Abstract
The circular (circ)RNAs are a newly recognized group of noncoding (nc)RNAs. Research to characterize the functional features of circRNAs has uncovered distinctive profiles of conservation, stability, specificity and complexity. However, a new line of evidence has indicated that although circRNAs can function as ncRNAs, such as in the role of miRNA sponges, they are also capable of coding proteins. This discovery is no accident. In the last century, scientist detected the ability of translate in some virus and artificial circRNAs. Artificial circRNA translation products are usually nonfunctional, whereas natural circRNA translation products are completely different. Those new proteins have various functions, which greatly broadens the new ideas and research direction for our research. These series findings also raise questions about whether circRNA is still classified as non-coding RNA. Here, we summarize the evidence concerning translation potential of circRNAs, including synthetic and endogenous circRNA translation ability, and discuss the mechanisms of circRNA translation.
Keywords: ncRNAs, circRNAs, Translation, mRNA, miRNA
Introduction
The classic “central dogma of molecular biology” suggests that the DNA constituent of our chromosomes is transcribed into RNA and subsequently translated into proteins. High-throughput sequencing technology has not only verified the dynamic complexity of gene expression but also revealed the existence of delicate regulatory processes at the RNA level (Pan et al., 2008). The RNA form of genetic information serves as the intermediary between DNA and its protein products (Crick, 1970); as such, it is believed that levels of RNA are at the core of life’s complex functions (Licatalosi & Darnell, 2010). At the turn of the century, whole-genome sequencing indicated that while approximately 93% of the DNA in the human genome is transcribed into RNA, only approximately 2% of the DNA sequences encode proteins (Consortium, 2012). This finding suggested that there are large amounts of noncoding (nc)RNAs in mammalian cells.
Although the newly discovered ncRNAs were at first largely dismissed as “transcriptional noise”, focused investigations began to reveal functional roles in cell biology and many disease types. Researchers’ attention has now turned towards defining the roles of ncRNAs in regulating and modulating host gene expression (Meller, Joshi & Deshpande, 2015; Peschansky & Wahlestedt, 2014). The current collective data have allowed the two major groups of ncRNAs—the long (l)ncRNAs and small RNAs, grossly stratified according to size—to be further categorized according to function; these functional subcategories include ribosomal (r)RNA, transfer (t)RNA, small nuclear (sn)RNA, small nucleolar (sno)RNA, PIWI-interacting (pi)RNA, micro (mi)RNA, lncRNA, circular (circ)RNA and transcription initiation (ti)RNA (Cech & Steitz, 2014; Wright & Bruford, 2011). Among these, the miRNAs and lncRNAs have been extensively studied and confirmed to function in gene transcription through pivotal activities in a versatile regulation network (Guil & Esteller, 2015; Mondal & Kanduri, 2013).
In 2012, Salzman et al. (2012) found the massive presence of circRNAs in eukaryotic cells. Thereafter, the circRNAs have been shown particular stability and functional versatility in vivo. For instances, Hansen et al. (2013) firstly reported the natural circRNAs’ function as efficient miRNA sponges in both physiological and pathological processes. CircRNAs are products of hnRNA backsplicing and the resulting RNAs represent covalently closed circles, which are devoid of terminal RNA cap structures and poly(A) tails. However, the rapid development of genome-wide translation profiling and ribosome profiling has revealed that a small number of small open reading frames (sORFs) within circRNAs actually have peptide- or protein-coding potential. Therefore, circRNAs have now been demonstrated as capable of translating directly into protein, indicating an intriguing potential to directly function in many processes of life. In this review, we will discuss the most recent progress of the research into the translational capacity of circRNAs and towards defining the underlying mechanisms.
