Abstract
Breast cancer (BC) is the second leading cause of cancer-associated deaths among women worldwide. Increasing evidence has indicated that microRNAs (miRNAs) have demonstrated great potential for improving the diagnosis and therapy for BC. In the present study, miRNA-155 was detected in human BC tissues using reverse transcription-quantitative (RT-q)PCR. RT-qPCR and western blot assays were used to analyze the levels of transforming growth factor β receptor type II (TGFBR2) in human BC tissues. MCF-7 cells were cultured and treated with miR-155 inhibitor and an MTT assay was performed to determine the role of miR-155 on the proliferation of MCF-7 cells. Subsequently, TGFBR2 and epithelial-mesenchymal transition (EMT)-associated molecules were analyzed using RT-qPCR and western blot assays. The direct binding of miR-155 to TGFBR2 was validated using a dual luciferase assay. Higher levels of miR-155 and lower levels of TGFBR2 were expressed in human BC tissues compared with paired normal tissues. Furthermore, the expression levels of miR-155 were associated with the tumor size, TNM stage and metastasis status of BC. Transfection of MCF-7 cells with miR-155 inhibitors resulted in reduced cell proliferation and suppressed the EMT process, characterized by upregulated expression of the epithelial markers, E-cadherin and CK18, and downregulated expression of mesenchymal markers, fibronectin and smooth muscle actin α. Transfection of a miR-155 inhibitor also resulted in increased expression of TGFBR2, and miR-155 may have regulated TGFBR2 through direct binding to the 3'untranslated region of TGFBR2 as determined using a dual-luciferase assay. Based on the results of the present study, miR-155 may serve as a novel diagnostic biomarker and therapeutic target for patients with BC.
Keywords: breast cancer, miR-155, proliferation, epithelial-mesenchymal-transition, MCF-7
Introduction
Breast cancer (BC) is the second leading cause of cancer-associated deaths amongst women worldwide (1,2) with >450,000 BC-associated deaths each year in the world according to the statistical analysis from World Cancer Research Fund (3). China is one of the countries with the top 3 highest incidence of BC worldwide (4). Despite the wide use of curative surgery and adjuvant chemotherapy for patients with BC, distant metastasis and tumor recurrence remains a major issue for improving BC prognosis (2,5). Therefore, understanding the pathological mechanisms underlying BC development and progression and identifying the specific targets, may provide an effective strategy for diagnosis and therapy for treating patients with BC.
Metastasis accounts for 90% of all BC-associated deaths, during which epithelial-mesenchymal transition (EMT) is a pivotal process which allows cancer progression and metastasis (2,6). EMT involves a transformational change of epithelial cells to mesenchymal cells characterized by loss of adhesion, invasive and metastatic properties and acquisition of a cancer stem cell phenotype (2,7). Tumor cells undergoing EMT demonstrate gain of mesenchymal markers such as fibronectin (FN) and smooth muscle actin α (α-SMA) (5,8,9), and a downregulation of epithelial markers, such as E-cadherin and CK18 (9-11).
MicroRNAs (miRNAs/miRs) are small (18-22 nucleotides) single-stranded non-coding RNAs that regulate gene expression at the post-transcriptional level by recognizing specific target mRNAs resulting in their targeted degradation or inhibition of translation (12,13). Various miRNAs have been demonstrated to serve varying roles in numerous tumor progressing biological behaviors, including proliferation, apoptosis, differentiation, migration, invasion, metastasis and angiogenesis (14-16). miRNAs are involved in the occurrence and development of cancer, functioning either as tumor suppressors or oncogenes (17,18). Abnormal expression of miRNAs has been demonstrated in a number of different types of cancer and is closely associated with cancer progression (17,18). Previous studies have shown that the levels of miR-155 were significantly increased in BC tissue, and this was positively associated with the occurrence and metastasis of BC, suggesting the oncogenic function of miR-155 (19,20). However, the mechanisms underlying miR-155 function are not completely understood, and it is unknown whether miR-155 functions partially by mediating EMT.
Transforming growth factor β receptor type II (TGFBR2) is a crucial signaling in tumor epithelial cells. Lower levels of TGFBR2 expression have been detected in BC tissue and the reduced expression of TGFBR2 is associated with adverse pathological characteristics and poor prognosis of patients with BC (21,22). TGFBR2 acts as bona fide metastasis suppressor gene (23). TGFBR2 pathways regulate various processes like EMT, angiogenesis and immunomodulation (24). To the best of our knowledge, a direct interaction between miR-155 and TGFBR2 has not been previously demonstrated.
