Abstract
Neurodegeneration in Alzheimer’s disease (AD) is closely associated with accumulation of pathologic tau aggregates in the form of neurofibrillary tangles. We found that a p.Asp395Gly mutation in VCP was associated with dementia characterized neuropathologically by neuronal vacuoles and neurofibrillary tangles. Moreover, VCP appeared to exhibit tau disaggregase activity in vitro which was impaired by the p.Asp395Gly mutation. Additionally, intracerebral microinjection of pathologic tau led to increased tau aggregates in p.Asp395Gly VCP knock-in mice compared to injected wild-type mice. These findings suggest that p.Asp395Gly VCP is an autosomal dominant genetic mutation associated with neurofibrillary degeneration in part due to reduced tau disaggregation, raising the possibility that VCP may represent a therapeutic target for the treatment of AD.
One Sentence Summary:
p.Asp395Gly VCP exhibits impaired PHF-tau disaggregase activity and is associated with a neurodegenerative disease, vacuolar tauopathy.
Main Text:
Alzheimer’s disease (AD) and other tauopathies are progressive and fatal neurologic diseases characterized by tau protein aggregation and neurodegeneration (1-3). Autosomal dominant mutations that result in AD pathologies have been linked mechanistically to the production or aggregation of pathologic proteins (4). Less is known about which endogenous disaggregating factors play an etiologic role in neurodegenerative disease. Valosin-containing protein (VCP, also known as p97) is a member of the AAA+ protein family (ATPases Associated with diverse cellular Activities) (5, 6) and uses the energy from ATP hydrolysis to extract proteins from various macromolecular complexes (7). VCP is an essential gene and is expressed across CNS cell types (8, 9). We report here that the loss of VCP tau disaggregase function is associated with autosomal dominant dementia, suggesting that impaired tau turnover can promote neurofibrillary degeneration.
Results
p.Asp395Gly VCP is Associated with Autosomal Dominant Frontotemporal Degeneration
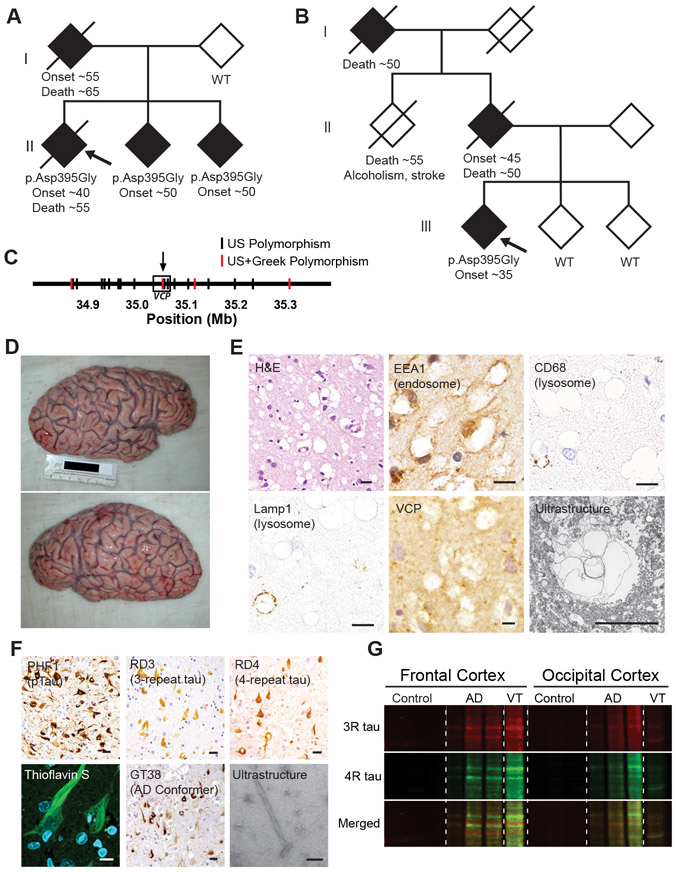
We identified two families from the United States and Greece (10) with an autosomal dominant inheritance pattern of frontotemporal degeneration (FTD) (Fig. 1A and B). Targeted, exome, and whole genome sequencing of the American family identified a VCP mutation (c.1184A>G, p.Asp395Gly, NM_007126.5) in three affected family members which was absent in the unaffected parent (Fig. 1A and Table S1). The Greek family also demonstrated an autosomal dominant inheritance pattern of FTD with p.Asp395Gly (c.1184A>G) VCP identified in the affected proband which was absent in both unaffected siblings (Fig. 1B and Table S1). Neither family exhibited muscle or bone disease. Whole genome sequencing revealed that VCP c.1184A>G was the only rare (allele frequency <0.1%) protein coding variant shared by the three affected American siblings and the Greek proband (Table S2). Haplotype data extracted from whole genome sequencing indicated that VCP c.1194A>G did not seem to originate from a common founder (Fig. 1C). MAPT mutations were absent in both families.
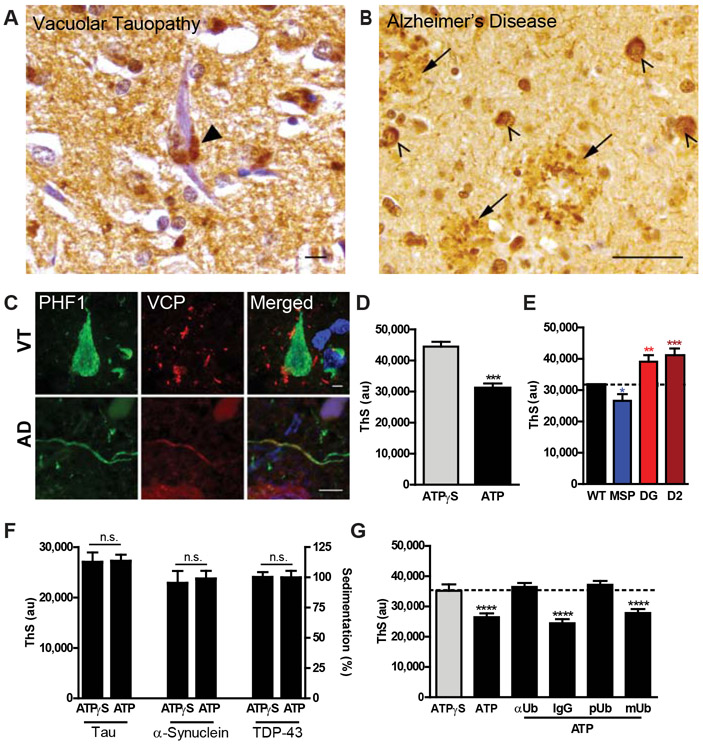
Fig. 1. p.Asp395Gly VCP is associated with vacuolar tauopathy.
(A and B) Family pedigrees for the kindred from the United States (A) or Greece (B) showing an autosomal dominant pattern of inheritance with shaded shapes denoting individuals with FTD. VCP genotype, approximate age of disease onset and death are listed, if available. Proband denoted by arrow. (C) Distinct VCP haplotypes suggest absence of a common founder mutation. Genotype data was extracted from whole genome sequencing where polymorphisms shared between the three affected American siblings but absent in their unaffected parent are shown in black, and polymorphisms which are additionally found in the affected Greek individual are highlighted in red. VCP designated by box with arrow showing location of the p.Asp395Gly VCP mutation. (D) Gross photographs of VT brain showing circumscribed frontal atrophy. (E) Vacuolar neuropathology. Representative hematoxylin and eosin (H&E), antibody immunostain, or electron microscopic ultrastructure images are shown. Scales bars are 10 μm. (F) Neuropathology of tau aggregates. Representative antibody immunostain, thioflavin S stain, or electron microscopic ultrastructure images are shown. Scale bars are 10 μm except for electron microscopy which is 200 nm. (G) Biochemical analysis of pathologic tau. Sarkosyl-insoluble frontal and occipital neocortex lysates from control, AD, or VT were immunoblotted for 3-repeat (RD3, red) and 4-repeat tau (anti-4R tau, green).
