Abstract
This paper briefly reviews the role of hypermethylation of host cell genes in cervical carcinogenesis and discusses potential clinical applications of methylation analysis in the management of high‐risk HPV (hrHPV) ‐positive women. We argue that methylation assays can be used: 1. for primary triage of hrHPV‐positive women to detect cervical cancer and advanced cervical intraepithelial neoplasia (CIN); 2. as secondary triage for women with minor cytological abnormalities to identify those with the highest risk of CIN3 or worse; 3. as exit test for women leaving the screening programme to identify cervical cancer and advanced CIN; and 4. to support management of CIN.
Tweetable abstract
This paper discusses potential clinical applications of DNA methylation analysis in the management of women with a high‐risk HPV infection.
Keywords: cervical cancer screening, cervical intraepithelial neoplasia, DNA methylation, human papillomavirus
Tweetable abstract
This paper discusses potential clinical applications of DNA methylation analysis in the management of women with a high‐risk HPV infection.
Introduction
Persistent infection with a high‐risk type of human papillomavirus (hrHPV) is a necessary cause for cervical cancer. 1 The development of invasive cervical cancer through premalignant lesions (cervical intraepithelial neoplasia [CIN] graded 1–3) is a slow process that can take up to 30 years from initial hrHPV infection, allowing detection and treatment of early‐stage disease. 2 Primary hrHPV DNA testing is currently preferred for cervical screening over cytology because of its superior sensitivity and high negative predictive value for high‐grade CIN or worse (CIN2+). 3 However, most hrHPV infections are productive infections, i.e. producing new virions, and do not give rise to cervical cancer, leading to a 3–5% lower specificity of the HPV test compared with cytology. 4 Therefore, a triage test is required to distinguish hrHPV‐positive women with clinically relevant cervical lesions from those with transient infections. New biomarkers, such as DNA methylation of specific host cell genes, are being evaluated as alternative triage methods to further improve risk stratification of hrHPV‐positive women.
Here we will briefly highlight the role of DNA methylation in cervical carcinogenesis and discuss the potential clinical applications of DNA methylation analysis in the detection and management of women with cervical neoplastic lesions. We will focus on methylation markers that have been clinically validated for the detection of cervical cancer, CIN3 and CIN2 in cervical scrapes and self‐collected cervico‐vaginal cells, and that are currently available within commercial or research methylation assays.
DNA methylation and cervical carcinogenesis
Cervical carcinogenesis is driven by the viral oncoproteins, E6 and E7. Their transcription and the HPV productive life cycle in differentiated epithelium relies on interactions with multiple cellular proteins and complex epigenetic remodelling of the viral chromatin, including both histone modifications and DNA methylation. 5 This epigenetic programme is disrupted during malignant transformation. The interaction of E6 and E7 with various epigenetic regulators, such as DNA methyltransferases, histone‐modifying enzymes and subunits of chromatin remodelling complexes, affect the transcription of host cell genes. 6
The best‐studied host cell modification in cervical cancer is DNA methylation, which involves the covalent binding of a methyl group to the 5′ position of a cytosine molecule in CpG dinucleotides. Besides global hypomethylation, the overall loss of methylation during carcinogenesis, resulting in chromosomal instability, 7 and the silencing of tumour suppressor genes by local hypermethylation of CpG‐rich promoter regions contribute to cancer development. 6 , 8 In vitro models have shown that methylation of (candidate) tumour suppressor genes occurs at the stage of immortalisation and increases with progression to a tumorigenic phenotype. 9 , 10 This progressive increase in DNA methylation seen in vitro is comparable to the observations in CIN and cervical cancer tissues, also demonstrating a gradual increase in methylation with progression to cancer (see Figure 1). 11 The functional relevance of methylation‐mediated gene silencing during HPV‐induced carcinogenesis has been demonstrated for a subset of currently known methylation targets, including miR124‐2, CADM1, MAL and PAX1. 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19
Figure 1.
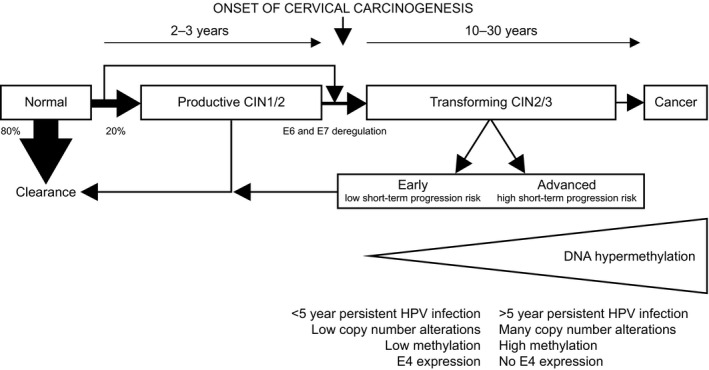
Schematic representation of the various outcomes of hrHPV infection of cervical epithelial cells and characteristics of early versus advanced transforming lesions. Most hrHPV infections are cleared by an effective immune response without causing cellular abnormalities (transient infections). Productive infections can give rise to productive cervical intraepithelial neoplasia (CIN, mainly CIN1 and a subset of CIN2), of which the majority regresses spontaneously within 1 or 2 years. Transforming infections are characterised by deregulated E6 and E7 expression and are associated with transforming CIN (the remaining subset of CIN2 and CIN3). Transforming CIN is a heterogeneous disease with variable duration of lesion existence and includes both progressive and regressive lesions. On routine haematoxylin & eosin‐stained sections these lesions cannot be distinguished. Current data support the division of transforming CIN lesions into early transforming lesions, characterised by a <5 year preceding HPV infection, few copy number alterations, low methylation levels and E4 expression and advanced transforming lesions, characterised by a ≥5 year preceding HPV infection, cancer‐like copy number alterations, high methylation levels and no E4 expression. As increases in genetic and epigenetic alterations are associated with disease progression, early transforming lesions are thought to have a low short‐term progression risk to cervical cancer, whereas advanced transforming lesions are thought to have a high short‐term progression risk. 8 , 47 , 50
Detection of DNA methylation
Detection of methylated host cell DNA can be performed by different molecular methods and on different sample types including cervical tissue, cervical scrapes, self‐collected cervico‐vaginal cells and even urine. 8 , 20 Most methods require a bisulphite conversion step, a chemical reaction of sodium bisulphite with DNA that converts unmethylated cytosines to uracil, whereas methylated cytosines remain unaffected. Quantitative methylation‐specific PCR is most commonly used to detect methylation of targeted sequences, as it requires a minimal amount of input DNA, is suitable for high‐throughput settings and is highly reproducible. Another sensitive method used for clinical application is pyrosequencing, a method providing an absolute level of methylation. 21
Available methylation assays
Although many methylation targets have been identified using both targeted and genome‐wide approaches, extensive validation for use in clinical practice and development into a commercial or research methylation assay has only been performed for some (Table 1). These include the marker panels CADM1 and MAL, later supplemented with miR124‐2 (PreCursor‐M®, Self‐Screen B.V., Amsterdam, the Netherlands), FAM19A4 and miR124‐2 (QIAsure Methylation Test®, Qiagen, Hilden, Germany), ASTN1, DLX1, ITG4, RXFP3, SOX17 and ZNF671 (GynTect®, Oncgnostics, Jena, Germany), EPB41L3 combined with viral target from four HPV types (HPV16L1, HPV16L2, HPV18L2, HPV31L1 and HPV33L2; S5 Classifier, research assay), POU4F3 (Confidence Marker™, Neumann Diagnostics, Budapest, Hungary) and PAX1 (Cervi‐M®, Ingenuity Healthcare, Mumbai, India). Other promising target genes include JAM3, C13ORF18, TERT, ANKRD18CP, CDH6, GFRA1, LHX8, PCDHA4, PCDHA13, SOX1 and ZNF582. 11 , 21 , 22 , 23 , 24
Table 1.