The aim of the present study was to assess the expression levels of miR-155 and TGFBR2 in BC tissue, determine whether miR-155 directly interacted with TGFBR2, and examine the effects of miR-155 on proliferation and invasion of MCF-7 cells. The present study may improve the understanding of the role of miR-155 in the pathological mechanisms underlying development and progression of BC.
Materials and methods
Tissue samples
A total of 30 pairs of BC and corresponding normal tissue were collected from patients who underwent surgery in Xinganmeng People's Hospital (Inner Mongolia, China) from January 1st 2016 to April 30th, 2017 and all the specimens were confirmed using routine pathological examinations and staged according to the tumor-node-metastasis (TNM) system (25,26). The patient (female) age range was 41-68 (median age=59). Radiotherapy and chemotherapy were not performed prior to surgery. Written informed consent was obtained from all the patients and the experiments were approved by The Ethics Committee of Xinganmeng People's Hospital (Inner Mongolia, China). Clinicopathological characteristics of patients collected, included age, sex, tumor size, pathological stage, lymph node metastasis, ER status, PR status and HER2 status. The characteristics and prevalence of each are listed in Table I.
Table I.
Expression of miR-155 in patients with breast cancer.
| Clinicopathological characteristic | n | miR-155 High | miR-155 Low | P-value |
|---|---|---|---|---|
| Age, years | 0.4642 | |||
| ≥50 | 16 | 7 | 9 | |
| <50 | 14 | 8 | 6 | |
| Tumor size, cm | 0.0303a | |||
| <3 | 13 | 4 | 9 | |
| ≥3 | 17 | 12 | 5 | |
| TNM stage | 0.0253a | |||
| I-II | 18 | 6 | 12 | |
| III-IV | 12 | 9 | 3 | |
| Lymph node metastasis | 0.0295a | |||
| No | 11 | 3 | 8 | |
| Yes | 19 | 13 | 6 | |
| Estrogen receptor status | 0.7125 | |||
| Positive | 15 | 8 | 7 | |
| Negative | 15 | 9 | 6 | |
| Progesterone receptor status | 0.7851 | |||
| Positive | 17 | 7 | 10 | |
| Negative | 13 | 6 | 7 | |
| Human epidermal growth factor receptor 2 status | 0.6956 | |||
| Positive | 16 | 8 | 8 | |
| Negative | 14 | 8 | 6 |
aP<0.05. TNM, tumor-node-metastasis stage.
Cell culture and treatment
MCF-7 BC cells were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific Inc.) and 1% (w/v) penicillin/streptomycin (Sigma-Aldrich; Merck KGaA) at 37˚C in a 5% CO2 incubator. Cells were passaged at 80-90% confluence, and the cells in the logarithmic growth phase were used for further experiments.
MCF-7 cells were divided into three groups according to the treatment as follows: No treatment (control group), miR-155 mimics group, miR-155 inhibitor (inhibitor group) and miR-155 negative control (NC) group. Cells were transfected with miR-155 mimics (5'-UUAAUGCUAAUCGUGAUAGGGG-3') miR-NC mimics (5'-CAGUACUUUUGUGUAGUACAA-3'), miR-155 inhibitor (5'-CCCCUAUCACGAUUAGCAUUAA-3') or NC (5'-CAGUACUUUUGUGUAGUACAA-3') (Guangzhou RiboBio Co., Ltd.) using Lipofectamine™ 2000 according to the manufacturer's protocols. The expression of miR-155 in each group was detected by reverse transcription quantitative (RT-q)PCR 48 h after transfection.
Cell proliferation assay
Cells in each group were seeded into a 96-well culture plate at a density of 1x105 cells/well and cultured for 12, 24 or 48 h. Cell proliferation was assessed using an MTT assay (Bio-Rad Laboratories, Inc.). According to the manufacturer's protocol, 10 µl MTT reagent (5 mg/ml) was added to each well. After incubation for 4 h at 37˚C, the medium was discarded and the cells were incubated with 150 µl DMSO (Bio-Rad Laboratories, Inc.). The plates were shaken on a microvibrator for 10 min and the optical density of each well was measured using an microplate reader (Bio-Rad Laboratories, Inc.) at 490 nm. The results are expressed as a percentage of the untreated control.