CircRNA biology
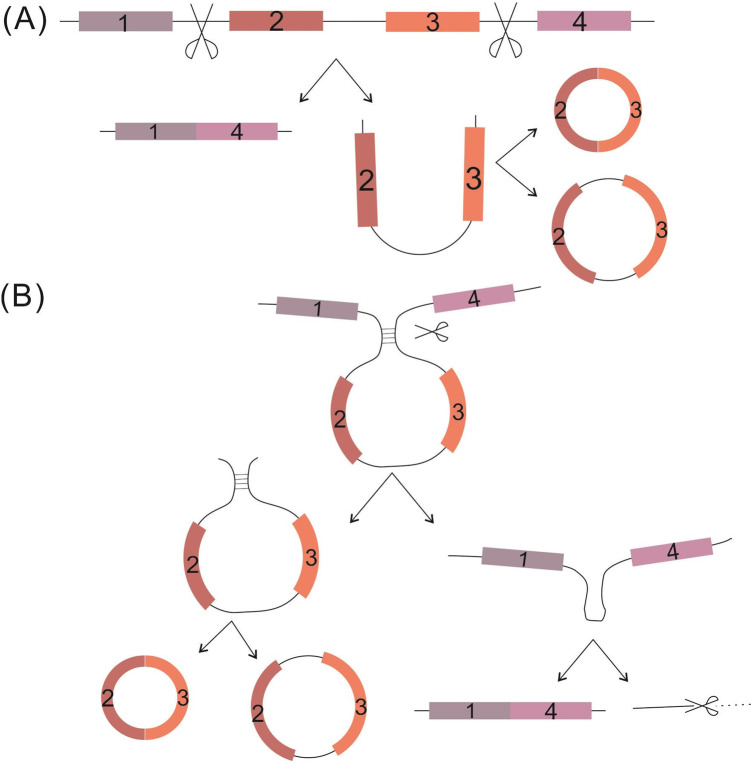
CircRNAs are single-stranded covalently closed circular RNA molecules generated from a broad array of genomic regions, ranging from intergenic, intronic and coding sequences to 5′- or 3′-untranslational sequences (Chen & Yang, 2015; Memczak et al., 2013). Two models of circRNA biosynthesis have been proposed, both involving back-splicing catalyzed by the spliceosomal machinery. The first of the two, the “exon skipping” model, begins with classical splicing to generate linear RNA. The downstream exon links to the upstream exon, with one or more exons being skipped; the skipped exons then further back-splice to form precursor circRNAs, which undergo further processing to become mature circRNAs. (B) The second of the two models, the “direct back-splicing” circularization model, is related mostly to complementary motifs; in this, the complementary pairing RNA back-splices to produce a precursor circRNA together with an exon-intron(s)-exon intermediate, and the latter is further processed to produce a linear RNA with skipped exons or which is targeted for degradation (Ashwal-Fluss et al., 2014; Jeck & Sharpless, 2014; Lasda & Parker, 2014) (Fig. 1).
Figure 1. Proposed circRNA formation models.
(A) The “exon skipping” model. (B) The “direct back-splicing” model. Black thin lines represent intron sequence; colored thick lines represent different exon sequences.
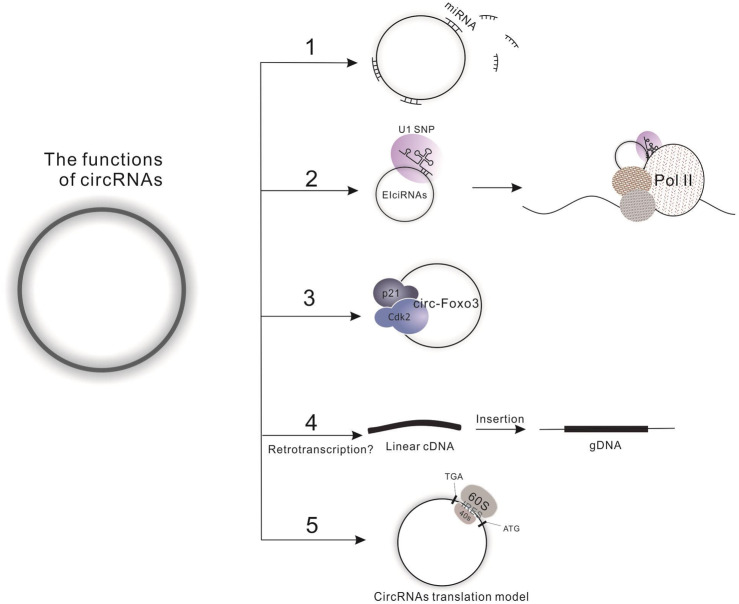
To date, four functions have been defined for the circRNAs. First, circRNAs harbor miRNA complementary sequences, facilitating their combination with and ability to adjust the biological function of a large number of miRNAs by functioning as molecular sponges. A specific example of this is the circMTO1, which acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression (Han et al., 2017). Furthermore, one circRNA may combine with several kinds of miRNAs; for instance, circHIPK3 has been reported to combine with 9 miRNAs (miR-29a, miR-29b, miR-124, miR-152, miR-193a, miR-338, miR-379, miR-584 and miR-654) to synergistically inhibit cell proliferation (Zheng et al., 2016). Second, circRNAs can directly regulate transcription, splicing and expression of a parental gene. The exon-intron circRNAs (EIciRNAs) are examples of this regulation, interacting with RNA polymerase II and enhancing transcription of their parental genes (Li et al., 2015). Third, circRNAs directly interact with proteins, such as the ternary complex circ-Foxo3-p21-CDK2, which serves to arrest the function of CDK2 and interrupt cell cycle progression (Du et al., 2016). However, studies indicate that one circRNA might simultaneously harbor more than one of the above functions, which is evidenced by the finding that circ-Amotl1 can act both as a sponge for miR-17 to promote cell proliferation, migration and wound healing and as a target for protein binding (c-Myc, Akt1 and PDK1) to promote the proliferation of tumor cells and enhancing cardiac repair (Yang et al., 2017a; Yang et al., 2017c; Zeng et al., 2017). Fourth, Dong et al. developed a computational pipeline (CIRCpseudo), and indicated that stabilized circRNAs could form circRNA pseudogenes by retrotranscribing and integrating into the genome (Dong et al., 2016). However, there is only one paper on circRNA’s formation of pseudogenes, which does not explain the specific mechanism of it. We need more evidence to prove this idea. More interestingly, the latest research is hinting at a potential fourth function of circRNAs: translation (Fig. 2), which opens a new field for researchers to explore the biological functions of circRNA-derived proteins. For detailed information on the biology of circRNA, please see the review written by Li X et al. (Barrett & Salzman, 2016; Chen, Chen & Chuang, 2015; Li, Yang & Chen, 2018).
