. Although the effect of this ubiquitination on effector binding is a matter of controversy, knockdown of either of these enzymes, as well as Rab7A [63, 64, 65], results in enlarged late endosomes (or lysosomes), suggesting that the ubiquitination/deubiquitination cycle is important for Rab7A function. Although ubiquitin is usually bound to a lysine residue of a specific substrate by the sequential action of E1, E2, and E3 enzymes, Legionella SidE family proteins atypically ubiquitinate a serine residue of Rab33B in a E1/E2‐independent manner [66, 67]. Determining whether and how this modification allows bacterial survival in host cells will require further investigation.
Inactivation, stabilization, and degradation
Although Rab proteins themselves are GTPases that can hydrolyze their bound GTP to GDP, Rab‐GAPs enable more rapid and regulated inactivation of Rabs by enhancing their intrinsic GTPase activity. For example, TBC1D4 [also known as AS160 (Akt substrate of 160 kDa)] inactivates Rab8, Rab10, and Rab13, thereby preventing surface expression of GLUT4 in unstimulated skeletal muscles and adipocytes. Insulin stimulation activates these Rabs, because TBC1D4 phosphorylation by Akt attenuates its GAP activity, thus enabling insulin‐dependent GLUT4 translocation to the plasma membrane (reviewed in Ref. [68]). Most Rab‐GAPs identified to date are members of the Tre‐2/Bub2/Cdc16 (TBC) domain‐containing protein family and are encoded by ~ 40 genes in mammals [69] (see Table S1). Each TBC protein is thought to exert GAP activity on specific Rab family proteins, although their corresponding substrates have not been fully clarified [6]. Recent research on TBC family proteins has delineated the roles of TBC1D6 (Rab26‐GAP), TBC1D9B (Rab11‐GAP), TBC1D25/OATL1 (Rab33B‐GAP), and RUTBC1 (Rab32/38‐GAP) in GPCR trafficking, epithelial transcytosis, autophagosome maturation, and melanogenesis, respectively [70, 71, 72, 73, 74]. Moreover, TBC1D15 (Rab7A‐GAP) has been shown to be recruited to mitochondria by binding to Fis1 and to drive untethering of mitochondria–lysosome contacts by inactivating Rab7A [75, 76, 77]. Interestingly, expression of constitutively active Rab7A or GAP activity‐deficient mutants of TBC1D15 increases contact duration and inhibits mitochondrial fission events, suggesting that spatiotemporal regulation of Rab7A is important to the dynamics of mitochondria–lysosome contacts, which may determine the place and timing of mitochondrial fission. On the other hand, the GAP activity‐independent functions of several TBC family proteins, including TBC1D12, TBC1D14, TBC1D23, and TBC1D32/BROMI, have also been determined [78, 79, 80, 81]; for example, TBC1D23 is involved in endosomal vesicle capture at the trans‐Golgi in a GAP activity‐independent manner [80]. Future studies should provide a more comprehensive understanding of the roles of TBC family proteins and the regulation of Rab inactivation.
GDI extracts inactivated Rabs from membranes and masks their hydrophobic prenyl moiety, thereby retaining them in the cytosol for next use. In addition to this general mechanism, stabilization and degradation systems that are specific to a subset of Rabs have recently been reported. RABIF (also known as Mss4) acts as a holdase (ATP‐independent chaperone) of Rab8, Rab10, and Rab13, and its knockout leads to rapid proteasomal degradation of these Rabs [82]. Furthermore, hydrophobic residues within the switch I region of inactive Rab8 are recognized by BAG6, which targets Rab8 to proteasomes for degradation [83]. BAG6 also recognizes Rab10 and Rab13. These findings suggest that Rab8, Rab10, and Rab13 (yeast Sec4 homologs) are relatively unstable (especially in their inactive form) and/or prone to aggregate during folding and thus require specific quality control machineries to maintain their protein levels and prevent cytotoxic aggregation.
New molecular mechanisms of Rab functions
Because the basic concept of the function of Rab proteins is that each family member decorates the surface of a specific organelle and recruits effectors that mediate membrane trafficking, effector identification is paramount to understanding the functions of each Rab. The first Rab effector was identified by a cross‐linking method, by using a membrane fraction from bovine brain and bacterially purified Rab3A loaded with GTPγS, a nonhydrolyzable GTP analog [84, 85]. GST pull‐down and yeast two‐hybrid screens that have used Rabs as bait have subsequently succeeded in identifying several effectors in early Rab studies [86, 87, 88, 89, 90]. Both methods are easy to perform and have continued to be used in recent works that have identified EHBP1L1 (a Rab8 effector for polarized transport) [91], Sec16A (a Rab10 effector for insulin secretion) [92], and RELCH (a Rab11 effector for cholesterol transport) [93]. They have also been applied to much larger scale analyses [94, 95, 96] that have provided clues to unveiling the roles of several Rabs. For example, Rab2 has been shown to interact with the HOPS complex (a tethering factor) and mediate autophagosome–lysosome fusion [97], while Rab18 has been demonstrated to bind to the NAG–RINT1–ZW10 complex (an interactor of ER‐associated Q‐SNAREs) and bridge ER–lipid droplet contacts [98].
Despite these efforts, there are still many Rabs to which few or no effectors have been assigned. One promising approach to further explore possibly weak and transient interactions between Rabs and their effectors is to use the recently developed proximity biotinylation techniques [99, 100]. Actually, APEX2 (an engineered form of a soybean ascorbate peroxidase)‐tagged Rab4, Rab5, Rab7A, and Rab21 have enabled identification and comparison of their interactors in living cells, and the results revealed that a Rab21–WASH complex interaction is required for trafficking of a subset of clathrin‐independent cargos [101]. However, two drawbacks of this method in terms of Rab‐effector identification need to be considered: (a) possible contamination by noninteractors that are merely localized on the same organelles, and (b) the difficulty of determining the nucleotide dependency of the interactions, because constitutively negative Rab mutants are often mislocalized or not membrane‐bound. The ‘mitoID’ method has recently been developed to overcome these drawbacks [102]. In brief, Rab (constitutively active/negative)‐BirA* (an Escherichia coli biotin ligase mutant) fusion proteins are ectopically targeted to mitochondria to eliminate background proteins identified in common. Many effectors of Rab2A, 6A, 7A, 8A, 9A, 10, 11A, 18, 30, and 33B, as well as several of their GEFs and GAPs, have been successfully identified by using this method. Although the mitoID method may be unsuited for effectors that require a specific lipid for membrane association [102], application to the other Rabs should be informative.
