Abstract
Recent studies implicate mitochondrial dysfunction in the development and progression of numerous chronic diseases, which may be partially due to modifications in mitochondrial DNA (mtDNA). There is also mounting evidence that epigenetic modifications to mtDNA may be an additional layer of regulation that controls mitochondrial biogenesis and function. Several environmental factors (eg, smoking, air pollution) have been associated with altered mtDNA methylation in a handful of mechanistic studies and in observational human studies. However, little is understood about other environmental contaminants that induce mtDNA epigenetic changes. Numerous environmental toxicants are classified as endocrine disrupting chemicals (EDCs). Beyond their actions on hormonal pathways, EDC exposure is associated with elevated oxidative stress, which may occur through or result in mitochondrial dysfunction. Although only a few studies have assessed the impacts of EDCs on mtDNA methylation, the current review provides reasons to consider mtDNA epigenetic disruption as a mechanism of action of EDCs and reviews potential limitations related to currently available evidence. First, there is sufficient evidence that EDCs (including bisphenols and phthalates) directly target mitochondrial function, and more direct evidence is needed to connect this to mtDNA methylation. Second, these and other EDCs are potent modulators of nuclear DNA epigenetics, including DNA methylation and histone modifications. Finally, EDCs have been shown to disrupt several modulators of mtDNA methylation, including DNA methyltransferases and the mitochondrial transcription factor A/nuclear respiratory factor 1 pathway. Taken together, these studies highlight the need for future research evaluating mtDNA epigenetic disruption by EDCs and to detail specific mechanisms responsible for such disruptions.
Keywords: mitochondria, epigenetics, environmental exposure, endocrine disruptors, methylation, oxidative stress, redox signaling, translational, lipids, gene expression
Mitochondria are subcellular organelles comprised of 2 separate and functionally distinct membranes that encapsulate the intermembrane space and matrix compartments (Nunnari and Suomalainen, 2012). As powerhouses of the cell, mitochondria are found in almost all human cell-types. Depending on the cell-type, the number of mitochondria in each cell and the copy number of mitochondrial DNA (mtDNA) also vary greatly. It was estimated that energy-intensive tissues (cardiac and skeletal muscle) have between 4000 and 6000 copies of mtDNA per cell, whereas liver, kidney, and lung tissues have between 500 and 2000 copies (D'Erchia et al., 2015). Mitochondria are responsible for up to 95% of cellular energy demands through oxidative phosphorylation (Tzameli, 2012). Beyond their role in cellular respiration, proper mitochondrial function is also critical for steroid synthesis, innate immunity, apoptosis, and nuclear epigenetic regulation (Shaughnessy et al., 2014). Another important role of mitochondria is to buffer cellular reactive oxygen species (ROS) levels. Therefore, mitochondrial dysfunction has been linked to aging and numerous chronic diseases.
Endocrine disrupting chemicals (EDCs) are a class of environmental contaminants that modulate hormone action. EDCs disrupt hormonal signals primarily by mimicking naturally occurring hormones, binding to hormone receptors, and then either activating or inhibiting important downstream pathways (Vandenberg et al., 2012). Well-characterized EDCs include phthalates, parabens, and bisphenols found in food packaging materials and personal care products, pesticides like DDT and atrazine, dioxins and polychlorinated biphenyls used as industrial solvents, various flame retardants, triclosan used in antibacterial soaps, and others (Gore et al., 2015). Numerous studies have demonstrated that mitochondria may be a target of some EDCs (Marroqui et al., 2018). Many EDCs are also known to influence regulation of the nuclear epigenome (Xin et al., 2015). Although several other environmental contaminants (eg, particulate matter) have been associated with mitochondrial epigenetic dysregulation (reviewed in the following sections), whether EDCs have similar effects on mitochondrial epigenetic endpoints is not entirely clear. We propose that substantially more needs to be understood about the ability of EDCs to alter the mitochondrial epigenome. Specifically, we recommend that this potential mechanism of toxicity or biomarker of EDC effect merits investigation in animal and human studies of EDC exposure. Therefore, this review will synthesize knowledge leading to this recommendation and the current limitations that need to be overcome to make progress in the field. We will discuss the function of mitochondria as it relates to health, disruption of mitochondrial endpoints by EDC exposures, currently available cross-sectional studies related to the mitochondrial epigenome and its relationship to human health, the impacts of non-EDC environmental contaminants on mitochondrial endpoints, and possible mechanisms by which EDCs could induce mitochondrial epigenetic dysregulation. Lastly, we will provide recommendations for incorporating mitochondrial epigenetic endpoints in research assessing the health impacts of EDCs.
Mitochondrial Dysfunction as a Biomarker of Health
Several rare diseases result directly from inherited mutations in mitochondrial or nuclear genes that regulate mitochondrial function (termed “primary mitochondrial diseases”) (Gorman et al., 2016; Rahman and Rahman, 2018). However, numerous chronic diseases are also characterized by systemic or organ-specific mitochondrial dysfunction. For example, given the mitochondria’s role in metabolic homeostasis, mitochondrial function has frequently been associated with metabolic disorders, including insulin resistance (Sangwung et al., 2020), nonalcoholic fatty liver disease (Mansouri et al., 2018), and obesity (de Mello et al., 2018). Although these studies have primarily evaluated mitochondria as a useful biomarker of chronic diseases, the field is beginning to consider mitochondria as a novel therapeutic target for the treatment of cardiovascular diseases (Bonora et al., 2019), reproductive disorders (Wolf et al., 2015), various cancers (Yang et al., 2016), neurodegeneration (Park et al., 2018), and metabolic diseases (Lee et al., 2019). Therefore, evaluating mitochondrial endpoints in response to various environmental and lifestyle factors and in diverse disease conditions could provide important clues into disease origins and even potential for treatment.
Mitochondrial Genome and Associations With Health Outcomes in Humans
As the hub of cellular respiration charged with producing the majority of the cell’s energy substrate (ATP), mitochondria have their own genome with unique structure and composition to sustain the cell’s energy requirements via oxidative phosphorylation within the electron transport chain (ETC) (St John, 2016). The human mitochondrial genome is a small circular double-stranded DNA molecule composed of 16 569 bp that encode 37 genes (Table 1). Thirteen of the approximately 100 protein subunits of the ETC are encoded by the mitochondrial genome, including 7 subunits of NADH dehydrogenase (Complex I), 1 subunit of cytochrome c reductase (Complex III), 3 subunits of cytochrome c oxidase (Complex IV), and 2 subunits of ATP synthase (Complex V) (Gao et al., 2017). The mitochondrial genome also encodes 22 species of transfer RNAs and 2 ribosomal RNAs for protein synthesis (Suzuki et al., 2011). In addition, the mitochondrial genome has 1 major noncoding region, the displacement loop (D-loop). Although the function of the D-loop is still unresolved, the regulatory elements within this region suggest a potential role for controlling mtDNA transcription and replication (eg, mtDNA copy number) (Chang and Clayton, 1985; Fernandez-Silva et al., 2003). Proper mtDNA replication and gene expression are crucial for cell viability, and disturbances in these processes have been shown to cause mitochondrial diseases in humans.