Figure 2. Functions of circRNAs.
(1) Molecular sponge for miRNA; (2) Regulation of transcription, splicing and expression of parental gene by binding to Pol II; (3) Interaction with proteins; (4) Direct translation of circRNAs.
CircRNA translation potential: a controversial issue explored unceasingly
It is commonly believed that mRNAs are the primary controller of cells, carrying out the necessary functions for life. Since the endogenous circRNAs appear to not be associated with polysomes, they presumably lack the potential for translation (Guo et al., 2014; Jeck et al., 2013). Although this notion has not been definitively disproven, it still attracts scientists’ interests in exploring the unknown, hoping to advance the field of research into circRNA translational potency forward from theory to practical knowledge.
Theoretical basis for direct translation of endogenous circRNAs
Molecular structure
Internal ribosome entry site (IRES).
It is well known that there are two translation modes, cap-dependent translation and cap-independent translation. The traditional cap-dependent translation accounts for a basal level of protein synthesis under normal growth conditions. In contrast, cap-independent translation contributes to cell proliferation or cellular adaptation/survival when traditional protein synthesis is severely inhibited; this second mode is mediated by the IRES. Therefore, IRES-mediated translation serves as an urgent breakdown maintenance mechanism during cell stress, ensuring basic protein needs are met (Lang, Kappel & Goodall, 2002; Riley, Jordan & Holcik, 2010); as such, this mechanism is often triggered in conditions of viral invasion, tumor or other human diseases (Faye & Holcik, 2015; Holcik & Sonenberg, 2005; Sonenberg & Hinnebusch, 2009). Thus, it is not surprising that the IRES itself was originally identified by researchers studying the virus parasitic mechanism (Baird et al., 2006); since then, comparative sequencing analysis has led to the identification of IRES components throughout the human genome (Weingarten-Gabbay et al., 2016). Functional studies have characterized the IRES in mRNA as dependent upon the molecule’s special structure, allowing the 40S subunit to avoid assembling directly at the 5′-untranslated sequence (Sonenberg & Hinnebusch, 2009).
In 2016, Chen et al. established a circRNA database, circRNADb (http://reprod.njmu.edu.cn/circrnadb) (Chen et al., 2016), the first of its kind, summarizing circRNA-encoded protein information based upon 32,914 human exonic circRNAs. Interestingly, their initial explorations of this dataset found ORFs in about half of the circRNAs and IRESs in about half of those; as such, those 7,170 circRNA sequences were considered to fit the characteristic requirements for protein translation capabilities. To date, four types of virus IRES structures are classified with the functional ability to hijack eukaryotic translation machinery, and all work with a common mechanical principle, leading to 80S ribosomal assembly and extension (Yamamoto, Unbehaun & Spahn, 2017). However, in eukaryotic mRNAs and circRNAs, the IRES-mediated ribosome assembly mechanism is less well known; only isolated examples of IRESs with known ITAF binding sites or resolved three-dimensional structure are available35,36.
RNA modification.
