Abstract
High mobility group protein B1 (HMGB1) is a nuclear protein that has been reported to contribute to tumor growth in humans. The present study identified a microRNA (miR/miRNA) that targets the 3' untranslated region (3'UTR) of the HMGB1 gene and assessed its effects on the proliferation of human cervical cancer cells and associated molecular mechanism. Western blotting was performed to determine HMGB1 levels in HeLa cells. TargetScan software was used to identify miRNA binding sites adjacent to the HMGB1. The viability of HeLa cells transfected with miR-142-3p mimics or inhibitors was determined using an MTT assay. The subcellular distribution (cytoplasmic or nuclear) of HMGB1 in HeLa cells was observed by western blotting. HMGB1 expression in HeLa and CaSKi cells was significantly higher compared with normal control cervical cells. TargetScan analysis indicated that miR-142-3p binds to the 3'UTR of HMGB1. Transfection with a miR-142-3p inhibitor increased cytoplasmic HMGB1 expression in HeLa cells, as shown by western blot analysis, while transfection with miR-142-3p mimics decreased the cytoplasmic expression of HMGB1 in HeLa cells. Therefore, miR-142-3p negatively regulated HMGB1 levels in cervical cancer cells. These findings indicated that miR-142-3p inhibited the proliferation of human cervical cancer cells, at least in part, by negatively regulating the cytoplasmic localization of HMGB1.
Keywords: microRNA-142-3p, high mobility group protein B1, cervical cancer, cytoplasmic distribution
Introduction
High mobility group protein B1 (HMGB1) is a member of a class of non-histone chromosomal binding proteins that is highly conserved and primarily present in the nucleus (1,2). Nuclear HMGB1 functions as a DNA chaperone and is involved in DNA replication, maintenance of DNA structure, genetic recombination, DNA repair and transcription (3). By contrast, extranuclear HMGB1 functions as a classical damage-associated molecular pattern (4). Inflammatory factors, including infection, cell necrosis and drugs can stimulate the displacement and release of HMGB1, which promotes autophagy and apoptosis (4). HMGB1 is highly homologous and human and rodent HMGB1 share >98% homology (5). HMGB1 has two main functional regions: The A- and B-boxes (5). The B-box has cytokine-like activity and the A-box inhibits the function of the B-box (5). HMGB1 has multiple receptors, including the receptor for advanced glycation end-products (RAGE), toll-like receptors (TLRs) and multi-ligand proteoglycans, including syndecan and plasminogen, although RAGE and TLR4/TLR2 are its main receptors (6).
HMGB1 is a multifunctional protein that exerts numerous biological functions in both normal and malignant tumor cells (7). HMGB1 was demonstrated to be involved in the development of various diseases, including sepsis, arthritis and various tumors, and has been shown to be of prognostic value (7). Additionally, HMGB1 is involved in the regulation of autophagy and apoptosis in tumor cells (4,8). HMGB1 is expressed in breast cancer (8,9), colon cancer (10,11), melanoma (12,13) and ovarian cancer (14). HMGB1 is associated with various characteristics of tumor cells, including proliferation, angiogenesis, escape from programmed cell death and resistance to growth factor inhibitors and biological behavior of tumors, including tissue invasion and distant spread (7,13). Furthermore, evidence has demonstrated that HMGB1 may induce tumorigenesis, metastasis and chemotherapy resistance and may be useful as a novel treatment target for lung cancer (7).
MicroRNAs (miRs or miRNAs) are a class of non-coding RNAs involved in post-transcriptional regulation of gene expression (15). Numerous miRNAs have been reported to be abnormally expressed in tumor cells (15). miRNAs are a class of evolutionarily conserved small non-coding RNAs that regulate numerous genes during tumorigenesis in humans and other mammals (15). Numerous long non-coding RNAs (lncRNAs) and miRNAs have been reported to regulate HMGB1. For example, the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) regulated HMGB1 by sponging miR-129-5p to promote the proliferation of SW480 and HCT116 colon cancer cells (15). Additionally, HMGB1 is a target of miR-1284 and was demonstrated to reverse the effects of miR-1284 on the proliferation and chemosensitivity of cervical cancer cells (16). The lncRNA urothelial carcinoma-associated 1 functions as an oncogene to promote the progression of lung cancer and lung cancer cell migration via the miRNA-193a/HMGB1 axis (17). In pancreatic cancer cells, the lncRNA zinc finger E-box-binding homeobox 2/antisense RNA 1 increases the proliferation and invasion of cancer cells by regulating the miR-204/HMGB1 axis (18). Additionally, miRNAs were reported to regulate cancer progression, invasion and metastasis. For instance, miR-216b expression decreased in colorectal cancer tissues compared with levels in corresponding adjacent normal tissues and miR-216b functioned as a tumor suppressor by inhibiting cancer development and progression partially through the HMGB1-mediated JAK2/STAT3 pathway (19). In non-small cell lung cancer cells, miR-449a inhibited the proliferation, migration and invasion of lung cancer cells via the HMGB1-mediated NF-κB signaling pathway (20).
However, the function and molecular mechanism of HMGB1 in cervical cancer remains to be elucidated. It has been demonstrated that miRNA-1284, acting as a regulator of HMGB1, suppressed cell proliferation and migration in osteosarcoma (21). Furthermore, the lncRNA MALAT1 was shown to promote the proliferation of osteosarcoma cells by inhibiting miR-142-3p or miR-129-5p and by targeting HMGB1(22). The present study investigated miRNAs that target HMGB1 in cervical cancer and explored the underlying molecular mechanism involved in the proliferation of cervical cancer cells, which may provide novel insights into the tumorigenesis of cervical cancer.