Table 1.
Genes Expressed Within the Human Mitochondrial Genome
| Gene Name | Gene Symbol | Strand | Function | Reference |
|---|---|---|---|---|
| Polypeptides | ||||
| Mitochondrially encoded NADH dehydrogenase 1 | MT-ND1 | H | Couples electron transfer from NADH to ubiquinone with transmembrane proton pumping contributing to the proton motive force used for ATP synthesis | Wirth et al. (2016) |
| Mitochondrially encoded NADH dehydrogenase 2 | MT-ND2 | H | ||
| Mitochondrially encoded NADH dehydrogenase 3 | MT-ND3 | H | ||
| Mitochondrially encoded NADH dehydrogenase 4 | MT-ND4 | H | ||
| Mitochondrially encoded NADH 4L dehydrogenase | MT-ND4L | H | ||
| Mitochondrially encoded NADH dehydrogenase 5 | MT-ND5 | H | ||
| Mitochondrially encoded NADH dehydrogenase 6 | MT-ND6 | L | ||
| Mitochondrially encoded cytochrome B | MT-CYB | H | As a component of Complex III, mediates electron transfer from ubiquinol to cytochrome c | Emmanuele et al. (2013) |
| Mitochondrially encoded cytochrome c oxidase I | MT-CO1 | H | Cytochrome c oxidase shuttles electrons from cytochrome c to molecular oxygen to capture energy in the membrane potential by the reduction of oxygen to water | Mick et al. (2011) |
| Mitochondrially encoded cytochrome c oxidase II | MT-CO2 | H | ||
| Mitochondrially encoded cytochrome c oxidase III | MT-CO3 | H | ||
| Mitochondrially encoded ATP synthase 6 | MT-ATP6 | H | Produces ATP through phosphorylation of ADP by using electrochemical energy generated by proton gradient across the inner membrane of mitochondria | Neupane et al. (2019) |
| Mitochondrially encoded ATP synthase 8 | MT-ATP8 | H | ||
| Ribosomal RNA | ||||
| Mitochondrially encoded 12S RNA | MT-RNR1 | H | Translation of mitochondrial mRNAs together with imported ribosomal proteins | Christian and Spremulli (2012) |
| Mitochondrially encoded 16S RNA | MT-RNR2 | H | ||
| Transfer RNA (tRNA) | ||||
| Mitochondrially encoded tRNA alanine | MT-TA | L | Codon reading during mitochondrial protein translation | Scaglia and Wong (2008) |
| Mitochondrially encoded tRNA arginine | MT-TR | H | ||
| Mitochondrially encoded tRNA asparagine | MT-TN | L | ||
| Mitochondrially encoded tRNA aspartic acid | MT-TD | H | ||
| Mitochondrially encoded tRNA cysteine | MT-TC | L | ||
| Mitochondrially encoded tRNA glutamic acid | MT-TE | L | ||
| Mitochondrially encoded tRNA glutamine | MT-TQ | L | ||
| Mitochondrially encoded tRNA glycine | MT-TG | H | ||
| Mitochondrially encoded tRNA histidine | MT-TH | H | ||
| Mitochondrially encoded tRNA isoleucine | MT-TI | H | ||
| Mitochondrially encoded tRNA leucine 1 (UUA/G) | MT-TL1 | H | ||
| Mitochondrially encoded tRNA leucine 2 (CUN) | MT-TL2 | H | ||
| Mitochondrially encoded tRNA lysine | MT-TK | H | ||
| Mitochondrially encoded tRNA methionine | MT-TM | H | ||
| Mitochondrially encoded tRNA phenylalanine | MT-TF | H | ||
| Mitochondrially encoded tRNA proline | MT-TP | L | ||
| Mitochondrially encoded tRNA serine 1 (UCN) | MT-TS1 | L | ||
| Mitochondrially encoded tRNA serine 2 (AGU/C) | MT-TS2 | H | ||
| Mitochondrially encoded tRNA threonine | MT-TT | H | ||
| Mitochondrially encoded tRNA tryptophan | MT-TW | H | ||
| Mitochondrially encoded tRNA tyrosine | MT-TY | L | ||
| Mitochondrially encoded tRNA valine | MT-TV | H |
Abbreviations: H, heavy strand; L, light strand.
Because each mitochondria can have multiple genome (DNA) copies, and each cell can have numerous mitochondria (Satoh and Kuroiwa, 1991), mtDNA copy number has been proposed as an estimate of mitochondrial cellular respiration capacity (Malik and Czajka, 2013). As such, mtDNA copy number reflects oxidative phosphorylation capacity in cells, with lower copy number linked with poor oxidative capacity and elevated oxidative stress (Liu et al., 2003; Wang et al., 2011). Given the many other roles of mitochondria in the cell, mtDNA copy number also likely reflects overall mitochondrial function. Metabolic disease is a simple example of such associations, as it is characterized by poor metabolic flexibility and oxidative capacity. For example, a study of 12 obese and 8 lean U.S. adults found that mtDNA copy number was lower in obese compared with lean individuals (1665 ± 213 vs 2514 ± 505, p < .05) (Ritov et al., 2005), whereas in 148 healthy Swedish volunteers, mtDNA copy number was shown to decrease significantly in adipose tissue with increasing BMI (r = −0.24, p = .004) (Kaaman et al., 2007). Similarly, a study in 94 healthy Korean young adults showed that mtDNA copy number in circulation was negatively associated with BMI (r = −0.22, p = .04), as well as waist circumference (r = −0.23, p = .03) and visceral fat area (r = −0.28, p = .01) after adjusting for age and sex (Lee et al., 2014). Analogous studies have shown associations between mtDNA copy number and various other diseases, including Parkinson’s disease (Dolle et al., 2016; Pyle et al., 2016), cardiovascular diseases (Ashar et al., 2017; Yue et al., 2018; Zhang et al., 2017), and numerous others (Al-Kafaji et al., 2020; Fazzini et al., 2019; Jędrak et al., 2017)—further confirming the role of mitochondrial homeostasis in human disease development and progression.
EDCs and Disruption of Mitochondrial Endpoints
There is mounting evidence that some EDCs are associated with disruptions to mitochondrial endpoints, and that such disruptions may be 1 potential cause of EDC toxicity (Chernis et al., 2020; Moon et al., 2012; Posnack et al., 2012). Although the precise mechanisms for these observations are not entirely clear, oxidative stress has been implicated as an important mediator between various EDC exposures and long-term deleterious health outcomes (Neier et al., 2015). Given the importance of mitochondria for cellular ROS homeostasis and the pro-oxidant nature of some EDCs (Marroqui et al., 2018), it is likely that EDCs act, in part, through ROS-mediated and mitochondrial pathways.