RT-qPCR
Total RNA was extracted from cells (48 h post-transfection) or tissue using TRIzol® reagent (Thermo Fisher Scientific, Inc.), and subsequently transcribed into cDNA using PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan) at 42˚C for 15 min. qPCR was performed using SYBR® Premix Ex Taq™ kit (Takara Bio, Inc.) on an ABI 7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with customized primer sets for TGFBR2, CK18, E-cadherin, FN and α-SMA. The relative expression of mRNA in each sample was normalized to the level of GAPDH using the 2-ΔΔCq method (27). The expression of miR-155 was examined using the Hairpin-it™ miRNAs qPCR Quantitation kit (Shanghai GenePharma Co., Ltd.) according to manufacturer's protocol, and U6 was used as an internal control for normalization. Primers were synthesized by GenScript and the sequences are listed in Table II.
Table II.
Sequences of the primers.
| Gene | Primer sequence |
|---|---|
| microRNA-155 | |
| Forward | 5'-UAAUACCGUCUUAAAACCGU-3' |
| Reverse | 5'-UUCUGGGAACGUGAAACCT-3' |
| U6 | |
| Forward | 5'-CTCGCTTCGGCAGCACA-3' |
| Reverse | 5'-AACGCTTCACGAATTTGCGT-3' |
| Transforming growth factor β receptor type II | |
| Forward | 5'-TCTGGGCTCCTGATTGCT-3' |
| Reverse | 5'-TGAGGCAGCTTTGTAAGT-3' |
| E-Cadherin | |
| Forward | 5'-GTGGCCCGGATGTGAGAAG-3' |
| Reverse | 5'-GGAGCCCTTGTCGGATGATG-3' |
| CK18 | |
| Forward | 5'-AAGAAAACCCGAAGAGG-3' |
| Reverse | 5'-CTGACTCAAGGTGCAGC-3' |
| Fibronectin | |
| Forward | 5'-TTGTTCGGTGGAGTAGACCC-3' |
| Reverse | 5'-GTGCCAGTGGTCTCTTGTTG-3' |
| Smooth muscle actin-α | |
| Forward | 5'-TTCCTTCGTGACTACTGCTGAG-3' |
| Reverse | 5'-CAATGAAAGATGGCTGGAAGAG-3' |
| GAPDH | |
| Forward | 5'-GCACCACCAACTGCTTAGC-3' |
| Reverse | 5'-GGCATGGACTGTGGTCATGAG-3' |
Western blotting
Cells (48 h post-transfection) from each group or tissue samples were collected and lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology), Shanghai, China) on ice. After centrifugation, the supernatant was obtained and the protein concentration was measured using a Pierce® BCA Protein assay kit (Thermo Fisher Scientific, Inc.). The protein extracts were mixed with loading buffer, and denatured in a boiling water bath for 10 min. Equivalent quantities 20 µg of protein were loaded onto a 10% SDS gel, resolved using SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with tris-buffered saline Tween 20 (TBST) containing 5% skimmed milk and subsequently incubated with primary antibodies against human TGFBR2 (cat. no. ab186838), CK18 (cat. no. ab133263), E-cadherin (cat. no. ab256580), FN (cat. no. ab2413), α-SMA (cat. no. ab32575; 1:200 for anti-CK18 antibody, 1:100 for all other antibodies) at 4˚C overnight. The membranes were washed and then incubated with Horseradish Peroxidase (HRP)-conjugated secondary antibodies (cat. no. ab6721; 1:5,000) at room temperature for 60 min. All the antibodies were purchased from Abcam. Target proteins were detected using Novex® ECL Chemiluminescent Substrate Reagent kit (Thermo Fisher Scientific, Inc.) using a ChemiDoc™ XRS+ imaging system (Bio-Rad Laboratories, Inc.). The density of protein bands was analyzed using Gel-Pro analyzer software 4.0 (Media Cybernetics, Inc.) and the data are expressed as the ratio of each target protein to the internal control, GAPDH (cat. no. ab181602, 1:10,000; Abcam) at 4˚C overnight.
