Abstract
In the mammalian central nervous system, nerve-glia antigen 2 (NG2) glia are considered the fourth glial population in addition to astrocytes, oligodendrocytes and microglia. The fate of NG2 glia in vivo has been carefully studied in several transgenic mouse models using the Cre/loxP strategy. There is a clear agreement that NG2 glia mainly serve as progenitors for oligodendrocytes and a subpopulation of astrocytes mainly in the ventral forebrain, whereas the existence of a neurogenic potential of NG2 glia is lack of adequate evidence. This mini review summarizes the findings from recent studies regarding the fate of NG2 glia during development. We will highlight the age-and-region-dependent heterogeneity of the NG2 glia differentiation potential. We will also discuss putative reasons for inconsistent findings in various transgenic mouse lines of previous studies.
Key Words: astrogliogenesis, cell fate, Cre/loxP system, development, differentiation, embryonic brain, neurogenesis, NG2 glia, oligodendrocyte lineage, oligodendrocyte precursor cells
Introduction
In the mammalian central nervous system (CNS), the nerve-glia antigen 2 (NG2) glycoprotein (also called chondrotin sulfate proteoglycan 4, CSPG4) is immunodetectable only in oligodendrocyte precursor cells (OPCs) and vascular pericytes (Nishiyama et al., 1996; Horner et al., 2002; Stallcup, 2002). OPCs in the adult CNS intensively co-express NG2 and platelet derived growth factor receptor α (PDGFRα), whereas the subcellular expression pattern of those two markers is different during the development (Nishiyama et al., 1996; Diers-Fenger et al., 2001; Dawson et al., 2003; Rivers et al., 2008). Despite their capacity to generate mature oligodendrocytes (OLs) throughout life, NG2-expressing OPCs also possess unique physiological properties such as promoting presynaptic specialization in neurons (Tanaka et al., 2009) and modulating neuroinflammation (Nakano et al., 2017; Zhang et al., 2019; Liu and Aguzzi, 2020). Thereby, to emphasize their status as the fourth glial population in addition to astrocytes, OLs and microglia, OPCs are also termed NG2 glia (Butt et al., 1999; Bergles et al., 2000; Nishiyama, 2001; Greenwood and Butt, 2003; Peters, 2004; Butt and Dinsdale, 2005; Ge et al., 2006; Kukley et al., 2007; Nishiyama et al., 2009; Vélez-Fort et al., 2010; Haberlandt et al., 2011). As the largest proliferative cell population in the adult CNS, NG2 glia are equally distributed over the whole brain (Figure 1Bb1 and b2) and spinal cord (Nishiyama et al., 2016). Their fate has been carefully studied in several transgenic mouse models using the Cre/loxP strategy in vivo (Doerflinger et al., 2003; Rivers et al., 2008; Zhu et al., 2008a, b, 2011; Guo et al., 2009, 2010; Kang et al., 2010; Hill et al., 2011; Simon et al., 2011; Clarke et al., 2012; Huang et al., 2014, 2018, 2019). Consensus has been achieved for the oligodendrogenic potential of postnatal NG2 glia, though debate still exists regarding the potential differentiation fate of NG2 glia to other cell types like astrocytes and neurons. For this mini review, we searched PubMed for literatures published up to 2020 by using the keyword combination: “NG2” OR “PDGFRα” OR “oligodendrocyte precursor cell” AND “Cre”. We will summarize results of fate-mapping studies of NG2 glia during development by using transgenic mice.
Figure 1.
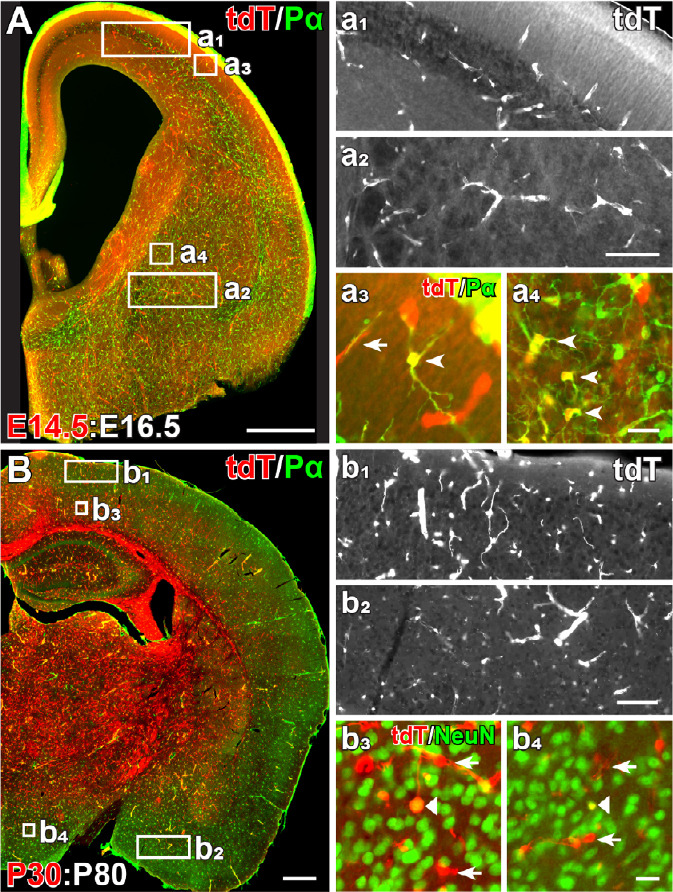
NG2 glia in embryonic and adult brain.