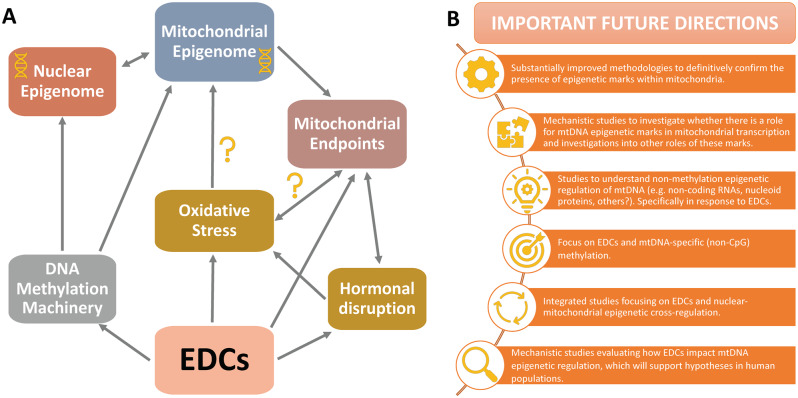
Two (and likely more) oxidative stress-related pathways could connect EDCs to mitochondrial disruption (Figure 1A). First, EDCs could induce oxidative stress, leading to mitochondrial damage and having deleterious consequences for numerous downstream mitochondria-related functions. Although mitochondrial oxidative phosphorylation of lipids generates damaging pro-oxidants (Taylor et al., 2014)—thereby exposing mitochondria to high endogenous pro-oxidants—mtDNA appears to be even more sensitive to oxidative damage than the nuclear compartment (Bulteau et al., 2006; Orrenius et al., 2007), as was first reported in a seminal study several decades ago (Yakes and Van Houten, 1997). Another potential mechanism is that EDCs directly damage mitochondria or target mitochondrial biogenesis, thereby decreasing the cell’s capacity to deal with intrinsic and extrinsic sources of ROS. Because of the challenges related to assessing direct effects on mitochondrial endpoints, this pathway has been less studied. Furthermore, because oxidative stress and mitochondrial function are so closely linked, EDC-mediated toxicity likely occurs through both mechanisms. Therefore, whereas studies have begun to concurrently assess oxidative stress and mitochondrial pathways in response to EDC exposures (Azevedo et al., 2020; Hornos Carneiro et al., 2020; Rodrigues-Pereira et al., 2020; Sammi et al., 2019), more research may be needed to clarify the precise mechanisms behind EDC-induced mito-toxicity. These mechanistic studies will be critical for driving research in human populations, which will require stable biomarkers of mitochondrial effects in response to EDC exposures.
Figure 1.
Evidence and plausible mechanisms supporting the role of endocrine disrupting chemicals (EDCs) in modulating mitochondrial DNA (mtDNA) epigenetic endpoints (A) and recommended future directions to move the field forward (B). Abbreviation: CpG, cytosines followed by guanine residues on DNA.
Given the hormone-modulating nature of EDCs, the interplay between hormones and mitochondrial function is also difficult to ignore. First, mitochondria serve critical roles in steroidogenesis, thus making them a likely target for EDC action. For example, the steroidogenic acute regulatory (StAR) protein—the acute control point for steroid synthesis—acts by relocating cholesterol from the outer- to innermitochondrial membrane (Miller, 2013). Several studies suggest that some EDCs target StAR and its downstream pathways. For example, tributyltin was shown to upregulate StAR mRNA expression in ovine ovarian theca cells and several other species, which authors suggested was a compensatory mechanism for tributyltin-induced cellular cholesterol efflux (Pu et al., 2019). One study also showed that a common pesticide inhibited steroidogenesis by disrupting StAR protein expression in a Leydig tumor cell line (Walsh et al., 2000), whereas another reported that Star mRNA expression was decreased in testes of mice exposed to dibutyl phthalate and 4-tert-octylphenol (Bunay et al., 2018). Second, numerous studies have confirmed the role of steroid hormones (estrogen, specifically) in regulating mitochondrial function, especially its role in oxidative stress (Klinge, 2020; Tower et al., 2020). Whether these endocrine mechanisms are directly related to the observed effects of EDCs on oxidative stress will need to be better understood, but these studies do suggest a role of EDCs in modulating critical mitochondrial functions.
The Mitochondrial Epigenome
There is recent evidence that the mitochondrial genome, like the nuclear genome, contains epigenetic marks, but as will be discussed later, there is controversy about the presence and potential functions of these modifications. The epigenome here is defined as heritable alterations to the genome that do not change the underlying DNA sequence but influence gene regulation. Epigenetic modifications of the nuclear genome include DNA methylation and hydroxymethylation, certain noncoding RNA (ncRNA), and chromatin packaging dictated in part by post-translational histone tail modifications (Bird, 1986; Strahl and Allis, 2000). In nuclear DNA, the histone code works in concert with DNA methylation to regulate gene expression by altering the accessibility of transcriptional machinery to a given gene. DNA methylation, specifically, is 1 form of epigenetic regulation by which methyl groups are added to DNA cytosine residues. In mammalian nuclear DNA, methylated cytosines are typically immediately followed by guanine residues (called CpG sites). These DNA modifications alter gene transcription by changing the physical conformation of the DNA, attracting methyl CpG-binding proteins, and altering the binding of transcription factors. As with other forms of epigenetic regulation, these modifications can be inherited across cell divisions and possibly across generations.
While the following sections will describe observational studies connecting mtDNA methylation to various exposures and health outcomes, the existence of methylation (specifically) within mtDNA has been somewhat controversial, with some studies reporting a complete lack of mtDNA methylation (Hong et al., 2013; Mechta et al., 2017). As has been recently described (Morris et al., 2020; Owa et al., 2018), this uncertainty may partly stem from the methodologies used to assess mtDNA methylation. Using these methodologies, the proportion of methylated CpG sites throughout the mitochondrial genome was reported to be much lower overall compared with the nuclear genome (< 5% compared with 70% of nuclear DNA CpG sites) (Hong et al., 2013). However, recent bisulfite sequencing studies have revealed extensive non-CpG methylation of mtDNA, with approximately 40% of cytosines being methylated (Patil et al., 2019). Of these non-CpG methylated cytosines, CpT and CpC dinucleotides are the most frequently methylated, followed by CpA and with CpG sites exhibiting the most infrequent methylation. Recently, GpC methylation (rather than CpG methylation) was also discovered and was shown to be inversely associated with mtDNA gene expression in vitro (van der Wijst et al., 2017). By using technologies that focus specifically on CpG methylation, it is possible that some studies have underestimated the true levels of mtDNA methylation (Morris et al., 2020), but substantially more evidence is needed to confirm whether this is truly the case. Although assessment of non-CpG methylation is needed in future studies of mtDNA methylation, researchers using methods that only assess CpG methylation (ie, pyrosequencing) can benefit from recent methodological advancements. Vos et al. modified the pyrosequencing protocol to avoid inaccurate results due to incomplete bisulfite conversion and/or amplification of nuclear mitochondrial sequences (NUMTs). First, they completely linearized mtDNA prior to conversion with BamHI. Then, they amplified regions of mtDNA genes that do not overlap with NUMTs (Vos et al., 2020). Such rigorous methods are essential going forward in mtDNA methylation studies.