Dual luciferase activity assay
The target of miR-155 was predicted by the online database Targetscan 7.2 [http://www.targetscan.org/vert_72/ (28)]. The 3'UTR of TGFBR2 containing the predicted miR-155 specific binding site was amplified by PCR from genomic DNA, and inserted into the pMIR-REPORT™ luciferase reporter vector (Thermo Fisher Scientific, Inc.) to obtain the wild-type luciferase reporter plasmid p-TGFBR2-wt. PCR was performed using the following thermocycling conditions: 94˚C for 2 min, followed by 35 cycles of 94˚C for 2 sec, 60˚C for 60 sec and 72˚C for 1 min. The PCR products were amplified using cDNA and fused to the firefly luciferase gene of the pGL3-control plasmid (Promega Corporation) with the restriction enzyme sites of KpnI and XhoI. Two site mutations were introduced to WT-TGFBR2-3'-UTR to construct the mutant (MUT) TGFBR2-3'-UTR using a Quick Site-directed mutation kit (Agilent Technologies, Inc.). The cells were co-transfected with pGL3 constructions including 200 ng pGL3-WT-TGFBR2 and 200 ng pGL3-Mut-TGFBR2, 10 nM miR-NC or 10 nM miR-155 mimics and 26 ng pRL-TK in 24-well plates using Lipofectamine® 2000 (Invitrogen, USA). At 24 h of transfection, luciferase activity (firefly and Renilla) was determined using the dual-luciferase reporter assay system (Promega, USA).
Statistical analysis
All data were analyzed using SPSS version 20.0 (IBM, Corp.). Data are presented as the mean ± standard deviation SD. Statistical significance between groups was analyzed using ANOVA analysis and a Bonferroni post-hoc test. A χ2 test was used to analyze the association between miR-155 expression and clinicopathological characteristics. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical features
As showed in Table I, overexpression of miR-155 was significantly associated with larger breast tumor sizes, TNM stage and lymph node metastasis. However, there was no significant difference between ages, ER status, PR status and HER2 status between the patients with high or low miR-155 expression levels.
miR-155 expression is increased in human BC tissue and is negatively associated with TGFBR2 expression
The levels of miR-151 and TGFBR2 were detected in all 30 pairs of human BC tissues and adjacent normal controls. RT-qPCR data showed that the levels of miR-151 were significantly increased in BC tissues compared with their paired normal tissues (Fig. 1A; P<0.01). Furthermore, human BC tissues exhibited a significant reduction in TGFBR2 expression, both at the mRNA and protein levels compared with the normal control tissues (Fig. 1A and B; P<0.01). These data are consistent with the previous studies, and suggested an inverse association between the expression levels of miR-155 and its target TGFBR2.
Figure 1.
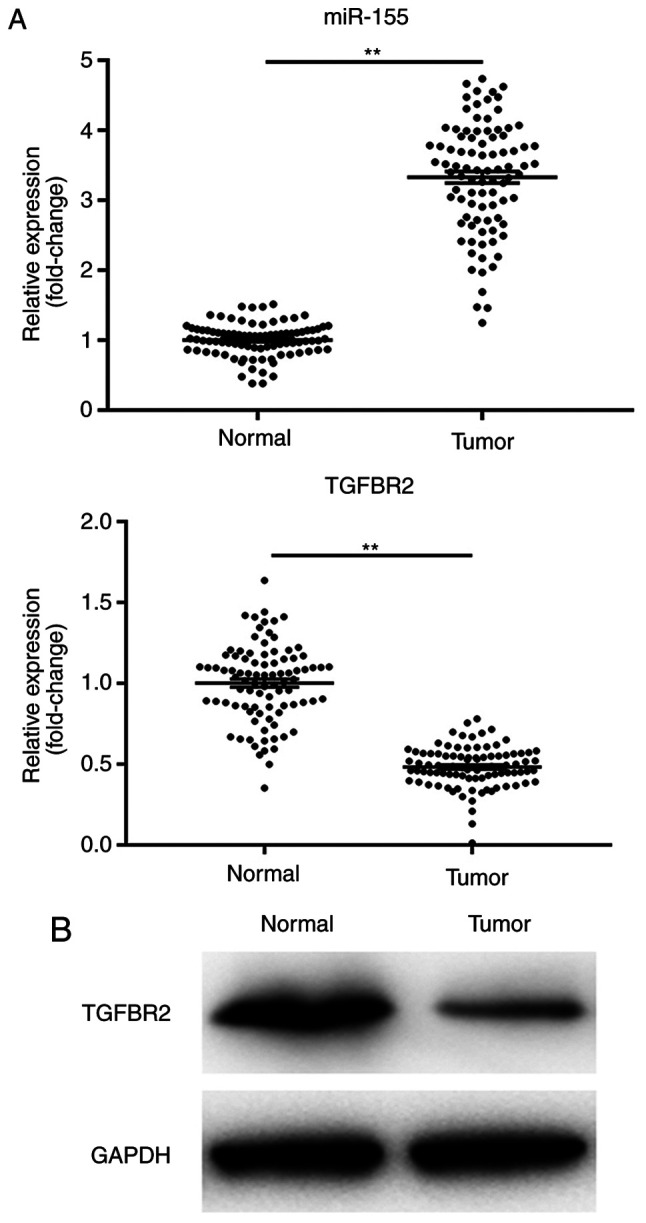
miR-155 expression is upregulated in BC tissue samples and is negatively associated with TGFBR2 expression. (A) Reverse transcription-quantitative PCR analysis of miR-155 and TGFBR2 mRNA expression levels in human breast cancer tissue and normal controls samples. Results are expressed relative to the value of normal controls that were assigned a value of 1. The line indicates the mean expression values. **P<0.01. (B) Representative western blots showing the expression of TGFBR2 protein in human BC tissues and normal controls. GAPDH was used as the loading control. BC, breast cancer; TGFBR2, transforming growth factor β receptor type II; miR-155, microRNA-155.