Neuropathologic Characterization of Vacuolar Tauopathy
An autopsy was performed on the American proband. The brain weighed 1,090 grams and exhibited severe circumscribed frontal atrophy consistent with frontotemporal lobar degeneration (Fig. 1D). Microscopic analysis revealed marked grey matter neuronal vacuolization (Fig. 1E). Vacuoles appeared to be immunoreactive for the endocytic marker, EEA1 (Fig. 1E), with less than 1% of vacuoles positive for lysosomal markers (CD68 and Lamp1, Fig. 1E). VCP staining showed granular staining in the neuropil but was negative in vacuoles (Fig. 1E). Vacuoles were negative for markers of endoplasmic reticulum, multivesicular bodies, and autophagosomes (calnexin, CD63, LC3B, Fig. S1A). Ultrastructural analysis revealed electron-lucent, membrane bound vacuoles (Fig. 1E). These findings suggested that these vacuoles may be endocytic in origin with rare vacuoles exhibiting lysosomal features.
Neuron loss affected the frontal, temporal, and parietal neocortices, together with abundant tau protein aggregates (Fig. 1F and Fig. S1B). TDP-43, Aβ, α-synuclein, and prion protein aggregates were absent (Fig. S1C and D). Tau protein aggregates were morphologically identical to the neurofibrillary tangles (NFTs) found in AD. Tau inclusions were strongly reactive with Thioflavin-S (ThS) and Gallyas silver stain, and were immunoreactive with antibodies specific to phosphorylated tau (PHF1), 3-repeat tau, and 4-repeat tau, and ubiquitin (Fig. 1F and Fig. S1E). Tau aggregates were also immunoreactive to GT-38, an antibody that recognizes a conformation specific to AD tau (Fig. 1F) (11). Accumulation of 3-repeat and 4-repeat sarkosyl-insoluble tau was confirmed biochemically, and VCP protein expression appeared normal (Fig. 1G and Fig S1F). Electron microscopy of biochemically extracted insoluble tau protein demonstrated paired helical filaments (PHF; Fig. 1F). Thus, the tau pathology of p.Asp395Gly VCP was characterized by accumulation of NFTs of similar composition to those found in AD.
Distribution of Neuronal Vacuolization and Neurofibrillary Degeneration
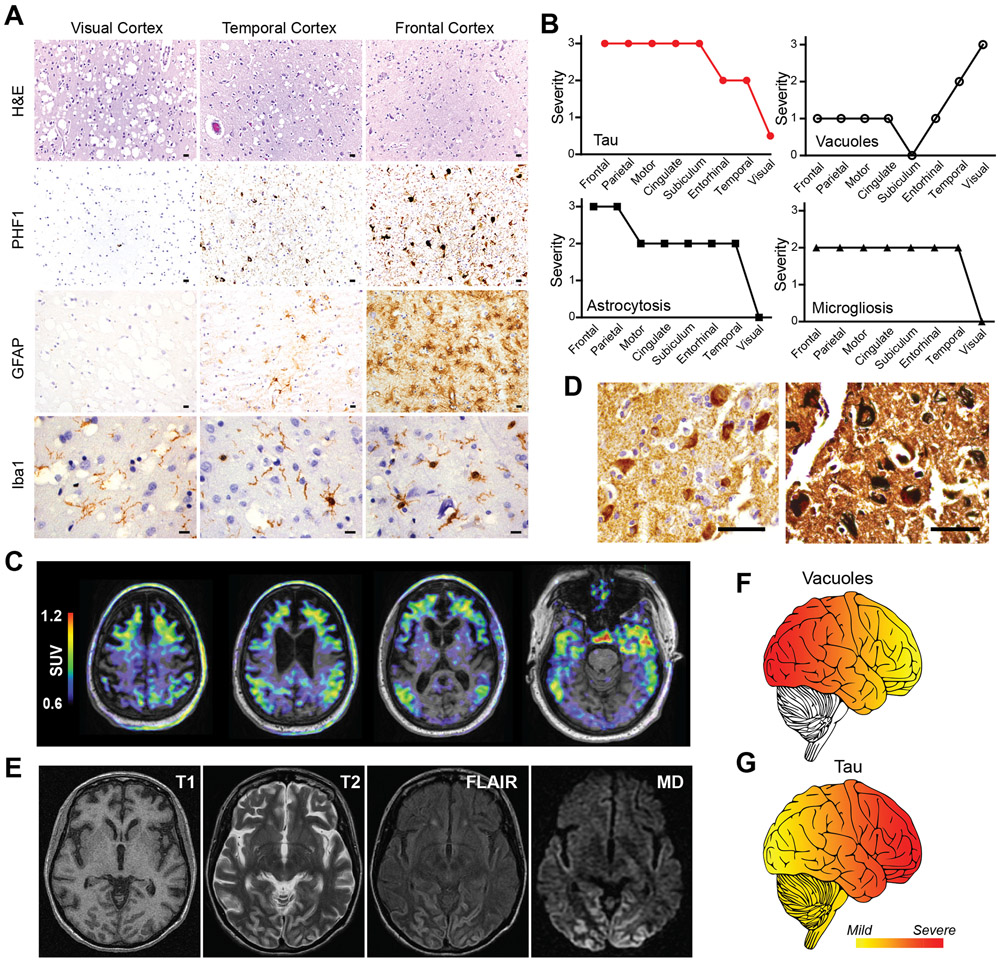
Neuronal vacuolization and neurofibrillary degeneration were inversely related (Fig. 2A). Regional analysis indicated that vacuoles were most prominent in the occipital neocortex with fewer vacuoles rostrally, in contrast with NFTs which were most abundant in the frontal neocortex with fewer along a caudal gradient (Fig. 2B and Fig. S1G). Reactive astrocytosis and microgliosis were seen in regions with neurofibrillary degeneration (Fig. 2A and B and Fig. S1G). White matter was unaffected by vacuoles or tauopathy. Thus, in contrast with NFTs, vacuoles appeared not to be directly associated with degeneration, thereby differentiating them from the vacuoles of prion disease, the rimmed vacuoles of inclusion body myopathy, or the non-specific microvacuolar superficial spongiosus common to many neurodegenerative diseases.
Fig. 2. Inverse regional relationship between vacuoles and neurofibrillary degeneration in vacuolar tauopathy.
(A) Rostral to caudal gradient of vacuoles versus neurofibrillary degeneration. Representative hematoxylin and eosin (H&E), PHF1 (phospho-tau), GFAP (astrocyte), and Iba1 (microglia) stained images from occipital/visual, temporal, and frontal neocortical regions are shown. Scale bars are 10 μm. (B) Regional scoring of tau accumulation, vacuoles, astrogliosis, and microgliosis. Cortical brain regions were scored for severity of tau (red), vacuoles (circles), astrogliosis (squares), and microgliosis (triangles). Severity scores ranged from 0 (none) to 3 (severe). (C) Neuroimaging of affected kindred. Overlaid T1-weighted MRI and tau PET images from affected p.Asp395Gly VCP carrier pseudocolored to standard uptake values (SUV). (D) Cerebral biopsy of Greek proband demonstrates neurofibrillary tangles. Representative images of AT8 (phospho-tau, left) and Gallyas silver (right) stained sections from a frontal neocortex biopsy. Scale bars are 50 μm (left) and 20 μm (right). (E) Neuroimaging of Greek proband. MRI neuroimaging images are shown including T1, T2, fluid-attenuated inversion recovery (FLAIR), and MD from diffuse tensor imaging. (F and G) Schematic showing the distribution of (F) vacuole or (G) tau pathology is illustrated (yellow=mild, red=severe) where vacuoles were most severe in caudal neocortical regions in contrast with tau aggregates which were most severe in rostral neocortical regions.
To confirm that p.Asp395Gly VCP was associated with tauopathy, MRI and 18F-flortaucipir (formerly 18F-AV-1451) PET imaging was performed on a living affected sibling which demonstrated frontotemporal atrophy coinciding with tau tracer retention in a frontotemporal distribution (Fig. 2C and Fig. S2). In addition, frontal cerebral biopsy of the affected Greek proband also confirmed the presence of tauopathy consisting of NFTs (Fig. 2D). Moreover, mean diffusivity (MD) from diffusion tensor imaging of the Greek proband demonstrated “cortical ribboning,” most prominently affecting the occipital lobe, which is a known feature of vacuolated cortex (Fig. 2E) (12). Thus, vacuolar pathology was most prominent in caudal neocortical regions (Fig. 2F) which showed minimal radiologic and pathologic evidence of neurodegeneration. In contrast, tau accumulation correlated with cerebral atrophy, neuron loss and reactive gliosis (Fig. 2G).