Vesicle tethering is thought to be an important step in the proper recognition of target membranes and efficient fusion to them [103]. Whereas many Rabs facilitate this process by recruiting tethering factors, including golgins, COG complex, HOPS complex, and exocyst complex, new modes of Rab‐mediated tethering beyond such a simple ‘effector recruiting’ model have been reported (Fig. 4). During lumenogenesisof epithelial cell, vesicles containing apical membrane proteins such as podocalyxin have been found to be transported and exocytosed to the apical membrane initiation site, thereby opening and expanding the lumen [104]. Rab35 has been shown to localize to the nascent apical membrane prior to the arrival of the apically directed vesicles and to capture them by direct interaction with the cytoplasmic tail of podocalyxin, not through the typical tethering factors [105] (Fig. 4A). Such trans interactions also occur between Rab5 and its effector EEA1 during the homotypic fusion of early endosomes. EEA1 is associated with early endosomes via its C‐terminal PI3P‐binding domain and extends its intermediate coiled‐coil region and N‐terminal Rab5‐binding domain outward from the membrane surface, enabling the capture of other Rab5‐bearing early endosomes. Although the predicted length of EEA1 (~ 200 nm) appears to be too long to allow subsequent fusion of the two membranes, a recent study using rotary shadowing electron microscopy and optical tweezers clearly answered to this problem [106]. Intriguingly, it was shown that while EEA1 alone adopted a fully extended conformation, upon binding to active Rab5 it becomes flexible enough to collapse into smaller structures (ranging from 20 to 200 nm). This causes an equilibrium shift and generates entropic force (~ 3 pN), which pulls the two vesicles together (i.e., tethering). This finding was the first evidence that an active Rab does not just recruit its effector, but induces a conformational change in its effector upon binding (Fig. 4B). Another example is the ability of Rab itself to serve as a tethering factor. The results of an in vitro liposome tethering assay using purified Vps21 (yeast Rab5) and chemically defined liposomes unexpectedly showed that a homotypic interaction of Vps21 is sufficient to promote liposome tethering in the absence of any effector molecules [107] (Fig. 4C). Similarly, purified Rab5A, loaded on liposomes at physiological density (equivalent to the previously reported Rab density of a single synaptic vesicle [108]), has been shown to efficiently drive liposome tethering in vitro [109]. In addition, homotypic tethering by Rab1A, 3A, 4A, 6A, 7A, 9A, 11A, and 33B at higher density and efficient heterotypic tethering by Rab1A and Rab9A have been demonstrated. Understanding the physiological significance of these intrinsic tethering activities of Rabs and their possible interplay with effector‐mediated tethering await further investigation.
Fig. 4.
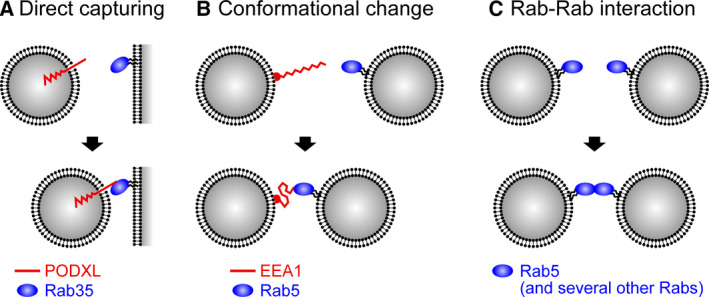
New mechanistic models of Rab‐mediated membrane tethering. (A) Podocalyxin (PODXL)‐containing vesicles are directly captured by Rab35 on the target membrane [105]. (B) Binding of EEA1 to Rab5 enables a conformational change in EEA1, which generates the pulling force that brings two endosomes together [106]. (C) Vesicle tethering can be induced by homo‐ and hetero‐typic interactions of Rabs in the absence of any effectors in vitro [109]. See the text for details.
Human diseases, mutant animals, and Rab knockout phenotypes
Finally, we will discuss the physiological roles of Rabs by focusing on their roles as revealed by studies on human diseases, mutant animals, and Rab‐KO phenotypes (summarized in Table 1 and Table S2). Lysosome‐related organelles (LROs) are typically nondegradative, cell‐type‐specific secretory organelles, such as the melanosomes in pigment cells, dense granules in platelets, and lamellar bodies in lung alveolar type II cells. Since these organelles play important roles in various biological phenomena (e.g., skin and hair pigmentation, blood clotting, and pulmonary surfactant secretion), defects in LRO biogenesis cause human diseases, including Hermansky–Pudlak syndrome (HPS), which is characterized by oculocutaneous albinism, a prolonged bleeding time, and often pulmonary fibrosis (reviewed in Ref. [110]). The mutated genes in the hypopigmentation pale ear (ep) mouse and light ear (le) mouse have turned out to be orthologs of the human HPS genes HPS1 and HPS4, respectively [111, 112, 113]. HPS1 and HPS4 have subsequently been shown to heterodimerize to form the BLOC‐3 complex, which exhibits GEF activity toward Rab32 and its paralog Rab38 [21]. Furthermore, double KO of Rab32/38 in mice results in severe coat color dilution and defects in hemostasis and lung morphology [114], indicating involvement of these Rabs in LRO biogenesis. The number of dense granules in Rab32/38‐KO mouse platelets is reduced, and their morphology is abnormal.
Bacteria‐containing vacuoles, in which infectious bacteria are retained in host cells, are also thought to be a kind of LROs that serve to isolate such bacteria. Interestingly, Rab32 has been shown to be recruited to Staphylococcus aureus‐ or Mycobacterium tuberculosis‐containing vacuoles in infected macrophages [115]. Other studies have demonstrated that Rab32 is also recruited to Salmonella Typhi‐containing vacuoles and that Rab32‐ or HPS4‐deficient mice are more susceptible to infection by the pathogen than wild‐type mice are [116, 117]. Not surprisingly, bacteria also evolved defenses against isolation in vacuoles. S. Typhimurium, but not S. Typhi, secretes GtgE and SopD2, a protease and GAP for Rab32, respectively, into the host cell cytosol that interfere with the Rab32‐dependent defense pathway [117]. Taken together, the results of these studies suggest that Rab32/38 play roles in diverse physiological processes through LRO biogenesis. Transport of LROs, such as melanosomes and dense granules, is also known to be regulated by Rab small GTPases, specifically by the Rab27 isoform (see review Ref. [118] for details).
As with Rab32, Rab20 has also been shown to have an antibacterial function. Rab20 is recruited to M. tuberculosis‐containing phagosomes in macrophages, and its recruitment is enhanced by the macrophage‐activating cytokine interferon‐γ [119]. In addition, whereas overexpression of Rab20 attenuates intracellular bacterial replication, KO of Rab20 results in inability of bacteria‐containing phagosomes to become acidic and proteolytic, thereby allowing bacterial replication. Consistent with these findings, after being infected with M. tuberculosis the lungs of Rab20‐KO mice were found to contain larger lesion areas than in infected wild‐type mice.
Other Rab‐KO mice recently reported include the following: Rab10‐KO mice, which are characterized by early embryonic lethality [120], Rab13‐KO mice, which have a smaller spleen and lymph nodes because of impaired lymphocyte migration [121], and Rab39A‐KO mice, which exhibit reduced cross‐presentation activity by antigen‐presenting cells [122]. Moreover, The International Mouse Phenotyping Consortium (IMPC) has generated over 8000 KO mouse lines to date, and the phenotypic data for more than half of the Rab family genes are already available on its website (https://www.mousephenotype.org/). The data available include the lethal phenotypes of Rab2A, 5C, 21, and 40C, which have not yet been reported in the literature. The CRISPR/Cas‐mediated genome editing technology has boosted the KO mouse production rate, and the IMPC now plans to generate KO mice for all of the rest of the genes (~ 10 000) in the mouse genome within 10 years.
The recent technical advances in genome editing have also led to acceleration of KO analyses in cultured cells. In particular, genome‐wide screening using a single guide RNA (sgRNA) lentiviral library has succeeded in identifying many positive and negative regulators, including Rabs, of particular phenotypes. For example, Rab34 has been found to be a positive regulator of Hedgehog (Hh) signaling, which is sensed by primary cilia and plays an essential role in the patterning of cell differentiation during development [123]. In the absence of Rab34, primary cilia formation by both cultured cells and in vivo was found to be inhibited [124], and another independent study by the IMPC has shown that Rab34‐KO mice exhibit typical ciliopathy phenotypes, including polydactyly and craniofacial malformation [125]. Thus, Rab34 is essential for ciliogenesis and Hh signaling, although the molecular mechanism has yet to be determined. Other cilia‐related Rabs, including Rab8 and Rab23, have also been reported (see review Ref. [126] for details). A recent advance has been the discovery that Fuzzy and Inturned, known regulators of the planar cell polarity pathway, form a Rab23‐GEF complex [29]. Although knockdown of this complex or Rab23 inhibited ciliogenesis in cultured cells, the number and length of nodal cilia were normal in Rab23‐KO mice [127]. Rab8 localizes on primary cilia and has been shown to be required for ciliogenesis in cultured cells and a zebrafish model [128, 129], but not in mice [130]. Taken together, Rab8 and Rab23 seem not to be essential for ciliogenesis itself, but to be likely to regulate ciliary functions in mammals.
