Abstract
Amyloidogenic peptides and proteins are rich sources of supramolecular assemblies. Sequences derived from well-known amyloids, including Aβ, human islet amyloid polypeptide, and tau have been found to assemble as fibrils, nanosheets, ribbons, and nanotubes. The supramolecular assembly of medin, a 50-amino acid peptide that forms fibrillary deposits in aging human vasculature, has not been heavily investigated. In this work we present an X-ray crystallographic structure of a cyclic β-sheet peptide derived from the 19–36 region of medin that assembles to form interpenetrating cubes. The edge of each cube is composed of a single peptide, and each vertex is occupied by a divalent metal ion. This structure may be considered a metal-organic framework (MOF) containing a large peptide ligand. This work demonstrates that peptides containing Glu or Asp that are preorganized to adopt β-hairpin structures can serve as ligands and assemble with metal ions to form MOFs.
Graphical Abstract
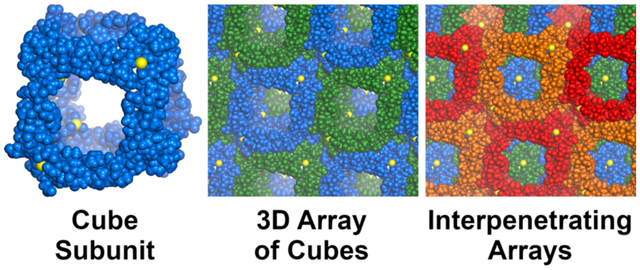
Introduction
Amyloidogenic peptides and proteins are rich sources of supramolecular assemblies. Sequences derived from Aβ (Alzheimer’s disease), human islet amyloid polypeptide (type II diabetes), and tau (Alzheimer’s disease and frontotemporal dementia) have been found to assemble as fibrils, nanosheets, ribbons, and nanotubes (Table 1).1–20 Other types of assemblies have also been observed, including square channels from sequences derived from transthyretin (senile systemic amyloidosis) and cylindrins from sequences derived from αB-crystallin (cataracts).21–22 Although these well characterized amyloidogenic peptides and proteins have yielded many novel supramolecular assemblies, the more recently discovered amyloidogenic peptide medin has not been as heavily studied and provides and exciting frontier for the discovery of interesting supramolecular assemblies.
Table 1.
Supramolecular Assemblies of Selected Sequences Derived from Amyloidogenic Peptides & Proteins
| Peptide | Sequence | Assembly | Citation |
|---|---|---|---|
| Aβ(10–35) | YEVHHQKLVFFAEDVGSNKGAIIGLM | fibrila,b | 1, 2, 3 |
| Aβ(16–22) | Ac-KLVFFAE-NH2 | fibril, helical ribbon, nanotubea,b,c,d | 4, 5, 6 |
| KLVF(N-Me)FAE, KLVFFAE | nanotubee | 7 | |
| Aβ(16–22), Italian mutant | KLVFFAK | nanosheetb,c | 8 |
| Aβ(16–20) | AAKLVFF | nanotubeb | 9, 10 |
| βAβAKLVFF | helical ribbond | 11 | |
| Aβ(19–20) | FF | nanotubeb | 12 |
| Ac-FF-NH2, NH2-FF-NH2, Boc-FF-NH2 | nanotubeb,c | 13 | |
| Fmoc-FF | flat ribbon, fibrilb,c,d | 13, 14 | |
| hIAPP(20–29) | SNNFGAILSS | flat ribbon, helical ribbonc,d | 15 |
| SNNFG(N-Bu)AI(N-Bu)LS(N-Bu)S | helical ribbonb | 16 | |
| hIAPP(21–29) | NNFGAILSS | helical ribbonb,c | 17 |
| hIAPP(23–27) | FGAIL | flat ribbonb,c | 18 |
| tau(306–311) | Ac-VQIVYK-NH2 | straight and twisted filamentb | 19 |
| transthyretin (106–112, 115–121) | TIAA(N-Me)LLS, SFSTTAV | square channele | 20 |
| αB-crystallin(90–100) | KVKVLGDVIEV | cylindrinb,e | 21 |
Solid-state NMR (ssNMR)
Electron microscopy (EM)
Atomic force microscopy (AFM)
Cryo-EM
X-ray crystallography
Medin, also referred to as aortic medial amyloid, is a 50-amino acid peptide that forms fibrillary deposits in aging human vasculature (Figure 1). These deposits have been implicated in the pathogenesis of thoracic aortic aneurysm and dissection. Westermark et al. found evidence of fibrillary medin in the aortic media of 97% of subjects over the age of 50.22 Although the amino acid sequence of medin was determined in 1999, the folding pattern of the monomer was not elucidated until 2017.23 Using 13C NMR spectroscopy, Madine et al. identified that the monomer of medin contains three β-strand regions consisting of residues 7–13, 21–25, and 30–36.24 The sequences 14–24 and 26–29 were suggested to form unstructured loops. Computational modeling using QUARK and ROSETTA consistently predicted the 21–35 region to fold as a β-hairpin.24
Figure 1.