The distribution of embryonic and adult NG2 glia in the brain demonstrated in NG2-CreERT2Huang KIxRosa26-tdTomato mice. In the embryonic brain, of the NG2 gene started in a small portion of OPCs in the ventral brain. When Cre activity was induced at E14.5 and analyzed at E16.5 (E14.5:E16.5), tdTomato (tdT) expression was found in PDGFRα+ OPCs (therefore termed NG2 glia) and vascular pericytes (A, a1–a4). Most tdT+ NG2 glia were detected in the ventral brain (A, a2, and a4, arrowheads) and few were found in the dorsal cortex (A, a1, and a3, arrowheads). Nevertheless, vascular tdT+ pericytes were observed in the whole brain (A, a1–a4, arrows). In adult brain at P30:P80, tdT+ NG2 glia were found equally distributed in the brain, irrespective of dorsal or ventral regions (B, b1, b2). Some NeuN+ tdT+ cells with typical neuronal morphology were sporadically located in the cortex (b3, triangle). However, only few tdT+ cells (~0.5%, 5 out of 946 cells analyzed from three mice) were NeuN immunopositive in the hypothalamus (b4, triangle). Scale bars: 500 μm in A and B; 100 μm in a2, and b2; 20 μm in a4, and b4 (unpublished data from Huang et al., 2014, 2019).
Cre/loxP Mouse Models for Fate-Mapping of NG2 Glia
The Cre recombinase, a 38 kDa protein found in the bacteriophage P1, catalyses site-specific homologous recombination of two particular 34 bp nucleotide sequences (loxP: locus of crossover of the bacteriophage P1, also floxed), allowing specific manipulation of floxed DNA strands (Sternberg and Hamilton, 1981; Abremski et al., 1983; Hoess and Abremski, 1985). To enable a temporal control of Cre activity (CreER), the enzyme was fused to mutated ligand-binding domain of the human estrogen receptor (ER), thereby trapping it by heat shock proteins within the cytosol and preventing its entry into the nucleus (Metzger et al., 1995; Feil et al., 1996; Zhang et al., 1996). In the presence of 4-OH tamoxifen (4-HT, a specific ligand of the mutated ER), CreER will be released from heat shock proteins and subsequently enter the nucleus to loxP sites. The CreER system underwent improvements. For example, the modified CreERT2 displays a much higher sensitivity to 4-HT than CreER and is highly recommended for inducible gene manipulation in the living mouse (Feil et al., 1997; Indra et al., 1999).
To map the fate of distinct cell populations, Cre-expressing animals are crossbred to reporter mice, where the expression of a transgenic reporter protein is driven by ubiquitously active promoters after Cre-mediated deletion of the floxed STOP cassette (lsl) (Soriano, 1999; Novak et al., 2000; Mao et al., 2001; Srinivas et al., 2001; Madisen et al., 2010). Therefore, Cre-expressing cells and their progeny will irreversibly turn on the reporter gene, allowing the tracing of defined Cre-expressing cell types and their progeny. Hence, fate-mapping studies are largely relying on the specificity of the promoter driving Cre-expression in the target cells.
By and large, Cre-expressing mice can be classified into two groups according to the strategies of transgene insertion: via non-homologous recombination (TgN) by injection of linearized vector DNA strands to oocytes (Gordon et al., 1980) and homologous recombination (TgH, also called knock-in, KI) in embryonic stem cells (Smithies et al., 1985; Thomas et al., 1986). Each strategy has advantages and disadvantages as discussed in previous reviews (Nishiyama et al., 2009; Richardson et al., 2011). Briefly, transgenic mice generated via non-homologous recombination are faster produced, however the transgene is controlled by just a short promoter sequence which may result in ectopic expression of the transgene. Although nowadays the bacterial or phage artificial chromosome (BAC or PAC) approaches greatly improved the specificity of the transgene by using much longer regulatory sequences. Mice generated by non-homologous recombination have still drawbacks due to the random insertion into the genome, which might cause unpredictable transgene expression or unintended gene deletion at the insertion site (Beil et al., 2012). The homologous knock-in strategy introduces the transgene into the endogenous targeted gene locus, ensuring the complete and true control of the transgene expression by all the regulatory elements of the targeted gene. However, it requires more effort and time-cost to generate knock-in mice and usually the knock-in strategy leads to the loss of at least one allele of the targeted gene (in heterozygous mice) unless Internal-Ribosome-Entry-Site strategy is successfully incorporated (Chan et al., 2011).
To achieve lineage tracing of NG2 glia in vivo, a series of transgenic mice has been generated in several research groups via different strategies. In these transgenic mice, the expression of Cre or CreER(T2) is driven by the promoters of either one of the two specific NG2 glia markers, (NG2/PDGFRα), or other genes that are active in the OL lineage such as Olig2, Sox10, or PLP (Table 1). Therefore, the expression pattern of the selected marker gene during development determines the cell populations with the recombined reporter gene. For example, in the developing forebrain at E15.5, Olig2 is expressed in OPCs and a sub-population of interneuronal progenitor cells. When Cre activity was induced at E15.5 in Olig2-CreERTTakebayashi KI mice, both OL lineage cells and interneurons expressing the chosen reporter could be found in the postnatal cortex (Miyoshi et al., 2007). In addition, transgene expression only represents the gene activity of the selected marker rather than the genuine protein expression. For example, the NG2 protein starts to be immunodetectable in OPCs in the rodent brain from about embryonic day 14 (E14), but NG2 gene activity can already be detected in a small portion of PDGFRα+ OPCs (3%) at E12.5 in NG2-EYFP KI mice (Karram et al., 2008; Huang et al., 2019). Although the proportion of EYFP+ PDGFRα+ OPCs increased drastically from E12.5 to birth in NG2-EYFP KI mice, the expression of EYFP could only be found in OPCs or pericytes, indicating that NG2 gene activity is strictly restricted to those two cell populations (Huang et al., 2019).
Table 1.