Materials and methods
Cell lines and reagents
Primary cervical epithelial cells (cat. no. ATCC® PCS-480-011™) were purchased from the American Type Culture Collection. Cells were cultured and passaged in complete expansion medium, which was prepared by adding the contents of the Cervical Epithelial Cell Growth kit (cat. no. ATCC® PCS-480-042) to one bottle of Cervical Epithelial Cell Basal medium (cat. no. ATCC® PCS-480-032), according to the manufacture's protocol. Human cervical cancer cell lines HeLa (cat. no. CBP60232; Nanjing Cobioer Co., Ltd.) and CaSKi (cat. no. CC-Y1086; EK Biosciences GmbH) cells were maintained and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1X streptomycin and penicillin (Gibco; Thermo Fisher Scientific, Inc.) in a cell incubator under a humidified atmosphere and 5% CO2 at 37˚C. MISSION® microRNA mimic miR-142-3p (has-miR-142-3p mimic; cat. no. HMI0219) and corresponding MISSION® miRNA, negative control 1 (negative control mimic; cat. no. HMC0002), MISSION® Lenti microRNA inhibitor, human miR-142-3p (miR-142-3p inhibitor; cat. no. HLTUD0219) and MISSION® Lenti microRNA inhibitor (negative control inhibitor; cat. no. HLTUD001C) were purchased from Sigma-Aldrich; Merck KGaA.
Specimens
Three pairs of clinical specimens (cervical cancer tissues and surrounding non-cancerous tissues; distance between tissues, 3 cm) were obtained from patients at Tianjin Central Hospital of Gynecology and Obstetrics (Tianjin, China). Specimen 1 was collected on April 4th, 2017, specimen 2 on July 8th, 2017 and specimen 3 on October 13th, 2017. The age distribution was 43-68 years old and the mean age was 6±2 years old.
Patients were included into the current study if they did not receive any cancer-related treatment prior to the surgery. Patients who underwent chemotherapy or had diabetes were excluded. The clinical specimens were immediately frozen in liquid nitrogen and stored at -80˚C. All patients provided written informed consent prior to enrollment. Studies involving humans were conducted in accordance with the Declaration of Helsinki and were approved by the Ethics Committee of Tianjin Central Hospital of Gynecology and Obstetrics.
Cell transfection
HeLa cells (2x105 cells/48-well) were transfected with the aforementioned has-miR-142-3p mimics, negative control mimics, hsa-miR-142-3p inhibitors and negative control inhibitors using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, a total of 0.5 µg has-miR-142-3p mimics, inhibitors, negative control mimics or negative control inhibitors was diluted in 100 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.). Opti-MEM was purchased from Thermo Fisher Scientific, Inc. A total of 2 µl Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) was diluted with 200 µl of Opti-MEM. Following 5 min at room temperature, 100 µl of diluted DNA and 100 µl of diluted Lipofectamine 2000® was mixed gently and incubated for 20 min at room temperature. The mixture was then added to the cells with Opti-MEM and free FBS and transfected for 6 h at 37˚C. Following transfection, Opti-MEM medium was changed into DMEM + FBS + antibiotics at 37˚C, as described above. Subsequently, HMGB1 expression was assessed and cell proliferation assays were performed.
Additionally, HMGB1 shRNA(h) (cat. no. sc-37982-SH) and control shRNA (cat. no. sc-108060) were purchased from Santa Cruz Biotechnology, Inc. Transfection into HeLa cells was performed as described above. The transfected cells were cultured for 24, 48 and 72 h for western blotting and MTT assays.
ELISA
A human HMGB1 ELISA kit (cat. no. JL13893-48T) was purchased from Shanghai Jianglai Biotech Co., Ltd. ELISA was performed to determine the concentration of HMGB1 in the cell culture supernatant of miR-142-3p mimic-transfected, mimic negative control-transfected and untreated HeLa cells. Cell culture supernatants in each group were collected and the concentration of HMGB1 was detected using the kit, according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from transfected HeLa cells using a PureLink RNA Mini kit (cat. no. 12183018A; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RNA was reverse transcribed into cDNA using a High-Capacity RNA-to-cDNA kit (cat. no. 4387406; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The MystiCq® microRNA qPCR assay primer for miR-142-3p (cat. no. MIRAP00174) was purchased from Sigma-Aldrich; Merck KGaA. Multiplex qPCR assay was performed using a TaqMan Gene Expression master mix (cat. no. 4369016; Invitrogen; Thermo Fisher Scientific, Inc.). The sequences used were as follows: U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCGT. miR-143-3p was commercially purchased from Sigma-Aldrich; Merck KgaA and, therefore, the sequences are not known. The thermocycling conditions were as follows for qPCR: denaturation for 1 min at 95˚C, 40 cycles of 15 sec at 95˚C and 1 min at 60˚C. Samples were stored at 4˚C. U6 was used as the reference gene. miR-142-3p levels were quantified according to the 2-ΔΔCq method (22).
MTT assay
HeLa cells (1.2x105 cells/well) were plated into 48-well plates and incubated for 8 h at 37˚C. Subsequently, cells were transfected with miR-142-3p mimics, mimic negative controls, miR-142-3p inhibitors or inhibitor negative controls for 24 and 48 h. Cell viability was determined using an MTT assay, as previously described (23). For the assay, 20 µl MTT stock solution (5 mg/ml) was added to the plate and incubated for 4 h. A total of 100 µl DMSO(cat. no. D8418; Sigma-Aldrich; Merck KGaA) was added to the culture medium and incubated for 10 min. The plates were then read on a microplate reader at 490 nm and data were analyzed.