Another CRISPR screening revealed that loss of Rab10 or its chaperone RABIF (see the above section) protected cells from L. pneumophila toxicity by attenuating its intracellular replication [131]. Although the bacteria normally entered Rab10‐KO or RABIF‐KO cells, the Legionella‐containing vacuoles (LCVs) were unable to recruit the ER‐resident proteins to its membrane, indicating that L. pneumophila needs to hijack the host Rab10 function to convert the LCV membrane into an ER‐like membrane. Interestingly, the same as Rab33B (see the above section), Rab10 is also ubiquitinated by the bacterial proteins SidC and SdcA, and these proteins are required for Rab10 recruitment to LCVs. Both Rab10 and RABIF have also been identified as positive regulators of the surface translocation of GLUT4 [82], thereby validating the usefulness of genome‐wide screenings as a means of identifying functional partners of Rabs (e.g., GEFs, GAPs, effectors, and chaperones).
In order to facilitate more detailed and comprehensive Rab‐KO analyses, a collection of KO cell lines for the entire Rab family has been generated using the epithelial cell line Madin–Darby canine kidney (MDCK) II [132] [available from RIKEN BioResource Research Center Cell Bank (https://cell.brc.riken.jp/en); Cat#: RCB5099–RCB5148], and subsequent analyses of these cells confirmed the roles of Rab2, Rab7, and Rab11 in Golgi integrity, lysosome homeostasis, and single lumen formation in epithelial cysts, respectively. In addition, Rab6‐KO cells lack the basement membrane of polarized MDCK II cells likely due to a secretory defect. Rab6 seems to be required for the post‐Golgi transport of a wide range of secretory cargos, and cargos have been shown to be mistargeted to lysosomes for degradation in Rab6‐KO cells. Since many of the Rab‐KO phenotypes have become visible only when close paralogs, for example, Rab6A and Rab6B, were simultaneously knocked out, future research using the collection is expected to reveal new phenotypes that have been missed by genome‐wide screenings.
Perspectives
As described above, biochemical analyses, including by mass spectrometry and proximity biotinylation, have continued to provide mechanistic insights into Rab functions by identifying novel effectors, GEFs, and GAPs, as well as post‐translational modifiers. In addition, electron microscopy techniques have been useful not only to study intracellular organelles but also for the structural analysis of purified proteins without crystallization. This allowed the study of high molecular weight Rab effectors, such as EEA1. In phenotypic studies, CRISPR/Cas technology has made it possible to accelerate KO analyses in both animals and cultured cells. Moreover, since many Rabs are involved in complicated neuronal disorders [133], pathogen defense pathways [134], and cancer [135], specific assays beyond the standard phenotyping pipeline are also needed. Such efforts will together provide a more detailed and comprehensive understanding of the physiological functions of the Rab family of GTPases in the future.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
YH, SH, and MF prepared the figures and tables. YH and MF wrote and edited the paper.
Supporting information
Fig. S1. Amino acid sequences of Rabs/Ypts that were used for the phylogenetic analysis in Fig. 2A.
Fig. S2. Amino acid sequences of the switch II region of human Rabs that were used for the phylogenetic analysis in Fig. 2B.
Table S1. Rab effectors, binding molecules, GEFs, and GAPs in mammals
Table S2. Mouse and human Rab genes.
Acknowledgements
This work was supported in part by Grant‐in‐Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (grant number 18K14692 to YH), Grant‐in‐Aid for Scientific Research(B) from the MEXT (grant number 19H03220 to MF), and by Japan Science and Technology Agency (JST) CREST (grant number JPMJCR17H4 to MF).
References
- 1. Fukuda M (2008) Membrane traffic in the secretory pathway: regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol Life Sci 65, 2801–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513–525. [DOI] [PubMed] [Google Scholar]
- 3. Hutagalung AH & Novick PJ (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91, 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pfeffer SR (2013) Rab GTPase regulation of membrane identity. Curr Opin Cell Biol 25, 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stenmark H & Olkkonen VM (2001) The Rab GTPase family. Genome Biol 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Itoh T, Satoh M, Kanno E & Fukuda M (2006) Screening for target Rabs of TBC (Tre‐2/Bub2/Cdc16) domain‐containing proteins based on their Rab‐binding activity. Genes Cells 11, 1023–1037. [DOI] [PubMed] [Google Scholar]
- 7. Diekmann Y, Seixas E, Gouw M, Tavares‐Cadete F, Seabra MC & Pereira‐Leal JB (2011) Thousands of Rab GTPases for the cell biologist. PLoS Comput Biol 7, e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klöpper TH, Kienle N, Fasshauer D & Munro S (2012) Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol 10, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawasaki M, Nakayama K & Wakatsuki S (2005) Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr Opin Struct Biol 15, 681–689. [DOI] [PubMed] [Google Scholar]
- 10. Srikanth S, Do KK, Gao Y, Woo JS, Ghosh S, Calmettes G, Paz A, Abramson J, Jiang M & Gwack Y (2016) A large Rab GTPase encoded by CRACR2A is a component of subsynaptic vesicles that transmit T cell activation signals. Sci Signal 9, ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamaguchi Y, Sakai E, Okamoto K, Kajiya H, Okabe K, Naito M, Kadowaki T & Tsukuba T (2018) Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation. Cell Mol Life Sci 75, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Huynh W, Skokan TD, Lu W, Weiss A & Vale RD (2019) CRACR2a is a calcium‐activated dynein adaptor protein that regulates endocytic traffic. J Cell Biol 218, 1619–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakayama K & Katoh Y (2018) Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin‐2 and dynein‐2 motors. J Biochem 163, 155–164. [DOI] [PubMed] [Google Scholar]
- 14. Mili S, Moissoglu K & Macara IG (2008) Genome‐wide screen reveals APC‐associated RNAs enriched in cell protrusions. Nature 453, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hinger SA, Cha DJ, Franklin JL, Higginbotham JN, Dou Y, Ping J, Shu L, Prasad N, Levy S, Zhang B et al (2018) Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Rep 25, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ioannou MS & McPherson PS (2016) Regulation of cancer cell behavior by the small GTPase Rab13. J Biol Chem 291, 9929–9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leung KF, Baron R & Seabra MC (2006) Thematic review series: lipid posttranslational modifications. Geranylgeranylation of Rab GTPases. J Lipid Res 47, 467–475. [DOI] [PubMed] [Google Scholar]
