Abstract
Adipose is a distributed organ that performs vital endocrine and energy homeostatic functions. Hypertrophy of white adipocytes is a primary mode of both adaptive and maladaptive weight gain in animals and predicts metabolic syndrome independent of obesity. Due to the failure of conventional culture to recapitulate adipocyte hypertrophy, technology for production of adult-size adipocytes would enable applications such as in vitro testing of weight loss therapeutics. To model adaptive adipocyte hypertrophy in vitro, we designed and built fat-on-a-chip using fiber networks inspired by extracellular matrix in adipose tissue. Fiber networks extended the lifespan of differentiated adipocytes, enabling growth to adult sizes. By micropatterning preadipocytes in a native cytoarchitecture and by adjusting cell-to-cell spacing, rates of hypertrophy were controlled independent of culture time or differentiation efficiency. In vitro hypertrophy followed a nonlinear, nonexponential growth model similar to human development and elicited transcriptomic changes that increased overall similarity with primary tissue. Cells on the chip responded to simulated meals and starvation, which potentiated some adipocyte endocrine and metabolic functions. To test the utility of the platform for therapeutic development, transcriptional network analysis was performed, and retinoic acid receptors were identified as candidate drug targets. Regulation by retinoid signaling was suggested further by pharmacological modulation, where activation accelerated and inhibition slowed hypertrophy. Altogether, this work presents technology for mature adipocyte engineering, addresses the regulation of cell growth, and informs broader applications for synthetic adipose in pharmaceutical development, regenerative medicine, and cellular agriculture.
Keywords: Adipocyte Hypertrophy, Cell Size, Obesity, Diabetes, Tissue Engineering, Extracellular Matrix
Introduction
The adipose organ is distributed into multiple subcutaneous and visceral depots and performs vital homeostatic functions including hormone secretion and energy metabolism1. The defining cellular constituent of adipose is the adipocyte,2 which stores energy by growing in size as it accumulates lipids in a process called hypertrophy3. In the contexts of vertebrate development or weight gain4,5, white adipocytes hypertrophy hundreds to thousands-fold, often coinciding with maladaptive metabolic changes implicated in diseases associated with obesity including diabetes6. Despite fundamental roles in cellular maturation and weight gain as well as suspected contributions to human pathophysiology, the specific biology of adipocyte hypertrophy and its application to drug development and tissue engineering remain elusive owing to the transience and fragility of primary adipocytes7 and the inability of in vitro adipocyte differentiation to recapitulate hypertrophy8.
Three-dimensional culture is a promising approach for supporting adipocyte hypertrophy and several materials and methods have already been developed for adipose tissue engineering. Existing platforms include decellularized adipose9-11, polyethyleneimine and elastin-like polypeptide surface coatings12, hydrogels13, silk fibroin or polyglycolic acid scaffolds14-16, microfiber meshes17, stirred or levitated beads18,19, hyaluronic acid sponges20, poly L-lactic acid or polycaprolactone electrospun fibers21,22, and freeze dried nanocellulose and alginate mixtures23. While each supports adipogenesis and some have been used to generate insulin sensitive adipocytes in vitro24,25, none of these platforms were specifically designed for adipocyte hypertrophy nor have they been used to produce adipocytes larger than those that can already be achieved with conventional methods.
Here we developed fat-on-a-chip technology and modeled adaptive weight gain by generating adult-size adipocytes in vitro from primary human precursors. We describe the limits of adipocyte hypertrophy for conventional methods and demonstrate inverse proportionality of the rate of hypertrophy with seeding density and consequent differentiation efficiency. By combining fiber networks and cell micropatterning inspired by extracellular matrix and cytoarchitecture in native adipose tissue, culture longevity was increased and seeding density was decoupled from differentiation efficiency, allowing control of adipocyte hypertrophy independent of differentiation efficiency or culture time. Using this technology, average adipocyte size was increased two orders of magnitude relative to conventional methods. By simulating meals or fasting, some functional responses were potentiated including nutrient uptake and release, as well as hormone secretion. In additional tests, gene expression in adipocytes on the chip was more similar to primary tissue than conventional controls, suggesting increased relevance to in vivo adipose biology. As proof of concept for using the chip for biological discovery and therapeutic development, network analysis of transcriptional changes and subsequent drug studies suggested that retinoic acid receptors regulate adipocyte enlargement and may be therapeutic targets for obesity.
Results
Limits of conventional differentiation on adipocyte hypertrophy
Our primary assumption in engineering fat-on-a-chip was that the distributed function of the adipose organ can be modeled with a discrete, tissue engineered system that recapitulates the cellular structure and regulatory functional responses of adipocytes in vivo. To design26 the chip, we considered features of native tissue and assembled a list of design criteria (Table 1). The primary criteria were the abilities to support adipogenesis of preadipocytes (10-15 μm diameter)27-30 and growth into the range of adult cell sizes (55-270 μm diameter) (Figure 1a)31-34. Since average adipocyte size increases over the course of several months during human development (second trimester to ~3 months postnatal)4, additional criteria included stability in cell culture to accommodate the time required for cell growth. To select an approach to achieve these initial design criteria, we quantified the limits of conventional two-dimensional in vitro adipogenesis in terms of cell size and culture longevity, which has been appreciated for decades35. Adipocyte cultures derived from multiple human preadipocyte sources (Supplementary Table 1; Figure S1) were tracked throughout their lifespan by lipid droplet staining at regular time points (Figure 1b). Per convention, differentiation was performed using confluent monolayers that inevitably aggregated and detached (Figure 1c) after approximately one month (37.6 ± 0.4 days; standard error; n = 93 culture wells from 3 separate experiments; Figure 1d). Once aggregated, culture utility is limited by diffusion gradients, cell death, imaging difficulty, and the logistics of suspension culture. Accordingly, culture detachment was considered the endpoint. By high content imaging analysis of stained intermediates, total cell number and adipocyte sizes were quantified (Figure 1e). In addition to aggregate detachment, individual adipocytes also detached throughout in vitro differentiation (Movie S1), attributed to the spherical morphogenesis and increasing buoyancy that accompanies adipogenesis and hypertrophy8. Consistently, total cell number decreased with time in culture, while adipocyte diameters grew on average to ~20 μm (Figures 1f and S1a-c) with no individual adipocytes greater than 65 μm (Figures 1g and S1d-f). These data suggested that both individual cell loss and culture aggregation prevent adipocytes differentiated in vitro from reaching adult sizes.
Table 1 ∣.
Design criteria for fat on a chip
| Design Criteria | Success Metric | Approach |
|---|---|---|
| Robust cell source | Primary cells, Commercially available | Primary human preadipocytes |
| Adipogenesis compatible | > 75% efficiency27,28 | Gelatin substrate29,30 |
| Adult cell size | > 55 μm diameter31,32 | Fiber network for 3D attachment |
| Developmental growth rate | Adult size in 2-6 months4 | Patterning for native cytoarchitecture34 |
| Stable in culture | > 2-6 months | Blend polycaprolactone (2-year in vivo half-life40) in fiber material |
| Biomimetic matrix topography | 100-1,000 nm fiber diameter37 | Pull spinning39 with custom collection mandrel |
| Simple installation and use | No special equipment, Well-plate compatibility | Press-fit installation with standard forceps |
| Controlled rates of hypertrophy | No genetic or culture media manipulation | Patterning for tuned cell-to-cell spacing |
Figure 1 ∣. Two-dimensional culture does not accommodate adult adipocyte sizes.
a. Illustration depicting the range of adipocyte sizes observed in humans. Fetal measurements refer to the end of the second trimester4. A comparison to adipocyte sizes achieved in typical in vitro differentiation is also shown. b. Fluorescence micrograph of conventional differentiation of human preadipocytes after 21 days of culture. Transcription factor C/EBPα (red) is shown with lipid droplets (yellow) and nuclei (blue). Scale bar is 100 μm. c. Brightfield micrographs of adipocyte differentiations that aggregated after 33 days of culture. Scale bar is 1 cm. d. Kaplan-Meier plot of 93 differentiations in separate wells from 3 separate experiments using subcutaneous primary human preadipocytes. Culture aggregation and detachment was considered the endpoint. e. Example of automated lipid droplet (gray) identification during high content image analysis. Droplets outlined in blue were analyzed while objects outlined in orange were excluded due to similarity with background. Scale bar is 100 μm. f. Cell number (black) and lipid droplet diameter (red) as a function of time for at least four replicate adipocyte differentiations per timepoint using ESC derived human preadipocytes. Means and standard errors are shown. g. Distribution of 205,403 lipid droplet diameters from all four differentiation replicates after 31 days of culture.