Fate-mapping studies of NG2 glia in different transgenic mice
| Short name | Type (TgH/TgN/ BAC/PAC) | Reporter | Region | Recombined NG2 glia (%) | Recombined cell types | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|
| OL Lineage | Astrocyte | Neuron | ||||||
| NG2-CreZhu | BAC | Z/EG | brain | ~86 | + | + | – | Zhu et al. (2008a, b) |
| Z/EG | spinal cord | ~70 | + | + | Not mentioned | |||
| NG2-CreERTZhu | BAC | Z/EG | brain | < 2 | + | + (from Embryonic) | + | Zhu et al. (2011) |
| Rosa26-EYFP | brain | ~45 | + | Not mentioned | – | |||
| Rosa26-tdTomato | brain | ~80 | + | - | + | Robins et al. (2013) | ||
| NG2-CreTsoa | BAC | Rosa26-LacZ | brain | ~99 | + | Not mentioned | + | Tsoa et al. (2014) |
| NG2-CreERT2Huang | TgH | Rosa26- tdTomato | brain | ~95 | + | + (from Embryonic) | + | Huang et al. (2014, 2018, 2019) |
| Rosa26-EYFP | brain | ~75 | + | Not mentioned | + | |||
| Rosa26-tdTomato | spinal cord | Not mentioned | + | – | + | |||
| PDGFRα-CreERT2Rivers | PAC | brain | ~47 | + | – | + (in PC) | Rivers et al. (2008) | |
| Rosa26-EYFP | spinal cord | ~40 | + | – | – | Zawadzka et al. (2010) | ||
| PDGFRα-CreERTKang | BAC | Rosa26-mGFP | brain | ~86 | + | – | + | Kang et al. (2010) |
| Rosa26-EYFP | brain | ~87 | + | – | + | |||
| Z/EG | brain | ~42 | + | – | + | |||
| PLP-CreERTDoerflinger | TgN | Rosa26-EYFP | brain | ~17 | + | + | + (in PC) | Guo et al. (2009, 2010) |
| spinal cord | ~27 | + | + | + | ||||
| Rosa26-EYFP | cerebellum | Not mentioned | + | + | – | Chung et al. (2013) | ||
| Olig2-CreERTTakebayashi | TgH | Rosa26-EYFP | cerebellum | Not mentioned | + | + | – | |
| Rosa26-EYFP | brain | Not mentioned | + | + | – | Dimou et al. (2008) | ||
| Z/EG | brain | Not mentioned | + | + | – | |||
+: Defined cell types were detected; –: defined cell types were not detected; BAC: bacterial artificial chromosome; NG2: nerve-glia antigen 2; TgH: transgene homologous recombination; TgN: transgene non-homologous recombination; PAC: phage artificial chromosome; PC: piriform cortex.
The sensitivity of the reporter can also lead to different results. The higher sensitivity of the reporter could reveal a broader range of Cre-expressing patterns (Madisen et al., 2010; Van Hove et al., 2020). For instance, compared to the Rosa26-EYFP reporter (Srinivas et al., 2001), a strong and ubiquitous CAG promoter and the WPRE (mRNA stabilizer woodchuck hepatitis virus posttranscriptional regulatory element) cassette were inserted into the construct of Rosa26-tdTomato reporter to improve expression efficiency (Madisen et al., 2010). Therefore, in 4-week old NG2-CreERT2Huang KI mice, the Rosa26-tdTomato reporter could label more NG2 glia by at least 20% than the Rosa26-EYFP reporter (Huang et al., 2014). However, reporter with high sensitivity might also generate unexpected recombined cells, which could be due to transient low or even ectopic activity of the selected gene driving Cre expression (Tognatta et al., 2017; Van Hove et al., 2020).
Spatiotemporally Controlled Generation of Oligodendrocytes from NG2 Glia
In the developing brain, OPCs arise in three waves sequentially from ventral to dorsal origin (Kessaris et al., 2006). The first OPCs are derived from Nkx2.1+ progenitors at E12.5 from the medial ganglionic eminence and anterior entopeduncular area in the ventral brain. By E16.5, the second wave of OPCs from Gsh2+ progenitors joined from the lateral and caudal ganglionic eminences (LGE and CGE) in the ventral brain. The ventrally derived OPCs quickly migrate to the dorsal cortex, in which those OPCs will be largely eliminated within the first postnatal weeks. The third wave of OPCs is generated from dorsal Emx1+ progenitors postnatally and contribute to ~80% of the OLs in the dorsal brain (Kessaris et al., 2006; Tripathi et al., 2011). Studies in NG2-EYFP KI mice suggested that at E12.5, the NG2 gene was already active in a small portion of OPCs, which was further confirmed by using NG2-CreERT2Huang KI mice (Huang et al., 2019). When NG2-CreERT2Huang KI embryos received tamoxifen at E12.5 and were analyzed 2 days later at E14.5 (E12.5:E14.5), only in the ventral brain reporter+ OPCs were detected. At E12.5:P0 (analysis at postnatal day 0), reporter+ OPCs could also be found in the dorsal cortex, indicating the migration of OPCs from the ventral to dorsal brain. A similar distribution pattern of reporter+ OPCs was revealed at E14.5:E16.5 (tamoxifen at E12.5 and analysis 2 days later, Figure 1Aa1–a4) and E14.5:P0 (Huang et al., 2019). However, at E14.5:P10 reporter+ OPCs (and OLs) migrating to the dorsal brain were greatly eliminated, consistent with previous studies (Huang et al., 2019). When Cre activity was induced embryonically (from E12.5 to E17.5) in NG2-CreERT2Huang KI or NG2-CreERTZhu BAC mice, some reporter positive cells were found to express mature OL markers and displayed OL morphology after birth which suggest that embryonic NG2 glia give rise to OLs (Zhu et al., 2011; Huang et al., 2019). Quantitative analysis revealed that embryonic NG2 glia generate more OLs proportionally in the white matter (WM) such as internal capsule than in the gray matter (GM) such as dorsal and ventral cortex (Huang et al., 2019). This region-dependent manner of OL differentiation from NG2 glia was confirmed in postnatal brains of several mouse lines (such as NG2-CreERT2Huang KI, NG2-CreERTZhu BAC, NG2-CreTsoa BAC, PDGFRα-CreERTKang BAC, PDGFRα-CreERT2Rivers PAC and Olig2-CreERTTakebayashi KI). Whenever Cre activity was induced, WM NG2 glia always displayed higher oligodendrogliogenic potential than their GM counterparts in terms of a greater proportion of reporter+ OLs within a confined period. In addition, NG2 glia quickly generate OLs within the early postnatal weeks, and such generation of OLs seems to continue throughout life although the rate declines significantly with age in both GM and WM of the brain (Rivers et al., 2008; Zhu et al., 2008a; Kang et al., 2010; Huang et al., 2014; Tsoa et al., 2014). Recent studies further demonstrated that newly generated mature OLs in the adult brain actively participated in myelin modelling (Hill et al., 2018; Hughes et al., 2018).