- 18. Blümer J, Rey J, Dehmelt L, Mazel T, Wu Y‐W, Bastiaens P, Goody RS & Itzen A (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol 200, 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerondopoulos A, Bastos RN, Yoshimura SI, Anderson R, Carpanini S, Aligianis I, Handley MT & Barr FA (2014) Rab18 and a Rab18 GEF complex are required for normal ER structure. J Cell Biol 205, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanza P, Evans RD, Briggs DA, Cantero M, Montoliu L, Patel S, Sviderskaya EV, Itzen A, Figueiredo AC, Seabra MC et al (2019) Nucleotide exchange factor Rab3GEP requires DENN and non‐DENN elements for activation and targeting of Rab27a. J Cell Sci 132, jcs212035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerondopoulos A, Langemeyer L, Liang JR, Linford A & Barr FA (2012) BLOC‐3 mutated in Hermansky‐Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 22, 2135–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novick P (2016) Regulation of membrane traffic by Rab GEF and GAP cascades. Small GTPases 7, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfeffer SR (2017) Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 28, 712–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas LL, van der Vegt SA & Fromme JC (2019) A steric gating mechanism dictates the substrate specificity of a Rab‐GEF. Dev Cell 48, 100–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marat AL, Dokainish H & McPherson PS (2011) DENN domain proteins: regulators of Rab GTPases. J Biol Chem 286, 13791–13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato M, Sato K, Liou W, Pant S, Harada A & Grant BD (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME‐4, a coated‐pit protein. EMBO J 27, 1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barr F & Lambright DG (2010) Rab GEFs and GAPs. Curr Opin Cell Biol 22, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishida M, Oguchi ME & Fukuda M (2016) Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct Funct 79, 61–79. [Google Scholar]
- 29. Gerondopoulos A, Strutt H, Stevenson NL, Sobajima T, Levine TP, Stephens DJ, Strutt D & Barr FA (2019) Planar cell polarity effector proteins Inturned and Fuzzy form a Rab23 GEF complex. Curr Biol 29, 3323–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lüningschrör P, Binotti B, Dombert B, Heimann P, Perez‐Lara A, Slotta C, Thau‐Habermann N, Von Collenberg CR, Karl F, Damme M et al (2017) Plekhg5‐regulated autophagy of synaptic vesicles reveals a pathogenic mechanism in motoneuron disease. Nat Commun 8, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailly E, McCaffrey M, Touchot N, Zahraoui A, Goud B & Bornens M (1991) Phosphorylation of two small GTP‐binding proteins of the Rab family by p34cdc2. Nature 350, 715–718. [DOI] [PubMed] [Google Scholar]
- 32. van der Sluijs P, Hull M, Huber LA, Mâle P, Goud B & Mellman I (1992) Reversible phosphorylation–dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J 11, 4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Francavilla C, Papetti M, Rigbolt KTG, Pedersen AK, Sigurdsson JO, Cazzamali G, Karemore G, Blagoev B & Olsen JV (2016) Multilayered proteomics reveals molecular switches dictating ligand‐dependent EGFR trafficking. Nat Struct Mol Biol 23, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin X, Zhang J, Chen L, Chen Y, Xu X, Hong W & Wang T (2017) Tyrosine phosphorylation of Rab7 by Src kinase. Cell Signal 35, 84–94. [DOI] [PubMed] [Google Scholar]
- 35. Heo JM, Ordureau A, Swarup S, Paulo JA, Shen K, Sabatini DM & Harper JW (2018) RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK‐PARKIN pathway. Sci Adv 4, eaav0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laviolette LA, Mermoud J, Calvo IA, Olson N, Boukhali M, Steinlein OK, Roider E, Sattler EC, Huang D, Teh BT et al (2017) Negative regulation of EGFR signalling by the human folliculin tumour suppressor protein. Nat Commun 8, 15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanafusa H, Yagi T, Ikeda H, Hisamoto N, Nishioka T, Kaibuchi K, Shirakabe K & Matsumoto K (2019) LRRK1 phosphorylation of Rab7 at S72 links trafficking of EGFR‐containing endosomes to its effector RILP. J Cell Sci 132, jcs228809. [DOI] [PubMed] [Google Scholar]
- 38. Shinde SR & Maddika S (2016) PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun 7, 10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steger M, Tonelli F, Ito G, Davies P, Trost M, Vetter M, Wachter S, Lorentzen E, Duddy G, Wilson S et al (2016) Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, Karayel O, Tonelli F, Martinez TN, Lorentzen E, Pfeffer SR et al (2017) Systematic proteomic analysis of LRRK2‐mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6, e31012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berndsen K, Lis P, Yeshaw WM, Wawro PS, Nirujogi RS, Wightman M, Macartney T, Dorward M, Knebel A, Tonelli F et al (2019) PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. eLife 8, e50416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dhekne HS, Yanatori I, Gomez RC, Tonelli F, Diez F, Schüle B, Steger M, Alessi DR & Pfeffer SR (2018) A pathway for parkinson's disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife 7, e40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eguchi T, Kuwahara T, Sakurai M, Komori T, Fujimoto T, Ito G, Yoshimura SI, Harada A, Fukuda M, Koike M et al (2018) LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci USA 115, E9115–E9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madero‐Pérez J, Fdez E, Fernández B, Lara Ordóñez AJ, Blanca Ramírez M, Gómez‐Suaga P, Waschbüsch D, Lobbestael E, Baekelandt V, Nairn AC et al (2018) Parkinson disease‐associated mutations in LRRK2 cause centrosomal defects via Rab8a phosphorylation. Mol Neurodegener 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bae E‐J, Kim D‐K, Kim C, Mante M, Adame A, Rockenstein E, Ulusoy A, Klinkenberg M, Jeong GR, Bae JR et al (2018) LRRK2 kinase regulates α‐synuclein propagation via RAB35 phosphorylation. Nat Commun 9, 3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, MacCabe BD, Marder KS, Honig LS, Clark LN, Small SA et al (2013) RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson's disease risk. Neuron 77, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K et al (2014) Unbiased screen for interactors of leucine‐rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc Natl Acad Sci USA 111, 2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Z, Bryant N, Kumaran R, Beilina A, Abeliovich A, Cookson MR & West AB (2018) LRRK2 phosphorylates membrane‐bound Rabs and is activated by GTP‐bound Rab7L1 to promote recruitment to the trans‐Golgi network. Hum Mol Genet 27, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Purlyte E, Dhekne HS, Sarhan AR, Gomez R, Lis P, Wightman M, Martinez TN, Tonelli F, Pfeffer SR & Alessi DR (2018) Rab29 activation of the Parkinson's disease‐associated LRRK2 kinase. EMBO J 37, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lai Y, Kondapalli C, Lehneck R, Procter JB, Dill BD, Woodroof HI, Gourlay R, Peggie M, Macartney TJ, Corti O et al (2015) Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J 34, 2840–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walther DJ, Peter JU, Winter S, Höltje M, Paulmann N, Grohmann M, Vowinckel J, Alamo‐Bethencourt V, Wilhelm CS, Ahnert‐Hilger G et al (2003) Serotonylation of small GTPases is a signal transduction pathway that triggers platelet α‐granule release. Cell 115, 851–862. [DOI] [PubMed] [Google Scholar]
- 52. Paulmann N, Grohmann M, Voigt JP, Bert B, Vowinckel J, Bader M, Skelin M, Jevšek M, Fink H, Rupnik M et al (2009) Intracellular serotonin modulates insulin secretion from pancreatic β‐cells by protein serotonylation. PLoS Biol 7, e1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS & Itzen A (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949. [DOI] [PubMed] [Google Scholar]