Statistical analyses have estimated that RNA molecules may contain more than 100 distinct modifications (Gilbert, Bell & Schaening, 2016). Approximately 16 species of modifications in mRNA have been recognized to date, and the vast majority of these involve the m6A, ψ and m5C chemical modifications (Cantara et al., 2011). The m6A modification is related to mRNA stability, splicing processing, polypeptide translation and miRNA processing and is correlated with stem cell fate and biological rhythms (Hoernes, Huttenhofer & Erlacher, 2016; Roundtree et al., 2017; Squires et al., 2012). The pseudouridylation modification (ψ) serves three main functions, namely, changing the codon, enhancing the transcript stability and regulating the stress response. To date, only the m6A modification has been verified in circRNAs, wherein it plays a role in promoting translation (Yang et al., 2017b). However, research on the m5C modification on ncRNAs has been very limited, though the ncRNA and mRNA have been found to hold thousands of m5C modification sites in recent years (Hoernes, Huttenhofer & Erlacher, 2016; Roundtree et al., 2017; Squires et al., 2012). Therefore, it is speculated that more modification types will be found in both circRNAs and mRNAs with continued research. Such modifications will likely function not only in terms of translation but also in adjusting the functions of circRNAs as ncRNAs. For detailed introduction about circRNAs translation by non-canonical initiation mechanisms, please see the reviews written by Diallo et al. (2019) and Zhang et al. (2020).
Analogous to similar ncRNAs
Recent studies demonstrate that many lncRNAs are able to translate into functional polypeptides. In 2013, Magny et al. (2013) found a putative noncoding RNA 003 in 2L (pncr003:2L), including two potentially functional smORFs in the fly’s heart, which could translate into bioactive peptides and synergistically regulate cardiac calcium uptake. In 2015, Anderson et al. (2015) discovered an annotated lncRNA that translates for a conserved micropeptide- myoregulin (MLN) that functions as a regulator of skeletal muscle physiology. One year afterward, Nelson et al. (2016) found that a peptide (named dwarf open reading frame (DWORF)) is encoded by a putative lncRNA. This peptide is mutually exclusive with the other three inhibitors (phospholamban, sarcolipin, and myoregulin) to competitively combine with the SEARC pump to adjust the reuptake of the Ca2+ in muscle. Then, Matsumoto et al. (2017) identified a functional novel polypeptide encoded by a lncRNA. This peptide can negatively regulate mTORC1 activation by interacting with the lysosomal v-ATPase in late endosome/lysosome. With deep research, increasingly more lncRNAs with the capacity of translating proteins (peptides) will be explored. As a special type of lncRNAs, we have reason to speculate that the biological significance of coding ability of circRNAs is still to be uncovered.
Experimental exploration for endogenous circRNA translation in eukaryotic cells
Early exploration findings
The first indications of a translational role for circRNAs emerged from studies of virus nucleic acids. One of the first observations of a circRNA behaving as a translational template was made with the single-stranded circular RNA genome of the hepatitis δ virus, a satellite virus of the hepatitis B virus; encapsulation of the former by hepatitis B virions was found to result in the production of a single viral protein of 122 amino acids, in a noncanonical manner (Kos et al., 1986). In 1995, Chen & Sarnow (1995) demonstrated that synthetic circRNAs containing IRES elements were able to correctly translate into polypeptides in rabbit reticulocyte lysate, but those without IRES could not. Furthermore, they speculated that this type of RNA can translate along the RNA circles for multiple consecutive rounds. In 1998, Perriman et al. used plasmids for creating RNA cyclase ribozymes to produce desired circular RNAs that were inserted into the green fluorescent protein (GFP) ORF (finite GFP encoding) and stop codon-devoid GFP reading frame (infinite GFP encoding) (Perriman & Ares Jr, 1998). The authors showed that both circRNAs can directly translate along with GFP in Escherichia coli strains and in the meantime, the infinite GFP-encoding RNA could be translated into an extremely long repeating poly-GFP. These findings validated Chen’s previous prediction in 1995 (Chen & Sarnow, 1995). In 1999, Li & Lytton (1999) observed that a circRNA containing NCX1 exon 2 might translate for a protein. It is a pity that they could not detect a protein corresponding exactly to what they predicted from the circular transcript; however, when the circRNAs were made into linear RNAs and transfected into HEK-293 cells, the linear versions of circRNAs were shown to result in the proteins of the expected size of ∼70 kDa, and the transfected cells possessed Na/Ca exchange activity.