Results from the constitutive NG2-CreZhu BAC mice showed that spinal NG2 glia could generate OLs (Zhu et al., 2008a). When Cre activity was induced in neonatal NG2-CreERT2Huang KI mice, a great portion of spinal NG2 glia quickly differentiated into OLs although the differentiation rate (the percentage of reporter+ OLs in all reporter+ glia) in WM was still higher than in GM (e.g., at P1:P13, ~70% and ~90% in the GM and WM, respectively) (Huang et al., 2018). However, the pattern of OL differentiation from NG2 glia in the adult spinal cord is different. When Cre activity was induced at P30 or P136 in NG2-CreERT2Huang KI mice, or at P70 in PDGFRα-CreERTKang BAC mice, spinal NG2 glia differentiated into OLs with a similar rate in WM and GM (Kang et al., 2010; Huang et al., 2018). In addition, although the rate of postnatal NG2 glia differentiating into OLs declined decreased with age, embryonic spinal NG2 glia did not show higher oligodendrogliogenic potential than neonatal NG2 glia (e.g., in the NG2-CreERT2Huang KI mice at E17.5:P10, ~50% and ~90% in the GM and WM respectively) (Huang et al., 2018).
Taken together, these studies suggest that NG2 glia differentiate into OLs in an age- and region-dependent manner, and it will be important to investigate the precise mechanisms that regulate the heterogeneous differentiation potential of NG2 glia to generate OLs.
Restricted Astrogliogenic Potential of NG2 Glia
The first in vivo evidence indicating NG2 glia generating astrocytes came from NG2-CreZhu BAC mice with the Z/EG reporter (Zhu et al., 2008a). In these mice, NG2 glia-derived astrocytes were mainly found in the GM of the ventral brain, where they contributed to about one third of the total S100B+ astrocytes. In addition, a few NG2 glia-derived astrocytes could be detected in other GM regions such as dorsal cortex and hippocampus, but never in WM areas. In the follow-up study, NG2-CreERTZhu BACxZ/EG mice were treated with tamoxifen either at E16.5 or postnatally to determine if NG2 glia could give rise to astrocytes all the time. However, only mice treated with tamoxifen embryomically displayed reporter+ astrocytes mostly in the ventral forebrain after birth, indicating that only a subpopulation of embryonic NG2 glia could differentiate into astrocytes (Zhu et al., 2011). These results were further confirmed by studies using NG2-CreERT2Huang KI mice crossed to the more sensitive Rosa26-tdTomato reporter. Only when CreERT2 activity was induced in embryos (at E12.5, E14.5 or E17.5), a significant number of reporter+ astrocytes could be detected in the postnatal forebrain, and the distribution pattern was similar to NG2-CreZhu BAC and NG2-CreERTZhu BAC mice (Zhu et al., 2008a, 2011; Huang et al., 2014, 2019). It was suggested that presumably those reporter+ astrocytes resulted from the transient activation of NG2 gene in certain radial glial cells (Richardson et al., 2011; Tognatta et al., 2017). However, when NG2-CreERT2Huang KIxRosa26-tdTomato embryos were analyzed just 2 days post tamoxifen administration, e.g. at E12.5:E14.5 (Huang et al., 2019) or E14.5:E16.5 (Figure 1Aa3 and a4), all the recombined cells were either PDGFRα+ OPCs or vascular pericytes, suggesting that reporter+ astrocytes detected after birth have to be derived from bona fide embryonic NG2 glia rather than radial glial cells. A recent single-cell RNA-Seq study of PDGFRα-CreERTKang BAC mice at E13.5:P3 demonstrated that the data set from reporter+ cells highly correlated with OLs and astrocytes, indicating again the astrogliogenic potential of embryonic NG2 glia (Marques et al., 2018).
In the spinal cord of NG2-CreZhu BACxZ/EG mice, reporter+ astrocytes were also found in the GM (Zhu et al., 2008b). However, in NG2-CreERT2Huang KIxRosa26-tdTomato mice, whenever Cre activity was induced (from E14.5 to adult), reporter+ cells were always restricted to the Sox10+ OL lineage cells, suggesting that spinal NG2 glia do not generate astrocytes (Huang et al., 2018). Such discrepancy between those two mouse lines might be attributed to early astrogliogenic progenitors in the spinal cord with transiently activated OL genes before E14.5, such as 2′,3′-cyclic nucleotide 3′ phosphodiesterase (Cnp) (Tognatta et al., 2017).