Overview of commercial or research host cell DNA methylation assays and their performance for the detection of CIN3 or worse in cervical scrapes or self‐collected specimens in different clinical settings
| Marker (panel) | Assay | Reference | HPV | Sample type | n | CIN3 (%) | ICC (%) | Sens. | Spec. | NPV | PPV | Comment | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Screening populations | |||||||||||||
| FAM19A4/miR124‐2 | QMSP | Bonde, subm. | HPV+ | Cervical scrapes | 2384 | 228 (9.6%) | 20 (0.8%) | 77.2 | 78.3 | 96.9 | 28.3 | End point CIN3; European screening populations | |
| FAM19A4/miR124‐2 | QMSP | [70] | HPV+ | Cervical scrapes | 979 | 109 (11.1%) | 6 (0.6%) | 71.3 | 78.3 | 94.8 | 33.0 | Cross‐sectional data | |
| — | — | 87.2 | — | 14‐year NPV | |||||||||
| FAM19A4/miR124‐2 | QMSP | [69] | HPV+ | Cervical scrapes | 1025 | 192 (18.7%) | 19 (1.9%) | — | — | 83.7 | 60.2 | 14‐year NPV and PPV | |
| CADM1/MAL | QMSP | [63] | HPV+ | Cervical scrapes | 234 | 38 (16.2%) | 3 (1.3%) | 68.4 | 75.5 | 92.5 | 35.1 | Consecutive series from screening cohort | |
| EPB41L3/HPV16L1/HPV16L2/HPV18L2/HPV31L1/HPV33L2* | PSQ | [34] | HPV+ | Cervical scrapes | 257 | 44 (17.1%) | 8 (3.1%) | 93.2 | 41.8 | — | 24.8 | Case–control within screening cohort; end point CIN3 | |
| EPB41L3/HPV16L1/HPV16L2/HPV18L2/HPV31L1/HPV33L2* | PSQ | [65] | HPV+ | Cervical scrapes | 341 | 18 (5.3%) | 1 (0.3%) | 84 | 63 | — | — | Selection of women from screening cohort; CIN2 (n = 20) lesions were excluded from analyses | |
| Non‐attendee population | |||||||||||||
| FAM19A4/miR124‐2 | QMSP | [28] | HPV+ | Self‐samples (brushes) | 254 | 68 (26.8%) | 4 (1.6%) | 69.4 | 76.4 | 86.3 | 53.8 | Performance data from validation set; brush‐ and lavage‐collected samples from (mostly) same patients | |
| Self‐samples (lavage) | 389 | 72 (18.5%) | 6 (1.5%) | 70.5 | 67.8 | 90.2 | 35.5 | ||||||
| CADM1/MAL | QMSP | [33] | HPV+ | Cervical scrapes | 364 | 56 (15.4%) | 6 (1.6%) | 69.4 | 71.2 | 91.9 | 33.1 | ||
| Gynaecological referral populations/Selection of archived material | |||||||||||||
| FAM19A4/miR124‐2 | QMSP | [30] | HPV+/− | Cervical scrapes | 538 | 66 (12.3%) | 6 (1.1%) | 77.8** | 69.3** | 98.3 ** | 36.4 ** | Women with abnormal cytology referred for colposcopy | |
| CADM1/MAL/miR124‐2 | QMSP | [27] | HPV+ | Cervical scrapes | 247 | 16 (6.5%) | 79 (32.0%) | 94.7 | 78.9 | — | — | Selection of archived material | |
| EPB41L3/HPV16L1/HPV16L2/HPV18L2/HPV31L1/HPV33L2** | PSQ | [68] | HPV+ | Cervical scrapes | 1493 | 373 (25.0%) | — | 90 | 49 | — | 51 | Women with abnormal cytology referred for colposcopy; end point CIN2/3 (n = 556) | |
| ASTN1/DLX1/ITGA4/RXFP3/SOX17/ZNF671 | QMSP | [37] | HPV+/− | Cervical scrapes | 280 | 50 (17.9%) | 5 (1.8%) | 64.8** | 94.6** | 91.7** | 74.5** | Residual samples from routine screening (NILM cytology) and from patients with CIN1+ on histology | |
| ASTN1/DLX1/ITGA4/RXFP3/SOX17/ZNF671 | QMSP | [39] | HPV+/− | Cervical scrapes | 306 | 88 (28.8%) | 5 (1.6%) | 67.7** | 82.6** | — | — | Women referred for colposcopy | |
| HPV+/− | 64.5 | 88.7 | — | — | |||||||||
| POU4F3 | QMSP | [40] | HPV+ | Cervical scrapes | 1287 | 65 (5.1%) | 12 (0.9%) | 89.6 | 60.9 | — | — | Mixed screening population and women referred for cone biopsy or hysterectomy; ICC cases included AIS and CIS | |
| PAX1 | QMSP | [44] | HPV+/− | Cervical scrapes | 449 | 87 (19.4%) | 71 (15.8%) | 69.6** | 81.8** | — | — | ||
| PAX1 | QMSP | [42] | HPV+/− | Cervical scrapes | 346 | 42 (12.1%) | 30 (8.7%) | 64** | 91** | — | — | Women with low‐ or high‐grade cytology referred for colposcopy and controls from routine screening (NILM cytology) | |
| HPV+/− | 57 | 96 | — | — | |||||||||
| PAX1 | QMSP | [43] | HPV+/− | Cervical scrapes | 419 | 32 (7.6%) | 4 (1.0%) | 86** | 85** | — | — | Mixed screening population and women referred with abnormal cytology | |
| PAX1 | QMSP | [45] | HPV+/− | Cervical scrapes | 321 | 24 (7.5%) | 1 (0.3%) | 52.0** | 90.3** | — | — | Women with ASC‐US cytology referred for colposcopy | |
AIS, adenocarcinoma in situ; ASC‐US, atypical squamous cells of unknown significance; CIN, cervical carcinoma in situ; CIS, cervical intraepithelial neoplasia, graded 1–3; HPV, human papillomavirus; ICC, invasive cervical cancer; n, samples size; NILM, negative for intraepithelial lesion or malignancy; NPV, negative predictive value; PPV, positive predictive value; PSQ, pyrosequencing; QMSP, quantitative methylation‐specific polymerase chain reaction; Sens., sensitivity; Spec., specificity.