These atypical neuropathologic features, together with the absence of the p.Asp395Gly variant of VCP from multiple genetic databases (1000 Genomes Project, the Alzheimer’s Disease Genetic Consortium, the Genome Aggregation Database - gnomAD), suggested that this mutation results in a unique neurodegenerative disease which we have named vacuolar tauopathy (VT). Despite its uniqueness, the similarities in morphology and composition of tau aggregates in VT and AD suggested that VCP may be involved in regulating the formation or dissolution of NFTs.
p.Asp395Gly VCP is a Hypomorph Mutation
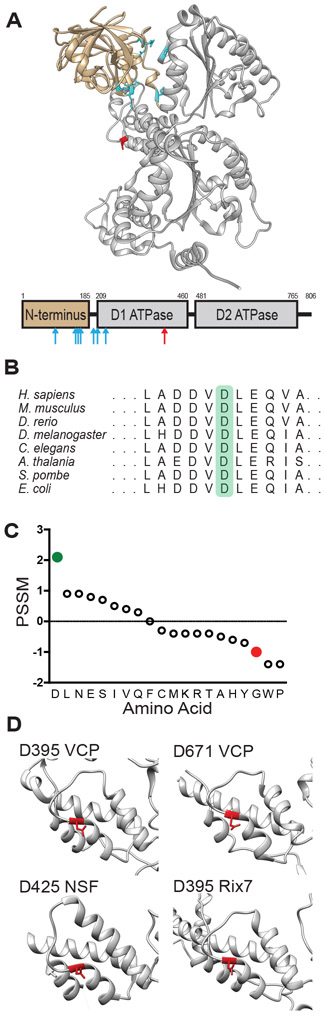
Previously identified VCP mutations cause multisystem proteinopathy (MSP), a pleiotropic degenerative disease that affects the muscle, bone, and nervous system which sometimes manifests as frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) (13-15). MSP mutations with autopsy confirmed FTLD-TDP uniformly lie in the cleft between the N-terminal and D1 ATPase domains (Fig. 3A, cyan). Furthermore, these MSP mutations increase ATPase activity (16, 17) and unfolding activity (18, 19) suggesting that MSP is associated with gain-of-function VCP mutations.
Fig. 3. In silico analysis of D395 VCP.
(A) Structure of VCP monomer. N terminal domain is tan, D1 and D2 ATPase domains are gray, MSP mutations are cyan, and VT mutation is red (PDB 5C19). The primary structure of VCP is underneath with the same color scheme, mutations marked by arrows. (B) Conservation of D395 VCP across species (highlighted in green). (C) PSSM scores for conservation in VCP homologues for amino acids including aspartic acid (D, green) and glycine (G, red). (D) Common aspartic acid N-cap in AAA+ proteins. The structures of several AAA+ proteins are shown with D395, or its equivalent, highlighted in red (VCP PDB 5C19, NSF PDB 1NSF, Rix7 PDB 6MAT).
In contrast, p.Asp395Gly VCP appeared to be associated with tauopathy in the absence of muscle or bone disease, and the mutation is found within the lid subdomain of the D1 ATPase domain (Fig. 3A, red). Amino acid position 395 is the N-cap of a short α-helix. Because the N-cap of an α-helix does not have amino acids above for hydrogen bonding, the helix is stabilized by the N-cap’s side chain hydrogen bonding within the interior of the α-helix. Aspartic acid (D) is among the most stabilizing of N-cap amino acids while glycine (G) is destabilizing (20, 21). Asp395 is conserved from humans to plants to bacteria (Fig. 3B). Furthermore, position specific scoring matrix (PSSM) scores for position 395 were calculated for VCP and its homologues (Fig. 3C). The positive PSSM score suggested that D has been evolutionary selected and is functional at this position. The gnomAD database contains a single instance of the relatively conservative p.Asp395Glu variant which is of uncertain significance given its rarity and the lack of accompanying phenotype data. In contrast, the negative PSSM score associated with glycine (G) at this position has been evolutionarily selected against and may be detrimental to VCP function. Indeed, multiple in silico analyses predicted that p.Asp395Gly has a deleterious effect on VCP function (Fig. S3A). In addition to being conserved among VCP homologues, this residue is found in the same corresponding lid subdomain location of several AAA+ ATPase domains (Fig. 3D) including the D2 ATPase domain of VCP, NSF, and Rix7. Overall, these analyses suggested that Asp395 may be important for VCP function.
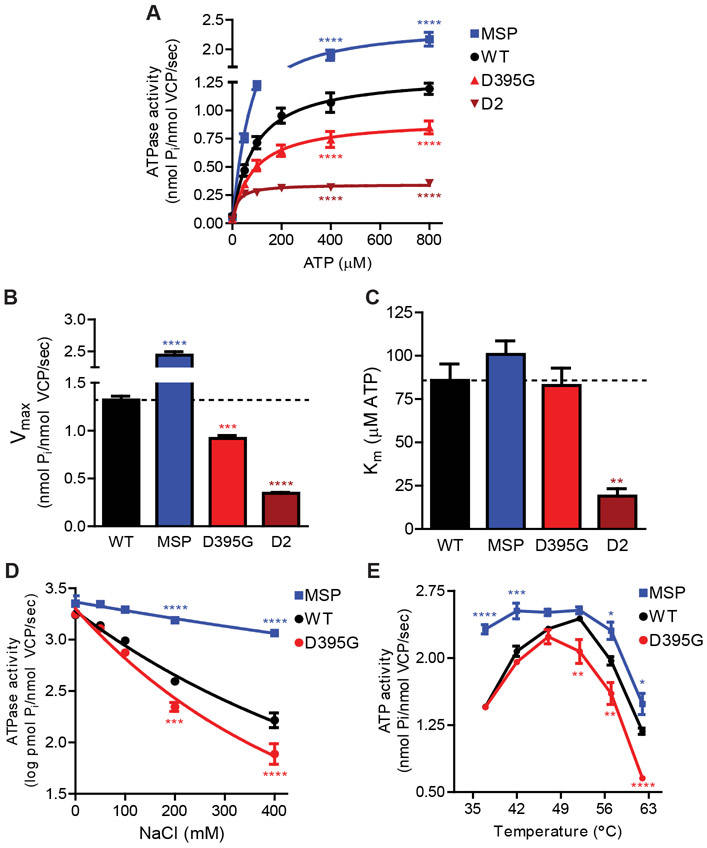
To determine the effect of p.Asp395Gly on VCP activity, Michaelis-Menten enzyme kinetics was performed on purified recombinant VCP proteins (Fig. 4A and Fig. S3B, UniProtKB: Q01853). As expected, the MSP mutation p.Asp232Glu increased VCP ATPase activity (MSP, blue) compared to wild type (black), while the D2 mutation p.Glu578Gln (Walker B mutation in the D2 ATPase domain) that inactivates ATPase hydrolysis decreased VCP ATPase activity (maroon). The p.Asp395Gly mutation decreased VCP ATPase activity (red), resulting in a 30.3% reduction in Vmax (Fig. 4B) and no significant change in Km (Fig. 4C).
Fig. 4. ATPase activity of mutant VCP proteins.
(A) Michaelis-Menten kinetic ATPase analysis. Recombinant WT, MSP, p.Asp395Gly, and D2 VCP proteins were assessed for ATPase activity over a range of ATP concentrations (n=3). ATPase activity shown as mean +/− SEM (two-way ANOVA: mutation p<0.0001; ATP p<0.0001; interaction p<0.0001; Bonferroni posthoc shown for only 400 and 800 μM ATP ****p<0.0001). (B) Vmax values and (C) Km from Michaelis-Menten kinetics (n=3). Data shown as mean +/− SEM (one-way ANOVA: Vmax p<0.0001; Km p=0.0005; Bonferroni posthoc **p<0.01 ***p<0.001 ****p<0.0001). (D) Salt denaturation of recombinant VCP proteins. VCP ATPase activity was assessed across NaCl concentrations (n=5). ATPase activity shown as mean +/− SEM (two-way ANOVA: mutation p<0.0001; NaCl p<0.0001; interaction p<0.0001; Bonferroni posthoc shown for only 200 and 400 mM NaCl ***p<0.001 ****p<0.0001). (E) Heat activation and denaturation of recombinant VCP proteins. VCP ATPase activity was assessed across temperature (n=3). ATPase activity shown as mean +/− SEM (two-way ANOVA: mutation p<0.0001; temperature p<0.0001; interaction p<0.0001; Bonferroni posthoc *p<0.05 **p<0.01 ***p<0.001 ****p<0.0001).