Binding prediction of miRNAs and HMGB1
To investigate how HMGB1 is regulated in cervical cancer cells, the 3'-untranslated region (3'UTR) of HMGB1 was scanned using the online search tool TargetScan 7.1 (www.targetscan.org/vert_72/) to search for possible complementary sites to human miRNAs and predict the possible miRNAs that may bind to and interact with HMGB1.
Isolation of nuclear and cytoplasmic proteins
HeLa cells were transfected with miR-142-3p inhibitors and inhibitor negative controls for 48 h. Following transfection, total protein in the cytoplasm or nucleus of transfected HeLa cells was extracted using a cell nuclear protein and cytoplasmic protein extraction kit (cat. no. P0028; Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Lamin B1 was used as a control for nuclear proteins and α-tubulin was used as a control for cytoplasmic proteins.
Western blotting
HeLa cells were transfected with miR-142-3p mimics, mimic negative controls, miR-142-3p inhibitors and inhibitor negative controls and cultured for 24 or 48 h. Subsequently, transfected cells were washed twice with ice-cold PBS and cell lysates were prepared using RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology). Total protein concentration of the prepared lysates was determined using a BCA kit. Next, 30 µg of total protein was separated by 10% SDS-PAGE at 120 V for 2 h and at 80 V for 15 min. The separated proteins were transferred to PVDF membranes, blocked with 5% non-fat milk for 1 h at room temperature and incubated with the following primary antibodies: Anti-HMGB1 (cat. no. ab79823; Abcam), anti-GAPDH (cat. no. ab70699; Abcam), anti-lamin B1 (cat. no. 66095-1-Ig; ProteinTech Group, Inc.) and rabbit monoclonal anti-α tubulin (cat. no. ab179513; Abcam) at 4˚C for 8 h. Dilutions of all primary antibodies were 1:5,000. Following primary antibody incubation, membranes were incubated with goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) and goat anti-mouse IgG-HRP (cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) secondary antibodies at room temperature for 1 h. The dilution of all secondary antibodies was 1:10,000. Bands were visualized using a Pierce™ ECL Western Blotting Substrate (cat. no. 32209; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The densitometry of the bands was analyzed by ImageJ software (version no. 1.51k; National Institutes of Health).
Statistical analysis
Data were analyzed using SPSS software (version no. 20.0; IBM Corp.). Multiple comparisons were analyzed using ANOVA, followed by Tukey's post-hoc test. An independent samples t-test was used to analyze independent samples. A paired t-test was used to analyze differences between paired experimental specimens. Data are presented as the mean ± SEM. P<0.05 was considered to indicate a statistically significant difference.
Results
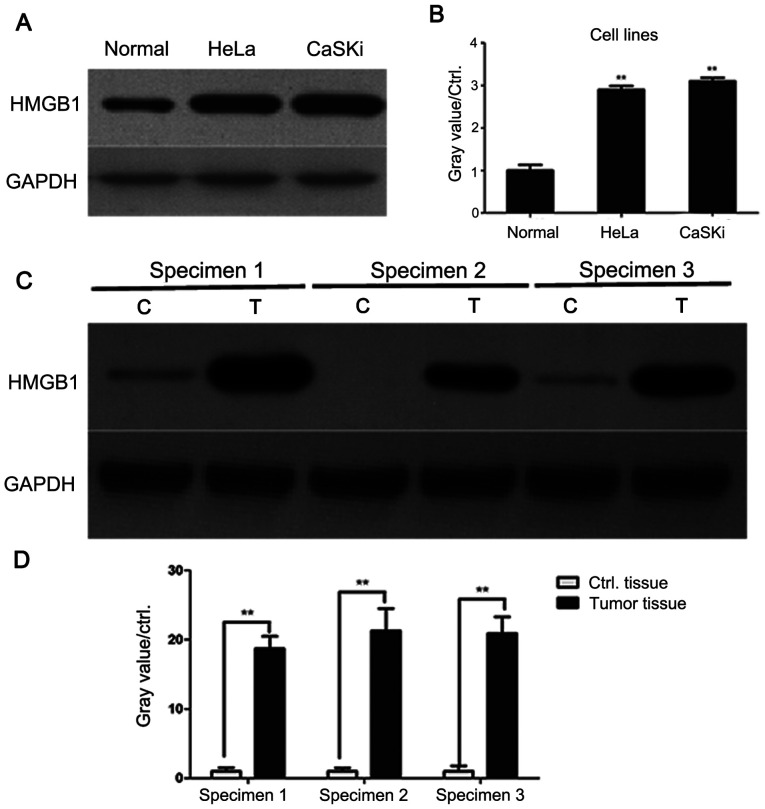
HMGB1 levels are higher in cervical cancer cells compared with normal cells. The expression levels of HMGB1 in human primary cervical epithelial cells and the human cervical cancer cell lines HeLa and CaSKi were examined. As presented in Fig. 1A and B, HMGB1 levels were significantly higher in HeLa and CaSKi cells compared with primary cervical epithelial cells, indicating that HMGB1 may be involved in cervical cancer.
Figure 1.
HMGB1 is highly expressed in cervical cancer cell lines. (A) HMGB1 levels were significantly higher in HeLa and CaSKi cells compared with primary cervical epithelial cells. (B) The gray values, representing HMGB1 levels in HeLa, CaSKi and primary cervical epithelial cells, were calculated using ImageJ software and presented in a histogram. HMGB1 expression levels in primary cervical epithelial cells were used as the control. Experiments were performed in triplicate. **P<0.01 vs. primary cervical epithelial cells. (C) Three sets of paired clinical specimens (cervical cancer and paracancer tissues) were collected and the levels of HMGB1 in the tissues were detected by western blotting. (D) Gray values representing HMGB1 levels in paired human cervical cancer and paracancer tissues are presented in a histogram. Data are presented as the mean ± standard deviation of two experiments. **P<0.01 vs. controls. T, human cervical cancer tissues; C, adjacent control tissues; HMGB1, high mobility group protein B1; ctrl, control.