Transcriptional regulation of mtDNA, much like that of bacterial genomes, is heavily influenced by nucleoid packaging, which involves proteins such as mitochondrial transcription factor A (TFAM) (Gilkerson et al., 2013). In terms of specific epigenetic marks within mtDNA, there is evidence to date for the presence of mtDNA methylation (Vos et al., 2020; Weinhouse, 2017), short and long ncRNA (Dong et al., 2017), and post-translational modifications to nucleoid proteins (King et al., 2018). Although substantially more data are needed to understand the potential regulation of these marks, studies suggest that mtDNA methylation patterns are dependent on isoform 3 of DNMT1 as well as DNMT3A in a local context and DNMT3B in a global context (Dou et al., 2019; Patil et al., 2019; Saini et al., 2017). It is important to note, however, that whereas the role of epigenetic marks in nuclear transcriptional regulation is relatively well established, the precise roles of epigenetic modifications within mtDNA are unclear. Several reviews have addressed this current limitation in the field and suggest that whereas epigenetic marks have been observed within mtDNA, there is currently little mechanistic insight connecting these marks to mitochondrial gene transcription (Mposhi et al., 2017; van der Wijst and Rots, 2015). The reason that some studies have suggested a role of mtDNA methylation in mitochondrial gene regulation is because mtDNA methylation is nonrandom and strand-specific; there is more methylation on the L-strand, which is specifically related to transcription. In addition, unique methylation patterns have been observed within the regulatory D-loop region, which controls mitochondrial gene transcription (van der Wijst and Rots, 2015). However, these studies provide correlations between methylation and transcription but lack causal evidence; substantially more data are needed to understand the specific roles that DNA methylation (and other epigenetic marks) plays within the mitochondria.
Proposed Roles of the Mitochondrial Epigenome in Health and Disease
Beyond measuring mtDNA copy number, recent studies have also suggested that mtDNA methylation may be an important marker of mitochondrial function (Iacobazzi et al., 2013; Stimpfel et al., 2018) and may in fact regulate mtDNA copy number (Lee et al., 2015; Sun et al., 2016; Tong et al., 2017). Therefore, similar to alterations in mtDNA copy number (and to nuclear DNA methylation), observational studies in humans have also begun to correlate mtDNA methylation with health outcomes, including several cancers (Ferreira et al., 2015), obesity (Zheng et al., 2015), metabolic health (Zheng et al., 2016), severe nonalcoholic liver disease (Pirola et al., 2013), Alzheimer’s disease (Stoccoro et al., 2017), cardiovascular disease (Baccarelli and Byun, 2015), and even pregnancy outcomes (Novielli et al., 2017). However, it is important to reiterate that additional studies may be needed to confirm whether these previous associations remain when using updated methods that more accurately assess mtDNA methylation.
Because the field is relatively new, the value of mtDNA methylation (or other epigenetic disruption) as a biomarker of disease remains to be established. For example, future studies are needed to determine whether epigenetic marks are better predictors of human diseases than are other mitochondrial biomarkers. Additional studies are also needed to understand whether perturbations in the mitochondrial epigenome are heritable not only across cell divisions but also across generations. There is substantial evidence of crosstalk in mitochondrial-nuclear epigenetic regulation, such that molecules generated as part of the mitochondrial energetics cascade regulate the nuclear epigenome, and proteins required for mtDNA transcriptional regulation are expressed by the nuclear genome (reviewed extensively in Weinhouse [2017]). Therefore, the disruption of the mitochondrial epigenome by environmental conditions may provide important clues into the regulation of the nuclear genome, and this process also needs to be better understood. Finally, it will be critical to investigate associations of nonmethylation epigenetic regulators within the mitochondria with various health outcomes, along with mtDNA methylation. Despite these current limitations, and assuming proper methodologies are in place to adequately establish mitochondria-specific epigenetic patterns, mitochondrial epigenetics may provide a novel approach for understanding organelle-specific epigenetic regulation of important physiological conditions in response to numerous systemic and environmental stressors.
Perturbation of the Mitochondrial Epigenome by Environmental Exposures
Similar to the nuclear epigenome (Martin and Fry, 2018), environmental health and toxicology studies are beginning to consider the mitochondrial epigenome as a target endpoint and potential mechanism of toxicity in response to numerous exposures. This field is still in its infancy, and studies to date have reported associations in human or animal studies between mtDNA and exposures to smoking, ambient particulate matter, metals, BDE-47, olive oil, and doxorubicin (reviewed previously by Lambertini and Byun [2016] and Sharma et al. [2019]). One example is a study of placental mtDNA methylation in 96 mother-newborn pairs from Rhode Island and foreskin from 62 infants from Kentucky that found that maternal smoking status was positively associated with D-loop methylation in both placenta (p = .001) and foreskin (p = .04) (Armstrong et al., 2016). Placental D-loop methylation was also higher among mothers who smoked or had high air pollution exposure compared with nonsmoking mothers with lower air pollution exposure (p < .05) using a method optimized for mtDNA methylation analysis (Vos et al., 2020). Similarly, in 381 Belgian women, higher exposure to airborne particulate matter with aerodynamic diameter ≤ 2.5 µm (PM2.5) was positively associated with mtDNA methylation of placental MT-RNR1 (p = .01) and D-loop (p = .06) (Janssen et al., 2015). Interestingly, authors also observed that higher PM2.5 exposure was associated with lower mtDNA copy number (p = .001), and established that mtDNA methylation explained a large proportion of this relationship (MT-RNR1: 54% and D-loop: 27%), potentially supporting the proposed role of mtDNA methylation in mitochondrial biogenesis (Janssen et al., 2015). In another example, mtDNA methylation of D-loop and ND6 was reduced in 221 highly arsenic exposed adults from West Bengal compared with 101 relatively unexposed adults. In turn, compared with the unexposed group, the exposed group had higher expression of ND4 and ND6, as well as increased mtDNA copy number (van der Wijst et al., 2017). While the precise mechanisms behind the findings in these observational studies are not well understood, these results connecting the environment to the mitochondrial epigenome need further exploration.
EDCs and Potential for Mitochondrial Epigenetic Disruption
While evidence is growing for environmental lability of the mitochondrial epigenome, studies have been limited to a handful of environmental exposures and have assessed primarily mtDNA methylation, generally without considering non-CpG methylation. To our knowledge, BDE-47, a lipophilic polybrominated diphenyl ether, is the only EDC studied to date with respect to mtDNA epigenetics. In a perinatal exposure study of the flame retardant BDE-47, exposed rat offspring exhibited decreased methylcytosine in mtDNA at postnatal day 41 in the brain compared with control offspring (Byun et al., 2015).