miR-155 inhibitor downregulates the expression of miR-155
MCF-7 cells were transfected with miR-155 NC inhibitor or miR-155 inhibitor. As shown in Fig. 2, the expression levels of miR-155 in cells transfected with miR-155 inhibitor was significantly lower compared with the control and NC group (P<0.01).
Figure 2.
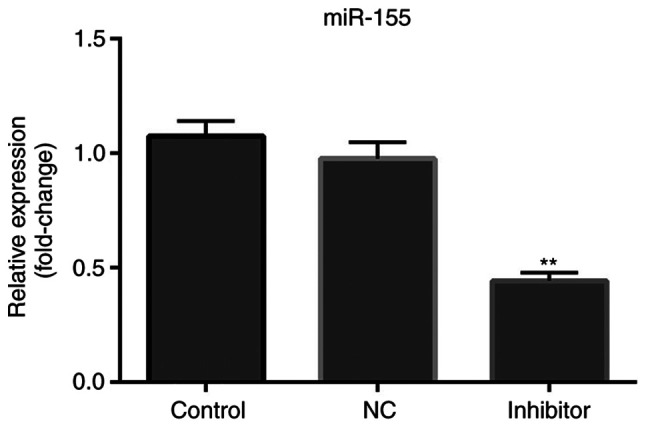
miR-155 inhibitor reduces the expression of miR-155. Expression of miR-155 was significantly decreased following transfection of miR-155 inhibitor compared with the control and NC groups. **P<0.01 vs. the control group. miR-155, microRNA-155.
miR-155 inhibitor suppresses proliferation of MCF-7 cells
miR-155 inhibitor or miR-155 NC inhibitor were transfected into MCF-7 cells and MTT assays were performed to assess the effects of miR-155 on cell proliferation. As shown in Fig. 3, miR-155 levels were lower in the miR-155 inhibitor transfected cells, compared with the NC-transfected or control cells. miR-155 inhibitor resulted in a slight decrease in cell proliferation 12 and 24 h after transfection (P>0.05), and a significant reduction in proliferation after 48 h (P<0.01).
Figure 3.
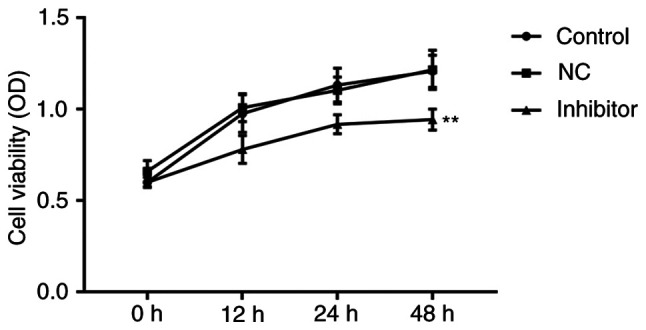
miR-155 inhibitor suppresses the proliferation of MCF-7 cells. An MTT assay was used to examine cell proliferation in each group (control, NC, miR-155 inhibitor). Data represent the optical density measured at 490 nm, and are presented as the mean ± standard deviation of three separate experiments. **P<0.01 vs. the control group. miR-155, microRNA-155.