Subsequently, three pairs of cervical cancer and paracancer tissues were collected. The levels of HMGB1 were assessed by western blotting. As demonstrated by Fig. 1C and D, HMGB1 protein levels were significantly higher in human cervical cancer tissues compared with paracancer tissues. These data showed that HMGB1 may serve an oncogenic role during the tumorigenesis of cervical cancer.
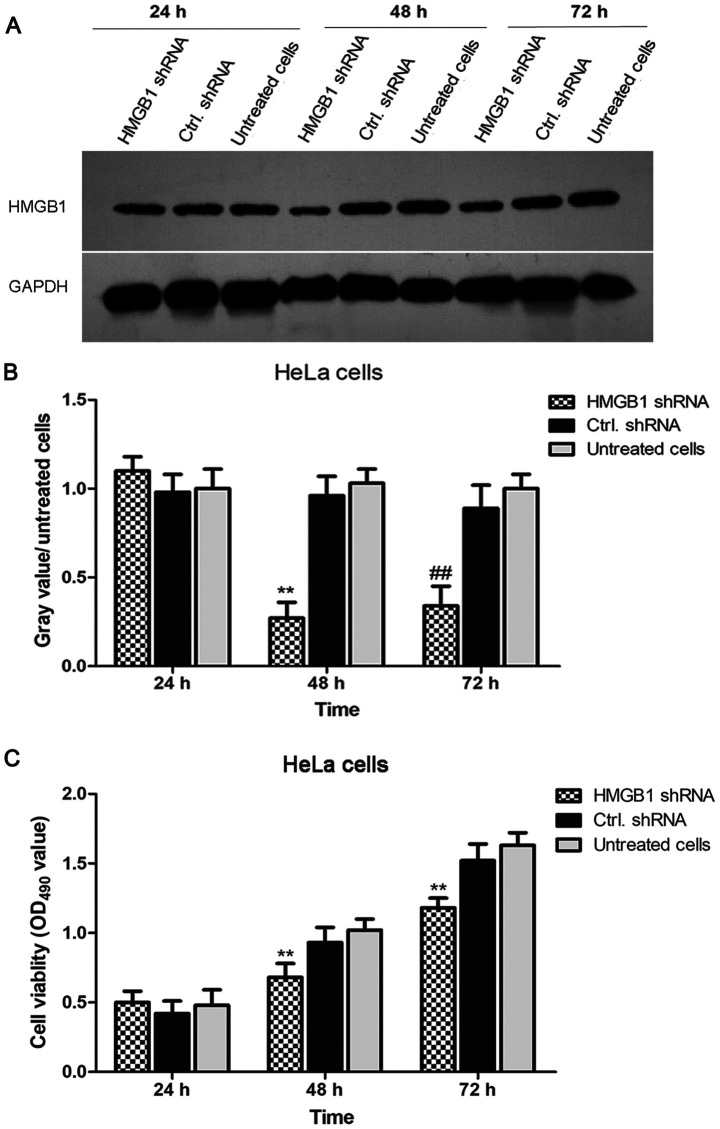
Knockdown of HMGB1 suppresses the viability of HeLa cells
To investigate the function of HMGB1 in the proliferation of cervical cancer cells, HMGB1 expression in HeLa cells was knocked down by RNA interference. As shown in Fig. 2A and B, the expression levels of HMGB1 were significantly decreased in HMGB1 shRNA-transfected HeLa cells compared with the levels in negative control shRNA-transfected HeLa cells at 24, 48 and 72 h. Subsequently, cell viability in HMGB1 shRNA-transfected, negative control shRNA-transfected and untreated HeLa cells was determined using an MTT assay. The results demonstrated that the viability of HMGB1 shRNA-transfected HeLa cells was significantly lower compared with negative control shRNA-transfected HeLa cells (Fig. 2C). These results indicated that HMGB1 shRNA effectively decreased HMGB1 expression in HeLa cells and the cell viability of HeLa cells was significantly decreased by knockdown of HMGB1.
Figure 2.
Knockdown of HMGB1 suppresses the viability of HeLa cells. (A) HMGB1 expression levels were significantly decreased in HMGB1 shRNA-transfected HeLa cells compared with the levels in negative control shRNA-transfected HeLa cells. GAPDH was used as an internal reference gene. (B) The gray value of each band is presented in a histogram. Experiments were performed in duplicate. **P<0.01 vs. shRNA-transfected HeLa cells after 48 h; ##P<0.01 vs. control shRNA-transfected HeLa cells after 72 h. (C) Cell viability was determined using an MTT assay. The results demonstrated that transfection with miR-142-3p inhibitor increased cytoplasmic levels of HMGB1 in HeLa cells. Data are presented as the mean ± standard deviation from three experiments. **P<0.01 vs. negative control shRNA-transfected cells at the respective time-points. HMGB1, high mobility group protein B1; shRNA, short hairpin RNA; OD, optical density; ctrl, control.