So far, postnatal fate-mapping studies in healthy mice of several transgenic lines (NG2-CreERTZhu BAC, NG2-CreERT2Huang KI, PDGFRα-CreERT2Rivers PAC, and PDGFRα-CreERTKang BAC) showed no incidence for astrogliogenesis from NG2 glia in the CNS after birth. Exceptions were from Olig2-CreERTTakebayashi KI and PLP-CreERTDoerflinger TgN mice in the brain and cerebellum (Takebayashi et al., 2002; Doerflinger et al., 2003; Dimou et al., 2008; Guo et al., 2009; Chung et al., 2013). However, the reporter+ astrocytes in postnatal Olig2-CreERTTakebayashi KI mice are mainly due to the direct gene activation of Olig2 in a subpopulation of astrocytes (Cai et al., 2007). In PLP-CreERTDoerflinger TgN mice, recombined astrocytes were detected within three days after tamoxifen administration, suggesting the potential direct/ectopic expression of Cre in astrocytes (Doerflinger et al., 2003; Guo et al., 2009; Tognatta et al., 2017). Thereby, we conclude that only a sub-group of embryonic NG2 glia in the forebrain can generate astrocytes after birth.
Controversial Evidence for a Neurogenic Potential of NG2 Glia
In the early embryonic ventral brain areas (medial ganglionic eminence and anterior entopeduncular area), Nkx2.1+ progenitor cells generate OPCs as well as interneurons, raising the possibility that some embryonic NG2 glia could be neurogenic (Petryniak et al., 2007; Nishiyama et al., 2016). However, in NG2-CreZhu BACxZ/EG mice, no evidence was found for neurogenesis from NG2 glia (Zhu et al., 2008a). On the contrary, in NG2-CreTsoa BAC mice with a more sensitive reporter (Rosa26-LacZ), a significant number of reporter+ interneurons were detected in the postnatal brain, which were assumed as progenies of the NG2+/Olig2+ progenitor cells immuno-positive for Cre at E14.5 (Tsoa et al., 2014). In the recent work on NG2-CreERT2Huang KI mice, with the sensitive reporter Rosa26-tdTomato, NG2 glia from E12.5 or E14.5 did not generate neurons. Moreover, when Cre activity was induced at later embryonic stages (after E16.5) in NG2-CreERTZhu BAC or NG2-CreERT2Huang KI mice, no reporter+ neurons were produced in the postnatal brain (Zhu et al., 2011; Huang et al., 2019). Considering that aberrant transgene activity may also occur in BAC transgenic mice, the reporter+ neurons in NG2-CreTsoa BAC mice thereby might be derived from certain neuronal progenitor cells with very low expression level of Cre beneath the detection threshold of immunostaining. Therefore, it is likely that at embryonic stages (at least after E12.5) the NG2 gene is restrictively activated in OPCs and pericytes. Considering NG2 becomes immuno-detectable earliest at age E13.5 (Nishiyama et al., 1996; Diers-Fenger et al., 2001), we conclude that embryonic NG2 glia do not generate neurons.
Also a neurogenic potential of postnatal NG2 glia has been studied intensively. Although reporter+ neurons can be detected in various mouse lines after postnatal Cre activity induction, the existence of neurogenic NG2 glia is still under debate. EdU incorporation experiments demonstrated that virtually all NG2 glia could be labelled by EdU after long-term treatment, indicating that all NG2 glia underwent proliferation (Clarke et al., 2012; Young et al., 2013). Therefore, neurons derived from NG2 glia should have been labelled by any of the thymidine analogues EdU or BrdU. However, the reporter+ neurons detected in NG2-CreERT2Huang KIxRosa26-tdTomato, PDGFRα-CreERTKang BACxRosa26-EYFP and PDGFRα-CreERT2Rivers PACxRosa26-EYFP mice were EdU/BrdU negative even after one-month treatment, indicating that those reporter+ neurons were derived from postmitotic cells rather than NG2 glia (Rivers et al., 2008; Kang et al., 2010; Huang et al., 2014). On the contrary, in the hypothalamus of NG2-CreERTZhu BACxtdTomato mice some reporter+ cells were detected with immunoreactivity to the mature neuronal maker NeuN or neuronal lineage maker HuC/D after adult Cre induction, accumulating to a significant number 60 days later (~4% and ~9% out of all reporter+ cells were NeuN+ and HuC/D+ respectively). In addition, a few HuC/D+ reporter+ cells demonstrated the incorporation of BrdU (Robins et al., 2013). Given that a small number of neuronal proteins including HuC/D were detected in OL lineage cells (Chittajallu et al., 2004; Clarke et al., 2012), immunostainings for OL lineage markers (such as Sox10 and Olig2) on those BrdU+ HuC/D+ reporter+ cells would help to better assess their cell types.
The number of reporter+ neurons in the brains of NG2-CreERT2Huang KIxRosa26-tdTomato, NG2-CreERTZhu BACxZ/EG and PDGFRα-CreERTKang BACxZ/EG mice did not accumulate over time, further confirming that those neurons were generated from postmitotic cells (Kang et al., 2010; Zhu et al., 2011; Huang et al., 2014). However, the reporter+ neurons in the piriform cortex of PDGFRα-CreERT2Rivers PACxRosa26-EYFP and PLP-CreERTDoerflinger TgNxRosa26-EYFP mice showed increased numbers of recombined neurons with very low rate (Rivers et al., 2008; Guo et al., 2009, 2010; Clarke et al., 2012). A recent study suggests that these reporter+ neurons might be derived from dormant doublecortin (DCX)+ progenitors (Rotheneichner et al., 2018). Likewise, very few reporter+ neurons (less than two neurons per cross-section) in the spinal cord of NG2-CreERT2Huang KIxRosa26-tdTomato mice, without incorporation of BrdU, increased in their number after tamoxifen administration (Huang et al., 2018). It is conceivable that those reporter+ neurons could be generated from certain mitotically quiescent progenitors, as also indicated by the quiescent neural progenitor cells in the adult olfactory bulb (Fuentealba et al., 2015).