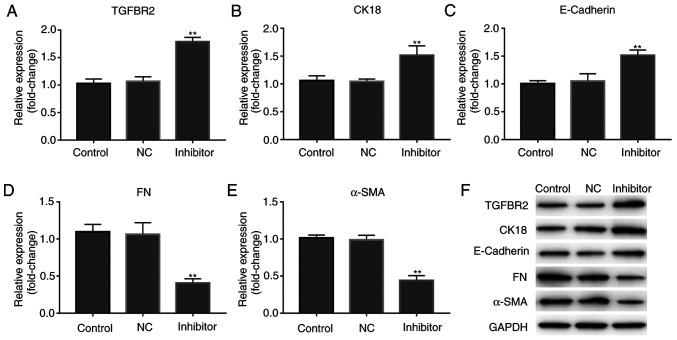
miR-155 inhibitor enhances the expression of TGFBR2 and mediates the expression of EMT-associated molecules
As there was an inverse association between miR-155 and TGFBR2 expression levels in human BC tissues, MCF-7 cells were transfected with miR-155 inhibitor, and the expression of TGFBR2 was shown to be increased. As shown in Fig. 4A and F, miR-155 inhibitor resulted in a significant increase in TGFBR2 expression, both at the mRNA and protein expression levels (P<0.01).
Figure 4.
miR-155 inhibitor enhances the expression of TGFBR2 and mediates the expression of EMT-related molecules. Reverse transcription-quantitative PCR analysis of the effect of miR-155 inhibitor on the expression of (A) TGFBR2, (B) CK18, (C) E-cadherin, (D) FN and (E) α-SMA. Results are expressed relative to the value of the controls that were assigned a value of 1. **P<0.01 vs. the control group. (F) Representative western blots showing the expression of TGFBR2, CK18, E-cadherin, FN and α-SMA in each cell group. GAPDH was used as the loading control. miR-155, microRNA-155; TGFBR2, transforming growth factor β receptor type II; EMT, epithelial-mesenchymal transition; FN, fibronectin; SMA-α, smooth muscle actin α.
Subsequently, whether miR-155 mediated expression of EMT-associated markers was assessed. As shown in Fig. 4B-F, miR-155 inhibitor resulted in alterations in the expression of epithelial markers, including a significant increase in E-cadherin and CK18 expression levels, and significant decrease in the expression of the mesenchymal markers, FN and α-SMA in MCF-7 cells. These data suggest that miR-155 inhibitor suppressed the metastatic properties of MCF-7 cells.
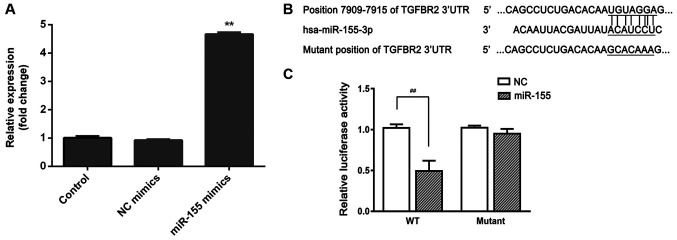
TGFBR2 is a direct target of miR-155
To confirm whether the observed reduction in TGFBR2 mRNA and protein levels was the result of direct binding between miR-155 and the 3'UTR of TGFBR2, first, cells were transfected miR-155 mimics or its negative control. As shown in Fig. 5A, and the expression of miR-155 was significantly increased in cells transfected with miR-155 mimics, suggesting successful transfection. Subsequently, the segment of TGFBR2 3'UTR containing the putative binding site for miR-155 was cloned and inserted into a firefly luciferase reporter vector to obtain p-TGFBR2-wt or p-TGFBR2-mut (Fig. 5B). When co-transfected in HEK-293T cells, miR-155 significantly repressed the luciferase activity of p-TGFBR2-wt, whereas luciferase expression was significantly higher in cells transfected with p-TGFBR2-mut and miR-155 (Fig. 5C). These data suggest that miR-155 binds directly binds to TGFBR2 3'UTR and thereby reduced mRNA and protein expression levels of TGFBR2.
Figure 5.
TGFBR2 is a direct target of miR-155. (A) Reverse transcription-quantitative PCR was performed to determine the expression levels of miR-155 in miR-155 mimics transfected cells. **P<0.01 vs. the control group. (B) Sequence alignment showing the relative position of the miR-155 binding site in the 3'UTR of TGFBR2 and the mutated nucleotides are indicated. The sequences were used to construct luciferase reporter plasmids. (C) Each reporter construct (p-TGFBR2-wt or p-TGFBR2-mut) was co-transfected with either miR-155 mimic or NC in 293T cells and a dual luciferase assay was performed after 24 h. Luciferase activity was normalized to Renilla and presented as relative to NC which was arbitrarily set at 1. Data are presented as the mean ± standard deviation of three independent experiments. ##P<0.01. miR-155, microRNA-155; TGFBR2, transforming growth factor β receptor type II; UTR, untranslated region; NC negative control; NC: miR-NC mimics.