Combination of cellular and viral targets.
Performance evaluated independent of HPV result.
Methylation in cervical cancer samples
Published data consistently show high methylation levels of most markers in nearly all cervical cancer specimens (tissue, scrapes or self‐samples), resulting in high accuracy for cervical cancer. A recent study evaluating a world‐wide series of over 500 cases of cervical cancer, including rare histotypes and HPV‐negative carcinomas, showed that 98.3% tested positive for FAM19A4/miR124‐2 methylation. 25 Methylation positivity was independent of histotype, International Federation of Obstetrics and Gynecology stage, HPV status and genotype, sample type and geographical region. This high cross‐sectional sensitivity for cervical cancer was also shown among hrHPV‐positive women from a Dutch cervical screening cohort. 26 Smaller series of cervical carcinomas were also evaluated for FAM19A4 alone, EPB41L3 (with and without viral markers), CADM1/MAL/miR124‐2, ASTN1/DLX1/ITGA4/RXFP3/SOX17/ZNF671 and POU4F3, all showing detection rates well over 90%. 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 For PAX1, positivity rates of at least 80% have been reported. 41 , 42 , 43 , 44 , 45 , 46
Methylation in CIN lesions
Host cell DNA methylation analysis of cervical samples is a promising tool to identify hrHPV‐positive women with clinically relevant cervical lesions. Methylation levels of promoter regions of specific host cell genes increase with increasing CIN grade and are extremely high in cervical cancer. As genetic and epigenetic alterations necessary for cervical cancer progression accumulate over time, high methylation levels in these regions are likely to be associated with advanced cervical lesions with a longer duration of lesion existence. 8 In support of this hypothesis is the finding that CIN2/3 lesions associated with a long‐term hrHPV infection (≥5 years), used as a proxy for duration of lesion existence, and a DNA copy number profile similar to cervical cancer had significantly higher methylation levels compared with CIN2/3 lesions associated with a short‐term hrHPV infection (<5 years) and few to no copy number alterations. 47 , 48 , 49 Additional arguments were provided by van Zummeren et al., who showed that CIN lesions expressing the HPV E4 protein, indicating a productive HPV infection, were associated with low methylation levels, and Louvanto et al., who showed that high methylation levels are predictive of progression of untreated CIN2 lesions in women up to age 30 years. 50 , 51 Taken together, these available data argue that methylation assays detect advanced CIN lesions in need of treatment.
Use of methylation analysis in cervical screening
Primary triage of hrHPV‐positive women
Cervical screening programmes based on primary hrHPV testing require triage of hrHPV‐positive women, but consensus on the optimal triage test is currently lacking. Triage strategies require a good balance between safety, i.e. high sensitivity for cervical cancer and CIN3 (CIN3+), and screening‐related burden, i.e. minimal over‐referral of women without clinically relevant cervical lesions. Although the optimal balance varies between regions depending on locally accepted risk thresholds, available resources and population characteristics, the following aspects are generally required for triage tests within cervical screening programmes: 1. high reproducibility; 2. high cross‐sectional sensitivity for CIN3+, leading to a high negative predictive value (~98%); 3. high specificity, leading to high positive predictive value of at least 20% (the Netherlands) or 5–10% (USA) and 4. long‐term safety: longitudinal data evaluating the long‐term CIN3+ risk of triage‐negative women are required to determine appropriate screening intervals with a chosen triage strategy. 52
Methods for triage of hrHPV‐positive women as used in current cervical screening programmes include cytology, HPV16/18 genotyping and repeat hrHPV testing. Although these triage strategies have acceptable clinical performance, there are some important drawbacks. Cytology triage cannot be performed on self‐collected specimens and its use is limited by its subjective nature, leading to suboptimal sensitivity and the need for repeat testing. Additional staining of cytology slides for p16INK4a and ki‐67 has been shown to mostly improve the specificity for CIN3+. 53 , 54 However, as with sole cytology, interpretation remains observer‐dependent and validation of this triage strategy is still ongoing. Triage by HPV16/18 genotyping or repeat hrHPV testing has the benefit of objective test results, but both methods have to be combined with cytology to have sufficient clinical performance. 55 A good alternative method for triage of hrHPV‐positive women is methylation analysis, with major benefits including high reproducibility, objectivity and applicability on both clinician‐collected and self‐collected cervical specimens. 28 , 56 These characteristics can facilitate full molecular screening with highly reproducible results.
Cross‐sectional performance of methylation markers for primary triage of hrHPV‐positive women
The cross‐sectional clinical performance of different methylation markers for the detection of CIN3+ has been studied using samples from different settings: cervical screening, screening non‐attendees and gynaecological referral populations (Table 1). Data from individual markers included in these assays are shown in the supplementary material (Table S1). As negative and positive predictive values for CIN3+ are influenced by the composition of the population tested, we will focus on the sensitivity and specificity of the different markers. CIN2 is not taken as an end point because it reflects a heterogeneous group of lesions and the diagnosis is moderately reproducible. 57 , 58 It should be noted that only marker panel FAM19A4/miR124‐2 has been evaluated in large, population‐based cervical screening populations, and that most studies used either smaller, selected series from screening populations, or samples from gynaecological referral populations (indicated in Table 1 and the Supplementary material, Table S1).