We hypothesized that p.Asp395Gly may result in local protein destabilization which may manifest as enhanced sensitivity to denaturation. Thus, we tested recombinant VCP proteins for ATPase activity in the presence of increasing salt concentration (Fig. 4D and Fig. S3C). MSP mutant protein was relatively resistant to salt denaturation while p.Asp395Gly mutant protein displayed increased sensitivity to salt denaturation. We also tested the ATPase activity of VCP with increasing temperature. VCP is known to heat activate prior to heat inactivating at higher temperatures due to denaturation (Fig. 4E, black and Fig. S3C) (22). While MSP mutant protein heat inactivated at a similar temperature compared to WT VCP protein, p.Asp395Gly mutant protein heat inactivated at lower temperatures. Overall, these data supported the hypothesis that p.Asp395Gly is a hypomorph mutation.
ATP- and Polyubiquitin-Dependent PHF-tau Disaggregase
Recent studies have demonstrated that VCP can unfold protein substrates in an ATP-dependent manner (18, 19, 23-26). Immunohistochemistry of VT brain demonstrated granular VCP staining adjacent to but excluded from NFTs (Fig. 5A and C). In contrast, AD tissues showed wild-type VCP localized to NFTs, dystrophic neurites within neuritic plaques, and tau-positive neuropil threads (Fig. 5B and C), raising the possibility that VCP interacts with pathologic tau in AD.
Fig. 5. ATP- and polyubiquitin- dependent tau disaggregase activity of VCP.
(A) VT neocortex showing granular VCP immunostaining (arrowhead) adjacent to a NFT. Scale bar is 10 μm. (B) AD neocortex showing VCP immunostaining within NFTs (arrows) and dystrophic neurites in neuritic plaques (arrowheads). Scale bar is 50 μm. (C) Localization of VCP and tau in human VT and AD tissues. Double immunofluorescence of VCP (green) and PHF1 (phospho-tau, red) in VT (top) and AD (bottom). Scale bars are 5 μm. (D) Recombinant VCP activity against pathologic PHF-tau. ThS fluorescence after incubation of human AD tissue derived pathologic tau with recombinant VCP, UFD1, NPLOC4, and either ATPγS or ATP (n=5). ThS signal shown as mean +/− SEM (two-tailed t-test, ***p=0.0003). (E) Mutant VCP activities against pathologic PHF-tau. ThS fluorescence after incubation of pathologic tau with recombinant WT, MSP, p.Asp395Gly (DG), or D2 VCP (n=5). ThS signal shown as beta +/− SEM (mixed effects model, MSP *p=0.0272; **DG p=0.0036; ***D2 p=0.0005). (F) VCP activity against recombinant protein aggregates. Recombinant tau, α-synuclein, or TDP-43 aggregates were incubated with VCP, VCP cofactors, and ATPγS or ATP. ThS fluorescence signal shown as a mean +/− SEM for tau (n=3) and α-synuclein (n=3). Sedimentation data shown as mean percent of average of ATPγS samples +/− SEM for TDP-43 (n=5). Pairwise testing by two-tail t-test resulted in non-significance (n.s.). (G) Polyubiquitin-dependence of VCP activity against pathologic PHF-tau. ThS fluorescence after addition of anti-ubiquitin monoclonal antibody (αUb), nonspecific isotope control (IgG), recombinant polyubiquitin (pUb), or recombinant monoubiquitin (mUb) to VCP, VCP cofactors, pathologic tau, and ATP (black with underline, n=5). Reactions with only VCP, VCP cofactors, pathologic tau, and either ATP or ATPγS (black and gray bar, respectively) were run in parallel as controls. ThS fluorescence signal shown as beta +/− SEM (mixed effects model: ATP, αUb, pUb ****p<0.0001).
Because VCP colocalizes with tau aggregates in AD and the partial loss-of-function p.Asp395Gly mutation is associated with tau accumulation, we hypothesized that VCP may be a tau disaggregase. To assess this, we treated pathologic tau aggregates purified from human AD brain tissue (27) with recombinant VCP protein together with VCP cofactors (UFD1L and NPLOC4, Fig. S3B and D) and either ATP or ATPγS, a nonhydrolyzable ATP analog. Tau aggregate levels were measured using ThS which undergoes a fluorescence shift when bound to pathologic tau fibrils (28). VCP exhibited a 30.1% ATP-dependent reduction in ThS signal (Fig. 5D). Furthermore, the gain-of-function MSP mutant protein (blue) led to a further decrease of ThS signal, while the loss-of-function p.Asp395Gly (red) and D2 mutant (maroon) proteins led to increased ThS signal (Fig. 5E; 16.3% decrease vs 23.0% and 30.0% increases, respectively).
Recent structural evidence suggests that VCP is a ubiquitin-dependent segregase that requires at least five conjugated polyubiquitins (25, 26). We predicted VCP would not have activity against recombinant protein aggregates due to the absence of polyubiquitination. In vitro aggregated recombinant tau, α-synuclein, or TDP-43 were incubated with VCP, VCP cofactors, and either ATP or ATPγS which indeed demonstrated that VCP had no activity against recombinant protein aggregates based on ThS signal or sedimentation analysis (Fig. 5F and Fig. S3E). Rather, VCP appeared to require polyubiquitin for substrate recognition, because addition of monoclonal antibodies specific to ubiquitin (αUb) was able to block VCP activity against AD tissue-derived pathologic tau while nonspecific isotype control antibodies had no effect (Fig. 5G). Similarly, addition of recombinant polyubiquitin (pUb) blocked VCP activity while recombinant mono-ubiquitin (mUb) did not block VCP activity (Fig. 5G). These data suggested that polyubiquitin mediates substrate recognition by VCP.
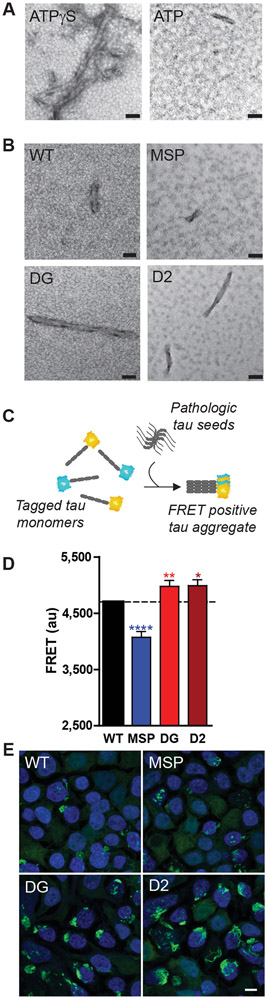
To provide orthogonal evidence for VCP activity against PHF tau, disaggregase reactions were examined by negative staining electron microscopy which revealed that VCP reduced the abundance of pathologic tau fibrils in an ATP-dependent manner (Fig. 6A) which was partially mitigated by the p.Asp395Gly mutation (Fig. 6B). These data suggested that VCP exhibits an ATP-dependent disaggregase activity against pathologic tau aggregates derived from human AD brain. Furthermore, tau disaggregase activity of VCP mutant proteins appeared to correlate with ATPase activity in that the gain-of-function MSP mutation increased VCP tau disaggregase activity while loss-of-function VT and D2 mutations decreased VCP tau disaggregase activity.
Fig. 6. VCP activity against pathologic tau fibrils and cellular tau aggregates.