This relative lack of information related to EDCs and epigenetic disruption is surprising given that several studies have concluded that EDCs target mitochondrial endpoints (discussed above). Furthermore, numerous EDCs have been shown to disrupt the nuclear epigenome and its regulators, including epigenetic disruption in the brain (reviewed by Walker and Gore [2017]) and disruption that persists inter- and transgenerationally (reviewed by Van Cauwenbergh et al. [2020]). There are 2 types of mechanisms by which EDCs are thought to induce epigenetic changes in nuclear DNA (reviewed by Alavian-Ghavanini and Rüegg [2018]). EDCs can regulate DNA methylation and chromatin state locally through inhibition or activation of nuclear receptors which recruit epigenetic machinery to their target genes. For example, bisphenol A (BPA) induced changes to DNA methylation of Fkbp5 in mice that were dependent on estrogen receptor-beta (Kitraki et al., 2015). An in vitro approach comparing the ability of various EDCs to occupy an ER-regulated promoter and ultimately induce local chromatin changes showed that some EDCs, including BPA and diethylstilbestrol, did induce promoter occupancy, albeit with lower affinities than estradiol (Ashcroft et al., 2011). In addition to gene-specific changes, some EDCs may induce widespread epigenetic changes indirectly through deregulation of epigenetic machinery (eg, DNMTs) or enzymes involved in the 1-carbon metabolism pathway (Laing et al., 2016). This deregulation occurs through activation or inhibition of nuclear receptors that control expression of epigenetic machinery genes. These proposed mechanisms, supported by indirect and some direct evidence, may also need to be investigated in relation to DNA methylation changes in mtDNA. Furthermore, as previously mentioned, the regulation of the nuclear and mitochondrial epigenomes appears to be closely intertwined (Weinhouse, 2017). Although the current review primarily focuses on mitochondria-specific epigenetic modifications, it is important to consider that these potential disruptions in response to environmental factors may have critical consequences for nuclear epigenetic regulation and vice versa. Therefore, whereas there is a relative lack of information related to EDC-induced epigenetic disruption of mtDNA, some knowledge can be gleaned by understanding the impacts of EDCs on the nuclear epigenome.
For example, DNA methylation relies on DNMTs, which are disrupted in the nuclear compartment by BPA, phthalates, and several other EDCs (reviewed extensively in Xin et al. [2015]). As previously discussed, DNMTs transcribed in the nucleus also appear to be required for mtDNA methylation, and their disruption in the nucleus may have important consequences for the mitochondrial epigenome. Another potential mechanism involves TFAM, the primary mitochondrial transcription factor that also has a role in mtDNA methylation (Rebelo et al., 2009). The transcription of TFAM is regulated by the nuclear respiratory factor 1 (Gleyzer et al., 2005), and this oxidative balance-related transcription factor is disrupted in response to numerous EDCs, including BPA and phthalates (reviewed in Marroqui et al. [2018]). These examples provide the impetus to better understand the impacts of EDCs on mitochondrial epigenetic endpoints. As discussed previously, the evidence linking mtDNA epigenetic changes to mitochondrial function remains to be concretely established. However, if EDCs do in fact target the mitochondrial epigenome, substantially more data are needed to understand the impacts of these disruptions for disease initiation and progression.
FUTURE DIRECTIONS AND CONCLUSIONS
Although direct evidence for EDC-induced mtDNA epigenetic disruption is exceedingly limited due to a lack of studies in this area, there is potential for EDCs to target mtDNA epigenetic endpoints. First, EDCs are known to target mitochondrial endpoints, including mitochondrial copy number and biogenesis, which have been shown to be regulated by mtDNA methylation. Second, there is evidence from cross-sectional studies for associations between mitochondrial epigenetic disruption and other environmental exposures—specifically smoking and air pollution, which, like EDCs, involve mitochondrial/pro-oxidant pathways. Finally, EDCs are known epigenetic disruptors, and whereas this evidence comes primarily from studies evaluating nuclear DNA, some mechanisms of nuclear epigenetic disruption may be relevant for mitochondrial epigenetic disruption. Therefore, we recommend that substantially more research is needed to investigate potential roles of EDCs in modifying mtDNA epigenetic endpoints and to determine how these potential changes impact human health. Ultimately, this knowledge will inform risk assessment and prevention/intervention strategies to mitigate the toxic effects of EDCs.
As discussed above, it is important to mention that such progress will also require more direct evidence for the fundamental function of mtDNA epigenetic disruption in mitochondrial and cellular health. Beyond that, given that so little is currently understood about mitochondrial epigenetic disruption by environmental toxicants, we also propose that more evidence is needed for mechanisms by which EDCs influence the mitochondrial epigenome (Figure 1B). Specifically, substantially more data are needed to understand whether the pro-oxidant and inflammatory pathways often associated with EDC exposures drive mitochondrial epigenetic disruption, or whether these pathways are activated in response to epigenetic shifts that cause mitochondrial dysfunction. Going forward, studies must also take into account the rapidly evolving knowledge about the unique mitochondrial epigenome when selecting methods for evaluation. For example, recent evidence shows that mtDNA methylation is more likely to occur in CpC, CpT, and GpC sites instead of CpG sites. Thus, next generation or targeted bisulfite sequencing-based methods should be used that quantify mtDNA methylation at all cytosines whenever possible. In addition, new mitochondria-specific approaches that assess nonmethylation epigenetic mechanisms, such as post-translational modifications of nucleoid proteins and the potential role of ncRNAs should be considered to gain a holistic understanding of environmental impacts of mtDNA regulation. Given the importance of mitochondria in human health and disease, such progress would provide valuable biomarkers that better characterize the mechanisms of action of EDCs and numerous other environmental toxicants.
FUNDING
United States Department of Agriculture National Institute of Food and Agriculture and Michigan AgBioResearch (Z.Z. and R.S.S.).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Contributor Information
Zheng Zhou, Department of Animal Sciences, Michigan State University, East Lansing, Michigan 48824.
Jaclyn M Goodrich, Department of Environmental Health Sciences, University of Michigan, Ann Arbor, Michigan 48109.
Rita S Strakovsky, Department of Food Science and Human Nutrition; Institute for Integrative Toxicology, Michigan State University, East Lansing, Michigan 48824.