The performance of FAM19A4 and/or miR124‐2 methylation has been studied extensively, showing consistently good sensitivity and specificity for the detection of CIN3+ in both clinician‐collected cervical scrapes (sensitivity 68.2–86.7%, specificity 60.6–79.3%) and self‐collected specimens (sensitivity 65.3–70.5%, specificity 67.8–81.3%). 28 , 29 , 49 , 59 , 60 , 61 , 62 Methylation analysis of CADM1 and/or MAL, and later with the addition of miR124‐2, has shown good performance for the detection of CIN3+ in cervical scrapes (sensitivity 68.0–94.7%, specificity 50.7–78.9%) and self‐collected specimens (sensitivity 64.9–71.6%, specificity 55.3–70.0%). 27 , 33 , 61 , 63 , 64 Good sensitivity has also been described for EPB41L3, particularly when combined with methylation analysis of defined HPV genes of the L1 and L2 region. Depending on the setting and the inclusion of viral methylation markers, CIN3 or CIN3+ sensitivities ranging from 67.0 to 93.2% and specificities ranging from 41.8 to 85.0% have been reported. 34 , 35 , 36 , 65 , 66 , 67 , 68 In a small series of self‐collected samples from a non‐attendee population, EPB41L3 showed good sensitivity and specificity for CIN3+. 36 The five‐gene marker panel consisting of DLX1, ITGA4, RXFP3, SOX17 and ZNF671, later adjusted with the addition of a sixth marker ASTN1, has been evaluated for the detection of CIN3+ among women visiting gynaecological outpatient departments (with or without hrHPV testing), showing sensitivities and specificities ranging from 64.5 to 76.2% and 76.0 to 94.6%, respectively. 37 , 38 , 39 POU4F3 has been evaluated in a mixed screening and referral population for triage of hrHPV‐positive women, showing CIN3+ sensitivity of 89.6% and specificity of 60.9%. 40 Although PAX1 has been evaluated in several Asian referral populations, only one study reported its performance as a triage marker for the detection of CIN3+ (sensitivity 57%, specificity of 96%), whereas the others reported the performance of the marker without previous hrHPV testing (sensitivity 44.1–86.0%, specificity 81.8–96.0%). 41 , 42 , 43 , 44 , 45 , 46
Longitudinal performance of methylation markers for primary triage of hrHPV‐positive women
Longitudinal CIN3+ risk data after a negative triage test are needed to determine the safety of the chosen screening interval. Longitudinal data on the performance of methylation triage of hrHPV‐positive women is only available for the marker panel FAM19A4/miR124‐2. Long‐term follow‐up data from a Dutch cervical screening cohort showed that the cervical cancer risk among hrHPV‐positive women after 14 years is lower following a negative FAM19A4/miR124‐2 methylation result compared with a negative cytology result and that the CIN3+ risk is similar following a negative methylation or cytology result. 26 , 69 This lower long‐term CIN3+ risk after a negative FAM19A4/miR124‐2 methylation test was confirmed in another study from the Netherlands. 70
Secondary triage for CIN3 or worse of women with ASC‐US/LSIL cytology
Current HPV‐based cervical screening programmes often use cytology for triage of hrHPV‐positive women, referring all women with abnormal cytology (≥atypical squamous cells of unknown significance [ASC‐US]) at baseline or after 6 months for colposcopy. Although this triage strategy significantly improves the specificity of the primary hrHPV test, over‐referral of women without clinically relevant lesions remains an issue, particularly among young women with low‐grade cytological abnormalities (ASC‐US or low‐grade squamous intraepithelial lesion [LSIL]). The latest monitoring report from the Dutch HPV‐based national screening programme illustrates this issue of over‐referral: 21% of hrHPV‐positive women (aged ≥30 years) had ASC‐US or LSIL cytology at baseline and at least 48% of all hrHPV‐positive women did not have clinically relevant disease (CIN1 or less). 71
Recent data from the VALID‐screen study, a multicentre study for the validation of FAM19A4/miR124‐2 methylation in European countries, indicate that the referral rate could be lowered by approximately 34% by secondary triage of hrHPV‐positive women with ASC‐US/LSIL cytology using FAM19A4/miR124‐2 methylation, while still detecting all cervical carcinomas and at least 70% of CIN3 lesions (Bonde et al., submitted). These data suggest that FAM19A4/miR124‐2 methylation analysis could be used to distinguish among women with ASC‐US/LSIL cytology those who require immediate colposcopy referral versus re‐testing for hrHPV and methylation within 3 years follow up in HPV‐based as well as in cytology‐based screening programmes. Similar results have been published for PAX1, although without prior hrHPV testing, detecting at least 50% of CIN3 and all cervical carcinomas among women with minor cytological abnormalities. 43 , 45
Exit test for women leaving the screening programme
A significant proportion of incident cervical cancers occur in elderly women outside the screening age. Because of the decreased balance between efficiency and harms, most countries screen women up to age 60 or 65 years. In this age‐group, the relatively low specificity of the hrHPV test is less problematic because of the lower HPV prevalence and fewer consequences of overtreatment. At the same time, some cervical cancers may be missed by hrHPV testing or by cytology triage testing, because of a higher frequency of hrHPV‐negative lesions and lower sensitivity of cytology testing at an older age. 72 , 73 With the high sensitivity of methylation analysis for cervical carcinomas, it would be an interesting option to combine hrHPV testing with methylation analysis at the final screening round, referring all women who test positive for either test for gynaecological examination (co‐testing). Vink et al. 25 showed that 90% of hrHPV‐negative cervical carcinomas were positive for FAM19A4/miR124‐2 methylation, suggesting that such a strategy may lead to earlier detection of women with cervical cancer and advanced CIN lesions.
Management of hrHPV‐positive women of childbearing age
Women with a positive cervical screening result, whether performed within a population‐based screening programme or because of specific complaints, should be referred for colposcopy. Clinical management of these women is based on the CIN grade of a colposcopy‐directed biopsy. All CIN3 and most CIN2 lesions are treated surgically, whereas CIN1 lesions are generally managed with a watch‐and‐wait policy. Conservative management can also be considered in young women with CIN2, as spontaneous regression rates are high: up to 60% of CIN2 and 32% of CIN3 lesions regress spontaneously. 74 , 75 , 76 Approximately 5% of untreated CIN2 and 12–31% of untreated CIN3 lesions ultimately progress to cancer, depending on the duration of lesion existence. 2 , 74 , 76 Current histopathological assessment of cervical biopsies cannot distinguish lesions with a low risk of progression to cervical cancer from those with a high progression risk. This leads to overtreatment and associated morbidity, most notably an increased risk of adverse pregnancy outcomes in subsequent pregnancies. 77 , 78 , 79 As most women with CIN2 and CIN3 lesions are of reproductive age, biomarkers to improve risk stratification and prevent overtreatment are needed. Several potential biomarkers have been evaluated, such as HPV genotyping, HPV viral load and immunohistochemical staining of cervical biopsies by p16ink4a and Ki‐67, but none of these markers has been shown to have clinical prognostic value or to be able to predict progression at the individual patient level. 80 Methylation analysis has emerged as a promising prognostic marker because of its specific sensitivity for advanced CIN lesions and cervical cancer.
The first data from a prospective longitudinal study on the value of methylation analysis in predicting the outcome of CIN lesions were recently published by Louvanto et al. 52 In this study, women under the age of 30 years and diagnosed with a CIN2 lesion were managed conservatively with 6‐monthly follow‐up visits up to 2 years. High methylation of host cell marker EPB41L3 combined with viral methylation markers (S5 classifier) were associated with progression to CIN3. A similar study is ongoing in the Netherlands, evaluating the value of FAM19A4/miR124‐2 methylation in the prediction of regression or non‐regression in untreated women. 81 This study also includes women with CIN3 and women aged up to 55 years. If their prognostic value is confirmed, methylation assays could be used to guide clinical management of women diagnosed with a CIN lesion.