(A) Electron microscopy of human AD tissue derived PHF tau fibrils after treatment with recombinant VCP, UFD1, NPLOC4, and either ATPγS or ATP (n=3). Scales bars are 50 nm. (B) Electron microscopy of VCP tau disaggregase reactions with recombinant WT VCP protein versus MSP, p.Asp395Gly (DG), and D2 mutant VCP proteins (n=3). Scales bars are 50 nm. (C) Schematic of tau seeding in tau biosensor cells. (D and E) Biosensor cells expressing WT or mutant VCP were transduced with human AD tissue derived pathologic tau (n=5). (D) FRET signal was assessed by flow cytometry and shown as beta +/− SEM (mixed effects model: MSP ****p<0.0001; **DG p=0.0095, *D2 p=0.0126). (E) Representative confocal fluorescence microscopy images of biosensor cells with intracellular tau aggregates (green) and DAPI nuclear counterstain (blue). Scale bar is 10 μm.
To confirm that VCP affects tau aggregates, we used a biosensor cell line engineered to develop intracellular aggregates upon seeding with proteopathic tau (29). When introduced to these cells, pathologic tau aggregates nucleate the aggregation of tau reporter proteins resulting in a FRET signal as a measure of intracellular tau aggregation (Fig. 6C). Biosensor cells overexpressing wild-type or mutant forms of VCP were transduced with AD brain derived tau. Relative to wild-type VCP, biosensor cells expressing p.Asp395Gly VCP exhibited higher FRET signal and increased intracellular tau aggregation, similar to what was observed with the ATPase deficient D2 mutant VCP (Fig. 6D). In contrast, biosensor cells expressing MSP mutant VCP exhibited reduced FRET signal and reduced intracellular tau aggregation (Fig. 6D). Confocal microscopy of biosensor cells confirmed that cells expressing p.Asp395Gly VCP exhibited increased intracellular tau aggregates (Fig. 6E).
Tau Accumulation in p.Asp395Gly Knock-in Mice
To confirm our in vitro findings and to test p.Asp395Gly VCP’s role in tau pathogenesis, we generated a p.Asp395Gly knock-in mutant mouse line (DG) using CRISPR-Cas9 (Fig. S4A) (30). While DG knock-in mice exhibited decreased litter size, non-Mendelian breeding, and mildly decreased one-year survival (Fig. S4B to D), no overt neurodegeneration was observed. RNA-sequencing revealed essentially no significantly altered transcripts in DG mice (Fig. S4E to G), and biochemical and immunohistochemical analysis did not reveal changes in VCP, tau, and other proteostasis related factors (p62, EEA1, LC3, and ubiquitin; Fig. S4H and I). Thus, mutant p.Asp395Gly VCP did not result in any overt CNS developmental phenotype, and appeared to be insufficient to initiate tau aggregation which was consistent with our hypothesis that VCP acts as a disaggregase, downstream of tau aggregation which does not occur spontaneously in mice.
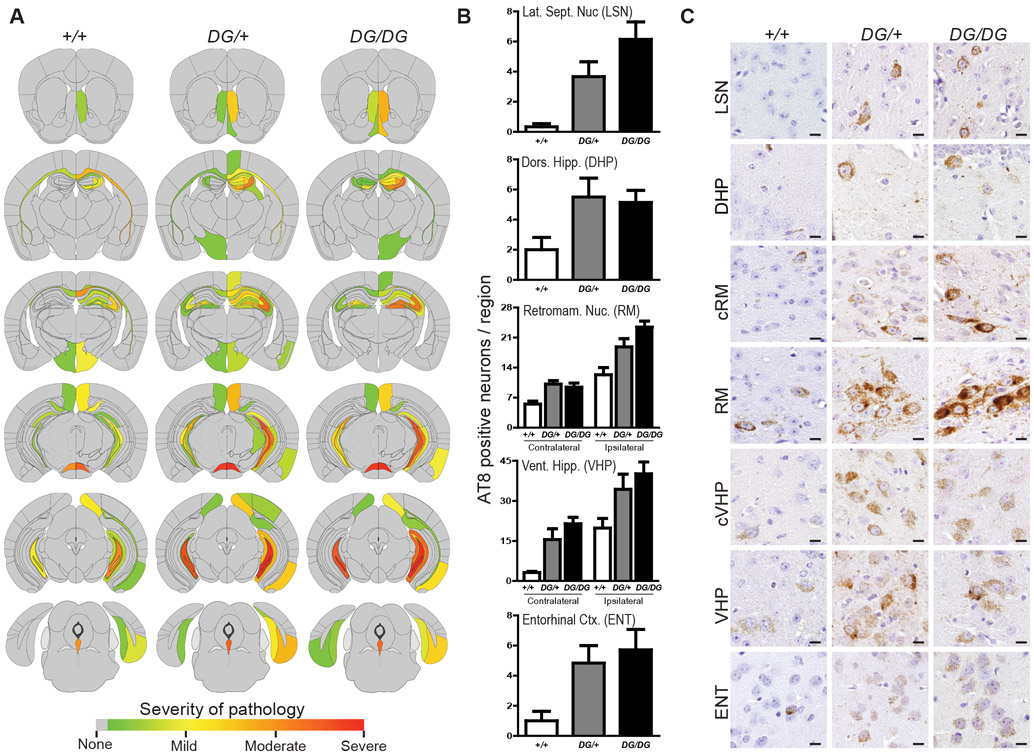
To initiate tau aggregation, three-month old +/+, DG/+, and DG/DG mice (wild-type, heterozygous, and homozygous p.Asp395Gly, respectively) were stereotaxically microinjected intracerebrally with pathologic AD tau in the dorsal hippocampus and overlying cortex and assessed three months post injection for tau pathology. Semiquantitiative scores of AT8-positive tau aggregates were used to generate brain heat maps to visualize the distribution and severity of tau pathology. While the distribution of tau pathology was similar across all genotypes (Fig. 7A), DG/+ and DG/DG mice appeared to exhibit more tau pathology in several brain regions. Selected brain regions that are neuroanatomically connected to the injection site were then quantitatively assessed for AT8-positive tau aggregates. DG/+ and DG/DG mice demonstrated an increase in the number of affected neurons across all examined regions (Fig. 7B). More severe pathology was present in both anterior and posterior regions as well as in connected regions contralateral to the injection site (Fig. 7B and C). A mixed effects linear regression model demonstrated that DG/+ and DG/DG mice exhibited significantly more AT8-positive neurons relative to wild-type mice (DG/+ p=0.0005; DG/DG p<0.0001; see Fig. S5A and B). Iba1 and GFAP immunohistochemical staining showed minimal to no reactive gliosis in response to accumulation of tau aggregation, as this paradigm specifically models tau aggregation without confounding downstream neurodegeneration (Fig. S5C). Moreover, injecting VCP mutant mice with control human brain lysates or AD lysates that were immunodepleted of tau did not induce tau aggregation (Fig. S5D). These data indicated that p.Asp395Gly VCP exacerbates tau pathology in vivo and were consistent with our in vitro data suggesting that VCP may function as a pathologic tau disaggregase.
Fig. 7. p.Asp395Gly VCP exacerbates tau pathology in vivo.
(A) Semiquantitative analysis of AT8-positive phospho-tau pathology. Six-month old wild-type (+/+), heterozygous p.Asp395Gly (DG/+), and homozygous p.Asp395Gly (DG/DG) knock-in mice analyzed three months after injection of pathologic tau in the right dorsal hippocampus and overlying cortex (n=6-7 mice per genotype). Semiquantitative scores were averaged across mice of the same genotype and mapped onto coronal brain maps (grey=no pathology, red=severe pathology). (B) Regional quantitative analysis of AT8-positive phospho-tau pathology. The number of AT8-positive neurons was counted across different brain regions, shown as beta +/− SEM (mixed effects model: DG/+ p=0.0005; DG/DG p<0.0001). (C) Tau pathology in p.Asp395Gly knock-in mice. Representative AT8 immunostained images are shown. Scale bars are 10 μm. LSN lateral septal nucleus, DHP dorsal hippocampus, RM retromammillary nucleus, VHP ventral hippocampus, ENT entorhinal cortex, c contralateral.