REFERENCES
- Alavian-Ghavanini A., Rüegg J. (2018). Understanding epigenetic effects of endocrine disrupting chemicals: From mechanisms to novel test methods. Basic Clin. Pharmacol. Toxicol. 122, 38–45. [DOI] [PubMed] [Google Scholar]
- Al-Kafaji G., Bakheit H. F., Alharbi M. A., Farahat A. A., Jailani M., Ebrahin B. H., Bakhiet M. (2020). Mitochondrial DNA copy number in peripheral blood as a potential non-invasive biomarker for multiple sclerosis. Neuromol. Med. 22, 304–313. [DOI] [PubMed] [Google Scholar]
- Armstrong D. A., Green B. B., Blair B. A., Guerin D. J., Litzky J. F., Chavan N. R., Pearson K. J., Marsit C. J. (2016). Maternal smoking during pregnancy is associated with mitochondrial DNA methylation. Environ. Epigenet. 2, dvw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar F. N., Zhang Y., Longchamps R. J., Lane J., Moes A., Grove M. L., Mychaleckyj J. C., Taylor K. D., Coresh J., Rotter J. I., et al. (2017). Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol. 2, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft F. J., Newberg J. Y., Jones E. D., Mikic I., Mancini M. A. (2011). High content imaging-based assay to classify estrogen receptor-α ligands based on defined mechanistic outcomes. Gene 477, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo L. F., Hornos Carneiro M. F., Dechandt C. R. P., Cassoli J. S., Alberici L. C., Barbosa F. Jr (2020). Global liver proteomic analysis of Wistar rats chronically exposed to low-levels of bisphenol A and S. Environ. Res. 182, 109080. [DOI] [PubMed] [Google Scholar]
- Baccarelli A. A., Byun H. M. (2015). Platelet mitochondrial DNA methylation: A potential new marker of cardiovascular disease. Clin. Epigenet. 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. (1986). CpG-rich islands and the function of DNA methylation. Nature 321, 209–213. [DOI] [PubMed] [Google Scholar]
- Bonora M., Wieckowski M. R., Sinclair D. A., Kroemer G., Pinton P., Galluzzi L. (2019). Targeting mitochondria for cardiovascular disorders: Therapeutic potential and obstacles. Nat. Rev. Cardiol. 16, 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau A. L., Szweda L. I., Friguet B. (2006). Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp. Gerontol. 41, 653–657. [DOI] [PubMed] [Google Scholar]
- Bunay J., Larriba E., Patino-Garcia D., Cruz-Fernandes L., Castaneda-Zegarra S., Rodriguez-Fernandez M., Del Mazo J., Moreno R. D. (2018). Editor's highlight: Differential effects of exposure to single versus a mixture of endocrine-disrupting chemicals on steroidogenesis pathway in mouse testes. Toxicol. Sci. 161, 76–86. [DOI] [PubMed] [Google Scholar]
- Byun H. M., Benachour N., Zalko D., Frisardi M. C., Colicino E., Takser L., Baccarelli A. A. (2015). Epigenetic effects of low perinatal doses of flame retardant BDE-47 on mitochondrial and nuclear genes in rat offspring. Toxicology 328, 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. (1985). Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. U.S.A. 82, 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernis N., Masschelin P., Cox A. R., Hartig S. M. (2020). Bisphenol AF promotes inflammation in human white adipocytes. Am. J. Physiol. Cell Physiol. 318, C63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian B. E., Spremulli L. L. (2012). Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta 1819, 1035–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mello A. H., Costa A. B., Engel J. D. G., Rezin G. T. (2018). Mitochondrial dysfunction in obesity. Life Sci. 192, 26–32. [DOI] [PubMed] [Google Scholar]
- D'Erchia A. M., Atlante A., Gadaleta G., Pavesi G., Chiara M., De Virgilio C., Manzari C., Mastropasqua F., Prazzoli G. M., Picardi E., et al. (2015). Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion 20, 13–21. [DOI] [PubMed] [Google Scholar]
- Dolle C., Flones I., Nido G. S., Miletic H., Osuagwu N., Kristoffersen S., Lilleng P. K., Larsen J. P., Tysnes O. B., Haugarvoll K., et al. (2016). Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat. Commun. 7, 13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Yoshitomi T., Hu J. F., Cui J. (2017). Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenet. Chromatin 10, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou X., Boyd-Kirkup J. D., McDermott J., Zhang X., Li F., Rong B., Zhang R., Miao B., Chen P., Cheng H., et al. (2019). The strand-biased mitochondrial DNA methylome and its regulation by DNMT3A. Genome Res. 29, 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuele V., Sotiriou E., Rios P. G., Ganesh J., Ichord R., Foley A. R., Akman H. O., Dimauro S. (2013). A novel mutation in the mitochondrial DNA cytochrome b gene (MTCYB) in a patient with mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes syndrome. J. Child Neurol. 28, 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzini F., Lamina C., Fendt L., Schultheiss U. T., Kotsis F., Hicks A. A., Meiselbach H., Weissensteiner H., Forer L., Krane V., et al. (2019). Mitochondrial DNA copy number is associated with mortality and infections in a large cohort of patients with chronic kidney disease. Kidney Int. 96, 480–488. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P., Enriquez J. A., Montoya J. (2003). Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 88, 41–56. [DOI] [PubMed] [Google Scholar]
- Ferreira A., Serafim T. L., Sardao V. A., Cunha-Oliveira T. (2015). Role of mtDNA-related mitoepigenetic phenomena in cancer. Eur. J. Clin. Invest. 45, 44–49. [DOI] [PubMed] [Google Scholar]
- Gao D., Zhu B., Sun H., Wang X. (2017). Mitochondrial DNA methylation and related disease. Adv. Exp. Med. Biol. 1038, 117–132. [DOI] [PubMed] [Google Scholar]
- Gilkerson R., Bravo L., Garcia I., Gaytan N., Herrera A., Maldonado A., Quintanilla B. (2013). The mitochondrial nucleoid: Integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb. Perspect. Biol. 5, a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleyzer N., Vercauteren K., Scarpulla R. C. (2005). Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 25, 1354–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A. C., Chappell V. A., Fenton S. E., Flaws J. A., Nadal A., Prins G. S., Toppari J., Zoeller R. T. (2015). Executive summary to EDC-2: The endocrine society's second scientific statement on endocrine-disrupting chemicals. Endocr. Rev. 36, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman G. S., Chinnery P. F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D. R., Zeviani M., Turnbull D. M. (2016). Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080. [DOI] [PubMed] [Google Scholar]
- Hong E. E., Okitsu C. Y., Smith A. D., Hsieh C. L. (2013). Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell. Biol. 33, 2683–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornos Carneiro M. F., Shin N., Karthikraj R., Barbosa F. Jr, Kannan K., Colaiacovo M. P. (2020). Antioxidant CoQ10 restores fertility by rescuing bisphenol A-induced oxidative DNA damage in the Caenorhabditis elegans germline. Genetics 214, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobazzi V., Castegna A., Infantino V., Andria G. (2013). Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol. Genet. Metab. 110, 25–34. [DOI] [PubMed] [Google Scholar]
- Janssen B. G., Byun H. M., Gyselaers W., Lefebvre W., Baccarelli A. A., Nawrot T. S. (2015). Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: An ENVIRONAGE birth cohort study. Epigenetics 10, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jędrak P., Krygier M., Tońska K., Drozd M., Kaliszewska M., Bartnik E., Sołtan W., Sitek E. J., Stanisławska-Sachadyn A., Limon J., et al. (2017). Mitochondrial DNA levels in Huntington disease leukocytes and dermal fibroblasts. Metab. Brain Dis. 32, 1237–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaman M., Sparks L. M., van Harmelen V., Smith S. R., Sjolin E., Dahlman I., Arner P. (2007). Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 50, 2526–2533. [DOI] [PubMed] [Google Scholar]