Use of methylation analysis for cervical screening in women living with HIV
The disease burden of both cervical cancer and HIV is disproportionally high in low‐ and middle‐income countries: >85% of global cervical cancer cases and >95% of all HIV infections occur in these regions. 82 , 83 Mainly because of HIV‐related immunodeficiency, women living with HIV are at increased risk for HPV infection, CIN lesions and cervical cancer. 84 , 85 , 86 Cervical screening is extremely relevant in women living with HIV, but because of poverty, lack of resources and infrastructure, and limited access to health care, implementation of organised screening programmes in low‐ and middle‐income countries has proven to be challenging. 87 Full molecular screening using methylation analysis could be used to improve cervical screening in low‐ and middle‐income countries, and women living with HIV in particular.
Studies on host cell methylation analysis in cervical scrapes of women living with HIV show promising results. The clinical performance of host cell methylation analysis for the detection of CIN3+ in these women has only been evaluated for some markers, including FAM19A4/miR124‐2, EPB41L3 and CADM1/MAL/miR124‐2. 88 , 89 , 90 , 91 Similar to HIV‐uninfected women, methylation levels increase with cervical disease severity and are extremely high in cervical cancer in women living with HIV. 88 , 89 , 90 , 92 These data suggest that also in women living with HIV, high methylation levels are associated with clinically relevant cervical lesions and a high cervical cancer progression risk.
FAM19A4/miR124‐2 methylation analysis was evaluated in cervical scrapes from a South African screening cohort of women living with HIV, showing good sensitivity and specificity for the detection of CIN3+ when used as a triage test of hrHPV‐positive women (sensitivity 72.9%, specificity 76.1%). 91 As women living with HIV have a high CIN3+ risk, direct referral of HP16/18‐positive women and triage of only those with a non‐16/18 hrHPV infection with FAM19A4/miR124‐2 methylation analysis is an interesting option to limit the number of tests performed while sensitivity and specificity are maintained (79.7% and 74.8%, respectively). The marker panel CADM1/MAL/miR124‐2 was evaluated in this same cohort and a Kenyan cohort of women living with HIV, showing similar clinical performance when used as a triage marker (sensitivity 73.8% and 72%, specificity 81.5% and 70%, respectively). 88 , 89 EPB41L3 was evaluated in cervical scrapes of women living with HIV from South Africa and Burkina Faso who were included in a prospective cervical screening study, showing a specificity of 66.2% at the threshold corresponding to 69.7% sensitivity for CIN2+, independent of hrHPV status. 90
Future directions
Currently available data justify further validation of methylation triage testing on cervical scrapes and self‐collected cervico‐vaginal specimens of hrHPV‐positive women in prospective implementation studies within cervical screening programmes in both high‐ and low‐resource settings, including a health‐economic evaluation of costs and benefits. Once similar data are available for methylation triage on self‐collected material and, potentially, urine, full molecular screening without the need for a visit to a clinician can be envisioned. 28 , 61 Given its high sensitivity for invasive cervical cancer and advanced CIN lesions, methylation analysis may eventually be considered as a primary cervical screening tool, either with or without hrHPV testing. Currently, methylation assays can be used following a positive HPV test, but are not yet available as point‐of‐care tests. However, the development of high‐throughput variants will change this. Future development of methylation assays into point‐of‐care tests would be ideal for low‐resource settings in order to facilitate same‐day screening and treatment and minimise loss to follow up.
Disclosure of interests
RDMS, DAMH and CJLMM are minority shareholders of Self‐screen B.V., a spin‐off company of VUmc, which develops, manufactures and licences the high‐risk HPV assay and methylation marker assays for cervical cancer screening and holds patents on these tests. CJLMM is part‐time CEO of Self‐Screen, has stock in Qiagen and MDXHealth, has received speakers fees from GSK, Qiagen and SPMSD/Merck, and served occasionally on the scientific advisory boards (expert meeting) of these companies. DAMH has been on the speakers bureau of Qiagen and serves occasionally on the scientific advisory boards of Pfizer and Bristol‐Myers Squibb. WWK and GGK declare no conflicts of interests. Completed disclosure of interests forms are available to view online as supporting information.
Contribution to authorship
WWK was responsible for the literature search and data acquisition. Data interpretation and writing of initial draft was by WWK and CJLMM. Manuscript writing and approval was by WWK, RDMS, DAMH, GGK and CJLMM.
Details of ethics approval
Not applicable.
Funding
CJLMM was in part supported by the European Research Council (ERC Advanced Masscare 322986).
Supporting information
Table S1. Overview of individual and combinations of host cell methylation markers included in commercial or research assays and their performance for the detection of CIN3 or worse in cervical scrapes or self‐collected specimens in different clinical settings.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Kremer WW, Steenbergen RDM, Heideman DAM, Kenter GG, Meijer CJLM. The use of host cell DNA methylation analysis in the detection and management of women with advanced cervical intraepithelial neoplasia: a review. BJOG 2021; 128:504–514.
Linked article: This article is commented on by SJ Bowden et al., p. 515 in this issue. To view this mini commentary visit https://doi.org/10.1111/1471-0528.16448.