Discussion
We report a genetic mutation that impairs tau aggregate turnover resulting in neurodegeneration, distinct from previously described genetic mutations linked to production or aggregation propensity of tau. We posit that neurons in VT are at increased susceptibility to tau aggregation resulting in downstream neurodegeneration. This also represents a remarkable instance of convergent allelic heterogeneity where different mutations of a single gene (VCP) result in different underlying diseases (VT vs. MSP) which can manifest with the same clinical phenotype (FTD). Moreover, our data suggest that VCP can function as a tau disaggregase and that a mutation in the lid subdomain can impair VCP ATPase and tau disaggregase activity. As the lid subdomain is highly conserved across VCP homologues, understanding the structure and function of the lid subdomain may further our understanding of the dynamic processes that underlie VCP disaggregase activity. Finally, while much attention has been afforded to the biosynthetic and aggregation pathways that lead to abnormal protein inclusions in AD and related disorders, these findings indicate that disaggregation pathways may play a causal role in neurodegeneration and suggests that modulating VCP activity may represent a new therapeutic target for the treatment of AD and other tauopathies.
Materials and Methods:
Cloning and Mutagenesis
N-terminal histidine tagged wild-type VCP was obtained from Addgene (plasmid #12373, gift from Axel Brunger). Untagged NPLOC4 (pCOLA-Duet, Novagen, NcoI/EcoR1) and N-terminal histidine tagged UFD1 (pET-Duet, Novagen, NcoI/HindIII) were obtained from Hermann Schindelin. Full-length human tau was inserted in pRK172 vector using NdeI/EcoR1, full-length human α-synuclein was inserted in pRK172 vector using NdeI/HindIII, and full-length human TDP-43 with a C-terminal histidine tag was inserted in pCOLD vector (CNDR, University of Pennsylvania). QuikChange II site-directed mutagenesis kit (Agilent Technology) was used to generate desired mutations. Plasmid sequences were confirmed with Sanger sequencing.
Recombinant Protein Purification
For VCP, plasmids were transformed into Escherichia coli Rosetta (DE3) cells (Novagen) following manufacturer’s instructions. Cells were grown in Terrific Broth supplemented with kanamycin and induced at OD600 ~0.600 with 1 mM IPTG for 18 hours at 15 °C. Cells were collected by centrifugation at 4,000xg for 15 minutes at 4 °C and then resuspended in lysis buffer (100 mM Tris pH 7.4, 500 mM KCl, 5 mM MgCl2, 1 mM ATP, 5% glycerol, 2 mM β-mercaptoethanol, 20 mM imidazole, 1 mg/mL lysoszyme, EDTA-free Complete Protease Inhibitor (Roche)) for 30 minutes at 4°C. Following sonication, lysates were cleared by centrifugation at 27,500xg for 30 minutes at 4°C. 2.5 mL of Ni-NTA Agarose (Qiagen) was added to the supernatant and nutated for 30 minutes at 4°C. The resin was washed three times with 20 mL of wash buffer 1 (50 mM HEPES pH 7.4, 150 mM KCl, 5 mM MgCl2, 1 mM ATP, 5% glycerol, 2 mM β-mercaptoethanol, EDTA-free Complete Protease Inhibitor (Roche)) and then three additional times with 20 mL of wash buffer 2 (1 M KCl in wash buffer 1). VCP was eluted with 350 mM imidazole in wash buffer 1. Elute was concentrated using an Amicon Ultra-15 centrifugal unit (Millipore) and loaded onto a precalibrated Superose-6 gel filtration column (GE Healthcare) at 4 °C. 0.5 mL fractions were collected in 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM MgCl2, 5% glycerol, 1 mM β-mercaptoethanol. Protein concentrations was determined using the Bradford Assay (Bio-Rad). Fractions were then concentrated to 5 mg/mL using an Amicon Ultra – 0.5 mL centrifugal unit (Millipore), snap frozen, and stored in liquid nitrogen. Additionally, expression plasmid was isolated from each protein preparation and confirmed by Sanger sequencing. Each preparation’s purity was assessed by gel electrophoresis.
For UFD1L and NPLOC4, plasmids were expressed similarly to VCP. Cultures were combined and then lysed in lysis buffer (50 mM Tris pH 7.4, 500 mM KCl, 5 mM MgCl2, 5% glycerol, 2 mM β-mercaptoethanol, and 20 mM imidazole). Purification was identical to VCP except resin was washed with lysis buffer then eluted with lysis buffer plus 300 mM imidazole. Elute was loaded onto a Superdex 200 Increase 10/300 GL (GE Healthcare) column precalibrated with 20 mM HEPES pH 7.4, 250 mM KCl, 1 mM MgCl2, 5% glycerol, and 1 mM β-mercaptoethanol.
For tau, plasmids were expressed in Escherichia coli BL21 (DE3) cells (Agilent Technology). After induction, bacterial pellets were resuspended in 100 mM MES pH 7.5, 750 mM NaCl, 2 mM NaF, 1 mM EGTA, 0.5 mM MgSO4, 1 mM PMSF, and protease inhibitors and then heated to 100°C for 10 minutes and centrifuged at 70,000xg for 30 minutes. Supernatants were then dialyzed into 20 mM piperazine-N,N’-bis(2-ethanesulfonic acid) pH 6.5, 10 mM Nacl, 1 mM EGTA, 1 mM MgSO4, 2 mM DTT, 0.1 mM PMSF and then separated on a HiTrap Sepharose HP IEX cation-exchange column (GE Healthcare) and eluted with a 0 to 400 mM NaCl gradient.
For α-synuclein, plasmids were expressed in Escherichia coli BL21 (DE3) cells (Agilent Technologies). Bacterial pellets were then resuspended in 50 mM Tris pH 7.4, 750 mM NaCl, 1 mM EDTA, and protease inhibitors, and then heated to 100°C for 10 minutes, and centrifuged at 70,000xg for 30 minutes. Supernatants were separated using a Superdex 200 gel filtration column (GE Healthcare). Fractions containing α-synuclein were concentrated using a Centriprep-10 (Millipore), dialyzed in 10 mM Tris pH 7.5, and subsequently separated on a Mono Q column (Amersham Pharmacia Biotech) and eluted with a 0 to 500 mM NaCl gradient.
For TDP-43, plasmids were expressed in Escherichia coli BL21 (DE3)-RIL cells (Agilent Technologies). Bacterial pellets were lysed in 20 mM sodium phosphate pH 7.8, 4 M urea, 500 mM NaCl. Solubilized protein was then bound onto a Ni-NTA column (Qiagen) and eluted with 20 mM sodium phosphate pH 7.8, 4 M urea, 500 mM NaCl and 200 mM imidazole prior to dialyzing into 20 mM Tris pH 8.0, 1M NaCl, 10 mM MgCl2, 1 mM CaCl2, and 50% glycerol.
Recombinant poly- and mono-ubiquitin were purchased (rPeptide).
Recombinant Protein Aggregation
Recombinant proteins were aggregated as previously described (31, 32). Recombinant α-synuclein at 5 mg/ml in PBS pH 7.0 was incubated with agitation at 37°C for 5 days. Recombinant tau at 40 μM was mixed with 40 μM low molecular weight heparin and 2 mM DTT in PBS pH 7.0 and incubated with agitation at 37°C for 5 days. Recombinant TDP-43 protein at 1 mg/ml in 20mM Tris pH 8.0, 1 M NaCl, 10 mM MgCl2, 1 mM CaCl2 and 9% glycerol was incubated with agitation at 37°C overnight, centrifuged at 100,000xg for 60 min at 4°C, and resuspended in PBS.
ATPase Activity Assay
ATPase activity was determined using the EnzCheck phosphatase assay kit (Molecular Probes). ATPase activity was measured in a 96 well flat bottom plate (Fisher Scientific) by a microplate reader (Spark 20M, Tecan). 50 nM of VCP was incubated in 100 μL ATPase reaction buffer (25 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM MgCl2) containing 0, 50, 100, 200, 400, or 800 μM ATP. The absorbance at 360 +/− 5 nm was measured at 45 second intervals for a total of 30 minutes at 37 °C. The amount of inorganic phosphate released was quantified based on a phosphate standard curve (0 to 150 μM) performed on the same plate. Initial enzyme velocity was determined by fitting the data to a linear regression model. Enzyme velocities were then plotted against ATP concentration for Michaelis-Menten kinetic analysis. Samples were run in duplicate. NaCl inactivation assays were conducted in a similar fashion but in the presence of 0, 50, 100, 200, or 400 mM NaCl with 800 μM ATP. For heat activation/inactivation, 50 nM of VCP was incubated in 100 μL ATPase reaction buffer containing 50 mM NaCl and 800 μM ATP. Reactions were incubated for 20 minutes at 37, 42, 47, 52, 57, or 62°C in a Veriti Thermal Cycler (Applied Biosystems). After 20 minutes, the reaction was stopped by incubation at 4 °C for 2 minutes and phosphate levels were determined using the EnzCheck phosphatase assay kit based on a phosphate standard curve performed on the same plate.