- King G. A., Hashemi Shabestari M., Taris K. H., Pandey A. K., Venkatesh S., Thilagavathi J., Singh K., Krishna Koppisetti R., Temiakov D., Roos W. H., et al. (2018). Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res. 46, 3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitraki E., Nalvarte I., Alavian-Ghavanini A., Rüegg J. (2015). Developmental exposure to bisphenol A alters expression and DNA methylation of Fkbp5, an important regulator of the stress response. Mol. Cell. Endocrinol. 417, 191–199. [DOI] [PubMed] [Google Scholar]
- Klinge C. M. (2020). Estrogenic control of mitochondrial function. Redox Biol. 31, 101435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing L. V., Viana J., Dempster E. L., Trznadel M., Trunkfield L. A., Uren Webster T. M., van Aerle R., Paull G. C., Wilson R. J., Mill J., et al. (2016). Bisphenol A causes reproductive toxicity, decreases dnmt1 transcription, and reduces global DNA methylation in breeding zebrafish (Danio rerio). Epigenetics 11, 526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini L., Byun H. M. (2016). Mitochondrial epigenetics and environmental exposure. Curr. Environ. Health Rep. 3, 214–224. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Park A., Oh K. J., Lee S. C., Kim W. K., Bae K. H. (2019). The role of adipose tissue mitochondria: Regulation of mitochondrial function for the treatment of metabolic diseases. Int. J. Mol. Sci. 20, 4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Lee D. C., Im J. A., Lee J. W. (2014). Mitochondrial DNA copy number in peripheral blood is independently associated with visceral fat accumulation in healthy young adults. Int. J. Endocrinol. 2014, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Johnson J., Gough D. J., Donoghue J., Cagnone G. L., Vaghjiani V., Brown K. A., Johns T. G., St John J. C. (2015). Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis. 6, e1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. S., Tsai C. S., Kuo C. L., Chen H. W., Lii C. K., Ma Y. S., Wei Y. H. (2003). Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic. Res. 37, 1307–1317. [DOI] [PubMed] [Google Scholar]
- Malik A. N., Czajka A. (2013). Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 13, 481–492. [DOI] [PubMed] [Google Scholar]
- Mansouri A., Gattolliat C. H., Asselah T. (2018). Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology 155, 629–647. [DOI] [PubMed] [Google Scholar]
- Marroqui L., Tuduri E., Alonso-Magdalena P., Quesada I., Nadal A., Dos Santos R. S. (2018). Mitochondria as target of endocrine-disrupting chemicals: Implications for type 2 diabetes. J. Endocrinol. 239, R27–45. [DOI] [PubMed] [Google Scholar]
- Martin E. M., Fry R. C. (2018). Environmental influences on the epigenome: Exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333. [DOI] [PubMed] [Google Scholar]
- Mechta M., Ingerslev L. R., Fabre O., Picard M., Barres R. (2017). Evidence suggesting absence of mitochondrial DNA methylation. Front. Genet. 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D. U., Fox T. D., Rehling P. (2011). Inventory control: Cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. L. (2013). Steroid hormone synthesis in mitochondria. Mol. Cell. Endocrinol. 379, 62–73. [DOI] [PubMed] [Google Scholar]
- Moon M. K., Kim M. J., Jung I. K., Koo Y. D., Ann H. Y., Lee K. J., Kim S. H., Yoon Y. C., Cho B. J., Park K. S., et al. (2012). Bisphenol A impairs mitochondrial function in the liver at doses below the no observed adverse effect level. J. Korean Med. Sci. 27, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M. J., Hesson L. B., Youngson N. A. (2020). Non-CpG methylation biases bisulphite PCR towards low or unmethylated mitochondrial DNA: Recommendations for the field. Environ. Epigenet. 6, dvaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mposhi A., Van der Wijst M. G., Faber K. N., Rots M. G. (2017). Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. (Landmark Ed.)22, 1099–1113. [DOI] [PubMed] [Google Scholar]
- Neier K., Marchlewicz E. H., Dolinoy D. C., Padmanabhan V. (2015). Assessing human health risk to endocrine disrupting chemicals: A focus on prenatal exposures and oxidative stress. Endocr. Disruptors (Austin) 3, e1069916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane P., Bhuju S., Thapa N., Bhattarai H. K. (2019). ATP synthase: Structure, function and inhibition. Biomol. Concepts 10, 1–10. [DOI] [PubMed] [Google Scholar]
- Novielli C., Mando C., Tabano S., Anelli G. M., Fontana L., Antonazzo P., Miozzo M., Cetin I. (2017). Mitochondrial DNA content and methylation in fetal cord blood of pregnancies with placental insufficiency. Placenta 55, 63–70. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Suomalainen A. (2012). Mitochondria: In sickness and in health. Cell 148, 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S., Gogvadze V., Zhivotovsky B. (2007). Mitochondrial oxidative stress: Implications for cell death. Annu. Rev. Pharmacol. Toxicol. 47, 143–183. [DOI] [PubMed] [Google Scholar]
- Owa C., Poulin M., Yan L., Shioda T. (2018). Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: Sources and avoidance of false-positive detection. PLoS One 13, e0192722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Davis R. L., Sue C. M. (2018). Mitochondrial dysfunction in Parkinson's disease: New mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. Rep. 18, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V., Cuenin C., Chung F., Aguilera J. R. R., Fernandez-Jimenez N., Romero-Garmendia I., Bilbao J. R., Cahais V., Rothwell J., Herceg Z. (2019). Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 47, 10072–10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola C. J., Gianotti T. F., Burgueno A. L., Rey-Funes M., Loidl C. F., Mallardi P., Martino J. S., Castano G. O., Sookoian S. (2013). Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 62, 1356–1363. [DOI] [PubMed] [Google Scholar]
- Posnack N. G., Swift L. M., Kay M. W., Lee N. H., Sarvazyan N. (2012). Phthalate exposure changes the metabolic profile of cardiac muscle cells. Environ. Health Perspect. 120, 1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y., Pearl S., Gingrich J., Jing J., Martin D., Murga-Zamalloa C. A., Veiga-Lopez A. (2019). Multispecies study: Low-dose tributyltin impairs ovarian theca cell cholesterol homeostasis through the RXR pathway in five mammalian species including humans. Arch. Toxicol. 93, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A., Anugrha H., Kurzawa-Akanbi M., Yarnall A., Burn D., Hudson G. (2016). Reduced mitochondrial DNA copy number is a biomarker of Parkinson's disease. Neurobiol. Aging 38, 216.e7–216.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman J., Rahman S. (2018). Mitochondrial medicine in the omics era. Lancet 391, 2560–2574. [DOI] [PubMed] [Google Scholar]
- Rebelo A. P., Williams S. L., Moraes C. T. (2009). In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 37, 6701–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritov V. B., Menshikova E. V., He J., Ferrell R. E., Goodpaster B. H., Kelley D. E. (2005). Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54, 8–14. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pereira P., Macedo S., Gaspar T. B., Canberk S., Selmi-Ruby S., Maximo V., Soares P., Miranda-Alves L. (2020). Relevant dose of the environmental contaminant, tributyltin, promotes histomorphological changes in the thyroid gland of male rats. Mol. Cell. Endocrinol. 502, 110677. [DOI] [PubMed] [Google Scholar]