References
- 1. zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002;2:342–50. [DOI] [PubMed] [Google Scholar]
- 2. Vink MA, Bogaards JA, van Kemenade FJ, de Melker HE, Meijer CJ, Berkhof J. Clinical progression of high‐grade cervical intraepithelial neoplasia: estimating the time to preclinical cervical cancer from doubly censored national registry data. Am J Epidemiol 2013;178:1161–9. [DOI] [PubMed] [Google Scholar]
- 3. Ronco G, Meijer CJ, Segnan N, Kitchener H, Giorgi‐Rossi P, Peto J, et al. HPV‐based screening for prevention of invasive cervical cancer – Authors' reply. Lancet 2014;383:1295. [DOI] [PubMed] [Google Scholar]
- 4. Koliopoulos G, Arbyn M, Martin‐Hirsch P, Kyrgiou M, Prendiville W, Paraskevaidis E. Diagnostic accuracy of human papillomavirus testing in primary cervical screening: a systematic review and meta‐analysis of non‐randomized studies. Gynecol Oncol 2007;104:232–46. [DOI] [PubMed] [Google Scholar]
- 5. Burley M, Roberts S, Parish JL. Epigenetic regulation of human papillomavirus transcription in the productive virus life cycle. Semin Immunopathol 2020;42:159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res 2017;772:36–50. [DOI] [PubMed] [Google Scholar]
- 7. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics 2009;1:239–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer 2014;14:395–405. [DOI] [PubMed] [Google Scholar]
- 9. Henken FE, Wilting SM, Overmeer RM, van Rietschoten JG, Nygren AO, Errami A, et al. Sequential gene promoter methylation during HPV‐induced cervical carcinogenesis. Br J Cancer 2007;97:1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schutze DM, Kooter JM, Wilting SM, Meijer CJ, Quint W, Snijders PJ, et al. Longitudinal assessment of DNA methylation changes during HPVE6E7‐induced immortalization of primary keratinocytes. Epigenetics 2015;10:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state‐of‐the‐science. Gynecol Oncol 2009;112:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szalmas A, Konya J. Epigenetic alterations in cervical carcinogenesis. Semin Cancer Biol 2009;19:144–52. [DOI] [PubMed] [Google Scholar]
- 13. Overmeer RM, Henken FE, Snijders PJ, Claassen‐Kramer D, Berkhof J, Helmerhorst TJ, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high‐grade CIN and cervical SCC. J Pathol 2008;215:388–97. [DOI] [PubMed] [Google Scholar]
- 14. Overmeer RM, Henken FE, Bierkens M, Wilting SM, Timmerman I, Meijer CJ, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol 2009;219:327–36. [DOI] [PubMed] [Google Scholar]
- 15. Wilting SM, Miok V, Jaspers A, Boon D, Sorgard H, Lando M, et al. Aberrant methylation‐mediated silencing of microRNAs contributes to HPV‐induced anchorage independence. Oncotarget 2016;7:43805–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation‐mediated silencing and tumour suppressive function of hsa‐miR‐124 in cervical cancer. Mol Cancer 2010;9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lando M, Fjeldbo CS, Wilting SM, Snoek BC, Aarnes E‐K, Forsberg MF, et al. Interplay between promoter methylation and chromosomal loss in gene silencing at 3p11‐p14 in cervical cancer. Epigenetics 2015;10:970–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer 2008;123:161–7. [DOI] [PubMed] [Google Scholar]
- 19. Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI, et al. Paired Box‐1 (PAX1) activates multiple phosphatases and inhibits kinase cascades in cervical cancer. Sci Rep 2019;9:9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snoek BC, Splunter APV, Bleeker MCG, Ruiten MCV, Heideman DAM, Rurup WF, et al. Cervical cancer detection by DNA methylation analysis in urine. Sci Rep 2019;9:3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorincz AT. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol 2016;60:501–12. [DOI] [PubMed] [Google Scholar]
- 22. Luttmer R, De Strooper LM, Steenbergen RD, Berkhof J, Snijders PJ, Heideman DA, et al. Management of high‐risk HPV‐positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn 2016;16:961–74. [DOI] [PubMed] [Google Scholar]
- 23. Kelly H, Benavente Y, Pavon MA, De Sanjose S, Mayaud P, Lorincz AT. Performance of DNA methylation assays for detection of high‐grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta‐analysis. Br J Cancer 2019;121:954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li N, He Y, Mi P, Hu Y. ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: a meta‐analysis of related studies in Chinese population. Medicine 2019;98:e14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vink FJ, Meijer C, Clifford GM, Poljak M, Ostrbenk A, Petry KU, et al. FAM19A4/miR124‐2 methylation in invasive cervical cancer: a retrospective cross‐sectional worldwide study. Int J Cancer 2019;147:1215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Strooper LMA, Berkhof J, Steenbergen RDM, Lissenberg‐Witte BI, Snijders PJF, Meijer C, et al. Cervical cancer risk in HPV‐positive women after a negative FAM19A4/mir124‐2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow‐up. Int J Cancer 2018:143:1541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Strooper LM, van Zummeren M, Steenbergen RD, Bleeker MC, Hesselink AT, Wisman GB, et al. CADM1, MAL and miR124‐2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol 2014;67:1067–71. [DOI] [PubMed] [Google Scholar]
- 28. De Strooper LM, Verhoef VM, Berkhof J, Hesselink AT, de Bruin HM, van Kemenade FJ, et al. Validation of the FAM19A4/mir124‐2 DNA methylation test for both lavage‐ and brush‐based self‐samples to detect cervical (pre)cancer in HPV‐positive women. Gynecol Oncol 2016;141:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bu Q, Wang S, Ma J, Zhou X, Hu G, Deng H, et al. The clinical significance of FAM19A4 methylation in high‐risk HPV‐positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer 2018;18:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leeman A, Del Pino M, Marimon L, Torne A, Ordi J, Ter Harmsel B, et al. Reliable identification of women with CIN3+ using hrHPV genotyping and methylation markers in a cytology‐screened referral population. Int J Cancer 2019;144:160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hesselink AT, Heideman DA, Steenbergen RD, Gok M, van Kemenade FJ, Wilting SM, et al. Methylation marker analysis of self‐sampled cervico‐vaginal lavage specimens to triage high‐risk HPV‐positive women for colposcopy. Int J Cancer 2014;135:880–6. [DOI] [PubMed] [Google Scholar]
- 32. Verhoef VM, Heideman DA, van Kemenade FJ, Rozendaal L, Bosgraaf RP, Hesselink AT, et al. Methylation marker analysis and HPV16/18 genotyping in high‐risk HPV positive self‐sampled specimens to identify women with high grade CIN or cervical cancer. Gynecol Oncol 2014;135:58–63. [DOI] [PubMed] [Google Scholar]
- 33. Verhoef VM, van Kemenade FJ, Rozendaal L, Heideman DA, Bosgraaf RP, Hesselink AT, et al. Follow‐up of high‐risk HPV positive women by combined cytology and bi‐marker CADM1/MAL methylation analysis on cervical scrapes. Gynecol Oncol 2015;137:55–9. [DOI] [PubMed] [Google Scholar]
- 34. Cook DA, Krajden M, Brentnall AR, Gondara L, Chan T, Law JH, et al. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer 2018;144:2587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boers A, Wang R, van Leeuwen RW, Klip HG, de Bock GH, Hollema H, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigenetics 2016;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boers A, Bosgraaf RP, van Leeuwen RW, Schuuring E, Heideman DA, Massuger LF, et al. DNA methylation analysis in self‐sampled brush material as a triage test in hrHPV‐positive women. Br J Cancer 2014;111:1095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitz M, Eichelkraut K, Schmidt D, Zeiser I, Hilal Z, Tettenborn Z, et al. Performance of a DNA methylation marker panel using liquid‐based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 2018;18:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansel A, Steinbach D, Greinke C, Schmitz M, Eiselt J, Scheungraber C, et al. A promising DNA methylation signature for the triage of high‐risk human papillomavirus DNA‐positive women. PLoS One 2014;9:e91905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitz M, Wunsch K, Hoyer H, Scheungraber C, Runnebaum IB, Hansel A, et al. Performance of a methylation specific real‐time PCR assay as a triage test for HPV‐positive women. Clin Epigenetics 2017;9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kocsis A, Takacs T, Jeney C, Schaff Z, Koiss R, Jaray B, et al. Performance of a new HPV and biomarker assay in the management of hrHPV positive women: Subanalysis of the ongoing multicenter TRACE clinical trial (n > 6000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. Int J Cancer 2017;140:1119–33. [DOI] [PubMed] [Google Scholar]