- 54. Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE & Roy CR (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goody PR, Heller K, Oesterlin LK, Müller MP, Itzen A & Goody RS (2012) Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J 31, 1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oesterlin LK, Goody RS & Itzen A (2012) Posttranslational modifications of Rab proteins cause effective displacement of GDP dissociation inhibitor. Proc Natl Acad Sci USA 109, 5621–5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Modica G, Skorobogata O, Sauvageau E, Vissa A, Yip CM, Kim PK, Wurtele H & Lefrancois S (2017) Rab7 palmitoylation is required for efficient endosome‐to‐TGN trafficking. J Cell Sci 130, 2579–2590. [DOI] [PubMed] [Google Scholar]
- 58. Piper RC, Dikic I & Lukacs GL (2014) Ubiquitin‐dependent sorting in endocytosis. Cold Spring Harb Perspect Biol 6, a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lachance V, Degrandmaison J, Marois S, Robitaille M, Génier S, Nadeau S, Angers S & Parent JL (2014) Ubiquitylation and activation of a Rab GTPase is promoted by a β2AR‐HACE1 complex. J Cell Sci 127, 111–123. [DOI] [PubMed] [Google Scholar]
- 60. Shin D, Na W, Lee JH, Kim G, Baek J, Park SH, Choi CY & Lee S (2017) Site‐specific monoubiquitination downregulates Rab5 by disrupting effector binding and guanine nucleotide conversion. eLife 6, e29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song P, Trajkovic K, Tsunemi T & Krainc D (2016) Parkin modulates endosomal organization and function of the endo‐lysosomal pathway. J Neurosci 36, 2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sapmaz A, Berlin I, Bos E, Wijdeven RH, Janssen H, Konietzny R, Akkermans JJ, Erson‐Bensan AE, Koning RI, Kessler BM et al (2019) USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7. Nat Commun 10, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vanlandingham PA & Ceresa BP (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 284, 12110–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Girard E, Chmiest D, Fournier N, Johannes L, Paul JL, Vedie B & Lamaze C (2014) Rab7 is functionally required for selective cargo sorting at the early endosome. Traffic 15, 309–326. [DOI] [PubMed] [Google Scholar]
- 65. Kuchitsu Y, Homma Y, Fujita N & Fukuda M (2018) Rab7 knockout unveils regulated autolysosome maturation induced by glutamine starvation. J Cell Sci 131, jcs215442. [DOI] [PubMed] [Google Scholar]
- 66. Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X & Luo ZQ (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I & Dikic I (2016) Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649. [DOI] [PubMed] [Google Scholar]
- 68. Jaldin‐Fincati JR, Pavarotti M, Frendo‐Cumbo S, Bilan PJ & Klip A (2017) Update on GLUT4 vesicle traffic: a cornerstone of insulin action. Trends Endocrinol Metab 28, 597–611. [DOI] [PubMed] [Google Scholar]
- 69. Fukuda M (2011) TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep 31, 159–168. [DOI] [PubMed] [Google Scholar]
- 70. Wei Z, Zhang M, Li C, Huang W, Fan Y, Guo J, Khater M, Fukuda M, Dong Z, Hu G et al (2019) Specific TBC domain‐containing proteins control the ER‐Golgi‐plasma membrane trafficking of GPCRs. Cell Rep 28, 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gallo LI, Liao Y, Ruiz WG, Clayton DR, Li M, Liu Y‐J, Jiang Y, Fukuda M, Apodaca G & Yin X‐M (2014) TBC1D9B functions as a GTPase‐activating protein for Rab11a in polarized MDCK cells. Mol Biol Cell 25, 3779–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Itoh T, Kanno E, Uemura T, Waguri S & Fukuda M (2011) OATL1, a novel autophagosome‐resident Rab33B‐GAP, regulates autophagosomal maturation. J Cell Biol 192, 839–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nottingham RM, Ganley IG, Barr FA, Lambright DG & Pfeffer SR (2011) RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem 286, 33213–33222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Marubashi S, Shimada H, Fukuda M & Ohbayashi N (2016) RUTBC1 functions as a GTPase‐activating protein for Rab32/38 and regulates melanogenic enzyme trafficking in melanocytes. J Biol Chem 291, 1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Onoue K, Jofuku A, Ban‐Ishihara R, Ishihara T, Maeda M, Koshiba T, Itoh T, Fukuda M, Otera H, Oka T et al (2013) Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J Cell Sci 126, 176–185. [DOI] [PubMed] [Google Scholar]
- 76. Yamano K, Fogel AI, Wang C, van der Bliek AM & Youle RJ (2014) Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 2014, e01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wong YC, Ysselstein D & Krainc D (2018) Mitochondria‐lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oguchi ME, Noguchi K & Fukuda M (2017) TBC1D12 is a novel Rab11‐binding protein that modulates neurite outgrowth of PC12 cells. PLoS One 12, e174883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Longatti A, Lamb CA, Razi M, Yoshimura S‐I, Barr FA & Tooze SA (2012) TBC1D14 regulates autophagosome formation via Rab11‐ and ULK1‐positive recycling endosomes. J Cell Biol 197, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shin JJH, Gillingham AK, Begum F, Chadwick J & Munro S (2017) TBC1D23 is a bridging factor for endosomal vesicle capture by golgins at the trans‐Golgi. Nat Cell Biol 19, 1424–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ko HW, Norman RX, Tran J, Fuller KP, Fukuda M & Eggenschwiler JT (2010) Broad‐minded links cell cycle‐related kinase to cilia assembly and Hedgehog signal transduction. Dev Cell 18, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gulbranson DR, Davis EM, Demmitt BA, Ouyang Y, Ye Y, Yu H & Shen J (2017) RABIF/MSS4 is a Rab‐stabilizing holdase chaperone required for GLUT4 exocytosis. Proc Natl Acad Sci USA 114, E8224–E8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Takahashi T, Minami S, Tsuchiya Y, Tajima K, Sakai N, Suga K, Hisanaga S, Ohbayashi N, Fukuda M & Kawahara H (2019) Cytoplasmic control of Rab family small GTPases through BAG6. EMBO Rep 20, e46794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shirataki H, Kaibuchi K, Yamaguchi T, Wada K, Horiuchi H & Takai Y (1992) A possible target protein for smg‐25A/rab3A small GTP‐binding protein. J Biol Chem 267, 10946–10949. [PubMed] [Google Scholar]
- 85. Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M & Takai Y (1993) Rabphilin‐3A, a putative target protein for smg p25A/rab3A p25 small GTP‐binding protein related to synaptotagmin. Mol Cell Biol 13, 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stenmark H, Vitale G, Ullrich O & Zerial M (1995) Rabaptin‐5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83, 423–432. [DOI] [PubMed] [Google Scholar]
- 87. Ren M, Zeng J, De Lemos‐Chiarandini C, Rosenfeld M, Adesnik M & Sabatini DD (1996) In its active form, the GTP‐binding protein rab8 interacts with a stress‐activated protein kinase. Proc Natl Acad Sci USA 93, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Díaz E, Schimmöller F & Pfeffer SR (1997) A novel Rab9 effector required for endosome‐to‐TGN transport. J Cell Biol 138, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Echard A, Jollivet F, Martinez O, Lacapère JJ, Rousselet A, Janoueix‐Lerosey I & Goud B (1998) Interaction of a Golgi‐associated kinesin‐like protein with Rab6. Science 279, 580–585. [DOI] [PubMed] [Google Scholar]
- 90. Christoforidis S, McBride HM, Burgoyne RD & Zerial M (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625. [DOI] [PubMed] [Google Scholar]