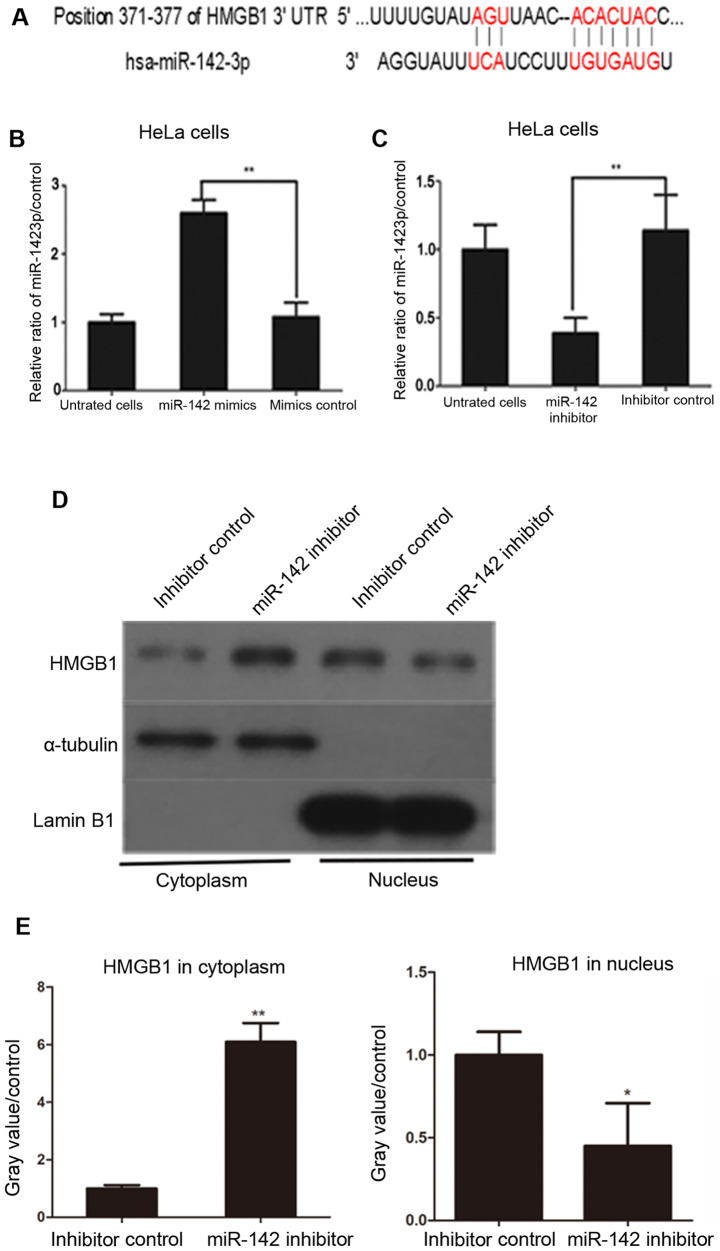
HMGB1 is predicted to be a target of miR-142-3p
As presented in Fig. 3A, a possible miR-142-3p binding sequence was found at position 371-377 of the HMGB1 3'UTR.
Figure 3.
HMGB1 is predicted to be a target of miR-142-3p and is negatively regulated by miR-142-3p. (A) Possible miRNA binding sites in HMGB1 were predicted by TargetScan, which identified miR-142-3p as a possible regulatory miRNA for HMGB1. The red color denotes the binding site of HMGB1 mRNA and miR-142-3p. (B) Human cervical cancer cells were transfected with miR-142-3p mimics or negative control mimics for 48 h. The relative expression of miR-142-3p was determined by RT-qPCR. **P<0.01 vs. inhibitor controls. (C) HeLa cells were transfected with miR-142-3p inhibitors or negative inhibitor controls for 48 h. The relative expression of miR-142-3p was determined by RT-qPCR. **P<0.01 vs. inhibitor controls. (D) Western blot analysis. The results showed that transfection with miR-142-3p inhibitor increased cytoplasmic levels of HMGB1 in HeLa cells. (E) Histogram of HMGB1 expression in the cytoplasm and nucleus. Data are presented as the mean ± standard deviation from three experiments. *P<0.05 vs. negative control inhibitor **P<0.01 vs. negative control inhibitor. HMGB1, high mobility group protein B1; miR, microRNA; 3'UTR, 3' untranslated region; RT-qPCR, reverse transcription-quantitative PCR; ctrl, control.
Transfection with miR-142-3p inhibitor increases cytoplasmic HMGB1 expression in HeLa cells
Western blotting was used to investigate the association between miR-142-3p and HMGB1 in human cervical cancer cells. miR-142-3p mimics were transfected into HeLa cells and cultured for 48 h. miR-142-3p expression significantly increased in cells transfected with miR-142-3p mimics compared with cells transfected with mimic negative controls (Fig. 3B). HeLa cells transfected with miR-142-3p inhibitor exhibited decreased expression of miR-142-3p compared with inhibitor control-transfected HeLa cells (Fig. 3C). Next, the effect of miR-142-3p on the expression and distribution of HMGB1 in cervical cancer cells was assessed.
HMGB1 protein levels in the cytoplasm and nucleus of miR-142-3p inhibitor- and negative control inhibitor-transfected HeLa cells were assessed at 48 h post-transfection. The results demonstrated that transfection with miR-142-3p inhibitors significantly increased cytoplasmic levels of HMGB1 in HeLa cells compared with negative control inhibitor-transfected cells (Fig. 3D and E). By contrast, nuclear levels of HMGB1 in miR-142-3p inhibitor-transfected cells were lower compared with control cells (Fig. 3D and E). These results suggested that overexpression of miR-142-3p inhibited the expression of HMGB1 in HeLa cells.