- Saini S. K., Mangalhara K. C., Prakasam G., Bamezai R. N. K. (2017). DNA methyltransferase1 (DNMT1) isoform3 methylates mitochondrial genome and modulates its biology. Sci. Rep. 7, 1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammi S. R., Foguth R. M., Nieves C. S., De Perre C., Wipf P., McMurray C. T., Lee L. S., Cannon J. R. (2019). Perfluorooctane sulfonate (PFOS) produces dopaminergic neuropathology in Caenorhabditis elegans. Toxicol. Sci. 172, 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwung P., Petersen K. F., Shulman G. I., Knowles J. W. (2020). Mitochondrial dysfunction, insulin resistance, and potential genetic implications. Endocrinology 161, bqaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Kuroiwa T. (1991). Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 196, 137–140. [DOI] [PubMed] [Google Scholar]
- Scaglia F., Wong L. J. (2008). Human mitochondrial transfer RNAs: Role of pathogenic mutation in disease. Muscle Nerve 37, 150–171. [DOI] [PubMed] [Google Scholar]
- Sharma N., Pasala M. S., Prakash A. (2019). Mitochondrial DNA: Epigenetics and environment. Environ. Mol. Mutagen. 60, 668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy D. T., McAllister K., Worth L., Haugen A. C., Meyer J. N., Domann F. E., Van Houten B., Mostoslavsky R., Bultman S. J., Baccarelli A. A., et al. (2014). Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ. Health Perspect. 122, 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpfel M., Jancar N., Virant-Klun I. (2018). New challenge: Mitochondrial epigenetics? Stem Cell Rev. Rep. 14, 13–26. [DOI] [PubMed] [Google Scholar]
- St John J. C. (2016). Mitochondrial DNA copy number and replication in reprogramming and differentiation. Semin. Cell Dev. Biol. 52, 93–101. [DOI] [PubMed] [Google Scholar]
- Stoccoro A., Siciliano G., Migliore L., Coppede F. (2017). Decreased methylation of the mitochondrial D-loop region in late-onset Alzheimer's disease. J. Alzheimers Dis. 59, 559–564. [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Sun X., Lee W., Vaghjiani V., St John J. C. (2016). Analysis of mitochondrial DNA copy number and its regulation through DNA methylation of POLGA. Methods Mol. Biol. 1351, 131–141. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Nagao A., Suzuki T. (2011). Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329. [DOI] [PubMed] [Google Scholar]
- Taylor E. M., Jones A. D., Henagan T. M. (2014). A review of mitochondrial-derived fatty acids in epigenetic regulation of obesity and type 2 diabetes. J. Nutr. Health Food Sci. 2, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Zhang L., Gao J., Wen S., Zhou H., Feng S. (2017). Methylation of mitochondrial DNA displacement loop region regulates mitochondrial copy number in colorectal cancer. Mol. Med. Rep. 16, 5347–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Pomatto L. C. D., Davies K. J. A. (2020). Sex differences in the response to oxidative and proteolytic stress. Redox Biol. 31, 101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I. (2012). The evolving role of mitochondria in metabolism. Trends Endocrinol. Metab. 23, 417–419. [DOI] [PubMed] [Google Scholar]
- Van Cauwenbergh O., Di Serafino A., Tytgat J., Soubry A. (2020). Transgenerational epigenetic effects from male exposure to endocrine-disrupting compounds: A systematic review on research in mammals. Clin. Epigenet. 12, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg L. N., Colborn T., Hayes T. B., Heindel J. J., Jacobs D. R. Jr, Lee D. H., Shioda T., Soto A. M., vom Saal F. S., Welshons W. V., et al. (2012). Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 33, 378–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst M. G., Rots M. G. (2015). Mitochondrial epigenetics: An overlooked layer of regulation? Trends Genet. 31, 353–356. [DOI] [PubMed] [Google Scholar]
- van der Wijst M. G., van Tilburg A. Y., Ruiters M. H., Rots M. G. (2017). Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 7, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos S., Nawrot T. S., Martens D. S., Byun H. M., Janssen B. G. (2020). Mitochondrial DNA methylation in placental tissue: A proof of concept study by means of prenatal environmental stressors. Epigenetics 1–11. doi: 10.1080/15592294.2020.1790923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. M., Gore A. C. (2017). Epigenetic impacts of endocrine disruptors in the brain. Front. Neuroendocrinol. 44, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh L. P., McCormick C., Martin C., Stocco D. M. (2000). Roundup inhibits steroidogenesis by disrupting steroidogenic acute regulatory (StAR) protein expression. Environ. Health Perspect. 108, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C., Lee W. C., Liao S. C., Lee L. C., Su Y. J., Lee C. T., Chen J. B. (2011). Mitochondrial DNA copy number correlates with oxidative stress and predicts mortality in nondiabetic hemodialysis patients. J. Nephrol. 24, 351–358. [DOI] [PubMed] [Google Scholar]
- Weinhouse C. (2017). Mitochondrial-epigenetic crosstalk in environmental toxicology. Toxicology 391, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth C., Brandt U., Hunte C., Zickermann V. (2016). Structure and function of mitochondrial Complex I. Biochim. Biophys. Acta 1857, 902–914. [DOI] [PubMed] [Google Scholar]
- Wolf D. P., Mitalipov N., Mitalipov S. (2015). Mitochondrial replacement therapy in reproductive medicine. Trends Mol. Med. 21, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F., Susiarjo M., Bartolomei M. S. (2015). Multigenerational and transgenerational effects of endocrine disrupting chemicals: A role for altered epigenetic regulation? Semin. Cell Dev. Biol. 43, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes F. M., Van Houten B. (1997). Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 94, 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Karakhanova S., Hartwig W., D'Haese J. G., Philippov P. P., Werner J., Bazhin A. V. (2016). Mitochondria and mitochondrial ROS in cancer: Novel targets for anticancer therapy. J. Cell. Physiol. 231, 2570–2581. [DOI] [PubMed] [Google Scholar]
- Yue P., Jing S., Liu L., Ma F., Zhang Y., Wang C., Duan H., Zhou K., Hua Y., Wu G., et al. (2018). Association between mitochondrial DNA copy number and cardiovascular disease: Current evidence based on a systematic review and meta-analysis. PLoS One 13, e0206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guallar E., Ashar F. N., Longchamps R. J., Castellani C. A., Lane J., Grove M. L., Coresh J., Sotoodehnia N., Ilkhanoff L., et al. (2017). Association between mitochondrial DNA copy number and sudden cardiac death: Findings from the Atherosclerosis Risk in Communities study (ARIC). Eur. Heart J. 38, 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. D., Brooke J., Smith C., Almeida F. A., Cheng Z. (2016) Mitochondrial epigenetic changes and progression from metabolically healthy obesity to metabolically unhealthy obesity: A cross-sectional study. Lancet Diabetes Endocrinol 4, S16. [Google Scholar]
- Zheng L. D., Linarelli L. E., Liu L., Wall S. S., Greenawald M. H., Seidel R. W., Estabrooks P. A., Almeida F. A., Cheng Z. (2015). Insulin resistance is associated with epigenetic and genetic regulation of mitochondrial DNA in obese humans. Clin. Epigenet. 7, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]