Over a decade later, Wang & Wang (2015) reported on their construction of an efficient back-splicing circRNA, which could be translated into functional GFP proteins in human and Drosophila cell lines. Furthermore, due to the nuclease resistance characteristics of circRNAs, when the cell was transfected with circRNA, protein production was prolonged for several days. In the same year, Abe et al. (2015) provided evidence that circRNAs were translated into infinite FLAG proteins in rabbit reticulocyte lysate and HeLa cells with an infinite ORF in the absence of any particular translation initiation element such as a poly-A tail, internal ribosome entry, or a cap structure. This series of experiments proves that artificial circRNAs with stop codon mutations have a rolling circle amplification (RCA) mechanism to code for long repeating poly proteins. In 2014, AbouHaidar et al. (2014) reported a small new virusoid with covalently closed circular (CCC) RNA (220 nt) associated with rice yellow mottle virus that could translate into a 16-kDa highly basic protein. This example is the only one that codes proteins among all known viroids and virusoids. This unique natural supercompact “nano genome” even overlaps its initiation and termination codons to UGAUGA (AbouHaidar et al., 2014).
Nevertheless, all these scattered reports, however, are limited to viruses, bacteria, or synthetic circRNA (Granados-Riveron & Aquino-Jarquin, 2016) (Table 1), and the translation ability of endogenous circRNAs still requires further exploration.
Table 1. The published circRNAs with translation potential.
| CircRNA source | Research model | Translation product | Functions | Reference |
|---|---|---|---|---|
| CircRNAs in viruses and bacteria | Hepatitis δ virus | Protein of 122 amino acids | The hepatitis delta antigen (HDAg) | Kos et al. (1986) |
| A virusoid associated with rice yellow mottle virus | 16-kDa highly basic protein | Unknown | AbouHaidar et al. (2014) | |
| Escherichia coli: 795-nt circular mRNA | GFP | Fluorescent | Perriman & Ares Jr (1998) | |
| HPV16-derived-circE7 | E7 protein | Influencing the development of cancer. | Zhao et al. (2019) | |
| Artificial circRNAs or synthetic modified RNA | HEK-293 cells | GFP | Fluorescent | Wang & Wang (2015) |
| Rabbit reticulocyte lysate and HeLa cells | FLAG protein (EGF, IGF-1, IGF-2) | Human growth factors | Abe et al. (2015) | |
| Rabbit reticulocyte lysate | 23-kDa product | Unknown | Chen & Sarnow (1995) | |
| HEK293 cells | GFP, Firefly luciferase, human erythropoietin | Convenient for the author to test | Wesselhoeft, Kowalski & Anderson (2018) | |
| Endogenous circRNAs | Drosophila: circMbl3 | 37.04-kDa protein | Unknown | Pamudurti et al. (2017) |
| Human: circ-ZNF609 | circ-ZNF609-encoded protein | Unknown | Legnini et al.(2017) | |
| Human: circ-FBXW7 | FBXW7-185aa | Cooperates with FBXW7 to control c-Myc stability | Yang et al. (2018) | |
| Human: circ-SHPRH | SHPRH-146aa | Guarding against full-length SHPRH protein degradation | Zhang et al. (2018a) | |
| Human: CircPINTexon2 | PINT87aa | Inhibiting oncogenes transcriptional elongation | Zhang et al. (2018b) | |
| Human: circPPP1R12A | circPPP1R12A-73aa | Promoting the colon cancer pathogenesis and metastasis | Zheng et al. (2019) | |
| Human: circβ-catenin | β-catenin-370aa | Stabilizing full-length β-catenin | Liang et al. (2019) | |
| Drosophila: circSfl | circlSfl protein | Extending the lifespan of fruit flies | Weigelt et al. (2020) |
Solid evidence for endogenous circRNA direct translation
In 2013, Jeck et al. (2013) reported that circRNAs are abundant, conserved and associated with ALU repeats, but there are no detectable levels of exonic circRNAs in the ribosome-bound fraction (via ribosome profiling). One year later, Dudekula et al. (2016) raised doubts about this conclusion when they reported their findings from a bioinformatic analysis; IRES regions in circRNAs represented predicted binding sites for RNA binding proteins, including some known to modulate IRES-driven translation.
In 2017, it was finally proved that endogenous circRNAs are capable of directly translating into proteins. By using ribosome footprinting and immunoprecipitation of Drosophila brain tissues, Pamudurti et al. demonstrated that circRNA sequences could be bound by ribosomes including the termination codon (Pamudurti et al., 2017). They focused on circ-Mbl from the Mbl gene among all of the ribo-circRNAs and repeatedly verified that circ-Mbl could translate into protein. Through the construction of an overexpression vector, the substitution of the ORF with a split Cherry molecule and target mass spectrometry from the Drosophila brain circ-Mbl was immunoprecipitated. In the same year, through a screening study of circRNAs related to human, mouse (C2,C12) and a Duchenne muscular dystrophy disease model, Legnini et al. reported that the circ-ZNF609 combined with ribosomes and that its encoded protein was suggested to be involved in the myoblast growth process; however, the circ-ZNF609 was found to be translated at almost two orders of magnitude lower efficiency than that of the linear form (Legnini et al., 2017).