VCP Tau Disaggregase Assays
50 nM pathologic tau, 100 nM VCP, and 300 nM UFD1L+NPLOC4 were incubated with 5 mM ATPγS or ATP in 25 mM HEPES pH 7.4, 100 mM KCl, 2 mM MgCl2, 1 mM β-mercaptoethanol at 42°C for 30 min. For Thioflavin-S measurements, 1% Sarkosyl was added to reaction and centrifuged at 186,000 x g for 30 minutes at 4°C. The pellet was resuspended in 10 mM MOPS pH 6.5, 5 μM Thioflavin-S to measure emission at 480 nm. Disaggregase reactions were also absorbed onto carbon/formvar-coated copper grids (Electron Microscopy Sciences) for visualization with a Joel 1010 electron microscope in a blinded manner. Recombinant protein aggregates (100 nM recombinant tau, 100 nM α-synuclein, or 600 nM TDP-43) were incubated with 300 nM VCP and 900 nM UFD1L+NPLOC4, and 5 mM ATP. For tau and α-synuclein, ThS measurements were then performed as described above. TDP-43 was analyzed by sedimentation because TDP-43 aggregates are not strongly positive for ThS. TDP-43 aggregates were centrifuged at 186,000 x g for 30 minutes at 4°C. Pellets were immunoblotted for TDP-43 by SDS PAGE.
FRET Biosensor Experiments
Tau RD P301S FRET Biosensor (ATCC) were maintained in DMEM (Invitrogen) with 10% FBS (Atlanta Biologicals) and 1% Pen-Strep (Invitrogen) at 37°C. Plasmids containing wild-type or mutant VCP were transfected using Lipofectamine 2000 (Thermo Fisher). 24 hours later, pathologic tau lysates were added to biosensor cells using Lipofectamine 2000 (Thermo Fisher). 24 hours later, cells were analyzed by flow cytometry (LSR-II, BD Biosciences) to obtain median FRET fluorescence signal intensity.
Mice
Mice were bred and maintained in a pathogen-free facility at the University of Pennsylvania. Experiments were completed in accordance to the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. Using an established protocol (30), the University of Pennsylvania Transgenic and Chimeric Mouse facility generated knock in animals. CRISPR-Cas9 components were injected into a fertilized oocyte then implanted into a pseudopregnant C57BL/6J mice (Jackson Laboratory). Single-guide RNA (sgRNA:AACAGGTAGAGTGGTGATAA) and single-stranded oligo donor nucleotide (ssODN:GGAATACCTGATGCTACAGGACGTTTGGAGATTCTTCAGATCCATACCAAGAACATGAAACTGGCAGATGATGTGGGCTTGGAACAGGTAGAGTGGTGATAAGGCCTGACCAGAATTTAGGATAGCTCTGTTTTAGGAGGTTGCAAATTACAAACATAGCCTAAGAAAGAGGCTTCATACCAGAAATACAAACTTGGGGA) were used to generate D395G mutant mice (see Fig. S4A). Because human and mouse VCP are identical at the protein level, the mouse genome was only altered by 3 base pairs. The first mutation led to the p.Asp395Gly mutation and the other silent, cis, non-coding mutation occurs in an intron to destroy the PAM site to prevent re-cutting by the Cas9 enzyme. Therefore, wild-type, heterozygous, and homozygous mice are wild-type, heterozygous, or homozygous on both the p.Asp395Gly and PAM site, respectively. Two founders were confirmed by Sanger sequencing to have D395G and no off-target mutations in the top 5 predicted off-target sites. Founders were bred with wild-type C57BL/6J mice for three generations prior to mouse experiments. For litter analyses, p.Asp395Gly mice were compared to PAM mice. PAM mice are wild-type in terms of VCP protein sequence but only harbor the non-coding PAM site mutation. Wild-type, heterozygous, and homozygous PAM mice exhibited no observable phenotype. Other lifespan, biochemical, molecular and histologic analyses compared p.Asp395Gly mice to WT C57BL/6J mice.
For intracerebral injections, mice were anesthetized with ketamine-xylazine-acepromazine and placed in a stereotaxic frame (David Kopf Instruments). Mice were aseptically injected with human AD pathologic tau in the right dorsal hippocampus and overlying cortex (bregma: −2.5mm, lateral: +2mm, depth: −2.4 mm and −1.4 mm from skull) with a Hamilton syringe. Each injection site received 2.5 μL of inoculum at 0.4 μg tau/μL as previously described (27, 33).
RNA-Sequencing
Mice were sacrificed by transcardiac perfusion with PBS. Brain regions were dissected, weighed, and frozen on dry ice. For RNA-sequencing, RNA from anterior cortex were extracted using RNeasy RNA extraction kit (Qiagen) following manufacturer’s instruction. RNA integrity was determined using the RNA Nanochip 6000 kit and an Agilent 2100 bioanalyzer (Agilent). Biological replicates were barcoded, pooled, and sequenced in a single lane on a HiSeq 2000 for 150 x 2 base pair paired-end RNA sequencing at the University of Pennsylvania Next Generation Sequencing Core Facility. Reads were mapped to the mouse genome (mm10) using the RNA-Seq Unified Mapper (RUM) and gene expression differences were analyzed using EdgeR with statistical significance set at a false discovery rate (FDR) of <5%.
Immunoblotting
For human brain, samples were homogenized in a high salt buffer containing 10 mM Tris pH 7.5, 500 mM NaCl, 2 mM EDTA, 1 mM DTT, 10% sucrose containing protease and phosphatase inhibitors (PhosphoSTOP, Roche). Samples were then spun at 55,000xg for 30 minutes at 4°C and supernatant was collected for VCP immunoblotting. Pellets were homogenized in buffer containing 10 mM Tris, 500 mM NaCl, 2 mM EDTA, 1 mM DTT, and 30% sucrose and spun at 55,000xg for 30 minutes at 4°C to float and remove myelin. The resulting pellet was homogenized in buffer containing 2% sarkosyl and 50 mM Tris pH 7.6, followed by centrifugation at 55,000xg for 30 minutes at 4°C. Supernatant was collected and the sarkosyl-insoluble pellets were homogenized at room temperature in buffer containing 2% SDS and 50 mM Tris pH 7.6. Samples were spun at 55,000xg for 30 minutes at room temperature and supernatant was collected for tau immunoblotting.
For mouse brain, samples were sonicated using a probe sonicator (Sonics Vibra-cell) in ice-cold radio immunoprecipitation assay (RIPA) buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS containing protease and phosphatase inhibitors. Sonicated sample were then spun at 55,000xg for 30 minutes at 4°C and supernatant was collected. Pellets were further extracted with 2% SDS buffer containing protease and phosphatase inhibitors at room temperature. Samples were spun at 55,000xg for 30 minutes at room temperature and supernatant was collected.
Brain lysate protein concentrations were quantified using BCA analysis (Pierce, Thermo Fisher). 50 μg of protein was run on either an 8% or 14% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred using the Trans-Blot Turbo system (Biorad, Hercules) to either nitrocellulose or polyvinylidene difluoride (PVDF) membranes. Membranes were then blocked in 5% non-fat milk for 1 hour at room temperature and immunoblotted with primary antibodies overnight at 4 °C (Table S3). IR dye secondary antibodies (Licor) were used to detect protein on a Licor Odyssey.