Supporting adipocyte hypertrophy with fiber networks
Conventional two-dimensional culture fails to recapitulate the in vivo boundary conditions of the adipocyte microenvironment36. In vivo, adipocyte growth is supported by a three-dimensional network of nanofibers known as extracellular matrix37, to which adipocytes anchor themselves via integrins embedded in the cell membrane38. Motivated by the hypothesis that a three-dimensional network would similarly support adipocyte hypertrophy in vitro, biomimetic matrix topography was added as a design criterion (Table 1). We selected pull spinning, a nanofiber manufacturing process we developed previously39, as our approach to build the biomimetic adipose extracellular matrix. Pull spinning uses a bristle on a spinning mandrel to form solid fibers from droplets of a volatile polymer solution (Figure S2a). To allow cell adhesion and to enable comparison with conventional methods, gelatin was selected as raw material for fiber fabrication. Polycaprolactone, a biocompatible synthetic polymer with an in vivo half-life of 2 years40, was also added to the spinning solution to stabilize fiber networks for long-term culture. To enhance utility and to simplify adoption, another important design criterion was compatibility with standard infrastructure (Table 1). Since infrastructure is application dependent, drug discovery was selected as a specific test application and polycarbonate inserts were design for fiber collection and press-fit installation in standard multi-well culture plates (Figure 2a). Insert arrays allowed for reproducible manufacture of fiber networks (S2b-d). Consistent with our goal to mimic the native microenvironment, we obtained fibers with diameters (518 ± 0.06 nm; standard error; n = 300 fibers from 3 separate networks; Figure 2b) in the observed range of adipose extracellular matrix (100 to 1,000 nm)37.
Figure 2 ∣. Fiber networks prolong adipocyte cultures and support adipocyte hypertrophy in vitro.
a. Illustration of a fiber network collected on a multi-well insert (left) and preadipocyte seeding onto the network after installation into a multi-well culture plate (right). b. Scanning electron micrograph of a gelatin-polycaprolactone fiber network used for adipocyte differentiation. Scale bar is 5 μm. c. Phase contrast and fluorescence micrographs of adipocyte differentiations after 1 day (top) or 21 days (middle and bottom) of culture in conventional well plates (left) or fiber networks (right). Transcription factor C/EBPα (red) is shown with lipid droplets (yellow) and nuclei (blue). Scale bar is 100 μm. d. Efficiencies of adipocyte differentiation in conventional well plates (gray; 8 differentiations) or fiber networks (black; 5 differentiations) using ESC derived human preadipocytes. Means and standard errors are shown. e. Lipid droplet diameter as a function of time for two to eight replicate adipocyte differentiations per timepoint in conventional well plates (gray) or fiber networks (black). Means and standard errors are shown. Red vertical line indicates endpoint of conventional cultures.
Since efficiency in conventional two-dimensional adipocyte differentiation relies on culture confluence29,41, we tested a range of seeding densities to obtain culture confluence within fiber networks. Based on contact inhibition of cell proliferation42 and promotion of adipogenesis, we concluded that 50,000 cells/mm3 was sufficient for confluence (Figure S3a-b). Confluence was also confirmed throughout the fiber network by live cell cytoplasmic staining. Cell viability was 97% (Figure S3c-d) and less than 1% of cells expressed markers of proliferation (Ki-67) or apoptosis (Annexin V) after three days of culture (Figure S3e-f). Upon induction of adipogenesis, characteristic lipid droplet formation and upregulation of the adipogenic transcription factor C/EBPα was observed at typical efficiency (Figure 2c-d). These results confirmed the compatibility of fiber networks with in vitro adipogenesis. To test whether fiber networks could prolong culture lifespan and support adipocyte hypertrophy beyond the extents of conventional methods, adipocyte cultures on fiber networks were tracked for 83 days, beyond the one-month time point when two-dimensional cultures consistently failed (Figure 1). During this time, none of the cultures on fiber networks aggregated. In addition, adipocyte diameters (Figure 2e) surpassed the 20 μm average size observed for two-dimension culture (Figure 1) with some adipocytes as large as 220 μm (Figure S4a). These results demonstrated the fiber networks’ ability to prolong culture longevity and to support adipocyte hypertrophy to extents observed in adults.
Controlling rates of adipocyte hypertrophy by Micropatterning and Matrix Mimicry
While fiber networks supported hypertrophy of individual adipocytes, they did not dramatically increase the average rate of growth (Figure S4b). By extrapolation, more than a year would still be required to produce adipocytes with an average diameter in the range of adult humans (i.e. greater than 58 μm in diameter or 100 pL in volume; see Methods). Since adipocytes grow faster than this during development, we reasoned that additional constraints on hypertrophy remained. Anecdotally, larger adipocytes have been observed in culture regions with lower cell density43, so we tested whether subconfluence could be used to accelerate average rates of adipocyte hypertrophy. For proof of principle, we quantified the sizes of adipocytes differentiated from cultures with a range of cell densities and observed an inverse relationship between culture density and adipocyte size (Pearson’s r = −.96; p = 4.4 x 10−11; Figure S4c), with 41% of adipocytes being larger than 30 μm in diameter when differentiated at 66 cells per mm2 versus only 6% at confluence (526 cells per mm2; p = 1.1 x 10−16; n = 230,259 cells; D = .38; two-sample Kolmogorov-Smirnov test; Figure S4d). However, we also observed decreased differentiation efficiency in subconfluent cultures (Pearson’s r = .94; p = 1.1 x 10−9), since preadipocyte spreading suppresses adipogenesis41. Thus, an approach that decoupled differentiation efficiency from culture density was needed to take advantage of the accelerated hypertrophy observed at subconfluence.
Single-cell islands can be patterned by microcontact printing to restrict cell spreading and circular islands have been shown to promote adipogenesis44. By micropatterning individual preadipocytes in circles and subsequently covering the patterned cells with fiber networks (Figures 3a and S5), the efficiency of differentiation could be maintained independent of cell density (Figure 3b) while simultaneously prolonging culture longevity and supporting hypertrophy. Cell proliferation was suppressed on the islands, a majority of which were occupied by a single cell (Figure S5). Circular islands were arranged with equidistant spacing in a hexagonal packing architecture generally found in native tissue34. By changing cell density, this approach provided means to achieve the design goal to control hypertrophy independent of time in culture, media composition, or genetic manipulation (Table 1).
Figure 3 ∣. Preadipocyte patterning facilitates control over the rate of adipocyte hypertrophy.
a. Micropatterning and Matrix Mimicry (M&MM) method for in vitro adipocyte hypertrophy: 1. Schematic depicts printing of circular extracellular matrix islands (gray) onto a culture surface (black) using a protein coated stamp (brown). An example fluorescence micrograph is shown below. 2. Schematic depicts preadipocyte seeding on islands and culture in induction medium. A fluorescence micrograph of adipocytes differentiated on islands after 11 days of culture is shown below. Transcription factor C/EBPα (red) is shown with lipid droplets (yellow) and nuclei (blue). White dashed circles mark positions of fibronectin islands (gray). 3. Photograph shows installation of a fiber network into a 12-well plate. The inset depicts a fiber network on top of adipocytes after installation. 4. Fluorescence micrograph of adipocytes differentiated on islands and cultured to continue hypertrophy for 30 days. Scale bars are 50 μm. b. Efficiencies of adipocyte differentiation as a function of culture density on fibronectin islands (blue) or on well plates uniformly coated with fibronectin (black) for two to eight replicate differentiations and at least 300 cells per time point using ESC derived human preadipocytes. Means and standard errors are shown. Significant decreases (p < .05; Wilcoxon rank sum test) in efficiency relative to that of confluent 2D cultures (dotted horizontal line) are indicated by asterisks. c. Distributions of lipid droplet diameters within adipocytes differentiated on fibronectin islands with either 50 (gray) or 100 um (semi-transparent blue) cell-to-cell spacing after 92 days of culture (n = 207 and 164 cells, respectively) using subcutaneous primary human preadipocytes. Adipocyte size ranges for conventional in vitro differentiations (2D) and adult humans (Adult) are indicated by red double-headed arrows. The Kolmogorov-Smirnov statistic (D) for the distributions and the corresponding p-value (p) are also shown. d. Empirical adipocyte size measurements from developing and postnatal humans (in vivo, solid gray curve) or from in vitro differentiations by fiber networks alone (solid black curve) or by the M&MM method (solid blue curve) are plotted as a function of time together with extrapolation (dotted lines) as in Figure S3b. A red dashed horizontal line indicates the volume of a cell that is 58 μm in diameter (i.e. 100 pL).