Amino acid sequence of medin. Amyloidogenic regions predicted by TANGO are highlighted in red. Underlined residues are β-strand regions identified by 13C NMR spectroscopy.
Medin is predicted by TANGO to have three amyloidogenic regions (Figure 1).25 Westermark et al. studied the medin(1–25), medin(32–41), and medin(42–49) peptides. The researchers only observed the formation of fibrils from the latter two peptides by electron microscopy, and concluded that the amyloid forming motif of medin lies in its C-terminus. Gazit et al. also observed the formation of fibrils by the medin(42–49) peptide by electron microscopy.26 Middleton et al. subsequently used solid-state NMR spectroscopy and X-ray fiber diffraction to establish that the fibrils from medin(42–49) consist of parallel, in-register β-sheets that assemble in a face-to-back manner.27 To our knowledge, no additional structural information regarding the assembly of medin(32–41) has been elucidated.
Results
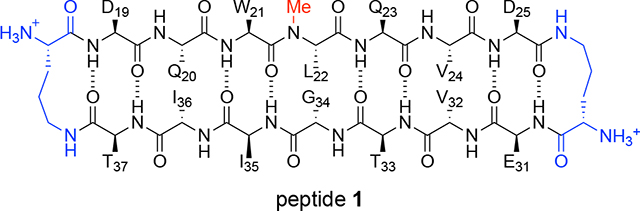
To gain further insights into the supramolecular assembly of medin, we set out to study the two β-strand regions in the medin monomer predicted to fold as a β-hairpin. To synthesize the β-hairpin mimic peptide 1, we connected two heptapeptide strands using δ-linked ornithine (δOrn) turn units (blue) to form a macrocycle.28,29 The two heptapeptide strands are derived from medin(19–25) DQWLQVD (top strand) and medin(31–37) EVTGIIT (bottom strand). N-Methylation (red) of the backbone was employed to attenuate uncontrolled aggregation.30 This strategy to make β-hairpin mimics has been successfully used by our lab in the past to identify other amyloid assemblies using X-ray crystallography.31
We used X-ray crystallography to study the structure and assembly of peptide 1. We began our crystallization efforts by screening peptide 1 in 576 conditions in a 96-well plate format using crystallization kits from Hampton Research. Cube-shaped crystals (Figure S4) grew in abundance in drops containing sodium acetate, calcium chloride, and 2-methyl-2,4-pentanediol. Further optimization of the crystallization conditions afforded monocrystals suitable for X-ray diffraction (optimized conditions: 0.1 M NaOAc, 0.02 M CaCl2, 30% MPD). The X-ray diffraction experiments were performed at 1.54 Å using an X-ray diffractometer. Despite the relatively high resolution of the acquired dataset (ca. 1.32 Å), we were unable to solve the structure by single wavelength anomalous diffraction (SAD). In an attempt to increase the magnitude of anomalous scattering, we crystallized peptide 1 in the optimized conditions substituting barium chloride for calcium chloride. The modified conditions afforded similar cube-shaped crystals that gave rise to enough measurable anomalous signal to facilitate SAD phasing and subsequent solution of the original dataset by isomorphous replacement. The structure was thus solved in the cubic I23 space group and refined to a final Rwork of 0.1772 (PDB 7JRH).
The X-ray crystallographic structure of peptide 1 reveals a three-dimensional network of large interpenetrating cubes, ca. 4.3 nm in size (Figure 2). Each cube is composed of molecules of peptide 1 located at the edges and coordinated to calcium ions at the vertices. In the structure, peptide 1 folds to form a hydrogen-bonded β-sheet. The elongated β-strand conformation of the top strand places the two aspartic acid residues (D19 and D25) at the opposite ends of the β-sheet, ca. 2.0 nm apart. The side chain carboxylate group of D19 binds one calcium ion, and the side chain of D25 binds another calcium ion. Each of the ions in turn binds either D19 or D25 residues of two other peptide molecules. This mode of coordination, in conjunction with the symmetry present in the crystal lattice, results in the formation of an assembly containing eight calcium ions and twelve molecules of peptide 1. Even though the arrangement of edges and vertices resembles a cube (Figure 2A and B), the point symmetry is actually tetrahedral, with four of the vertices arising from the coordination of D19 and the other four vertices arising from the coordination of D25.
Figure 2.