If neurogenic NG2 glia exist, they should be located in the same brain regions among different mouse lines. However, the distribution pattern of reporter+ neurons varies in different lines. In adult NG2-CreERT2Huang KIxRosa26-tdTomato mice, reporter+ neurons were detected mainly in the cortex where they form clusters in the amygdala, but they are scattered in the dorsal cortex (Figure 1Bb3), and could be found rarely in the hypothalamus (Figure 1Bb4, ~0.5% out of all reporter+ cells were NeuN+), and piriform cortex (Huang et al., 2014). In NG2-CreERTZhu BACxZ/EG mice, reporter+ neurons appeared sporadically in the neocortex (Zhu et al., 2011). In PDGFRα-CreERT2Rivers PACxRosa26-EYFP and PLP-CreERTDoerflinger TgNxRosa26-EYFP mice, reporter+ neurons were mainly located in the piriform cortex (Doerflinger et al., 2003; Rivers et al., 2008; Guo et al., 2009, 2010), while in PDGFRα-CreERTKang BACxZ/EG mice a few reporter+ neurons were found in the hypothalamus but never in the piriform cortex (Kang et al., 2010). In Olig2-CreERTTakebayashi KIxZ/EG mice, no reporter+ neurons were discovered (Dimou et al., 2008). It is therefore hard to find a conclusive distribution pattern of reporter+ neurons in common among those mice, suggesting they are derived from different cell populations.
Last, the appearance of morphologically mature reporter+ neurons happened shortly (within 3 days) post tamoxifen administration without proof of any intermediate transitional stage, suggesting that the reporter expression is due to direct recombination in mature neurons (Kang et al., 2010; Huang et al., 2014). Another explanation for this fast neuronal recombination could be independent from the neurogenic potential of NG2 glia. Recombined neurons could also be resulted from ectopic Cre activity acquired by endocytosis of exosomes potentially released by NG2 glia. In vitro studies have shown that mature OLs release exosomes carrying biomolecules such as myelin proteins (Bakhti et al., 2011; Fitzner et al., 2011; Frühbeis et al., 2019) and might be promoting neuronal long-term maintenance. Cre-carrying exosomes prepared from MOG-iCre mice lead to recombination of loxP sites as indicated by reporter gene activation (Frühbeis et al., 2013). In vivo evidence highlights the uptake of exosomes by neurons (Frühbeis et al., 2013), but the release from OLs in vivo remains to be elusive. Therefore, it is not too tempting to speculate that neurons could be recombined after engulfing Cre (mRNA or/and protein)-carrying exosomes secreted by NG2 glia. Still, whether NG2 glia can release exosomes in vivo remains to be elucidated. Altogether, to date, results from different transgenic mice did not provide adequate evidence for a neurogenic potential of NG2 glia.
Conclusion
In the past decades, the fate of NG2 glia in the CNS has been studied systematically during the development using a variety of transgenic mouse models. It is well established that NG2 glia are the most proliferative cells throughout life and could quickly generate OLs within the early postnatal weeks. The generation of OLs continues throughout life although the rate of OL differentiation declines with age in both GM and WM significantly. The astrogliogenic potential of NG2 glia is regionally restricted to a subgroup of embryonic NG2 glia mainly in the ventral forebrain. Meanwhile, few studies showed a potential neurogenesis from NG2 glia, but the findings were ambiguous. In conclusion, NG2 glia have the potential to generate OLs throughout life and astrocytes in the embryonic brain, but they do not generate neurons.
Acknowledgments
The authors are grateful to Prof. Dr. Frank Kirchhoff for his generous support and comments on the manuscript.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This work was supported by grants from the Deutsche Forschungsgemeinschaft DFG Sino-German joint project (Kl 503/14-1) to WH, DFG FOR 2289 to AS; from the Saarland University Medical Faculty HOMFOR2015 and HOMFORexzellenz2016 to AS and WH, respectively. WH was also supported by DFG SFB 894 and the European Commission EC-H2020 FET ProAct Neurofibres.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This work was supported by grants from the Deutsche Forschungsgemeinschaft DFG Sino-German joint project (Kl 503/14-1) to WH, DFG FOR 2289 to AS; from the Saarland University Medical Faculty HOMFOR2015 and HOMFORexzellenz2016 to AS and WH, respectively. WH was also supported by DFG SFB 894 and the European Commission EC-H2020 FET ProAct Neurofibres.