To demonstrate control over the rate of hypertrophy, we differentiated preadipocytes in patterns with either 50 or 100 μm cell-to-cell spacing. A clear separation in the distributions of adipocyte diameters in the two groups after 92 days in culture was observed (p = 1.1 x 10−16; n = 371 cells; D = .91; two-sample Kolmogorov-Smirnov test), with most adipocytes in the 100 μm spacing group in the range of adult cell sizes (Figures 3c and S5). Unlike the use of fibers alone, this combined approach, which we call Micropatterning and Matrix Mimicry (M&MM), required only two months for adipocytes to enter the range of adult cell sizes (i.e. average size greater than 58 μm in diameter or 100 pL in volume; Figure 3d). Collectively, these results demonstrated a positive relationship between cultured adipocyte spacing and the rate of adipocyte hypertrophy and defined an approach to control adipocyte hypertrophy on the chip independent of culture time.
Functional responses to simulated meals and fasting
Hormones released during feeding (e.g. insulin) and fasting (e.g. epinephrine) regulate nutrient uptake and release from adipocytes45. To demonstrate functional responses to a simulated meal by adipocytes grown on the M&MM chip, we first measured glucose uptake in the presence or absence of insulin and observed a two-fold increase when insulin was added (p = 0.048; n = 3-6 samples per condition; Wilcoxon rank sum test; Figure 4a). In addition, the lipolytic compound, forskolin promoted the release of glycerol (p = 4.2 x 10−4; n = 6-12 samples per condition; Wilcoxon rank sum test) and free fatty acids (p = 1.9 x 10−3; n = 6-12 samples per condition; Wilcoxon rank sum test) from adipocytes (Figure 4b). Moreover, insulin suppressed forskolin induced glycerol release (p = 3.0 x 10−5; n = 12-24 samples per condition; Wilcoxon rank sum test) but did not significantly reduce the induction of free fatty acid release by forskolin (p = 0.56; n = 12-24 samples per condition; Wilcoxon rank sum test). Although meals do not noticeably affect the secretion of adiponectin from adipocytes, fasting can increase plasma levels of adiponectin46. We observed elevated adiponectin secretion when glucose and insulin were removed from culture media to simulate fasting (p = 3.3 x 10−4; n = 5-10 samples per condition; Wilcoxon rank sum test; Figure 4c). These tests suggested that some adipocyte functions could be potentiated on the M&MM chip by simulated meals and fasting.
Figure 4 ∣. Adipocytes grown by Micropatterning and Matrix Mimicry (M&MM) respond to simulated meals and fasting.

a. Glucose uptake with (n = 6 samples from two differentiations) and without insulin (n = 3 samples from one differentiation; p = 0.048). b. Basal (n = 6 samples from two differentiations), forskolin stimulated (n = 12 samples from one differentiation; p = 4.2 x 10−4 and 1.9 x 10−3 for glycerol and free fatty acid, respectively), and insulin repressed (n = 24 samples from two differentiations; p = 3.0 x 10−5 and 0.56 for glycerol and free fatty acid, respectively) lipolytic release of free fatty acid (gray bars) and glycerol (black bars). c. Basal (n = 6 samples from three differentiations) and fasting (n = 5 samples from three differentiations; p = 3.3 x 10−4) adiponectin secretion. Means with standard error are shown. All measurements were taken using adipocytes differentiated from subcutaneous primary human preadipocytes after 40 days of culture and normalized by total protein. P-values were calculated with Wilcoxon rank sum tests.
Comparing gene expression during in vitro adipocyte hypertrophy to conventional differentiation and primary tissue
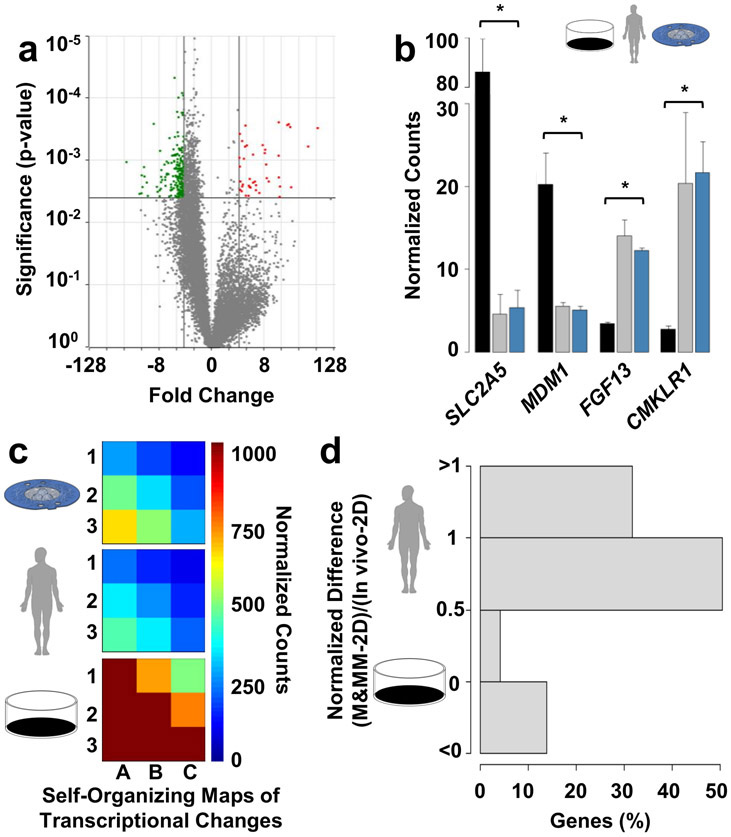
To further test the physiological relevance of the M&MM chip, we mapped transcriptional changes by sequencing RNA extracted from three replicate adipocyte differentiations cultured for 40 days on the M&MM chip or conventional 2D controls (48.7 ± 1.5 and 15.7 ± 0.6 μm diameter, respectively; mean ± standard error). Differential expression of 246 genes was observed (adjusted p-value < .05; ∣fold-change∣ > 3), of which 41 and 205 were up- or down-regulated with increased cell size, respectively (Figure 5a). By comparing these data to published datasets from primary tissue47 (see Materials and Methods), we observed that the transcriptional changes on the M&MM chip increased the similarity of in vitro differentiations with primary adipose tissue (Spearman’s ρ = .63 vs .53). For example, expression of the fructose transporter SLC2A5 and microtubule-binding protein MDM1 fell to match RNA levels in primary tissue (Figure 5b). Similarly, expression of fibroblast growth factor FGF13 and adipokine receptor CMKLR1 expression rose to in vivo levels (Figure 5b). Overall, 202 of the 246 differentially expressed genes were transcribed at levels more similar to primary tissue than conventional differentiation (p = 2.2 x 10−16; χ2 = 101.48; Chi-squared test for given probabilities; Figure 5c), most of which within 50% of in vivo levels (Figure 5d). Increased similarity with gene expression profiles of primary tissue suggests the biological relevance of the M&MM chip and its utility for molecular studies of adipocyte enlargement.
Figure 5 ∣. Micropatterning and Matrix Mimicry (M&MM) increases transcriptomic similarity of in vitro adipocyte differentiations with primary tissue.
a. Volcano plot of all 11,356 genes (gray dots) identified by RNA sequencing in three replicate adipocyte differentiations cultured by M&MM or two-dimensional controls for 40 days (48.7 ± 1.5 and 15.7 ± 0.6 μm mean diameter, respectively; standard error) using subcutaneous primary human preadipocytes. Genes that were up- (red; n = 41) or downregulated (green; n = 205) in larger adipocytes have positive and negative fold changes, respectively. b. RNA sequencing counts of exemplary down- (SLC2A5, MDM1) or upregulated (FGF13, CMKLR1) genes on the M&MM chip (blue) relative to control (black) adipocyte differentiations. RNA sequencing counts in five primary tissue samples are also shown for comparison (gray). Means and standard errors are shown. The same significance thresholds shown in panel a are indicated by asterisks. c. Comparison of self-organizing maps generated from M&MM (blue), control (black), or in vivo (gray) expression values of genes that were differentially expressed between the M&MM chip and conventional culture. d. Distribution of transcriptional similarity for all 246 genes that were differentially expressed on the M&MM chip. Expression equal to the levels of primary tissue (in vivo), conventional controls (2D), and their midpoint are indicated, as well as expression 50% beyond that range (150% 2D, 150% in vivo) or more in either direction.