- 91. Nakajo A, Yoshimura S‐I, Togawa H, Kunii M, Iwano T, Izumi A, Noguchi Y, Watanabe A, Goto A, Sato T et al (2016) EHBP1L1 coordinates Rab8 and Bin1 to regulate apical‐directed transport in polarized epithelial cells. J Cell Biol 212, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bruno J, Brumfield A, Chaudhary N, Iaea D & McGraw TE (2016) SEC16A is a RAB10 effector required for insulin‐stimulated GLUT4 trafficking in adipocytes. J Cell Biol 214, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sobajima T, Yoshimura S‐I, Maeda T, Miyata H, Miyoshi E & Harada A (2018) The Rab11‐binding protein RELCH/KIAA1468 controls intracellular cholesterol distribution. J Cell Biol 217, 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fukuda M, Kanno E, Ishibashi K & Itoh T (2008) Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 7, 1031–1042. [DOI] [PubMed] [Google Scholar]
- 95. Kanno E, Ishibashi K, Kobayashi H, Matsui T, Ohbayashi N & Fukuda M (2010) Comprehensive screening for novel Rab‐binding proteins by GST pull‐down assay using 60 different mammalian Rabs. Traffic 11, 491–507. [DOI] [PubMed] [Google Scholar]
- 96. Gillingham AK, Sinka R, Torres IL, Lilley KS & Munro S (2014) Toward a comprehensive map of the effectors of Rab GTPases. Dev Cell 31, 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fujita N, Huang W, Lin TH, Groulx JF, Jean S, Nguyen J, Kuchitsu Y, Koyama‐Honda I, Mizushima N, Fukuda M et al (2017) Genetic screen in Drosophila muscle identifies autophagy‐mediated T‐tubule remodeling and a Rab2 role in autophagy. eLife 6, e23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu D, Li Y, Wu L, Li Y, Zhao D, Yu J, Huang T, Ferguson C, Parton RG, Yang H et al (2018) Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J Cell Biol 217, 975–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Roux KJ, Kim DI, Raida M & Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rhee H‐W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA & Ting AY (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Del Olmo T, Lauzier A, Normandin C, Larcher R, Lecours M, Jean D, Lessard L, Steinberg F, Boisvert F & Jean S (2019) APEX2‐mediated RAB proximity labeling identifies a role for RAB21 in clathrin‐independent cargo sorting. EMBO Rep 20, e47192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gillingham AK, Bertram J, Begum F & Munro S (2019) In vivo identification of GTPase interactors by mitochondrial relocalization and proximity biotinylation. eLife 8, e45916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gillingham AK & Munro S (2019) Transport carrier tethering – how vesicles are captured by organelles. Curr Opin Cell Biol 59, 140–146. [DOI] [PubMed] [Google Scholar]
- 104. Bryant DM, Datta A, Rodríguez‐Fraticelli AE, Peränen J, Martín‐Belmonte F & Mostov KE (2010) A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 12, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Klinkert K, Rocancourt M, Houdusse A & Echard A (2016) Rab35 GTPase couples cell division with initiation of epithelial apico‐basal polarity and lumen opening. Nat Commun 7, 11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Murray DH, Jahnel M, Lauer J, Avellaneda MJ, Brouilly N, Cezanne A, Morales‐Navarrete H, Perini ED, Ferguson C, Lupas AN et al (2016) An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature 537, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lo SY, Brett CL, Plemel RL, Vignali M, Fields S, Gonen T & Merz AJ (2012) Intrinsic tethering activity of endosomal Rab proteins. Nat Struct Mol Biol 19, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P et al (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846. [DOI] [PubMed] [Google Scholar]
- 109. Segawa K, Tamura N & Mima J (2019) Homotypic and heterotypic trans‐assembly of human Rab family small GTPases in reconstituted membrane tethering. J Biol Chem 294, 7722–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Marks MS, Heijnen HFG & Raposo G (2013) Lysosome‐related organelles: unusual compartments become mainstream. Curr Opin Cell Biol 25, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Feng GH, Bailin T, Oh J & Spritz RA (1997) Mouse pale ear (ep) is homologous to human Hermansky‐Pudlak syndrome and contains a rare “AT‐AC” intron. Hum Mol Genet 6, 793–797. [DOI] [PubMed] [Google Scholar]
- 112. Gardner JM, Wildenberg SC, Keiper NM, Novak EK, Rusiniak ME, Swank RT, Puri N, Finger JN, Hagiwara N, Lehman AL et al (1997) The mouse pale ear (ep) mutation is the homologue of human Hermansky‐Pudlak syndrome. Proc Natl Acad Sci USA 94, 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Suzuki T, Li W, Zhang Q, Karim A, Novak EK, Sviderskaya EV, Hill SP, Bennett DC, Levin AV, Nieuwenhuis HK et al (2002) Hermansky‐Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light‐ear gene. Nat Genet 30, 321–324. [DOI] [PubMed] [Google Scholar]
- 114. Aguilar A, Weber J, Boscher J, Freund M, Ziessel C, Eckly A, Magnenat S, Bourdon C, Hechler B, Mangin PH et al (2019) Combined deficiency of RAB32 and RAB38 in the mouse mimics Hermansky‐Pudlak syndrome and critically impairs thrombosis. Blood Adv 3, 2368–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Seto S, Tsujimura K & Koide Y (2011) Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic 12, 407–420. [DOI] [PubMed] [Google Scholar]
- 116. Spanò S & Galán JE (2012) A Rab32‐dependent pathway contributes to Salmonella typhi host restriction. Science 338, 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Spanò S, Gao X, Hannemann S, Lara‐Tejero M & Galán JE (2016) A bacterial pathogen targets a host Rab‐family GTPase defense pathway with a GAP. Cell Host Microbe 19, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fukuda M (2013) Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic 14, 949–963. [DOI] [PubMed] [Google Scholar]
- 119. Schnettger L, Rodgers A, Repnik U, Lai RP, Pei G, Verdoes M, Wilkinson RJ, Young DB & Gutierrez MG (2017) A Rab20‐dependent membrane trafficking pathway controls M. tuberculosis replication by regulating phagosome spaciousness and integrity. Cell Host Microbe 21, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lv P, Sheng Y, Zhao Z, Zhao W, Gu L, Xu T & Song E (2015) Targeted disruption of Rab10 causes early embryonic lethality. Protein Cell 6, 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nishikimi A, Ishihara S, Ozawa M, Etoh K, Fukuda M, Kinashi T & Katagiri K (2014) Rab13 acts downstream of the kinase Mst1 to deliver the integrin LFA‐1 to the cell surface for lymphocyte trafficking. Sci Signal 7, ra72. [DOI] [PubMed] [Google Scholar]
- 122. Cruz FM, Colbert JD & Rock KL (2019) The GTPase Rab39a promotes phagosome maturation into MHC‐I antigen‐presenting compartments. EMBO J, e102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Pusapati GV, Kong JH, Patel BB, Krishnan A, Sagner A, Kinnebrew M, Briscoe J, Aravind L & Rohatgi R (2018) CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen Sonic Hedgehog. Dev Cell 44, 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Xu S, Liu Y, Meng Q & Wang B (2018) Rab34 small GTPase is required for Hedgehog signaling and an early step of ciliary vesicle formation in mouse. J Cell Sci 131, jcs213710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H et al (2016) High‐throughput discovery of novel developmental phenotypes. Nature 537, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Blacque OE, Scheidel N & Kuhns S (2018) Rab GTPases in cilium formation and function. Small GTPases 9, 76–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Fuller K, O'Connell JT, Gordon J, Mauti O & Eggenschwiler J (2014) Rab23 regulates Nodal signaling in vertebrate left‐right patterning independently of the Hedgehog pathway. Dev Biol 391, 182–195. [DOI] [PubMed] [Google Scholar]