Discussion
miRNAs exhibit notable potential for use in the diagnosis and treatment of BC (29,30). miR-155 has been reported to be strongly associated with BC progression (16,17). Combining previous studies (16,17) with the results of the present study, it was confirmed that the levels of miR-155 were significantly increased in human BC tissues compared with paired normal controls. These data suggest that miR-155 may function as an oncogene.
To determine the roles of miR-155 in the pathogenesis of BC, in vitro studies using MCF-7 human BC cells were performed. Cell proliferation and metastasis are essential events accounting for tumor progression. The results of the present study data showed that transfection of a miR-155 inhibitor resulted in reduced proliferation and survival of MCF-7 cells. miR-155 has also been shown to increase BC metastasis in vivo in previous studies (19,20). Thus, whether miR-155 modulated expression of EMT markers in MCF7 cells was assessed, as EMT is a major cause of tumor metastasis. A decrease in the expression of the mesenchymal markers (FN and α-SMA), and an increase in the expression epithelial markers (E-cadherin and CK18) was observed in MCF-7 cells following treatment with miR-155 inhibitor, suggesting that downregulation of miR-155 reduced EMT in MCF-7 cells.
Bioinformatics analysis showed that TGFBR2 was a potential direct target of miR-155. Lower levels of TGFBR2 expression were observed in human BC tissues, consistent with previous studies (21,22). Therefore, it was hypothesized that miRNA-155 could directly negatively regulate TGFBR2 mRNA and protein expression levels. To confirm this hypothesis, MCF-7 cells were transfected with miR-155 inhibitor. The miR-155 inhibitor induced enhanced expression of TGFBR2, suggesting that miR-155 inversely regulated TGFBR2 expression. Subsequently, direct binding of miR-155 to the TGFBR2 3'UTR was confirmed through the use of a dual luciferase assay.
Taken together, the results of the present study demonstrate that miR-155 promoted proliferation and metastasis of MCF-7 cells, and negatively regulated expression of its target, TGFBR2, by directly binding to its mRNA transcript. These results suggest that miR-155 may serve an important role in BC pathogenesis and potentially be used for development of novel diagnostic and therapeutic strategies for treatment of patients with BC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TH conceived and designed the study. XL performed the majority of the experiments and wrote the manuscript. YL assisted with the experiments. ZL participated in the analysis and data interpretation. All authors read and approved the final manuscrip.
Ethics approval and consent to participate
The present study was performed in accordance with standard guidelines and was approved by the Ethics Committee of Xinganmeng People’s Hospital (Inner Mongolia, China). Written informed consent was obtained from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zohrap N, Saatci Ö, Ozes B, Coban I, Atay HM, Battaloglu E, Şahin Ö, Bugra K. SIK2 attenuates proliferation and survival of breast cancer cells with simultaneous perturbation of MAPK and PI3K/Akt pathways. Oncotarget. 2018;9:21876–21892. doi: 10.18632/oncotarget.25082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Liu J, Ying X, Lin PC, Zhou BP. Twist-mediated epithelial-mesenchymal transition promotes breast tumor cell invasion via inhibition of hippo pathway. Sci Rep. 2016;6(24606) doi: 10.1038/srep24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. Cancer Genome Atlas Network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast Cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 5.Li CL, Yang D, Cao X, Wang F, Hong DY, Wang J, Shen XC, Chen Y. Fibronectin induces epithelial-mesenchymal transition in human breast cancer MCF-7 cells via activation of calpain. Oncol Lett. 2017;13:3889–3895. doi: 10.3892/ol.2017.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Moustafa AE, Achkhar A, Yasmeen A. EGF-receptor signaling and epithelial-mesenchymal transition in human carcinomas. Front Biosci (Schol Ed) 2012;4:671–684. doi: 10.2741/s292. [DOI] [PubMed] [Google Scholar]
- 7.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Lupia A, Peppicelli S, Witort E, Bianchini F, Carloni V, Pimpinelli N, Urso C, Borgognoni L, Capaccioli S, Calorini L, Lulli M. CD63 tetraspanin is a negative driver of epithelial-to-mesenchymal transition in human melanoma cells. J Invest Dermatol. 2014;134:2947–2956. doi: 10.1038/jid.2014.258. [DOI] [PubMed] [Google Scholar]