Retinoic acid receptors regulate transcriptional changes during in vitro adipocyte hypertrophy
To test the utility of the M&MM chip for drug target discovery and development, we organized differentially expressed genes (Figure 5a) into a minimal self-regulating network using known regulatory relationships among genes (Figure 6a). Increased stringency (adjusted p-value < .05; ∣fold-change∣ > 5) was used for gene selection relative to the analysis in Figure 5 to focus on genes with the largest expression changes and for practical limits in computational processing. While activator protein-1 and early growth response transcription factors were also differentially expressed, retinoic acid receptors were positioned at the top of the hierarchy and were the only upregulated transcription factors represented in the network. Although treatment with retinoic acid has been associated with browning of white adipose tissue in mice48 and inhibition of adipogenesis49, no clear role in adipocyte hypertrophy has been described and retinoic acid has even been reported to decrease cell size49,50.
Figure 6 ∣. Retinoic acid receptors regulate transcriptional changes that contribute to adipocyte hypertrophy in vitro.
a. Minimal self-regulating network of differentially expressed genes during in vitro adipocyte hypertrophy from Figure 5. A key of protein symbols is included at the bottom left. All symbols surrounded by a blue circle were in the set of differentially expressed genes used to seed the network. Up- or down regulation of seed nodes during size expansion is indicated by smaller red or blue circles to the upper left of each symbol. b-e. Phase contrast images of adipocytes after 47 days in culture and 32 days of treatment with either retinoic acid, synthetic agonist or antagonist, or vehicle. Scale bar is 50 μm. f. Quantification of lipid droplet diameter by image analysis of three separate experiments. Retinoic acid and agonist groups were significantly different than the vehicle control (respectively, p = 5.6 x 10−8 and 2.2 x 10−16, n = 1,362 and 1,730 cells, and D = .24 and .25; two-sample Kolmogorov-Smirnov test). g. Dose response of retinoic acid receptor antagonist on lipid droplet diameter after 40 days of culture and 20 days of treatment. The antagonist concentration of 10 μM was significantly different than the vehicle control (p = 3.6 x 10−2, n = 132 cells, and D = .29; two-sample Kolmogorov-Smirnov test). Subcutaneous primary human preadipocytes were used for all panels. Means with standard error are shown.
To test the hypothesis that retinoic acid receptors mediate the changes in gene expression that support adipocyte enlargement, we treated adipocyte cultures on the M&MM chip with either retinoic acid, a synthetic agonist or antagonist, or vehicle (i.e. dimethyl sulfoxide). Cultures were treated every 3-4 days using media supplemented with 1 μM of the respective compounds. After at least 30 days of treatment using three separate differentiations, retinoic acid (p = 5.6 x 10−8; n = 1,362 cells; D = .24; two-sample Kolmogorov-Smirnov test) or agonist (p = 2.2 x 10−16; n = 1,730 cells; D = .25; two-sample Kolmogorov-Smirnov test) significantly increased adipocyte size relative to the vehicle control (Figure 6b-f). Since treatment with 1 μM antagonist every 3-4 days did not significantly affect adipocyte size (p = 0.22; n = 1,819 cells; D = .05; two-sample Kolmogorov-Smirnov test), we measured the response to daily treatment and higher doses (Figure 6g). Relative to cells fed every 3-4 days in our initial drug study, cells fed daily were larger at baseline per comparison of vehicle treated controls in the two experiments. Adipocyte size decreased with increasing antagonist concentration and reached statistical significance at 10 μM (p = 3.6 x 10−2, n = 132 cells, and D = .29; two-sample Kolmogorov-Smirnov test). Through identification of transcriptional changes, transcriptional network analysis, and rational target selection for drug testing, our data suggest that the M&MM chip could be used for basic biological discovery, therapeutic target or lead compound validation prior to in vivo testing.
Discussion
The M&MM chip described here advances existing fat-on-a-chip technology51-61 by supporting in vitro adipocyte hypertrophy to adult cell sizes while extending culture lifespan and controlling growth rates independent of differentiation efficiency. These platform features complement previous ex vivo models that obtained different adipocyte sizes by varying donor age62, diet63, genetic background64, adipose depot65 or enzymatic digestion66. Even cell age, which differentially influences hypertrophy due to heterogeneity in lipid metabolism67, could not be controlled in studies that isolated adipocytes of different sizes from the same adipose depot of the same donor7,68,69. By decoupling cell size and cell age, the M&MM chip enables investigation of adipocyte-intrinsic mechanisms for size control and could be applied to the development of drugs to treat maladaptive adipocyte hypertrophy.
As proof of concept for applying the chip to drug development, we identified retinoid signaling intermediates among other candidates as potential regulators of adipocyte hypertrophy. Regulation by retinoids may help explain why retinoic acid is enriched more than 30-fold in adipose tissue relative to serum70,71. Of note, the concentration of retinoic acid receptor antagonist required to significantly decrease hypertrophy was ten-fold higher (10 μm) than the concentration of agonist (1 μm) required to increase it. While the network analysis and drug study provide evidence that M&MM technology can enable drug discovery, the general significance of retinoid signaling to adipocyte hypertrophy will need to be demonstrated in differentiations of preadipocytes from additional donors and adipose depots. Consistent with our findings, the potential therapeutic benefits of modulating retinoid signaling in obesity and diabetes have been demonstrated by independent studies72-74 and proposed mechanisms often consider a role in enhancing peroxisome proliferator-activated receptor-gamma signaling73. Retinoid signaling has also been implicated in hypertrophy of additional cell types, including hepatocytes75 and cardiomyocytes76, although changes in retinoic acid receptor transcription were not detected in our previous studies of cardiac hypertrophy77,78.
Further development of the M&MM chip could expand potential applications by mimicking additional features of in vivo adipose tissue. For example, adipocytes interact with multiple cell types and develop in vivo from aggregated mesenchymal cells interconnected via gap junctions79, which are transiently down-regulated during differentiation80. Previous studies found that Connexin43, a gap junction protein expressed on adipocyte membranes, was required for adipogenesis, although the importance of synchronized electrophysiology and metabolism enabled by gap junctions in adipose tissue is not known81. On the M&MM chip, adipocytes differentiated and hypertrophied without physical coupling to other cells, demonstrating gap junctions are neither required for adipogenesis nor hypertrophy. The gene encoding Connexin43, GJA1, is transcribed on the chip and may regulate differentiation independent of its role in functional gap junctions.
The design principles we employed to control adipocyte hypertrophy may also be applied to engineer three-dimensional tissues, such as adipose lobules. Lobules are subunits of adipose depots, encased in a dense network of matrix fibers called septa and supported internally by sparse, stromal fibers82. Others have proposed that lobule geometry emerges primarily from adipocyte-matrix interactions that govern adipogenesis and hypertrophy83,84. While extracellular matrix opposes adipose overgrowth85-87 and must be remodeled to accommodate hypertrophy34, our findings demonstrate that distances between neighboring preadipocytes during adipose formation will also dictate tissue growth. Since adipocyte sizes influence adipose mechanics88, future efforts to engineer biomimetic lobules should incorporate preadipocyte spacing, in addition to matrix scaffold mechanics, into tissue designs.