Thereafter, Yang et al. (2017b) explored circRNA translation ability by the same approach and found that control sequences without IRES were also capable of translating the target protein. These unexpected circRNA translation events were initiated by eIF4G2 and eIF3A and associated with the m6A modification. When the m6A modifications were “erased”, the target protein translation activity was substantially affected, to the point that it completely disappeared. Ribosome spectrum analysis confirmed that a multitude of endogenous circRNAs were bound by ribosomes, but whether these circRNAs harbored any IRESs was not examined. Finally, high-throughput sequencing analysis determined that approximately 13% of the total circRNAs carried the m6A modification. Months later, another independent study showed that circRNAs carry extensive m6A modifications and are expressed in cell type-specific patterns (Zhou et al., 2017). The writing and reading machinery of these m6A modifications were found to be similar to those of mRNAs (i.e., involving the METTL3/14 and YTH proteins) but were distinctive in their location patterns; the data also suggested that the m6A modification did not appear to promote degradation of circRNAs as it does for mRNAs. Ultimately, interpretation of these findings indicates that switching the state of m6A modifications may allow for functional control of circRNAs.
In 2018, Yang et al. (2018) reported that the circ-FBXW7 can translate for a new protein FBXW7-185aa during glioma tumorigenesis. Intriguingly, this protein cooperates with FBXW7, which is encoded in their parental genes, to control c-Myc stability and repress cell cycle acceleration and the consequent proliferation. This is the first study to provide definitive evidence of protein translation via circRNA synergy with the protein expression by parental genes and joint function of the proteins. Zhang et al. (2018a) further reported that circ-SHPRH, a circRNA containing an IRES-driven ORF, translates into a functional protein. For this process, circ-SHPRH utilizes overlapping genetic codes to create a UGA stop codon, causing translation of the SHPRH-146aa protein. The translated SHPRH-146aa functions as a protector of the full-length SHPRH protein, guarding against degradation by the ubiquitin proteasome and consequently inhibiting cell proliferation and tumorigenicity in human glioblastoma. In the same year, Zhang et al. (2018b) found that the 1084 nt CircPINTexon2 which generated by the circularization of exon 2 of LINC-PINT encodes an 87-aa peptide. This peptide (PINT87aa) suppresses glioblastoma cell proliferation in vitro and in vivo by directly interacting with polymerase associated factor complex (PAF1c) and inhibiting the transcriptional elongation of multiple oncogenes.
In 2019, Zheng et al. (2019) reported an upregulated circRNA (circPPP1R12A) in colon cancer tissues that could translate a 73-aa protein (circPPP1R12A-73aa). The circPPP1R12A-73aa promotes the proliferation, migration and invasion abilities of colon cancer via activating hippo-YAP signaling pathway. In addition, Liang et al. (2019) reported circ β-catenin originated from β-catenin gene locus could promote tumorigenesis. Knockdown of circ β-catenin repressed liver cancer cell growth and migration in vitro and in vivo by inhibiting Wnt/ β-catenin pathway. In terms of mechanism, circ β-catenin encoded a novel protein (β-catenin-370aa) which shared homologous N-terminus sequence with wild type β-catenin, but it contained a new C-terminus with 9 specific amino acids. The β-catenin-370aa might function as a decoy for GSK3 β, leading to escape from GSK3 β-induced β-catenin degradation (Liang et al., 2019).
Interestingly, Zhao et al. (2019) in the same year identified a virus-derived circRNA, HPV16-circE7, which could translate E7 oncoprotein. By constructing various mutant vectors (such as circE7_noATG) for comparison, the authors found that circE7 can also provide the template for E7 oncoprotein translation. Further studies have found that the initiation of circE7 translation may be related to m6A modification and capable of generating the E7 oncoprotein in a heat-shock regulated manner. Moreover, HPV16 circE7 is essential for the transformed growth of CaSki cervical carcinoma cells and could be regulated by keratinocyte differentiation (Zhao et al., 2019).