- 9.Jeong H, Ryu YJ, An J, Lee Y, Kim A. Epithelial-mesenchymal transition in breast cancer correlates with high histological grade and triple-negative phenotype. Histopathology. 2012;60:E87–E95. doi: 10.1111/j.1365-2559.2012.04195.x. [DOI] [PubMed] [Google Scholar]
- 10.Yang D, Ma M, Zhou W, Yang B, Xiao C. Inhibition of miR-32 activity promoted EMT induced by PM2.5 exposure through the modulation of the Smad1-mediated signaling pathways in lung cancer cells. Chemosphere. 2017;184:289–298. doi: 10.1016/j.chemosphere.2017.05.152. [DOI] [PubMed] [Google Scholar]
- 11.Na Y, Kaul SC, Ryu J, Lee JS, Ahn HM, Kaul Z, Kalra RS, Li L, Widodo N, Yun CO, Wadhwa R. Stress chaperone mortalin contributes to epithelial-mesenchymal transition and cancer metastasis. Cancer Res. 2016;76:2754–2765. doi: 10.1158/0008-5472.CAN-15-2704. [DOI] [PubMed] [Google Scholar]
- 12.Hujie G, Zhou SH, Zhang H, Qu J, Xiong XW, Hujie O, Liao CG, Yang SE. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/KLF11/Smads in hepatocellular carcinoma. Cancer Cell Int. 2018;18(10) doi: 10.1186/s12935-018-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Jiang Y, Miao R, Qu K, Zhang J, Liu C. MicroRNA-1271 functions as a metastasis and epithelial-mesenchymal transition inhibitor in human HCC by targeting the PTP4A1/c-Src axis. Int J Oncol. 2018;52:536–546. doi: 10.3892/ijo.2017.4224. [DOI] [PubMed] [Google Scholar]
- 14.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Sha HH, Li HJ. Functions and mechanisms of miR-186 in human cancer. Biomed Pharmacother. 2019;119(109428) doi: 10.1016/j.biopha.2019.109428. [DOI] [PubMed] [Google Scholar]
- 16.Iacona JR, Lutz CS. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. 2019;10(e1533) doi: 10.1002/wrna.1533. [DOI] [PubMed] [Google Scholar]
- 17.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: Molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–2540. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Chernyy V, Pustylnyak V, Kozlov V, Gulyaeva L. Increased expression of miR-155 and miR-222 is associated with lymph node positive status. J Cancer. 2018;9:135–140. doi: 10.7150/jca.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bašová P, Pešta M, Sochor M, Stopka T. Prediction potential of serum miR-155 and miR-24 for relapsing early breast cancer. Int J Mol Sci. 2017;18(E2116) doi: 10.3390/ijms18102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei CY, Tan QX, Zhu X, Qin QH, Zhu FB, Mo QG, Yang WP. Expression of CDKN1A/p21 and TGFBR2 in breast cancer and their prognostic significance. Int J Clin Exp Pathol. 2015;8:14619–14629. [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, Cantrell MA, Rack PG, Neal JT, Chan CWM, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20:769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricke F, Lee J, Michalak M, Warnken U, Hausser I, Suarez-Carmona M, Halama N, Schnölzer M, Kopitz J, Gebert J. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun Signal. 2017;15(14) doi: 10.1186/s12964-017-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, Li P, Hei Y, Li S, Wang J, Lv X, Zhang J. Long Non-coding RNA-ZNF281 promotes cancer cell migration and invasion in gastric cancer via downregulation of microRNA-124. Oncol Lett. 2020;19:1849–1855. doi: 10.3892/ol.2020.11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Zhao B, Jin F. The assessment of 8th edition AJCC prognostic staging system and a simplified staging system for breast cancer: The analytic results from the SEER database. Breast J. 2019;25:838–847. doi: 10.1111/tbj.13347. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14(52) doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Wang Y, Shang L, Qi L, Song M. Five common functional polymorphisms in microRNAs and susceptibility to breast cancer: An updated meta-analysis. Genet Test Mol Biomarkers. 2018;22:350–358. doi: 10.1089/gtmb.2017.0270. [DOI] [PubMed] [Google Scholar]
- 30.Sandiford OA, Moore CA, Du J, Boulad M, Gergues M, Eltouky H, Rameshwar P. Human Aging and Cancer: Role of miRNA in Tumor Microenvironment. Adv Exp Med Biol. 2018;1056:137–152. doi: 10.1007/978-3-319-74470-4_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.