The incorporation of specific matrix proteins is another chip feature that can be further developed and tuned based on application. Matrix proteins vary among adipose depots and regulate adipocyte function89. While gelatin was sufficient for in vitro hypertrophy, the ability to control protein composition in fiber networks could be leveraged to engineer specific adipose depots for pharmaceutical development. Depot- and disease-specific decellularized matrix have been used to demonstrate the functional importance of adipose matrix protein composition11,90 and can similarly be used for fiber fabrication in our platform. Control over scaffold turnover and remodeling also provides opportunities to model fibrosis. Inclusion of polycaprolactone in our fibers helped to maintain long-term culture integrity and has been included by others in scaffolds for mesenchymal stem cells91. Conversely, scaffolds composed primarily of polylactic acid are readily degraded during tissue remodeling and do not support long-term adipogenesis15. While alternative materials have been used as adipose scaffolds12-17,20,22,23 and may similarly promote culture longevity, manufacture of stable fiber networks from strictly native matrix constituents would provide more relevant substrates for matrix remodeling and will be an important future direction. Taken collectively, this technology enables disease modeling and drug testing prior to in vivo validation and informs tissue engineering approaches for adipose reconstruction, aesthetic medicine, food and other applications for which adipocytes could be exploited.
Materials and Methods
Chip Design and Fabrication
Micropatterning
To recreate adipocyte cytoarchitecture on the chip, patterning masks for photolithography were designed using custom Python scripts and fabricated by CAD/Art Services, Inc. (Bandon, OR, USA). Masters for single-cell patterns were subsequently fabricated on silicon wafers92. Briefly, SU-8 (MicroChem, 3005) was spin coated onto each wafer, soft baked, UV-crosslinked, baked, and developed in propylene glycol monomethyl ether acetate (Sigma, 484431) per manufacturer instructions. Polydimethylsiloxane (Sylgard 184, Dow, 2646340) was prepared per manufacturer instructions using a centrifugal mixer (Thinky, AR-100), poured on masters, placed in a vacuum for 1 hour to remove bubbles, and cured overnight at 60°C to make stamps93. Stamps were sterilized by sonication for 30 minutes in ethanol (VWR, 89125-188), air dried, and coated with 50 μg/mL fibronectin (Corning, 356008) in PBS for 1 hour. Coated stamps were then air dried and pressed onto culture surfaces to transfer patterned fibronectin. Culture surfaces were then treated with 1% Pluronics F127 (Sigma, P2443) in PBS for 5 minutes and rinsed before cell seeding.
Matrix Mimicry
To mimic extracellular matrix in adipose tissue, custom multi-well insert arrays and collection mandrels for fiber network fabrication were designed in SolidWorks software (Dassault Systemes, France). Insert arrays were cut from polycarbonate sheets (McMaster) with a ProtoLaser U3 UV engraver (LPKF, Germany), while collection mandrels were fabricated on an Objet Connex500 3D printer (Stratasys, Israel). Pull spinning39 was performed in a chemical fume hood using a rotating cylindrical bristle (length 0.531 cm, diameter 0.1 cm) press fit to a custom mandrel as previously described. As the mandrel rotates, the bristle contacts a droplet of polymer solution and draws the droplet into a single fiber. The mandrel was driven at 25,000 rpm by a motor and NE211 series control unit (NSK America Corporation, E3000) with a ceramic bearing spindle (NSK America Corporation, NR-3080S) and air supply of 0.3 MPa. Polymer solution was flowed at 0.3 mL/min into the bristle’s path using a needle (18G flat tip; BD Biosciences) positioned under the spinning mandrel and connected to a PHD Ultra syringe pump (Harvard Apparatus, 70-3310C). The gelatin (Sigma G2500-1KG) : polycaprolactone (Sigma 440744-500G) (1:4) spinning solution was 6% weight to volume in 1,1,1,3,3,3-hexafluoro-2-propanol (Oakwood Chemical 003409; Hazardous Material: TOXIC & CORROSIVE) and was mixed overnight at 200 rpm on a stir plate (IKA, RO10) before spinning. The collection mandrel was rotated at 60 rpm using a mounted drill (IKA, RW20DS1). Multi-well insert arrays were affixed to both sides of the collection mandrel for the consistent manufacture of 64 fiber networks per batch (Figure S1). Per batch, 5 mL of spinning solution were extruded to produce fiber networks that were approximately 30 μm thick. Networks were air dried for 20 minutes in a chemical fume hood to ensure complete solvent evaporation94, and sterilized by 20-minute UV treatment on each side prior to installation in well plates for culture.
Preadipocyte Culture and Adipogenesis
To grow adipocytes on the chip, cryopreserved subcutaneous (PromoCell, C-12977, lot numbers 394Z027.1 and 423Z037.1), visceral (Zen-Bio, OP-F-3, lot number OMM020216C), and diabetic visceral (Zen-Bio, MSND-F, lot number DMSNM072005) primary human preadipocytes (Table S1) were expanded in Dulbecco's Modification of Eagle's Medium (DMEM, VWR, 45000-304) with 2.5 ng/mL basic fibroblast growth factor (Aldevron), 7.5% fetal bovine serum (Thermo Fisher, 16140071), 1x GlutaMAX (Thermo Fisher, 35050-061), and 1x Penicillin:Streptomycin solution (VWR, 45000-652) on gelatin coated plates for four passages prior to differentiation. Unless varied experimentally as noted, preadipocytes were rinsed and seeded at 526 cells per mm2 in DMEM with 1 μm dexamethasone (Sigma, D1756), 400 nM insulin (Sigma, I9278), 0.5 μm rosiglitazone (Santa Cruz Biotechnology, sc-202795A), 7.5% KnockOut Serum Replacement (Thermo Fisher, 10828028) and 1x Penicillin:Streptomycin solution to induce adipogenesis95. Human embryonic stem cell (ESC) derived preadipocytes (Table S1) were generated and differentiated into white adipocytes as described previously96.
Chip Testing
Imaging
Cell number, proliferation, adipogenic differentiation efficiency, and lipid droplet diameters were quantified by high content image analysis on an Arrayscan XTI (Thermo Fisher, ASN00003L). Prior to imaging, samples were fixed in 4% paraformaldehyde in Dulbecco’s Phosphate-Buffered Saline (PBS, VWR, 45000-434) and stained with Hoechst 33342 Stain (Thermo Fisher, H1399), Alexa Fluor 568 Phalloidin (Thermo Fisher, A12380), and/or HCS LipidTOX Red Neutral Lipid Stain (Thermo Fisher, H34476) to label nuclei, microfilaments, and lipid droplets, respectively. Fixed samples were also immunostained with antibodies for either Ki-67 (Thermo Fisher, PA5-16785) or C/EBPα (Abcam, ab40761), or Vimentin (Abcam, ab92547) and α-Tubulin (Abcam, ab7291) with Alexa Fluor 488 (Thermo Fisher, A-21206) and/or Alexa Fluor 647 (Thermo Fisher, A-31571) secondary antibody to label proliferating or differentiated cells or intermediate filaments and microtubules, respectively. A 5x objective was used to scan all 36 fields of view without edge overlap from each 12-well plate culture. Nuclei and lipid droplets were automatically identified using fluorescence intensity thresholds from their respective channels. Nuclei number was used as a surrogate for cell number, except in Figure S3c where cells were stained with Trypan Blue and counted with an automated cell counter (Bio-Rad, TC10) after trypsinization. Average intensity within nuclear masks was used to calculate the percentage of Ki-67 or C/EBPα positive cells. Lipid droplet diameters larger than 10 μm were used as surrogates for adipocyte size. Cell viability and apoptosis were quantified by live cell staining and microscopy using Calcein AM (Thermo Fisher, C3100MP), 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher, D1306), and Annexin V (Thermo Fisher, A13203) per manufacturer instructions. An Olympus ix83 confocal microscope was used for z-stacks of adipocytes for 3D imaging. To quantify fiber diameters, fibers were collected on electron microscopy stubs, sputter coated using an Au target (Denton Vacuum, Moorestown, NJ), and imaged in a field emission scanning electron microscope (Carl Zeiss, Dresden, Germany). ImageJ software (National Institutes of Health, Bethesda, MD) was used for diameter quantification of 300 individual fibers (four random fields of view per sample from three samples).