- 128. Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC et al (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 129. Yoshimura SI, Egerer J, Fuchs E, Haas AK & Barr FA (2007) Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 178, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, Jung Y, Hagiwara H, Yoshihara Y, Yuzaki M, Harada R et al (2014) Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J Cell Sci 127, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jeng EE, Bhadkamkar V, Ibe NU, Gause H, Jiang L, Chan J, Jian R, Jimenez‐Morales D, Stevenson E, Krogan NJ et al (2019) Systematic identification of host cell regulators of Legionella pneumophila pathogenesis using a genome‐wide CRISPR screen. Cell Host Microbe 26, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Homma Y, Kinoshita R, Kuchitsu Y, Wawro PS, Marubashi S, Oguchi ME, Ishida M, Fujita N & Fukuda M (2019) Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J Cell Biol 218, 2035–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kiral FR, Kohrs FE, Jin EJ & Hiesinger PR (2018) Rab GTPases and membrane trafficking in neurodegeneration. Curr Biol 28, R471–R486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Asrat S, de Jesús DA, Hempstead AD, Ramabhadran V & Isberg RR (2014) Bacterial pathogen manipulation of host membrane trafficking. Annu Rev Cell Dev Biol 30, 79–109. [DOI] [PubMed] [Google Scholar]
- 135. Tzeng HT & Wang YC (2016) Rab‐mediated vesicle trafficking in cancer. J Biomed Sci 23, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Müller MP & Goody RS (2018) Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases 9, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sivars U, Aivazian D & Pfeffer SR (2003) Yip3 catalyses the dissociation of endosomal Rab–GDI complexes. Nature 425, 856–859. [DOI] [PubMed] [Google Scholar]
- 138. Shinde SR & Maddika S (2018) Post translational modifications of Rab GTPases. Small GTPases 9, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kapfhamer D, Valladares O, Sun Y, Nolan PM, Rux JJ, Arnold SE, Veasey SC & Bućan M (2002) Mutations in Rab3a alter circadian period and homeostatic response to sleep loss in the mouse. Nat Genet 32, 290–295. [DOI] [PubMed] [Google Scholar]
- 140. Geppert M, Bolshakov VY, Siegelbaum SA, Takei K, De Camilli P, Hammer RE & Südhof TC (1994) The role of Rab3A in neurotransmitter release. Nature 369, 493–497. [DOI] [PubMed] [Google Scholar]
- 141. Schlüter OM, Schmitz F, Jahn R, Rosenmund C & Südhof TC (2004) A complete genetic analysis of neuronal Rab3 function. J Neurosci 24, 6629–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Schlüter OM, Basu J, Südhof TC & Rosenmund C (2006) Rab3 superprimes synaptic vesicles for release: implications for short‐term synaptic plasticity. J Neurosci 26, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gilleron J, Bouget G, Ivanov S, Meziat C, Ceppo F, Vergoni B, Djedaini M, Soprani A, Dumas K, Jacquel A et al (2018) Rab4b deficiency in T cells promotes adipose Treg/Th17 imbalance, adipose tissue dysfunction, and insulin resistance. Cell Rep 25, 3329–3341. [DOI] [PubMed] [Google Scholar]
- 144. Bardin S, Miserey‐Lenkei S, Hurbain I, Garcia‐Castillo D, Raposo G & Goud B (2015) Phenotypic characterisation of RAB6A knockout mouse embryonic fibroblasts. Biol Cell 107, 427–439. [DOI] [PubMed] [Google Scholar]
- 145. Shafaq‐Zadah M, Gomes‐Santos CS, Bardin S, Maiuri P, Maurin M, Iranzo J, Gautreau A, Lamaze C, Caswell P, Goud B et al (2016) Persistent cell migration and adhesion rely on retrograde transport of β1 integrin. Nat Cell Biol 18, 54–64. [DOI] [PubMed] [Google Scholar]
- 146. Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer‐Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A et al (2003) Mutations in the small GTP‐ase late endosomal protein RAB7 cause Charcot‐Marie‐Tooth type 2B neuropathy. Am J Hum Genet 72, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kawamura N, Sun‐Wada GH, Aoyama M, Harada A, Takasuga S, Sasaki T & Wada Y (2012) Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat Commun 3, 1071. [DOI] [PubMed] [Google Scholar]
- 148. Sato T, Mushiake S, Kato Y, Sato K, Sato M, Takeda N, Ozono K, Miki K, Kubo Y, Tsuji A et al (2007) The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature 448, 366–369. [DOI] [PubMed] [Google Scholar]
- 149. Feng Q, Bonder EM, Engevik AC, Zhang L, Tyska MJ, Goldenring JR & Gao N (2017) Disruption of Rab8a and Rab11a causes formation of basolateral microvilli in neonatal enteropathy. J Cell Sci 130, 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yu S, Nie Y, Knowles B, Sakamori R, Stypulkowski E, Patel C, Das S, Douard V, Ferraris RP, Bonder EM et al (2014) TLR sorting by Rab11 endosomes maintains intestinal epithelial‐microbial homeostasis. EMBO J 33, 1882–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yu S, Yehia G, Wang J, Stypulkowski E, Sakamori R, Jiang P, Hernandez‐Enriquez B, Tran TS, Bonder EM, Guo W et al (2014) Global ablation of the mouse Rab11a gene impairs early embryogenesis and matrix metalloproteinase secretion. J Biol Chem 289, 32030–32043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sobajima T, Yoshimura SI, Iwano T, Kunii M, Watanabe M, Atik N, Mushiake S, Morii E, Koyama Y, Miyoshi E et al (2015) Rab11a is required for apical protein localisation in the intestine. Biol Open 4, 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lamers IJC, Reijnders MRF, Venselaar H, Kraus A, Jansen S, de Vries BBA, Houge G, Gradek GA, Seo J, Choi M et al (2017) Recurrent de novo mutations disturbing the GTP/GDP binding pocket of RAB11B cause intellectual disability and a distinctive brain phenotype. Am J Hum Genet 101, 824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bem D, Yoshimura SI, Nunes‐Bastos R, Bond FF, Kurian MA, Rahman F, Handley MTW, Hadzhiev Y, Masood I, Straatman‐Iwanowska AA et al (2011) Loss‐of‐function mutations in RAB18 cause Warburg micro syndrome. Am J Hum Genet 88, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Carpanini SM, McKie L, Thomson D, Wright AK, Gordon SL, Roche SL, Handley MT, Morrison H, Brownstein D, Wishart TM et al (2014) A novel mouse model of Warburg Micro syndrome reveals roles for RAB18 in eye development and organisation of the neuronal cytoskeleton. Dis Model Mech 7, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jenkins D, Seelow D, Jehee FS, Perlyn CA, Alonso LG, Bueno DF, Donnai D, Josifiova D, Mathijssen IMJ, Morton JEV et al (2007) RAB23 mutations in carpenter syndrome imply an unexpected role for Hedgehog signaling in cranial‐suture development and obesity. Am J Hum Genet 80, 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Eggenschwiler JT, Espinoza E & Anderson KV (2001) Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature 412, 194–198. [DOI] [PubMed] [Google Scholar]