In 2020, Weigelt et al. (2020) showed that circSfl was highly upregulated in all tissues by next-generation sequencing of wild-type and mutant flies. However, circSfl is lack of enrichment of miRNA binding sites in loop, which makes it unlikely acts as a miRNA sponge. Further study verified that circSfl is translated into a small protein that shares the N terminus with full-length Sfl protein. Furthermore, the protein encoded by circSfl and the protein encoded from the linear Sfl transcripts can positively extend the lifespan of female flies (Weigelt et al., 2020), indicating the unique role of circSfl in fly life.
Detailed information for the published coding circRNAs is summarized in Table 1.
Challenges and Perspectives
The field of RNA research has continually emphasized the structural and functional versatility of RNA molecules. This versatility has in turn inspired translational and clinical researchers to explore the utility of RNA-based therapeutic agents for a wide variety of medical applications. Several RNA therapeutics with diverse modes of action are currently being evaluated in large late-stage clinical trials, and many more are in the early clinical development stage, including strategies to modulate target gene expression, such as mRNA, siRNA and miRNA (Sullenger & Nair, 2016). For instance, mRNA-modified dendritic cells have shown promising and efficient results in clinical trials (Benteyn et al., 2015), and siRNA-based therapeutic agents such as bevacizumab (Avastin; off-label use) have shown success for the treatment of wet, age-related macular degeneration in clinical testing (Garba & Mousa, 2010). The circRNAs may regulate gene expression through different mechanisms, including direct translation (Lyu & Huang, 2017). Therefore, considering their stability and specific expression features, circRNAs with translation potential could represent strong candidates for development as clinical tools to therapeutically manipulate a wide variety of physiologic and pathologic processes. So far, Wesselhoeft et al. have pioneered the transformation of circRNA into robust and stable protein expression in eukaryotic cells. They also considered that circRNA is a promising alternative to linear mRNA (Wesselhoeft, Kowalski & Anderson, 2018). But using circRNA as a clinical tool to treat disease remains a challenge, requiring extensive and in-depth research.
In terms of the discovery and exploration of endogenous circRNA translation proteins, we speculate that there will be a large number of circRNAs with translational function gradually discovered. But these need to be supplemented by a large number of experiments, especially the function of those unknown proteins translated by circRNAs. For example, two cases of circRNA translation of small proteins have been found to act as molecular inhibitors or agonist of their mother protein (Yang et al., 2018; Zhang et al., 2018a). Therefore, studying the function of these small proteins may improve the mechanism of action of some molecules and even serve as a new target for clinical drugs.
The collective evidence to date implies that the translation of endogenous circular RNA into proteins or peptides may be a widespread phenomenon, though the coding potential of circRNAs previously had been largely disregarded. Therefore, further studies on the translational capacity of circRNAs should be encouraged and should focus on the existing problems, such as the functions and detailed mechanisms of circRNA modifications, the 5′ cap-independent translation of circRNAs and circRNA-derived protein or peptides. The resulting insights will also be helpful towards furthering our understanding of ncRNA functions in general.
Funding Statement
This work was supported by grants from the National Key Research and Development Project (No. 2016YFA0502203), the National Foundation of China (Nos. 81502728 and 81670534), and the Anhui Provincial Natural Science Foundation (No. 1408085MH149). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Bing Ni, Email: nibing@tmmu.edu.cn, nibingxi@126.com.
Jun Tang, Email: jiangzhuyan@stu.ahmu.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Qingqing Miao performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Bing Ni and Jun Tang conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
There is no original, experimental raw data in this literature review.