- 41. Huang TH, Lai HC, Liu HW, Lin CJ, Wang KH, Ding DC, et al. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer 2010;20:513–9. [DOI] [PubMed] [Google Scholar]
- 42. Lai HC, Ou YC, Chen TC, Huang HJ, Cheng YM, Chen CH, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a Taiwanese Gynecologic Oncology Group (TGOG) study. Cancer Med 2014;3:1062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kan YY, Liou YL, Wang HJ, Chen CY, Sung LC, Chang CF, et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer 2014;24:928–34. [DOI] [PubMed] [Google Scholar]
- 44. Liou YL, Zhang TL, Yan T, Yeh CT, Kang YN, Cao L, et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics 2016;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin CJ, Lai HC, Wang KH, Hsiung CA, Liu HW, Ding DC, et al. Testing for methylated PCDH10 or WT1 is superior to the HPV test in detecting severe neoplasms (CIN3 or greater) in the triage of ASC‐US smear results. Am J Obstet Gynecol 2011;204;21 e1‐7. [DOI] [PubMed] [Google Scholar]
- 46. Xu J, Xu L, Yang B, Wang L, Lin X, Tu H. Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. Int J Clin Exp Pathol 2015;8:1674–81. [PMC free article] [PubMed] [Google Scholar]
- 47. Bierkens M, Hesselink AT, Meijer CJ, Heideman DA, Wisman GB, van der Zee AG, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–9. [DOI] [PubMed] [Google Scholar]
- 48. Bierkens M, Wilting SM, van Wieringen WN, van Kemenade FJ, Bleeker MC, Jordanova ES, et al. Chromosomal profiles of high‐grade cervical intraepithelial neoplasia relate to duration of preceding high‐risk human papillomavirus infection. Int J Cancer 2012;131:E579–E585. [DOI] [PubMed] [Google Scholar]
- 49. De Strooper LM, Meijer CJ, Berkhof J, Hesselink AT, Snijders PJ, Steenbergen RD, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014;7:1251–7. [DOI] [PubMed] [Google Scholar]
- 50. Zummeren MV, Kremer WW, Leeman A, Bleeker MCG, Jenkins D, Sandt MV, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124‐2 genes in cervical cancer and precursor lesions. Mod Pathol 2018;31:1842–50. [DOI] [PubMed] [Google Scholar]
- 51. Louvanto K, Aro K, Nedjai B, Butzow R, Jakobsson M, Kalliala I, et al. Methylation in predicting progression of untreated high‐grade cervical intraepithelial neoplasia. Clin Infect Dis 2019;70:2582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Polman NJ, de Haan Y, Veldhuijzen NJ, Heideman DAM, de Vet HCW, Meijer C, et al. Experience with HPV self‐sampling and clinician‐based sampling in women attending routine cervical screening in the Netherlands. Prev Med 2019;125:5–11. [DOI] [PubMed] [Google Scholar]
- 53. Wentzensen N, Clarke MA, Bremer R, Poitras N, Tokugawa D, Goldhoff PE, et al. Clinical evaluation of human papillomavirus screening with p16/Ki‐67 dual stain triage in a large organized cervical cancer screening program. JAMA Int Med 2019;179:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luttmer R, Dijkstra MG, Snijders PJ, Berkhof J, van Kemenade FJ, Rozendaal L, et al. p16/Ki‐67 dual‐stained cytology for detecting cervical (pre)cancer in a HPV‐positive gynecologic outpatient population. Mod Pathol 2016;29:870–8. [DOI] [PubMed] [Google Scholar]
- 55. Polman NJ, Snijders PJF, Kenter GG, Berkhof J, Meijer C. HPV‐based cervical screening: rationale, expectations and future perspectives of the new Dutch screening programme. Prev Med 2019;119:108–17. [DOI] [PubMed] [Google Scholar]
- 56. Floore A, Hesselink A, Ostrbenk A, Alcaniz E, Rothe B, Pedersen H, et al. Intra‐ and inter‐laboratory agreement of the FAM19A4/mir124‐2 methylation test: results from an international study. J Clin Lab Anal 2019;33:e22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Darragh TM, Colgan TJ, Cox JT, Heller DS, Henry MR, Luff RD, et al. The lower anogenital squamous terminology standardization project for HPV‐associated Lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis 2012;16:205–42. [DOI] [PubMed] [Google Scholar]
- 58. Herrington CS. The terminology of pre‐invasive cervical lesions in the UK cervical screening programme. Cytopathology 2015;26:346–50. [DOI] [PubMed] [Google Scholar]
- 59. Luttmer R, De Strooper LM, Berkhof J, Snijders PJ, Dijkstra MG, Uijterwaal MH, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). Int J Cancer 2016;138:992–1002. [DOI] [PubMed] [Google Scholar]
- 60. Luttmer R, De Strooper LM, Dijkstra MG, Berkhof J, Snijders PJ, Steenbergen RD, et al. FAM19A4 methylation analysis in self‐samples compared with cervical scrapes for detecting cervical (pre)cancer in HPV‐positive women. Br J Cancer 2016;115:579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dankai W, Khunamornpong S, Siriaunkgul S, Soongkhaw A, Janpanao A, Utaipat U, et al. Role of genomic DNA methylation in detection of cytologic and histologic abnormalities in high risk HPV‐infected women. PLoS One 2019;14:e0210289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rogeri CD, Silveira HCS, Causin RL, Villa LL, Stein MD, de Carvalho AC, et al. Methylation of the hsa‐miR‐124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol Oncol 2018;150:545–51. [DOI] [PubMed] [Google Scholar]
- 63. De Strooper LM, Hesselink AT, Berkhof J, Meijer CJ, Snijders PJ, Steenbergen RD, et al. Combined CADM1/MAL methylation and cytology testing for colposcopy triage of high‐risk HPV‐positive women. Cancer Epidemiol Biomarkers Prev 2014;23:1933–7. [DOI] [PubMed] [Google Scholar]
- 64. Vasiljevic N, Scibior‐Bentkowska D, Brentnall AR, Cuzick J, Lorincz AT. Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high‐risk HPV positive women. Gynecol Oncol 2014;132:709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lorincz AT, Brentnall AR, Scibior‐Bentkowska D, Reuter C, Banwait R, Cadman L, et al. Validation of a DNA methylation HPV triage classifier in a screening sample. Int J Cancer 2016;138:2745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Leeuwen RW, Ostrbenk A, Poljak M, van der Zee AGJ, Schuuring E, Wisman GBA. DNA methylation markers as a triage test for identification of cervical lesions in a high risk human papillomavirus positive screening cohort. Int J Cancer 2019;144:746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eijsink JJ, Lendvai A, Deregowski V, Klip HG, Verpooten G, Dehaspe L, et al. A four‐gene methylation marker panel as triage test in high‐risk human papillomavirus positive patients. Int J Cancer 2012;130:1861–9. [DOI] [PubMed] [Google Scholar]
- 68. Brentnall AR, Vasiljevic N, Scibior‐Bentkowska D, Cadman L, Austin J, Cuzick J, et al. HPV33 DNA methylation measurement improves cervical pre‐cancer risk estimation of an HPV16, HPV18, HPV31 and EPB41L3 methylation classifier. Cancer Biomark 2015;15:669–75. [DOI] [PubMed] [Google Scholar]
- 69. Dick S, Kremer WW, De Strooper LMA, Lissenberg‐Witte BI, Steenbergen RDM, Meijer C, et al. Long‐term CIN3+ risk of HPV positive women after triage with FAM19A4/miR124‐2 methylation analysis. Gynecol Oncol 2019;154:368–73. [DOI] [PubMed] [Google Scholar]
- 70. Vink FJ, Lissenberg‐Witte BI, Meijer CJLM, Berkhof J, van Kemenade FJ, Siebers AG, et al. FAM19A4/miR124‐2 methylation analysis as a triage test for HPV‐positive women: cross‐sectional and longitudinal data from a Dutch screening cohort [published online ahead of print, 2020 Mar 25]. Clin Microbiol Infect 2020. 10.1016/j.cmi.2020.03.018 [DOI] [PubMed] [Google Scholar]
- 71. Landelijke Monitor Bevolkingsonderzoek Baarmoederhalskanker . Erasmus MC, PALGA; 2017. https://www.rivm.nl/documenten/landelijke‐evaluatie‐van‐bevolkingsonderzoek‐baarmoederhalskanker‐leba‐tm‐2017 (in Dutch).