C-Editors: Zhao M, Li JY; T-Editor: Jia Y
References
- 1.Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 2.Bakhti M, Winter C, Simons M. Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J Biol Chem. 2011;286:787–796. doi: 10.1074/jbc.M110.190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beil J, Fairbairn L, Pelczar P, Buch T. Is BAC transgenesis obsolete. State of the art in the era of designer nucleases? J Biomed Biotechnol. 2012;2012:308414. doi: 10.1155/2012/308414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergles DE, Roberts JDB, Somogyi P, Jahr CE. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature. 2000;405:187. doi: 10.1038/35012083. [DOI] [PubMed] [Google Scholar]
- 5.Butt A, Dinsdale J. Opposing actions of fibroblast growth factor-2 on early and late oligodendrocyte lineage cells in vivo. J Neuroimmunol. 2005;166:75–87. doi: 10.1016/j.jneuroim.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Butt AM, Duncan A, Hornby MF, Kirvell SL, Hunter A, Levine JM, Berry M. Cells expressing the NG2 antigen contact nodes of Ranvier in adult CNS white matter. Glia. 1999;26:84–91. [PubMed] [Google Scholar]
- 7.Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- 8.Chan HY, V S, Xing X, Kraus P, Yap SP, Ng P, Lim SL, Lufkin T. Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PLoS One. 2011;6:e28885–28885. doi: 10.1371/journal.pone.0028885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittajallu R, Aguirre A, Gallo V. NG2-positive cells in the mouse white and grey matter display distinct physiological properties. J Physiol. 2004;561:109–122. doi: 10.1113/jphysiol.2004.074252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung S, Guo F, Jiang P, Pleasure DE, Deng W. Olig2/Plp-positive progenitor cells give rise to Bergmann glia in the cerebellum. Cell Death Dis. 2013;4:e546–e546. doi: 10.1038/cddis.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke LE, Young KM, Hamilton NB, Li H, Richardson WD, Attwell D. Properties and fate of oligodendrocyte progenitor cells in the corpus callosum, motor cortex, and piriform cortex of the mouse. J Neurosci. 2012;32:8173–8185. doi: 10.1523/JNEUROSCI.0928-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 13.Diers-Fenger M, Kirchhoff F, Kettenmann H, Levine JM, Trotter J. AN2/NG2 protein-expressing glial progenitor cells in the murine CNS: Isolation, differentiation, and association with radial glia. Glia. 2001;34:213–228. doi: 10.1002/glia.1055. [DOI] [PubMed] [Google Scholar]
- 14.Dimou L, Simon C, Kirchhoff F, Takebayashi H, Götz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J Neurosci. 2008;28:10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 16.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 18.Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch U-K, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 19.Frühbeis C, Fröhlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Möbius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Krämer-Albers E-M. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frühbeis C, Kuo-Elsner WP, Barth K, Peris L, Tenzer S, Möbius W, Werner H, Nave KA, Fröhlich D, Krämer-Albers EM. Oligodendrocyte-derived exosomes promote axonal transport and axonal long-term maintenance. bioRxiv. 2019 doi: 101101/20191220884171. [Google Scholar]
- 21.Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge WP, Yang XJ, Zhang Z, Wang HK, Shen W, Deng QD, Duan S. Long-term potentiation of neuron-glia synapses mediated by Ca2+-permeable AMPA receptors. Science. 2006;312:1533–1537. doi: 10.1126/science.1124669. [DOI] [PubMed] [Google Scholar]
- 23.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci U S A. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwood K, Butt AM. Evidence that perinatal and adult NG2-glia are not conventional oligodendrocyte progenitors and do not depend on axons for their survival. Mol Cell Neurosci. 2003;23:544–558. doi: 10.1016/s1044-7431(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 25.Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29:7256–7270. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo F, Maeda Y, Ma J, Xu J, Horiuchi M, Miers L, Vaccarino F, Pleasure D. Pyramidal neurons are generated from oligodendroglial progenitor cells in adult piriform cortex. J Neurosci. 2010;30:12036–12049. doi: 10.1523/JNEUROSCI.1360-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haberlandt C, Derouiche A, Wyczynski A, Haseleu J, Pohle J, Karram K, Trotter J, Seifert G, Frotscher M, Steinhäuser C. Gray matter NG2 cells display multiple Ca2+-signaling pathways and highly motile processes. PLoS One. 2011;6:e17575. doi: 10.1371/journal.pone.0017575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill RA, Li AM, Grutzendler J. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat Neurosci. 2018;21:683–695. doi: 10.1038/s41593-018-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill RA, Natsume R, Sakimura K, Nishiyama A. NG2 cells are uniformly distributed and NG2 is not required for barrel formation in the somatosensory cortex. Mol Cell Neurosci. 2011;46:689–698. doi: 10.1016/j.mcn.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoess RH, Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985;181:351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- 31.Horner PJ, Thallmair M, Gage FH. Defining the NG2-expressing cell of the adult CNS. J Neurocytol. 2002;31:469–480. doi: 10.1023/a:1025739630398. [DOI] [PubMed] [Google Scholar]
- 32.Huang W, Bai X, Stopper L, Catalin B, Cartarozzi LP, Scheller A, Kirchhoff F. During development NG2 glial cells of the spinal cord are restricted to the oligodendrocyte lineage, but generate astrocytes upon acute injury. Neuroscience. 2018;385:154–165. doi: 10.1016/j.neuroscience.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Huang W, Guo Q, Bai X, Scheller A, Kirchhoff F. Early embryonic NG2 glia are exclusively gliogenic and do not generate neurons in the brain. Glia. 2019;67:1094–1103. doi: 10.1002/glia.23590. [DOI] [PubMed] [Google Scholar]
- 34.Huang W, Zhao N, Bai X, Karram K, Trotter J, Goebbels S, Scheller A, Kirchhoff F. Novel NG2-CreERT2 knock-in mice demonstrate heterogeneous differentiation potential of NG2 glia during development. Glia. 2014;62:896–913. doi: 10.1002/glia.22648. [DOI] [PubMed] [Google Scholar]