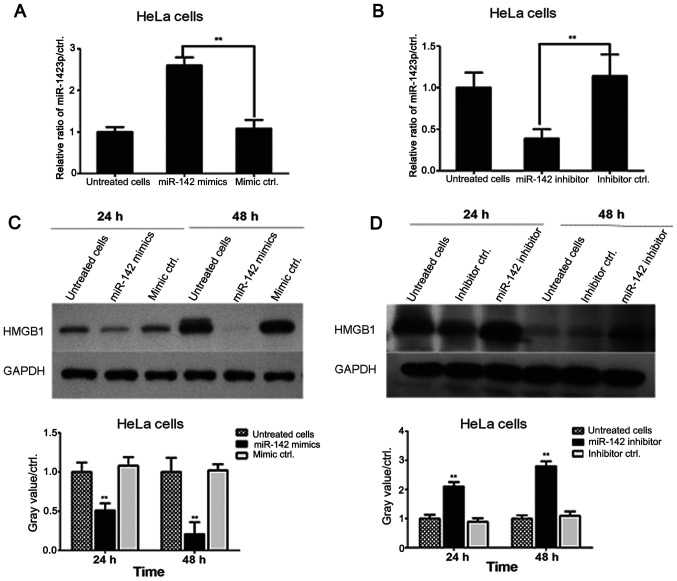
miR-142-3p negatively regulates the levels of HMGB1 in cervical cancer cells
miR-142-3p mimics were transfected into HeLa cells for 24 and 48 h. HMGB1 levels were assessed by western blotting. As shown in Fig. 4A, transfection of miR-142-3p mimics significantly downregulated HMGB1 expression levels in HeLa cells compared with the levels in mimic negative control-transfected cells at 24 and 48 h. Additionally, HeLa cells were transfected with either miR-142-3p inhibitors or inhibitor negative controls for 24 and 48 h, and the results demonstrated that transfection with miR-142-3p inhibitor significantly increased the levels of HMGB1 in cervical cancer cells compared with the levels in inhibitor negative control-transfected cells (Fig. 4B). These data revealed that miR-142-3p levels were negatively associated with HMGB1 expression in HeLa cells.
Figure 4.
miR-142-3p negatively regulates HMGB1 levels in cervical cancer cells. A. Human cervical cancer cells were transfected with hsa-miR-142-3p mimic or negative control mimics for 48 hours. The relative expression of hsa-miR-142-3p was determined by real time PCR. **p<0.01, compared with mimic control transfected HeLa cells. B. HeLa cells were transfected with hsa-miR-142-3p inhibitor or negative inhibitor control for 48 hours. The relative expression of hsa-miR-142-3p was determined by real time PCR. **p<0.01, compared with mimic control transfected HeLa cells. (C) Human cervical cancer cell lines were transfected with miR-142-3p and negative control mimics for 24 and 48 h and HMGB1 levels were determined by western blotting. GAPDH was used as an internal reference gene. The gray value of each HMGB1 band in miR-142-3p mimic- and negative control mimic-transfected cervical cancer cells was determined using ImageJ software and was presented in a histogram. Experiments were performed in duplicate. **P<0.01 vs. negative control mimic-transfected cells. (D) Cervical cancer cell lines were transfected with miR-142 inhibitor and inhibitor negative controls for 24 and 48 h. HMGB1 expression levels were assessed by western blotting. The gray values in miR-142-3p inhibitor- and inhibitor negative control-transfected HeLa cells are presented in a histogram. Data are presented as the mean ± standard deviation from two experiments. **P<0.01 vs. negative control inhibitor-transfected HeLa cells. HMGB1, high mobility group protein B1; RT-qPCR, reverse transcription-quantitative PCR; miR-142, microRNA-142; ctrl, control.
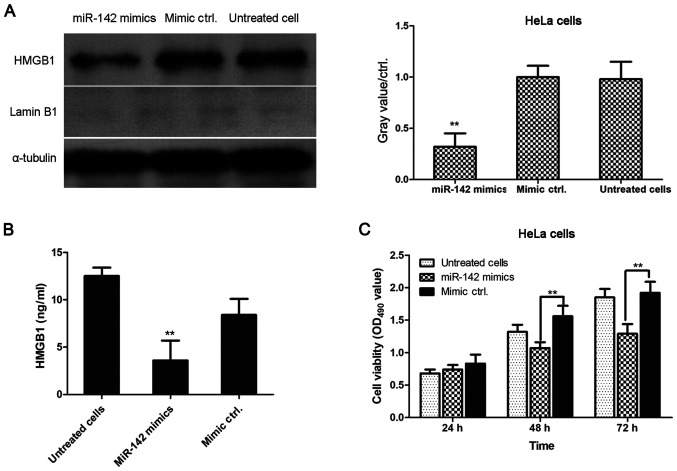
Transfection with miR-142-3p mimics decreases the levels of cytoplasmic HMGB1 in HeLa cells
As presented in Fig. 5A, HMGB1 expression was significantly lower in miR-142-3p mimic-transfected cells compared with mimic negative control-transfected cells. ELISA was performed to determine the concentration of HMGB1 in the cell culture supernatant of miR-142-3p mimic-transfected, mimic negative control-transfected and untreated HeLa cells. As demonstrated by Fig. 5B, miR-142-3p mimic-transfected cells secreted significantly lower levels of HMGB1 into the cell culture supernatant compared with mimic negative control-transfected cells. Additionally, the viability of transfected cells was assessed using an MTT assay. As shown in Fig. 5C, cell viability was significantly inhibited in miR-142-3p mimic-transfected HeLa cells compared with mimic negative control-transfected HeLa cells at 48 and 72 h. Cell viability was not significantly different between groups at 24 h.
Figure 5.
Transfection with miR-142-3p mimics decreases cytoplasmic levels of HMGB1 in HeLa cells. (A) Gray value of each HMGB1 band in miR-142-3p mimic- and negative control mimic-transfected cervical cancer cells and untreated cells is presented in a histogram. Experiments were performed in duplicate. **P<0.01 vs. negative control mimic-transfected cells. (B) ELISA was performed to measure the concentration of HMGB1 in the supernatant of miR-142-3p mimic-transfected, mimic negative control-transfected and untreated HeLa cells. **P<0.01 vs. mimic negative control-transfected HeLa cells. (C) HeLa cells were transfected with miR-142-3p mimics and mimic control and cultured for 24, 48 and 72 h, and cell viability was determined using an MTT assay. **P<0.01 vs. mimic control-transfected HeLa cells. Data are presented as the mean ± standard deviation from two experiments. HMGB1, high mobility group protein B1; miR-142, microRNA-142; ctrl, control.