Purification of Pathologic Tau
AD brains with histologically confirmed tau pathology were used to extract insoluble pathologic tau as previously reported.(27) ~12 grams of frontal cortical gray matter from AD or control cases were homogenized in high-salt buffer (10 mM Tris pH 7.4, 800 mM NaCl, 0.1% Sarkosyl, 10% sucrose) and centrifuged at 10,000xg for 10 min at 4°C. Supernatant was filtered and additional Sarkosyl was added to 1%. Samples were nutated for 1 hour at room temperature and then centrifuged at 300,000xg for 1 hour at 4°C. The 1% Sarkosyl-insoluble pellet containing pathological tau was resuspended in PBS. Immunodepletion of tau from AD lysates was described previously.(33) Anti-tau antibodies (R2295, Tau5, and K9JA) were conjugated to Dynabeads M-280 Toxylactivated (Fisher Scientific) according to manufacturer’s instructions. Tau immunodepletion was performed by adding 200 μL of pathologic tau lysate with anti-tau antibody-bead complexes and rotating overnight at 4°C. A magnet was used to remove the Dynabeads, and this process was performed six times to obtain lysates with a final tau concentration of 0.61 ng/μL tau.
Immunohistochemistry
Six μm formalin-fixed, paraffin-embedded tissue sections were stained using immunohistochemistry or immunofluorescence as previously described (34) using antibodies listed in Table S3. Semiquantitative analysis of mouse tau pathology was performed by grading coronal mouse sections as 0-3 (0=no pathology, 3=severe pathology) for each mouse. Averaged scores from each genotype were used to generate heat maps representing amount and distribution of tau pathology. Quantitative analysis was performed by manual counting of number of AT8-positive neurons in designated brain regions. Analysis was conducted blinded to genotype.
Human Subjects Research
Approval was obtained from the Attikon University General Hospital’s Bioethics Committee and the University of Pennsylvania IRB (702681, 298201, 824867). Informed consent was obtained from all participants or their next of kin surrogate. Consents did not include language permitting public sharing of genomic data.
Neuroimaging
Tau PET imaging was performed on an Ingenuity TF PET/CT scanner (Philips). Images were acquired over 30 minutes starting 75 minutes after intravenous injection of 370MBq (10 mCi) 18F-flortaucipir (previously called 18F-AV-1451). PET volumes were acquired at 2 mm3 isotropic resolution over six five-minute frames and after correction for signal attenuation and patient head motion the acquisition frames were summed across volumes. T1 magnetization-prepared rapid acquisition with gradient echo MRI was performed with isotropic 1 mm voxel size. PET and MRI images were aligned using MIM (MIM Software).
Statistics
Statistical analyses were performed using the GraphPad Prism 5.0 or Rstudio 3.0.1+. Student’s t-test, one- or two-way ANOVA with Bonferroni multiple comparisons correction posthoc tests, or linear mixed effects regression models were used in this study.
Supplementary Material
Acknowledgments:
We thank Tanya M. Bardakjian, Hannah Brown, Ethan Chung, Laynie Dratch, Stephanie Huang, Amber Wang, Elisabeth McCarty Wood, Bin Zhang, the National Prion Disease Pathology Surveillance Center (Mark Cohen and Katie Glisic), and the Center for Neurodegenerative Disease Research for their assistance, and the patients and families to whom this research is dedicated.
Funding: This study was supported by grants from the NIH (F30AG058317, R56AG063344, P30AG010124, P01AG017586, P01AG066597), the BrightFocus Foundation (A2018802S), and a Clinical Scientist Development Award from the Doris Duke Charitable Foundation. The NIH Undiagnosed Diseases Program is funded by the Common Fund, Office of the Director, NIH.
Footnotes
Competing interests: The authors declare no competing interests. ATN is currently affiliated with the Mayo Clinic, Rochester, MN. ZH is currently affiliated with the Chinese Academy of Sciences, Shanghai, China.
Data and materials availability: All transcriptome data are deposited in NCBI’s Gene Expression Omnibus (accession number GSE156831). Human genomics data is available upon request subject to a data use agreement to ensure maintenance of personal privacy. Requests for data or materials should be addressed to edward.lee@pennmedicine.upenn.edu.
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes:
- 1.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K, Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol 112, 389 (October, 2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs GG, Tauopathies. Handb Clin Neurol 145, 355 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Goedert M, Tau pathology and neurodegeneration. Lancet Neurol 12, 609 (June, 2013). [DOI] [PubMed] [Google Scholar]
- 4.Lee VM, Goedert M, Trojanowski JQ, Neurodegenerative tauopathies. Annu Rev Neurosci 24, 1121 (2001). [DOI] [PubMed] [Google Scholar]
- 5.van den Boom J, Meyer H, VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 69, 182 (January 18, 2018). [DOI] [PubMed] [Google Scholar]
- 6.Hanson PI, Whiteheart SW, AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6, 519 (July, 2005). [DOI] [PubMed] [Google Scholar]
- 7.Meyer H, Bug M, Bremer S, Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14, 117 (February 2, 2012). [DOI] [PubMed] [Google Scholar]
- 8.Uhlen M et al. , Tissue-based map of the human proteome. Science 347, 1260419 (January 23, 2015). [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929 (September 3, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos EM et al. , Frontotemporal dementia spectrum: first genetic screen in a Greek cohort. Neurobiol Aging 75, 224 e1 (March, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons GS et al. , Detection of Alzheimer Disease (AD)-Specific Tau Pathology in AD and NonAD Tauopathies by Immunohistochemistry With Novel Conformation-Selective Tau Antibodies. J Neuropathol Exp Neurol 77, 216 (March 1, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demaerel P, Heiner L, Robberecht W, Sciot R, Wilms G, Diffusion-weighted MRI in sporadic Creutzfeldt-Jakob disease. Neurology 52, 205 (January 1, 1999). [DOI] [PubMed] [Google Scholar]
- 13.Neumann M et al. , TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol 66, 152 (February, 2007). [DOI] [PubMed] [Google Scholar]
- 14.Watts GD et al. , Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36, 377 (April, 2004). [DOI] [PubMed] [Google Scholar]
- 15.Taylor JP, Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology 85, 658 (August 25, 2015). [DOI] [PubMed] [Google Scholar]
- 16.Manno A, Noguchi M, Fukushi J, Motohashi Y, Kakizuka A, Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Genes Cells 15, 911 (August, 2010). [DOI] [PubMed] [Google Scholar]
- 17.Halawani D et al. , Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol Cell Biol 29, 4484 (August, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blythe EE, Olson KC, Chau V, Deshaies RJ, Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc Natl Acad Sci U S A 114, E4380 (May 30, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blythe EE, Gates SN, Deshaies RJ, Martin A, Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4.UFD1L Binding and Substrate Processing. Structure 27, 1820 (December 3, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyu PC, Liff MI, Marky LA, Kallenbach NR, Side chain contributions to the stability of alpha-helical structure in peptides. Science 250, 669 (November 2, 1990). [DOI] [PubMed] [Google Scholar]
- 21.O'Neil KT, DeGrado WF, A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250, 646 (November 2, 1990). [DOI] [PubMed] [Google Scholar]
- 22.Song C, Wang Q, Li CC, ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J Biol Chem 278, 3648 (February 7, 2003). [DOI] [PubMed] [Google Scholar]
- 23.Olszewski MM, Williams C, Dong KC, Martin A, The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun Biol 2, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodnar NO et al. , Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Nat Struct Mol Biol 25, 616 (July, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twomey EC et al. , Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365, (August 2, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooney I et al. , Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science 365, 502 (August 2, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo JL et al. , Unique pathological tau conformers from Alzheimer's brains transmit tau pathology in nontransgenic mice. J Exp Med 213, 2635 (November 14, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedhoff P, Schneider A, Mandelkow EM, Mandelkow E, Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry 37, 10223 (July 14, 1998). [DOI] [PubMed] [Google Scholar]
- 29.Holmes BB et al. , Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci U S A 111, E4376 (October 14, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henao-Mejia J et al. , Generation of Genetically Modified Mice Using the CRISPR-Cas9 Genome-Editing System. Cold Spring Harb Protoc 2016, pdb prot090704 (February 1, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luk KD et al. , Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbons GS et al. , GFP-Mutant Human Tau Transgenic Mice Develop Tauopathy Following CNS Injections of Alzheimer's Brain-Derived Pathological Tau or Synthetic Mutant Human Tau Fibrils. J Neurosci 37, 11485–11494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Z et al. , Amyloid-beta plaques enhance Alzheimer's brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat Med 24, 29–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E et al. , Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathologica 134, 65–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.