- 158. Agler C, Nielsen DM, Urkasemsin G, Singleton A, Tonomura N, Sigurdsson S, Tang R, Linder K, Arepalli S, Hernandez D et al (2014) Canine hereditary ataxia in Old English Sheepdogs and Gordon Setters is associated with a defect in the autophagy gene encoding RAB24 . PLoS Genet 10, e1003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nam KT, Lee HJ, Smith JJ, Lapierre LA, Kamath VP, Chen X, Aronow BJ, Yeatman TJ, Bhartur SG, Calhoun BC et al (2010) Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. J Clin Invest 120, 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Jeong H, Lim KM, Kim KH, Cho Y, Lee B, Knowles BC, Roland JT, Zwerner JP, Goldenring JR & Nam KT (2019) Loss of Rab25 promotes the development of skin squamous cell carcinoma through the dysregulation of integrin trafficking. J Pathol 249, 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Dong W, He B, Qian H, Liu Q, Wang D, Li J, Wei Z, Wang Z, Xu Z, Wu G et al (2018) RAB26‐dependent autophagy protects adherens junctional integrity in acute lung injury. Autophagy 14, 1677–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Ménasché G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffraat N, Bianchi D, Fischer A, Le Deist F et al (2000) Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet 25, 173–176. [DOI] [PubMed] [Google Scholar]
- 163. Wilson SM, Yip R, Swing DA, O'Sullivan TN, Zhang Y, Novak EK, Swank RT, Russell LB, Copeland NG & Jenkins NA (2000) A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc Natl Acad Sci USA 97, 7933–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hume AN, Collinson LM, Rapak A, Gomes AQ, Hopkins CR & Seabra MC (2001) Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J Cell Biol 152, 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Stinchcombe JC, Barral DC, Mules EH, Booth S, Hume AN, Machesky LM, Seabra MC & Griffiths GM (2001) Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J Cell Biol 152, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bahadoran P, Aberdam E, Mantoux F, Buscà R, Bille K, Yalman N, De Saint‐Basile G, Casaroli‐Marano R, Ortonne JP & Ballotti R (2001) Rab27a: a key to melanosome transport in human melanocytes. J Cell Biol 152, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Tolmachova T, Abrink M, Futter CE, Authi KS & Seabra MC (2007) Rab27b regulates number and secretion of platelet dense granules. Proc Natl Acad Sci USA 104, 5872–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Mizuno K, Tolmachova T, Ushakov DS, Romao M, Åbrink M, Ferenczi MA, Raposo G & Seabra MC (2007) Rab27b regulates mast cell granule dynamics and secretion. Traffic 8, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Gomi H, Mori K, Itohara S & Izumi T (2007) Rab27b is expressed in a wide range of exocytic cells and involved in the delivery of secretory granules near the plasma membrane. Mol Biol Cell 18, 4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Roosing S, Rohrschneider K, Beryozkin A, Sharon D, Weisschuh N, Staller J, Kohl S, Zelinger L, Peters TA, Neveling K et al (2013) Mutations in RAB28, encoding a farnesylated small GTPase, are associated with autosomal‐recessive cone‐rod dystrophy. Am J Hum Genet 93, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Ying G, Boldt K, Ueffing M, Gerstner CD, Frederick JM & Baehr W (2018) The small GTPase RAB28 is required for phagocytosis of cone outer segments by the murine retinal pigmented epithelium. J Biol Chem 293, 17546–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Tucci A, Nalls MA, Houlden H, Revesz T, Singleton AB, Wood NW, Hardy J & Paisán‐Ruiz C (2010) Genetic variability at the PARK16 locus. Eur J Hum Genet 18, 1356–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kuwahara T, Inoue K, D'Agati VD, Fujimoto T, Eguchi T, Saha S, Wolozin B, Iwatsubo T & Abeliovich A (2016) LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci Rep 6, 29945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Alshammari MJ, Al‐Otaibi L & Alkuraya FS (2012) Mutation in RAB33B, which encodes a regulator of retrograde Golgi transport, defines a second Dyggve‐Melchior‐Clausen locus. J Med Genet 49, 455–461. [DOI] [PubMed] [Google Scholar]
- 175. Dupuis N, Lebon S, Kumar M, Drunat S, Graul‐Neumann LM, Gressens P & El Ghouzzi V (2013) A novel RAB33B mutation in Smith‐McCort dysplasia. Hum Mutat 34, 283–286. [DOI] [PubMed] [Google Scholar]
- 176. Wheeler DB, Zoncu R, Root DE, Sabatini DM & Sawyers CL (2015) Identification of an oncogenic RAB protein. Science 350, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA III & Pavan WJ (2002) Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci USA 99, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Oiso N, Riddle SR, Serikawa T, Kuramoto T & Spritz RA (2004) The rat Ruby (R) locus is Rab38: identical mutations in Fawn‐hooded and Tester‐Moriyama rats derived from an ancestral Long Evans rat sub‐strain. Mamm Genome 15, 307–314. [DOI] [PubMed] [Google Scholar]
- 179. Osanai K (2018) Rab38 mutation and the lung phenotype. Int J Mol Sci 19, 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, D'Elia E, Vecellio M, Russo S, Cogliati F, Larizza L et al (2010) Mutations in the small GTPase gene RAB39B are responsible for X‐linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet 86, 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wilson GR, Sim JCH, McLean C, Giannandrea M, Galea CA, Riseley JR, Stephenson SEM, Fitzpatrick E, Haas SA, Pope K et al (2014) Mutations in RAB39B cause X‐linked intellectual disability and early‐onset parkinson disease with α‐synuclein pathology. Am J Hum Genet 95, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Jiang Y, Sun Y, Hu J, Yu N, Liu H, Fan J, Ning X, Li Y, Liu B, Sun Y et al (2019) A germline mutation in Rab43 gene identified from a cancer family predisposes to a hereditary liver‐colon cancer syndrome. BMC Cancer 19, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Kretzer NM, Theisen DJ, Tussiwand R, Briseño CG, Grajales‐Reyes GE, Wu X, Durai V, Albring J, Bagadia P, Murphy TL et al (2016) RAB43 facilitates cross‐presentation of cell‐associated antigens by CD8α+ dendritic cells. J Exp Med 213, 2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Fitzgerald ML & Reed GL (1999) Rab6 is phosphorylated in thrombin‐activated platelets by a protein kinase C‐dependent mechanism: effects on GTP/GDP binding and cellular distribution. Biochem J 342, 353–360. [PMC free article] [PubMed] [Google Scholar]
- 185. Tzeng HT, Li TH, Tang YA, Tsai CH, Lu PJF, Lai WW, Chiang CW & Wang YC (2017) Phosphorylation of Rab37 by protein kinase C alpha inhibits the exocytosis function and metastasis suppression activity of Rab37. Oncotarget 8, 108556–108570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Pavarotti M, Capmany A, Vitale N, Colombo MI & Damiani MT (2012) Rab11 is phosphorylated by classical and novel protein kinase C isoenzymes upon sustained phorbol ester activation. Biol Cell 104, 102–115. [DOI] [PubMed] [Google Scholar]
- 187. Ong ST, Freeley M, Skubis‐Zegad J, Fazil MHUT, Kelleher D, Fresser F, Baier G, Verma NK & Long A (2014) Phosphorylation of Rab5a protein by protein kinase Cε is crucial for T‐cell migration. J Biol Chem 289, 19420–19434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Levin RS, Hertz NT, Burlingame AL, Shokat KM & Mukherjee S (2016) Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc Natl Acad Sci USA 113, E4776–E4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Sun L, Xu X, Chen Y, Zhou Y, Tan R, Qiu H, Jin L, Zhang W, Fan R, Hong W et al (2018) Rab34 regulates adhesion, migration, and invasion of breast cancer cells. Oncogene 37, 3698–3714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Amino acid sequences of Rabs/Ypts that were used for the phylogenetic analysis in Fig. 2A.
Fig. S2. Amino acid sequences of the switch II region of human Rabs that were used for the phylogenetic analysis in Fig. 2B.
Table S1. Rab effectors, binding molecules, GEFs, and GAPs in mammals
Table S2. Mouse and human Rab genes.