Discussion
HMGB1 is a highly conserved non-histone DNA-binding protein that is abnormally expressed in numerous types of human cancer (7,8). The present study revealed that HMGB1 levels were significantly higher in the two tested cervical cancer cell lines, HeLa and CaSKi, compared with primary cervical epithelial cells. This is consistent with previous findings that have demonstrated that HMGB1 is involved in cell proliferation and migration, and alters the protein expression of epithelial-mesenchymal transition-associated genes (7). These results indicated that HMGB1 may function as an oncogene in the progression of cervical cancer.
Subsequently, possible miRNAs involved in regulating the expression of HMGB1 were predicted using TargetScan. The results demonstrated that miR-142-3p may bind to position 371-377 of the HMGB1 3'UTR. Transfection with miR-142-3p mimics significantly decreased the levels of HMGB1 and the proliferation of miR-142-3p mimic-transfected HeLa cells was significantly decreased compared with mimic negative control-transfected HeLa cells. Conversely, transfection with miR-142-3p inhibitor increased the expression of HMGB1 in cervical cancer cells. This was consistent with the results reported in a study by Liu et al (24). Two candidate miRNAs, miR-142-3p and miR-129-5p, were screened and the results demonstrated that MALAT1 promoted the development of osteosarcoma by association with HMGB1 via both miR-142-3p and miR-129-5p. Additionally, previous studies have reported that miR-1284(21) and miR-22 (25,26) regulated HMGB1 and were involved in the regulation of cell proliferation and migration in cervical cancer.
Higher HMGB1 expression induced the proliferation and invasion of cervical cancer cells, which was consistent with the findings of Jiang et al (27). This previous study reported that HMGB1 suppressed the progression and development of cervical cancer. However, there were certain differences between the findings by Jiang et al (27) and the present study. While this previous study focused on the effects of miR-142 on the proliferation and invasiveness of cervical cancer cells, the present study demonstrated that miR-142 affected the subcellular distribution of HMGB1 in HeLa cells.
The molecular mechanism underlying the effects of HMGB1 on the proliferation of cervical cancer cells was explored. Western blotting showed that transfection with miR-142-3p inhibitor increased cytoplasmic HMGB1 expression in HeLa cells, while transfection with miR-142-3p mimics decreased cytoplasmic HMGB1 levels in HeLa cells. It has been reported that cytoplasmic HMGB1 translocation and HMGB1-induced cell autophagy contributed to cisplatin resistance by inhibiting the apoptosis of cervical cancer cells (28). Additionally, cytoplasmic HMGB1 expression was reported to be associated with the levels of tumor-infiltrating lymphocytes in breast cancer (29) and induced acute liver failure (30). These data indicated that the cytoplasmic translocation of HMGB1 likely promoted cancer progression. The present study demonstrated that miR-142-3p affected the subcellular distribution of HMGB1, as it inhibited the translocation of HMGB1 from the nucleus into the cytoplasm.
Since the current experiments were primarily performed in HeLa cells, future experiments will be performed in several other cervical cancer cell lines to support the results obtained. The levels of HMGB1 in three pairs of cervical cancer tissues and paracancer tissues were detected. The 5-year overall survival and disease-free survival rates of patients with different expression levels of HMGB1 and miR-142-3p have not been analyzed in the current study and will be investigated in future research.
In conclusion, the present results demonstrated that miR-142-3p negatively regulated the cytoplasmic localization of HMGB1, which inhibited the proliferation of human cervical cancer cells. Therefore, miR-142-3p may be a novel therapeutic target for human cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HD designed the current study and experiments, analyzed data and wrote the manuscript. JS performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Studies involving humans were conducted in accordance with the Declaration of Helsinki and were approved by the Ethics Committee of Tianjin Central Hospital of Gynecology and Obstetrics, Tianjin, China.