Adipocyte Growth Modeling
The exponential model of cell growth implies two assumptions for adipocytes: 1) cell growth is only limited by the amount of lipid synthesis machinery inside the cell, and 2) the amount of lipid synthesis machinery increases linearly with cell volume. The second assumption is supported by the observation that adipocyte water content increases linearly with respect to cell volume (despite occupying a nonlinearly decreasing fraction of cell volume)97 and the concentration of lipid synthesis enzymes is roughly constant during cell growth98. However, water content does not necessarily predict the amount of synthesis machinery, as in the case of chondrocyte hypertrophy where increasing water content outpaces organelle synthesis99. Moreover, growth patterns that are neither linear nor exponential have been reported repeatedly for eukaryotic cells100-102. Volumetric growth was fitted with the following equation:
Here V is the average cell volume, V0 is the initial average cell volume, t is the time in culture, τ is the only fitting parameter, which accounts for the growth rate constant. The model was tested with growth data from conventional 2D cell culture (i.e. Figure 1f), using spherical morphology to calculate volume. Spherical morphology is expected since lipid droplets are stiffer than the nucleus or surrounding cytoplasm103. The assumption of spherical adipocyte morphology was also confirmed by confocal microscopy (Figure S4d). Data was fitted and the 95% confidence interval for the fitting parameter was calculated using the nlinfit and the nlparci functions in MATLAB (MathWorks, USA), respectively. Deviation from the exponential model is likely due to the fact that the lipid droplet occupies an increasing majority of the cell volume, so that the second assumption of the exponential model is violated.
RNA Sequencing
RNA was extracted from ~ 175,000 cells per sample using a RNeasy Lipid Tissue Mini Kit (Qiagen, 74804). Extracted RNA was run on a BioAnalyzer RNA Pico chip (Agilent, 5067-1513) and all samples sequenced had RNA Integrity Numbers above 8. Isolation of mRNA and library preparation were performed using a KAPA mRNA HyperPrep kit (Roche, 08098115702) and libraries were sequenced on an Illumina NextSeq 500 sequencer. To compare transcriptomic data from our fat-on-a-chip with primary tissue, we downloaded FASTQ files with unprocessed RNA sequencing data from adipose tissue generated by the Human Protein Atlas project47. Starting from unprocessed sequencing results, the technical variables between our data and the previously published datasets were: 1) methods for RNA isolation (Trizol for this study vs tissue biopsy, Optimal Cutting Temperature compound embedding, and mechanical homogenization for the published study), and 2) the location and people who did the work. Remaining workflows were comparable: RNA purification (Qiagen), quality control (Agilent), cDNA library preparation and sequencing (Illumina). Biological differences were 1) the intended test variable (i.e. in vitro versus in vivo differentiation and hypertrophy), and 2) cell-type diversity (i.e. while adipocytes were the predominant cell type in both conditions, primary tissue also included contaminating blood vessels, nerves, and immune cells). Like the preadipocytes used for adipogenesis and sequencing data in this study, primary tissues in the published study were obtained from subcutaneous depots of female individuals. Unprocessed sequencing results from this study and from the previously published study were mapped to human genome build hg38 using the Spliced Transcripts Alignment to a Reference104 (version 2.5.3a) aligner and binned by GENCODE release 29 genes105. Gene counts in all datasets were normalized by the trimmed mean of M values method106 and considered differentially expressed if the absolute value of the change in a normalized gene count was greater than 3-fold with an adjusted107 p-value < .05 per ANOVA.
Glucose Uptake, Lipolysis, and Adiponectin Secretion
Functional assays were performed using the Glucose Uptake-Glo™ Assay (Promega, J1341), the Lipolysis Assay Kit, Dual Glycerol and Free Fatty Acids Detection (Zen-Bio, LIP-3-NC), and the Human Adiponectin ELISA (Ray Biotech, ELH-Adiponectin-2) per manufacturer instructions. DMEM, no glucose, no glutamine, no phenol red (Thermo Fisher, A1443001) was used as assay/starvation medium for glucose uptake (16 hours starvation prior to assay), lipolysis (4 hours starvation prior to assay), and for the starvation condition for adiponectin quantification (4 hours incubation time). Insulin and forskolin were used at 1 μM and 5 μM, respectively. Total protein content in each culture was quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher, 23227) after extraction in RIPA buffer (Sigma, R0278).
Retinoic Acid Receptor Modulation
Retinoic acid (Tocris, 695), Ch 55, Potent RAR agonist (Tocris, 2020), and AGN 193109, High affinity pan-RAR antagonist (Tocris, 5758) were dissolved in dimethyl sulfoxide and added to fresh culture media without rosiglitazone to feed every 3-4 days for a final concentration of 1 μM. Cultures were differentiated for 9-16 days prior to treatment. For dose-response studies, cultures were differentiated for 20 days and then treated every day for an additional 20 days with the indicated concentrations of retinoic acid receptor antagonist in fresh culture media.
Statistical Analysis
The self-organizing map of transcriptional changes was generated using Gene Expression Dynamics Inspector (Version 2.1)108 software with 20 and 80 first and second phase training iterations, respectively. To calculate the normalized expression difference for each gene on the M&MM chip versus conventional or in vivo controls, the differences between gene values on the M&MM chip and conventional culture were normalized by dividing by the difference between in vivo values and conventional culture. The minimal self-regulating network of differentially expressed genes was created using MetaCore software (Clarivate Analytics). Correlations and Chi-squared, Kolmogorov-Smirnov, and Wilcoxon tests for given probabilities were performed, respectively using the cor(), chisq.test(), ks.test(), and wilcox.test() functions in R (https://www.r-project.org/).
Supplementary Material
Acknowledgements
We thank Michael Rosnach, Danielle Gamboa, Brian Fountaine, and Grace Matthews for photographs and illustrations, Blakely O’Connor and Ilke Akartuna for photolithography training, members of the Disease Biophysics Group and the Cowan Lab for helpful discussions, and Lance Davidow, Kathleen Plaff, Gizem Rizki, Lee Rubin, Lee Barrett, Clifford Woolf, and the Human Neuron Core at Boston Children’s Hospital (BCH IDDRC, U54HD090255) for use, training, and troubleshooting of high-content imaging equipment. Benjamin Pope is a Good Ventures Fellow of the Life Sciences Research Foundation. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases through award UC4 DK104165. We acknowledge the financial support of the Harvard Stem Cell Institute and National Institutes of Health grants U01HL100408, U01HL107440, R01DK095384, and R01DK097768 to CAC. We also acknowledge the Wyss Institute for Biologically Inspired Engineering at Harvard University and the Harvard Center for Nanoscale Systems (CNS), which is a member of the National Nanotechnology Infrastructure Network (NNIN) under NSF award no. 1541959. This work was performed in part at the Harvard MRSEC (grant no. DMR 14-20570).
Footnotes
Conflicts of Interest
The authors are named inventors on pending patent applications PCT US16/62693 (A.L.G., B.D.P., K.K.P.) and PCT US18/56649 (B.D.P., K.K.P.) filed by Harvard University. K.K.P. is a principal at KK Parker & Associates, with consulting responsibilities to several biotechnology and pharmaceutical companies.