- 35.Hughes EG, Orthmann-Murphy JL, Langseth AJ, Bergles DE. Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat Neurosci. 2018;21:696–706. doi: 10.1038/s41593-018-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karram K, Goebbels S, Schwab M, Jennissen K, Seifert G, Steinhäuser C, Nave KA, Trotter J. NG2-expressing cells in the nervous system revealed by the NG2-EYFP-knockin mouse. Genesis. 2008;46:743–757. doi: 10.1002/dvg.20440. [DOI] [PubMed] [Google Scholar]
- 39.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kukley M, Capetillo-Zarate E, Dietrich D. Vesicular glutamate release from axons in white matter. Nat Neurosci. 2007;10:311. doi: 10.1038/nn1850. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Aguzzi A. NG2 glia are required for maintaining microglia homeostatic state. Glia. 2020;68:345–355. doi: 10.1002/glia.23721. [DOI] [PubMed] [Google Scholar]
- 42.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 44.Marques S, van Bruggen D, Vanichkina DP, Floriddia EM, Munguba H, Väremo L, Giacomello S, Falcão AM, Meijer M, Björklund ÅK, Hjerling-Leffler J, Taft RJ, Castelo-Branco G. Transcriptional convergence of oligodendrocyte lineage progenitors during development. Dev Cell. 2018;46:504–517. doi: 10.1016/j.devcel.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyoshi G, Butt SJB, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano M, Tamura Y, Yamato M, Kume S, Eguchi A, Takata K, Watanabe Y, Kataoka Y. NG2 glial cells regulate neuroimmunological responses to maintain neuronal function and survival. Sci Rep. 2017;7:42041–42041. doi: 10.1038/srep42041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishiyama A. NG2 cells in the brain: a novel glial cell population. Human Cell. 2001;14:77–82. [PubMed] [Google Scholar]
- 49.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 50.Nishiyama A, Boshans L, Goncalves CM, Wegrzyn J, Patel KD. Lineage, fate, and fate potential of NG2-glia. Brain Res. 2016;1638:116–128. doi: 10.1016/j.brainres.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup W. Co-localization of NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 52.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 53.Peters A. A fourth type of neuroglial cell in the adult central nervous system. J Neurocytol. 2004;33:345–357. doi: 10.1023/B:NEUR.0000044195.64009.27. [DOI] [PubMed] [Google Scholar]
- 54.Petryniak MA, Potter GB, Rowitch DH, Rubenstein JLR. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richardson WD, Young KM, Tripathi RB, McKenzie I. NG2-glia as multipotent neural stem cells: fact or fantasy. Neuron. 2011;70:661–673. doi: 10.1016/j.neuron.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robins SC, Trudel E, Rotondi O, Liu X, Djogo T, Kryzskaya D, Bourque CW, Kokoeva MV. Evidence for NG2-glia derived, adult-born functional neurons in the hypothalamus. PLoS One. 2013;8:e78236. doi: 10.1371/journal.pone.0078236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rotheneichner P, Belles M, Benedetti B, König R, Dannehl D, Kreutzer C, Zaunmair P, Engelhardt M, Aigner L, Nacher J, Couillard-Despres S. Cellular plasticity in the adult murine piriform cortex: continuous maturation of dormant precursors into excitatory neurons. Cereb Cortex. 2018;28:2610–2621. doi: 10.1093/cercor/bhy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon C, Götz M, Dimou L. Progenitors in the adult cerebral cortex: cell cycle properties and regulation by physiological stimuli and injury. Glia. 2011;59:869–881. doi: 10.1002/glia.21156. [DOI] [PubMed] [Google Scholar]
- 60.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature. 1985;317:230. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 61.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 62.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stallcup WB. The NG2 proteoglycan: past insights and future prospects. J Neurocytol. 2002;31:423–435. doi: 10.1023/a:1025731428581. [DOI] [PubMed] [Google Scholar]
- 64.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 65.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka Y, Tozuka Y, Takata T, Shimazu N, Matsumura N, Ohta A, Hisatsune T. Excitatory GABAergic activation of cortical dividing glial cells. Cereb Cortex. 2009;19:2181–2195. doi: 10.1093/cercor/bhn238. [DOI] [PubMed] [Google Scholar]
- 67.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 68.Tognatta R, Sun W, Goebbels S, Nave KA, Nishiyama A, Schoch S, Dimou L, Dietrich D. Transient Cnp expression by early progenitors causes Cre-Lox-based reporter lines to map profoundly different fates. Glia. 2017;65:342–359. doi: 10.1002/glia.23095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripathi RB, Clarke LE, Burzomato V, Kessaris N, Anderson PN, Attwell D, Richardson WD. Dorsally and ventrally derived oligodendrocytes have similar electrical properties but myelinate preferred tracts. J Neurosci. 2011;31:6809–6819. doi: 10.1523/JNEUROSCI.6474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsoa RW, Coskun V, Ho CK, de Vellis J, Sun YE. Spatiotemporally different origins of NG2 progenitors produce cortical interneurons versus glia in the mammalian forebrain. Proc Natl Acad Sci U S A. 2014;111:7444–7449. doi: 10.1073/pnas.1400422111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Hove H, Antunes ARP, De Vlaminck K, Scheyltjens I, Van Ginderachter JA, Movahedi K. Identifying the variables that drive tamoxifen-independent CreERT2 recombination: Implications for microglial fate mapping and gene deletions. Eur J Immunol. 2020;50:459–463. doi: 10.1002/eji.201948162. [DOI] [PubMed] [Google Scholar]
- 72.Vélez-Fort M, Maldonado PP, Butt AM, Audinat E, Angulo MC. Postnatal switch from synaptic to extrasynaptic transmission between interneurons and NG2 cells. J Neurosci. 2010;30:6921–6929. doi: 10.1523/JNEUROSCI.0238-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K, Richardson WD. Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron. 2013;77:873–885. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang SZ, Wang QQ, Yang QQ, Gu HY, Yin YQ, Li YD, Hou JC, Chen R, Sun QQ, Sun YF, Hu G, Zhou JW. NG2 glia regulate brain innate immunity via TGF-β2/TGFBR2 axis. BMC Med. 2019;17:204. doi: 10.1186/s12916-019-1439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008a;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- 77.Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, Nishiyama A. Age-dependent fate and lineage restriction of single NG2 cells. Development. 2011;138:745–753. doi: 10.1242/dev.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu X, Hill RA, Nishiyama A. NG2 cells generate oligodendrocytes and gray matter astrocytes in the spinal cord. Neuron Glia Biol. 2008b;4:19–26. doi: 10.1017/S1740925X09000015. [DOI] [PubMed] [Google Scholar]