Patient consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang K, Shan S, Wang S, Gu X, Zhou X, Ren T. HMGB1-containing nucleosome mediates chemotherapy-induced metastasis of human lung cancer. Biochem Biophys Res Commun. 2018;500:758–764. doi: 10.1016/j.bbrc.2018.04.150. [DOI] [PubMed] [Google Scholar]
- 2.Kumari T, Kumar B. High-mobility group box 1 protein (HMGB1) gene polymorphisms and cancer susceptibility: A comprehensive meta-analysis. Clin Chim Acta. 2018;483:170–182. doi: 10.1016/j.cca.2018.04.042. [DOI] [PubMed] [Google Scholar]
- 3.Osmanov T, Ugrinova I, Pasheva E. The chaperone like function of the nonhistone protein HMGB1. Biochem Biophys Res Commun. 2013;432:231–235. doi: 10.1016/j.bbrc.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Kang R, Xie Y, Zhang Q, Hou W, Jiang Q, Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res. 2017;27:916–932. doi: 10.1038/cr.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Wang F, Yang L, Yuan Y, Chen Y, Zhang G, Fan Z. HMGB1 a-box reverses brain edema and deterioration of neurological function in a traumatic brain injury mouse model. Cell Physiol Biochem. 2018;46:2532–2542. doi: 10.1159/000489659. [DOI] [PubMed] [Google Scholar]
- 6.Ying S, Xiao X, Chen T, Lou J. PPAR ligands function as suppressors that target biological actions of HMGB1. PPAR Res. 2016;2016(2612743) doi: 10.1155/2016/2612743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L, Yang L. The function and mechanism of HMGB1 in lung cancer and its potential therapeutic implications. Oncol Lett. 2018;15:6799–6805. doi: 10.3892/ol.2018.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang BF, Tzeng HE, Chen PC, Wang CQ, Su CM, Wang Y, Hu GN, Zhao YM, Wang Q, Tang CH. HMGB1 genetic polymorphisms are biomarkers for the development and progression of breast cancer. Int J Med Sci. 2018;15:580–586. doi: 10.7150/ijms.23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exner R, Sachet M, Arnold T, Zinn-Zinnenburg M, Michlmayr A, Dubsky P, Bartsch R, Steger G, Gnant M, Bergmann M, et al. Prognostic value of HMGB1 in early breast cancer patients under neoadjuvant chemotherapy. Cancer Med. 2016;5:2350–2358. doi: 10.1002/cam4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang CC, Gdynia G, Ehemann V, Roth W. The HMGB1 protein sensitizes colon carcinoma cells to cell death triggered by pro-apoptotic agents. Int J Oncol. 2015;46:667–676. doi: 10.3892/ijo.2014.2785. [DOI] [PubMed] [Google Scholar]
- 11.Yadav SS, Kumar M, Varshney A, Yadava PK. KLF4 sensitizes the colon cancer cell HCT-15 to cisplatin by altering the expression of HMGB1 and hTERT. Life Sci. 2019;220:169–176. doi: 10.1016/j.lfs.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Xie B, Cao K, Li J, Chen J, Tang J, Chen X, Xia K, Zhou X, Cheng Y, Zhou J, et al. Hmgb1 inhibits Klotho expression and malignant phenotype in melanoma cells by activating NF-κB. Oncotarget. 2016;7:80765–80782. doi: 10.18632/oncotarget.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parodi M, Pedrazzi M, Cantoni C, Averna M, Patrone M, Cavaletto M, Spertino S, Pende D, Balsamo M, Pietra G, et al. Natural killer (NK)/melanoma cell interaction induces NK-mediated release of chemotactic high mobility group box-1 (HMGB1) capable of amplifying NK cell recruitment. OncoImmunology. 2015;4(e1052353) doi: 10.1080/2162402X.2015.1052353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou LY, Shi LY, Xiao Y. Changes of HMGB1 expression on angiogenesis of ovarian cancer and its mechanism. J Biol Regul Homeost Agents. 2016;30:233–238. [PubMed] [Google Scholar]
- 15.Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233:6750–6757. doi: 10.1002/jcp.26383. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Li G. MiR-1284 enhances sensitivity of cervical cancer cells to cisplatin via downregulating HMGB1. Biomed Pharmacother. 2018;107:997–1003. doi: 10.1016/j.biopha.2018.08.059. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Zhou C. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. 2018;496:738–745. doi: 10.1016/j.bbrc.2018.01.097. [DOI] [PubMed] [Google Scholar]
- 18.Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y, Zhang G. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi: 10.1016/j.ijbiomac.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Liu X, He B, Pan Y, Sun H, Xu T, Hu X, Wang S. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7:2051–2069. [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR-449a suppresses tumor growth, migration, and invasion in non-small cell lung cancer by targeting a HMGB1-mediated NF-κB signaling pathway. Oncol Res. 2019;27:227–235. doi: 10.3727/096504018X15213089759999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv S, Guan M. miRNA-1284, a regulator of HMGB1, inhibits cell proliferation and migration in osteosarcoma. Biosci Rep. 2018;38(BSR20171675) doi: 10.1042/BSR20171675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.He R, Yang L, Lin X, Chen X, Lin X, Wei F, Liang X, Luo Y, Wu Y, Gan T, et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8:15632–15641. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K, Huang J, Ni J, Song D, Ding M, Wang J, Huang X, Li W. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16:578–587. doi: 10.1080/15384101.2017.1288324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 targets the 3'UTR of HMGB1 and inhibits the HMGB1-associated autophagy in osteosarcoma cells during chemotherapy. Tumour Biol. 2014;35:6021–6028. doi: 10.1007/s13277-014-1797-0. [DOI] [PubMed] [Google Scholar]
- 26.Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang Y, Wang Y, Zhao W, Wang W. miR-22 inhibits osteosarcoma cell proliferation and migration by targeting HMGB1 and inhibiting HMGB1-mediated autophagy. Tumour Biol. 2014;35:7025–7034. doi: 10.1007/s13277-014-1965-2. [DOI] [PubMed] [Google Scholar]
- 27.Jiang D, Wang H, Li Z, Li Z, Chen X, Cai H. MiR-142 inhibits the development of cervical cancer by targeting HMGB1. Oncotarget. 2017;8:4001–4007. doi: 10.18632/oncotarget.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Yu X, Song X, Li G, Mao X, Zhang Y. Inhibiting the cytoplasmic location of HMGB1 reverses cisplatin resistance in human cervical cancer cells. Mol Med Rep. 2017;15:488–494. doi: 10.3892/mmr.2016.6003. [DOI] [PubMed] [Google Scholar]
- 29.Lee HJ, Kim A, Song IH, Park IA, Yu JH, Ahn JH, Gong G. Cytoplasmic expression of high mobility group B1 (HMGB1) is associated with tumor-infiltrating lymphocytes (TILs) in breast cancer. Pathol Int. 2016;66:202–209. doi: 10.1111/pin.12393. [DOI] [PubMed] [Google Scholar]
- 30.Zhou RR, Zhao SS, Zou MX, Zhang P, Zhang BX, Dai XH, Li N, Liu HB, Wang H, Fan XG. HMGB1 cytoplasmic translocation in patients with acute liver failure. BMC Gastroenterol. 2011;11(21) doi: 10.1186/1471-230X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.