References
- Abe et al. (2015).Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling circle translation of circular RNA in living human cells. Scientific Reports. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbouHaidar et al. (2014).AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson et al. (2015).Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss et al. (2014).Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Baird et al. (2006).Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett & Salzman (2016).Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benteyn et al. (2015).Benteyn D, Heirman C, Bonehill A, Thielemans K, Breckpot K. mRNA-based dendritic cell vaccines. Expert Review of Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- Cantara et al. (2011).Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Research. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech & Steitz (2014).Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chen & Sarnow (1995).Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- Chen, Chen & Chuang (2015).Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdisciplinary Reviews-RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2016).Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Scientific Reports. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen & Yang (2015).Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biology. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium (2012).Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick (1970).Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- Diallo et al. (2019).Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC, Garmy-Susini B, Lacazette E. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. 2019;164:45–52. doi: 10.1016/j.biochi.2019.06.015. [DOI] [PubMed] [Google Scholar]
- Dong et al. (2016).Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L. CircRNA-derived pseudogenes. Cell Research. 2016;26:747–750. doi: 10.1038/cr.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du et al. (2016).Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudekula et al. (2016).Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye & Holcik (2015).Faye MD, Holcik M. The role of IRES trans-acting factors in carcinogenesis. Biochimica et Biophysica Acta/General Subjects. 2015;1849:887–897. doi: 10.1016/j.bbagrm.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Garba & Mousa (2010).Garba AO, Mousa SA. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmology and Eye Diseases. 2010;2:75–83. doi: 10.4137/OED.S4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, Bell & Schaening (2016).Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron & Aquino-Jarquin (2016).Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochimica et Biophysica Acta/General Subjects. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Guil & Esteller (2015).Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends in Biochemical Sciences. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2014).Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biology. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han et al. (2017).Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- Hansen et al. (2013).Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hoernes, Huttenhofer & Erlacher (2016).Hoernes TP, Huttenhofer A, Erlacher MD. mRNA modifications: dynamic regulators of gene expression? RNA Biology. 2016;13:760–765. doi: 10.1080/15476286.2016.1203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik & Sonenberg (2005).Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nature Reviews Molecular Cell Biology. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Jeck & Sharpless (2014).Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature Biotechnology. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck et al. (2013).Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos et al. (1986).Kos A, Dijkema R, Arnberg AC, Meide PHvander, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Lang, Kappel & Goodall (2002).Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Molecular Biology of the Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda & Parker (2014).Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini et al. (2017).Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2015).Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li & Lytton (1999).Li XF, Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. Journal of Biological Chemistry. 1999;274:8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- Li, Yang & Chen (2018).Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Liang et al. (2019).Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, Zhang JF. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biology. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi & Darnell (2010).Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature Reviews Genetics. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu & Huang (2017).Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biology. 2017;14:1000–1006. doi: 10.1080/15476286.2016.1227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magny et al. (2013).Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Matsumoto et al. (2017).Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- Meller, Joshi & Deshpande (2015).Meller VH, Joshi SS, Deshpande N. Modulation of chromatin by noncoding RNA. Annual Review of Genetics. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- Memczak et al. (2013).Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Noble Fle, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mondal & Kanduri (2013).Mondal T, Kanduri C. Maintenance of epigenetic information: a noncoding RNA perspective. Chromosome Research. 2013;21:615–625. doi: 10.1007/s10577-013-9385-5. [DOI] [PubMed] [Google Scholar]
- Nelson et al. (2016).Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti et al. (2017).Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Molecular Cell. 2017;66:9–21 e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan et al. (2008).Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Perriman & Ares Jr (1998).Perriman R, Ares Jr M. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4:1047–1054. doi: 10.1017/S135583829898061X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschansky & Wahlestedt (2014).Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, Jordan & Holcik (2010).Riley A, Jordan LE, Holcik M. Distinct 5′UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Research. 2010;38:4665–4674. doi: 10.1093/nar/gkq241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree et al. (2017).Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman et al. (2012).Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg & Hinnebusch (2009).Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires et al. (2012).Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Research. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger & Nair (2016).Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang & Wang (2015).Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt et al. (2020).Weigelt CM, Sehgal R, Tain LS, Cheng J, Esser J, Pahl A, Dieterich C, Gronke S, Partridge L. An insulin-sensitive circular RNA that Regulates lifespan in drosophila. Molecular Cell. 2020;79:268–279 e265. doi: 10.1016/j.molcel.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay et al. (2016).Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351: aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- Wesselhoeft, Kowalski & Anderson (2018).Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nature Communications. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright & Bruford (2011).Wright MW, Bruford EA. Naming ’junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Human Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Unbehaun & Spahn (2017).Yamamoto H, Unbehaun A, Spahn CMT. Ribosomal chamber music: toward an understanding of IRES mechanisms. Trends in Biochemical Sciences. 2017;42:655–668. doi: 10.1016/j.tibs.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2017c).Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, Gupta S, Yang W, Yang BB. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Molecular Therapy. 2017c;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2017a).Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, Dong J, Khorshidi A, Yang BB. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death and Differentiation. 2017a;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2017b).Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Research. 2017b;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2018).Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. Journal of the National Cancer Institute. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng et al. (2017).Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li RK, Wu Z, Backx PH, Yang BB. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2020).Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Molecular Cancer. 2020;19:105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018a).Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018a;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2018b).Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, Yang X, Huang N, Liu J, He K, Xie K, Zhang G, Huang S, Zhang N. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Communications. 2018b;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2019).Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, Zhan X, Laimins L, Wang RC. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nature Communications. 2019;10:2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2016).Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2019).Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, Wu C, Zhou Q, Hu W, Wu C, Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Molecular Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2017).Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Wittenberghe NVan, Xing Y, Giallourakis CC, Mullen AC. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Reports. 2017;20:2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
There is no original, experimental raw data in this literature review.