- 72. Gustafsson L, Sparen P, Gustafsson M, Pettersson B, Wilander E, Bergstrom R, et al. Low efficiency of cytologic screening for cancer in situ of the cervix in older women. Int J Cancer 1995;63:804–9. [DOI] [PubMed] [Google Scholar]
- 73. Aro K, Nieminen P, Louvanto K, Jakobsson M, Virtanen S, Lehtinen M, et al. Age‐specific HPV type distribution in high‐grade cervical disease in screened and unvaccinated women. Gynecol Oncol 2019;154:354–9. [DOI] [PubMed] [Google Scholar]
- 74. Ostor AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol 1993;12:186–92. [PubMed] [Google Scholar]
- 75. Tainio K, Athanasiou A, Tikkinen KAO, Aaltonen R, Cardenas J, Hernandes, et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: systematic review and meta‐analysis. BMJ 2018;360:k499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McCredie MRE, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol 2008;9:425–34. [DOI] [PubMed] [Google Scholar]
- 77. Kyrgiou M, Mitra A, Arbyn M, Paraskevaidi M, Athanasiou A, Martin‐Hirsch PP, et al. Fertility and early pregnancy outcomes after conservative treatment for cervical intraepithelial neoplasia. Cochrane Database Syst Rev 2015;9:CD008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kyrgiou M, Athanasiou A, Paraskevaidi M, Mitra A, Kalliala I, Martin‐Hirsch P, et al. Adverse obstetric outcomes after local treatment for cervical preinvasive and early invasive disease according to cone depth: systematic review and meta‐analysis. BMJ 2016;354:i3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Arbyn M, Kyrgiou M, Simoens C, Raifu AO, Koliopoulos G, Martin‐Hirsch P, et al. Perinatal mortality and other severe adverse pregnancy outcomes associated with treatment of cervical intraepithelial neoplasia: meta‐analysis. BMJ 2008;337:a1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koeneman MM, Kruitwagen RF, Nijman HW, Slangen BF, Van Gorp T, Kruse AJ. Natural history of high‐grade cervical intraepithelial neoplasia: a review of prognostic biomarkers. Expert Rev Mol Diagn 2015;15:527–46. [DOI] [PubMed] [Google Scholar]
- 81. Kremer WW, Berkhof J, Bleeker MC, Heideman DA, van Trommel NE, van Baal MW, et al. Role of FAM19A4/miR124‐2 methylation analysis in predicting regression or non‐regression of CIN2/3 lesions: a protocol of an observational longitudinal cohort study. BMJ Open 2019;9:e029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 83. Prevention gap report. UNAIDS; 2016. [Google Scholar]
- 84. De Vuyst H, Lillo F, Broutet N, Smith JS. HIV, human papillomavirus, and cervical neoplasia and cancer in the era of highly active antiretroviral therapy. Eur J Cancer Prev 2008;17:545–54. [DOI] [PubMed] [Google Scholar]
- 85. Sun XW, Kuhn L, Ellerbrock TV, Chiasson MA, Bush TJ, Wright TC Jr. Human papillomavirus infection in women infected with the human immunodeficiency virus. N Engl J Med 1997;337:1343–9. [DOI] [PubMed] [Google Scholar]
- 86. Chaturvedi AK, Madeleine MM, Biggar RJ, Engels EA. Risk of human papillomavirus‐associated cancers among persons with AIDS. J Natl Cancer Inst 2009;101:1120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Denny L, Quinn M, Sankaranarayanan R. Chapter 8: Screening for cervical cancer in developing countries. Vaccine 2006;8:S71–7. [DOI] [PubMed] [Google Scholar]
- 88. De Vuyst H, Franceschi S, Plummer M, Mugo NR, Sakr SR, Meijer CJ, et al. Methylation levels of CADM1, MAL, and MIR124‐2 in cervical scrapes for triage of HIV‐infected, high‐risk HPV‐positive women in Kenya. J Acquir Immune Defic Syndr 2015;70:311–8. [DOI] [PubMed] [Google Scholar]
- 89. Van Zummeren M, Kremer WW, Van Aardt MC, Breytenbach E, Richter KL, Rozendaal L, et al. Selection of women at risk for cervical cancer in an HIV‐infected South African population. AIDS 2017;31:1945–53. [DOI] [PubMed] [Google Scholar]
- 90. Kelly HA, Chikandiwa A, Warman R, Segondy M, Sawadogo B, Vasiljevic N, et al. Associations of human gene EPB41L3 DNA methylation and cervical intraepithelial neoplasia in women living with HIV‐1 in Africa. AIDS 2018;32:2227–36. [DOI] [PubMed] [Google Scholar]
- 91. Kremer WW, van Zummeren M, Breytenbach E, Richter KL, Steenbergen RDM, Meijer C, et al. The use of molecular markers for cervical screening of women living with HIV in South Africa. AIDS 2019;33:2035–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kremer WW, Van Zummeren M, Novianti PW, Richter KL, Verlaat W, Snijders PJ, et al. Detection of hypermethylated genes as markers for cervical screening in women living with HIV. J Int AIDS Soc 2018;21:e25165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Overview of individual and combinations of host cell methylation markers included in commercial or research assays and their performance for the detection of CIN3 or worse in cervical scrapes or self‐collected specimens in different clinical settings.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material