References
- 1.Rosen ED and Spiegelman BM, Cell, 2014, 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutkowski JM, Stern JH and Scherer PE, J. Cell Biol, 2015, 208, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haczeyni F, Bell-Anderson KS and Farrell GC, Obes. Rev. Off. J. Int. Assoc. Study Obes, 2018, 19, 406–420. [DOI] [PubMed] [Google Scholar]
- 4.Dauncey MJ and Gairdner D, Arch. Dis. Child, 1975, 50, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand SS, Tarnopolsky MA, Rashid S, Schulze KM, Desai D, Mente A, Rao S, Yusuf S, Gerstein HC and Sharma AM, PloS One, 2011, 6, e22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laforest S, Labrecque J, Michaud A, Cianflone K and Tchernof A, Crit. Rev. Clin. Lab. Sci, 2015, 52, 301–313. [DOI] [PubMed] [Google Scholar]
- 7.Björntorp P and Karlsson M, Eur. J. Clin. Invest, 1970, 1, 112–117. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HH, Kumar S, Barnett AH and Eggo MC, J. Endocrinol, 2000, 164, 119–128. [DOI] [PubMed] [Google Scholar]
- 9.Cheung HK, Han TTY, Marecak DM, Watkins JF, Amsden BG and Flynn LE, Biomaterials, 2014, 35, 1914–1923. [DOI] [PubMed] [Google Scholar]
- 10.Lin C-Y, Liu T-Y, Chen M-H, Sun J-S and Chen M-H, Biomed. Mater. Bristol Engl, 2016, 11, 035010. [DOI] [PubMed] [Google Scholar]
- 11.Baker NA, Muir LA, Washabaugh AR, Neeley CK, Chen SY-P, Flesher CG, Vorwald J, Finks JF, Ghaferi AA, Mulholland MW, Varban OA, Lumeng CN and O’Rourke RW, J. Clin. Endocrinol. Metab, 2017, 102, 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner PA, Harris LM, Purser CA, Baker RC and Janorkar AV, Biotechnol. Bioeng, 2014, 111, 174–183. [DOI] [PubMed] [Google Scholar]
- 13.Hilliou F, Pairault J, Dominice J and Redziniak G, Exp. Cell Res, 1988, 177, 372–381. [DOI] [PubMed] [Google Scholar]
- 14.Patrick CW, Chauvin PB, Hobley J and Reece GP, Tissue Eng., 1999, 5, 139–151. [DOI] [PubMed] [Google Scholar]
- 15.Mauney JR, Nguyen T, Gillen K, Kirker-Head C, Gimble JM and Kaplan DL, Biomaterials, 2007, 28, 5280–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanken H, Göhler F, Smeets R, Heiland M, Gröbe A, Friedrich RE, Busch P, Blessmann M, Kluwe L and Hartjen P, Vivo Athens Greece, 2016, 30, 567–572. [PubMed] [Google Scholar]
- 17.Kral JG and Crandall DL, Plast. Reconstr. Surg, 1999, 104, 1732–1738. [DOI] [PubMed] [Google Scholar]
- 18.Rubin J. Peter., Bennett JM, Doctor JS, Tebbets BM and Marra KG, Plast. Reconstr. Surg, 2007, 120, 414–424. [DOI] [PubMed] [Google Scholar]
- 19.Daquinag AC, Souza GR and Kolonin MG, Tissue Eng. Part C Methods, 2013, 19, 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halbleib M, Skurk T, de Luca C, von Heimburg D and Hauner H, Biomaterials, 2003, 24, 3125–3132. [DOI] [PubMed] [Google Scholar]
- 21.Kang X, Xie Y, Powell HM, James Lee L, Belury MA, Lannutti JJ and Kniss DA, Biomaterials, 2007, 28, 450–458. [DOI] [PubMed] [Google Scholar]
- 22.Shanti RM, Janjanin S, Li W-J, Nesti LJ, Mueller MB, Tzeng MB and Tuan RS, Ann. Plast. Surg, 2008, 61, 566–571. [DOI] [PubMed] [Google Scholar]
- 23.Krontiras P, Gatenholm P and Hägg DA, J. Biomed. Mater. Res. B Appl. Biomater, 2015, 103, 195–203. [DOI] [PubMed] [Google Scholar]
- 24.Brännmark C, Paul A, Ribeiro D, Magnusson B, Brolén G, Enejder A and Forslöw A, PloS One, 2014, 9, e113620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JH, Gimble JM, Vunjak-Novakovic G and Kaplan DL, Tissue Eng. Part C Methods, 2010, 16, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capulli AK, Tian K, Mehandru N, Bukhta A, Choudhury SF, Suchyta M and Parker KK, Lab. Chip, 2014, 14, 3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green H and Kehinde O, Cell, 1974, 1, 113–116. [Google Scholar]
- 28.Mackall JC, Student AK, Polakis SE and Lane MD, J. Biol. Chem, 1976, 251, 6462–6464. [PubMed] [Google Scholar]
- 29.Spiegelman BM and Ginty CA, Cell, 1983, 35, 657–666. [DOI] [PubMed] [Google Scholar]
- 30.Xiong C, Xie C-Q, Zhang L, Zhang J, Xu K, Fu M, Thompson WE, Yang L-J and Chen YE, Stem Cells Dev., 2005, 14, 671–675. [DOI] [PubMed] [Google Scholar]
- 31.Veilleux A, Caron-Jobin M, Noel S, Laberge PY and Tchernof A, Diabetes, 2011, 60, 1504–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wree A, Schlattjan M, Bechmann LP, Claudel T, Sowa J-P, Stojakovic T, Scharnagl H, Köfeler H, Baba HA, Gerken G, Feldstein AE, Trauner M and Canbay A, Metabolism, 2014, 63, 1542–1552. [DOI] [PubMed] [Google Scholar]
- 33.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Näslund E, Britton T, Concha H, Hassan M, Rydén M, Frisén J and Arner P, Nature, 2008, 453, 783–787. [DOI] [PubMed] [Google Scholar]
- 34.Pope BD, Warren CR, Parker KK and Cowan CA, Trends Cell Biol., , DOI: 10.1016/j.tcb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Björntorp P, Karlsson M, Pertoft H, Pettersson P, Sjöström L and Smith U, J. Lipid Res, 1978, 19, 316–324. [PubMed] [Google Scholar]
- 36.Spiegelman BM and Farmer SR, Cell, 1982, 29, 53–60. [DOI] [PubMed] [Google Scholar]
- 37.Young DA, Ibrahim DO, Hu D and Christman KL, Acta Biomater., 2011, 7, 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, DeYoung SM, Zhang M, Zhang M, Cheng A and Saltiel AR, Cell Metab., 2005, 2, 165–177. [DOI] [PubMed] [Google Scholar]
- 39.Deravi LF, Sinatra NR, Chantre CO, Nesmith AP, Yuan H, Deravi SK, Goss JA, MacQueen LA, Badrossamy MR, Gonzalez GM, Phillips MD and Parker KK, Macromol. Mater. Eng, 2017, 1600404. [Google Scholar]
- 40.Sun H, Mei L, Song C, Cui X and Wang P, Biomaterials, 2006, 27, 1735–1740. [DOI] [PubMed] [Google Scholar]
- 41.Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H and Kano K, Nat. Commun, 2014, 5, 3368. [DOI] [PubMed] [Google Scholar]
- 42.Eagle H and Levine EM, Nature, 1967, 213, 1102–1106. [DOI] [PubMed] [Google Scholar]
- 43.Hausman GJ, Novakofski JE, Ramsay T and Martin RJ, J. Anim. Sci, 1985, 60, 1553–1561. [DOI] [PubMed] [Google Scholar]
- 44.McBeath R, Pirone DM, Nelson CM, Bhadriraju K and Chen CS, Dev. Cell, 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- 45.Galton DJ and Bray GA, J. Clin. Invest, 1967, 46, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feizollahzadeh S, Rasuli J, Kheirouri S and Alizadeh M, Health Promot. Perspect, 2014, 4, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J and Ponten F, Science, 2015, 347, 1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 48.Mercader J, Ribot J, Murano I, Felipe F, Cinti S, Bonet ML and Palou A, Endocrinology, 2006, 147, 5325–5332. [DOI] [PubMed] [Google Scholar]
- 49.Noy N, Adipocyte, 2013, 2, 184–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arana A, Mendizabal JA, Alzón M, Soret B and Purroy A, J. Anim. Sci, 2008, 86, 3393–3400. [DOI] [PubMed] [Google Scholar]
- 51.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D and Yu H, Lab. Chip, 2009, 9, 3185–3192. [DOI] [PubMed] [Google Scholar]
- 52.Moraes C, Labuz JM, Leung BM, Inoue M, Chun T-H and Takayama S, Integr. Biol. Quant. Biosci. Nano Macro, 2013, 5, 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zambon A, Zoso A, Gagliano O, Magrofuoco E, Fadini GP, Avogaro A, Foletto M, Quake S and Elvassore N, Anal. Chem, 2015, 87, 6535–6543. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Schneider N, Platen A, Mitra I, Blazek M, Zengerle R, Schüle R and Meier M, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, E4143–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller PG and Shuler ML, Biotechnol. Bioeng, 2016, 113, 2213–2227. [DOI] [PubMed] [Google Scholar]
- 56.Brooks JC, Judd RL and Easley CJ, Methods Mol. Biol. Clifton NJ, 2017, 1566, 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loskill P, Sezhian T, Tharp KM, Lee-Montiel FT, Jeeawoody S, Reese WM, Zushin P-JH, Stahl A and Healy KE, Lab. Chip, 2017, 17, 1645–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanataweethum N, Zelaya A, Yang F, Cohen RN, Brey EM and Bhushan A, Biotechnol. Bioeng, 2018, 115, 1979–1987. [DOI] [PubMed] [Google Scholar]
- 59.Zhu J, He J, Verano M, Brimmo AT, Glia A, Qasaimeh MA, Chen P, Aleman JO and Chen W, Lab. Chip, 2018, 18, 3550–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, Kongsuphol P, Chiam SY, Zhang QX, Gourikutty SBN, Saha S, Biswas SK and Ramadan Q, Lab. Chip, 2019, 19, 241–253. [DOI] [PubMed] [Google Scholar]
- 61.Paek J, Park SE, Lu Q, Park K-T, Cho M, Oh JM, Kwon KW, Yi Y-S, Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR and Huh D, ACS Nano, 2019, 13, 7627–7643. [DOI] [PubMed] [Google Scholar]
- 62.Vann Bennett G and Cuatrecasas P, Science, 1972, 176, 805–806. [DOI] [PubMed] [Google Scholar]
- 63.Salans LB, Horton ES and Sims EAH, J. Clin. Invest, 1971, 50, 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salans LB, Bray GA, Cushman SW, Danforth E, Glennon JA, Horton ES and Sims EAH, J. Clin. Invest, 1974, 53, 848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldrick RB and McLoughlin GM, J. Clin. Invest, 1970, 49, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith U, J. Lipid Res, 1971, 12, 65–70. [PubMed] [Google Scholar]
- 67.Wang QA, Scherer PE and Gupta RK, J. Lipid Res, 2014, 55, 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Etherton TD, Aberle ED, Thompson EH and Allen CE, J. Lipid Res, 1981, 22, 72–80. [PubMed] [Google Scholar]
- 69.Blüher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB and Kahn CR, Dev. Cell, 2002, 3, 25–38. [DOI] [PubMed] [Google Scholar]
- 70.Perumal J, Sriram S, Lim HQ, Olivo M and Sugii S, Adipocyte, 2016, 5, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, Chen H, Mu D, Fan J, Song J, Zhong Y, Li D and Xia M, J. Clin. Endocrinol. Metab, 2016, 101, 1686–1692. [DOI] [PubMed] [Google Scholar]
- 72.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L and Kahn BB, Nature, 2005, 436, 356–362. [DOI] [PubMed] [Google Scholar]
- 73.Yamauchi T and Kadowaki T, Nihon Rinsho Jpn. J. Clin. Med, 2001, 59, 2245–2254. [PubMed] [Google Scholar]
- 74.Lin Y-W, Park SW, Lin Y-L, Burton FH and Wei L-N, Int. J. Obes 2005, , DOI: 10.1038/s41366-019-0379-z. [DOI] [Google Scholar]
- 75.Lenhard JM, Lancaster ME, Paulik MA, Weiel JE, Binz JG, Sundseth SS, Gaskill BA, Lightfoot RM and Brown HR, Diabetologia, 1999, 42, 545–554. [DOI] [PubMed] [Google Scholar]
- 76.Freire CMM, Azevedo PS, Minicucci MF, Oliveira Júnior SA, Martinez PF, Novo R, Chiuso-Minicucci F, Matsubara BB, Matsubara LS, Okoshi K, Novelli EL, Zornoff LAM and Paiva SAR, Nutr. Burbank Los Angel. Cty. Calif, 2011, 27, 824–828. [DOI] [PubMed] [Google Scholar]
- 77.Sheehy SP, Huang S and Parker KK, Circ. Cardiovasc. Genet, 2009, 2, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCain ML, Sheehy SP, Grosberg A, Goss JA and Parker KK, Proc. Natl. Acad. Sci, 2013, 110, 9770–9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burke S, Nagajyothi F, Thi MM, Hanani M, Scherer PE, Tanowitz HB and Spray DC, Microbes Infect., 2014, 16, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Azarnia R and Russell TR, J. Cell Biol, 1985, 100, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saez JC, Berthoud VM, Branes MC, Martinez AD and Beyer EC, Physiol. Rev, 2003, 83, 1359–1400. [DOI] [PubMed] [Google Scholar]
- 82.Estève D, Boulet N, Belles C, Zakaroff-Girard A, Decaunes P, Briot A, Veeranagouda Y, Didier M, Remaury A, Guillemot JC, Ledoux S, Dani C, Bouloumié A and Galitzky J, Nat. Commun, 2019, 10, 2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peurichard D, Delebecque F, Lorsignol A, Barreau C, Rouquette J, Descombes X, Casteilla L and Degond P, J. Theor. Biol, 2017, 429, 61–81. [DOI] [PubMed] [Google Scholar]
- 84.Peurichard D, Ousset M, Paupert J, Aymard B, Lorsignol A, Casteilla L and Degond P, J. Theor. Biol, 2019, 469, 127–136. [DOI] [PubMed] [Google Scholar]
- 85.Toni S, Morandi R, Busacchi M, Tardini L, Merlini L, Battistini NC and Pellegrini M, Front. Aging Neurosci, , DOI: 10.3389/fnagi.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chun T-H, Hotary KB, Sabeh F, Saltiel AR, Allen ED and Weiss SJ, Cell, 2006, 125, 577–591. [DOI] [PubMed] [Google Scholar]
- 87.Lackey DE, Burk DH, Ali MR, Mostaedi R, Smith WH, Park J, Scherer PE, Seay SA, McCoin CS, Bonaldo P and Adams SH, Am. J. Physiol. Endocrinol. Metab, 2014, 306, E233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alkhouli N, Mansfield J, Green E, Bell J, Knight B, Liversedge N, Tham JC, Welbourn R, Shore AC, Kos K and Winlove CP, Am. J. Physiol. Endocrinol. Metab, 2013, 305, E1427–1435. [DOI] [PubMed] [Google Scholar]
- 89.Roca-Rivada A, Belen Bravo S, Pérez-Sotelo D, Alonso J, Isabel Castro A, Baamonde I, Baltar J, Casanueva FF and Pardo M, Sci. Rep, 2015, 5, 12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grandl G, Müller S, Moest H, Moser C, Wollscheid B and Wolfrum C, Mol. Metab, 2016, 5, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Das P, Salerno S, Remigy J-C, Lahitte J-F, Bacchin P and De Bartolo L, Colloids Surf. B Biointerfaces, 2019, 184, 110493. [DOI] [PubMed] [Google Scholar]
- 92.Nall JR and Lathrop JW, IEEE Trans Electron Devices, 1957, 5, 117. [Google Scholar]
- 93.Adams WJ, Pong T, Geisse NA, Sheehy SP, Diop-Frimpong B and Parker KK, J. Comput.-Aided Mater. Des, 2007, 14, 19–29. [Google Scholar]
- 94.Badrossamay MR, McIlwee HA, Goss JA and Parker KK, Nano Lett., 2010, 10, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scott MA, Nguyen VT, Levi B and James AW, Stem Cells Dev., 2011, 20, 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahfeldt T, Schinzel RT, Lee Y-K, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, Clish CB, Deo RC, Shen T, Lau FH, Cowley A, Mowrer G, Al-Siddiqi H, Nahrendorf M, Musunuru K, Gerszten RE, Rinn JL and Cowan CA, Nat. Cell Biol, 2012, 14, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DiGirolamo M and Owens JL, Am. J. Physiol, 1976, 231, 1568–1572. [DOI] [PubMed] [Google Scholar]
- 98.Hames KC, Koutsari C, Santosa S, Bush NC and Jensen MD, Int. J. Obes, 2015, 39, 884–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buckwalter JA, Mower D, Ungar R, Schaeffer J and Ginsberg B, J. Bone Joint Surg. Am, 1986, 68, 243–255. [PubMed] [Google Scholar]
- 100.Anderson EC, Bell GI, Petersen DF and Tobey RA, Biophys. J, 1969, 9, 246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sinclair WK and Ross DW, Biophys. J, 1969, 9, 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cermak N, Olcum S, Delgado FF, Wasserman SC, Payer KR, A Murakami M, Knudsen SM, Kimmerling RJ, Stevens MM, Kikuchi Y, Sandikci A, Ogawa M, Agache V, Baléras F, Weinstock DM and Manalis SR, Nat. Biotechnol, 2016, 34, 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shoham N, Girshovitz P, Katzengold R, Shaked NT, Benayahu D and Gefen A, Biophys. J, 2014, 106, 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR, Bioinforma. Oxf. Engl, 2013, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML and Flicek P, Nucleic Acids Res., 2019, 47, D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Robinson MD and Oshlack A, Genome Biol., 2010, 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benjamini Y and Hochberg Y, J. R. Stat. Soc. Ser. B Methodol, 1995, 57, 289–300. [Google Scholar]
- 108.Eichler GS, Huang S and Ingber DE, Bioinformatics, 2003, 19, 2321–2322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.