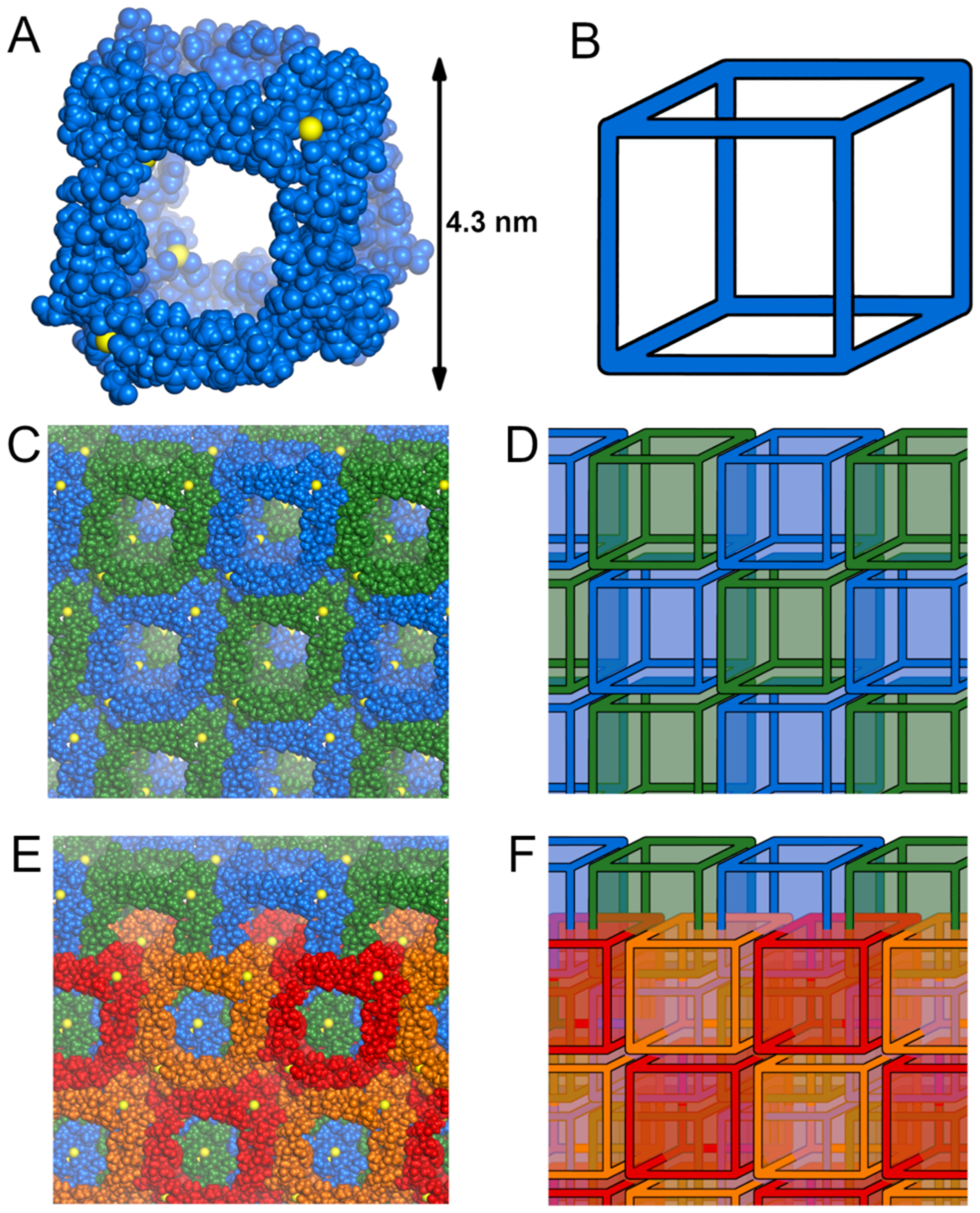
Three-dimensional assembly observed in the X-ray crystallographic structure of peptide 1. Individual cube (A), infinite array of cubes (C), and two interpenetrating arrays (E), along with their respective schematic representations (B, D, and F). The cubes are all symmetry-equivalent, and the coloring scheme has been chosen arbitrarily for clarity.
The cubes stack face-to-face in the crystal lattice in all three directions, forming an infinite 3-dimensional array (Figure 2C and D). Consistent with the body-centered lattice type, the complete crystal lattice contains two symmetry-equivalent mutually-interpenetrating arrays, related through a [0.5, 0.5, 0.5] Bravais translation vector (Figure 2E and F).32 Such a polycatenated structure belongs to the bcu-y class in knot theory — a 3D ‘chainmail’of linked cubes.33
The packing of the cubes, as shown in Figure 2C and D, is stabilized by hydrogen bonding and hydrophobic interactions between molecules of peptide 1 located at the edges of the cubes. The four molecules of peptide 1 that constitute the edges of every four adjacent cubes form a tetramer (Figure 3A). The tetramer can be interpreted as a dimer of dimers, in which each dimer is stabilized by six intermolecular hydrogen bonds involving side chains and main chains of T37, Q23, and I35 (Figure 3B). The dimers stack to form the tetramer. Packing of the hydrophobic side chains of I35, V32, and L22 — a total of twelve residues, three per monomer — help stabilize the tetramer. Eight hydrogen bonds between Q20 and V32 further stabilize the assembly.
Figure 3.
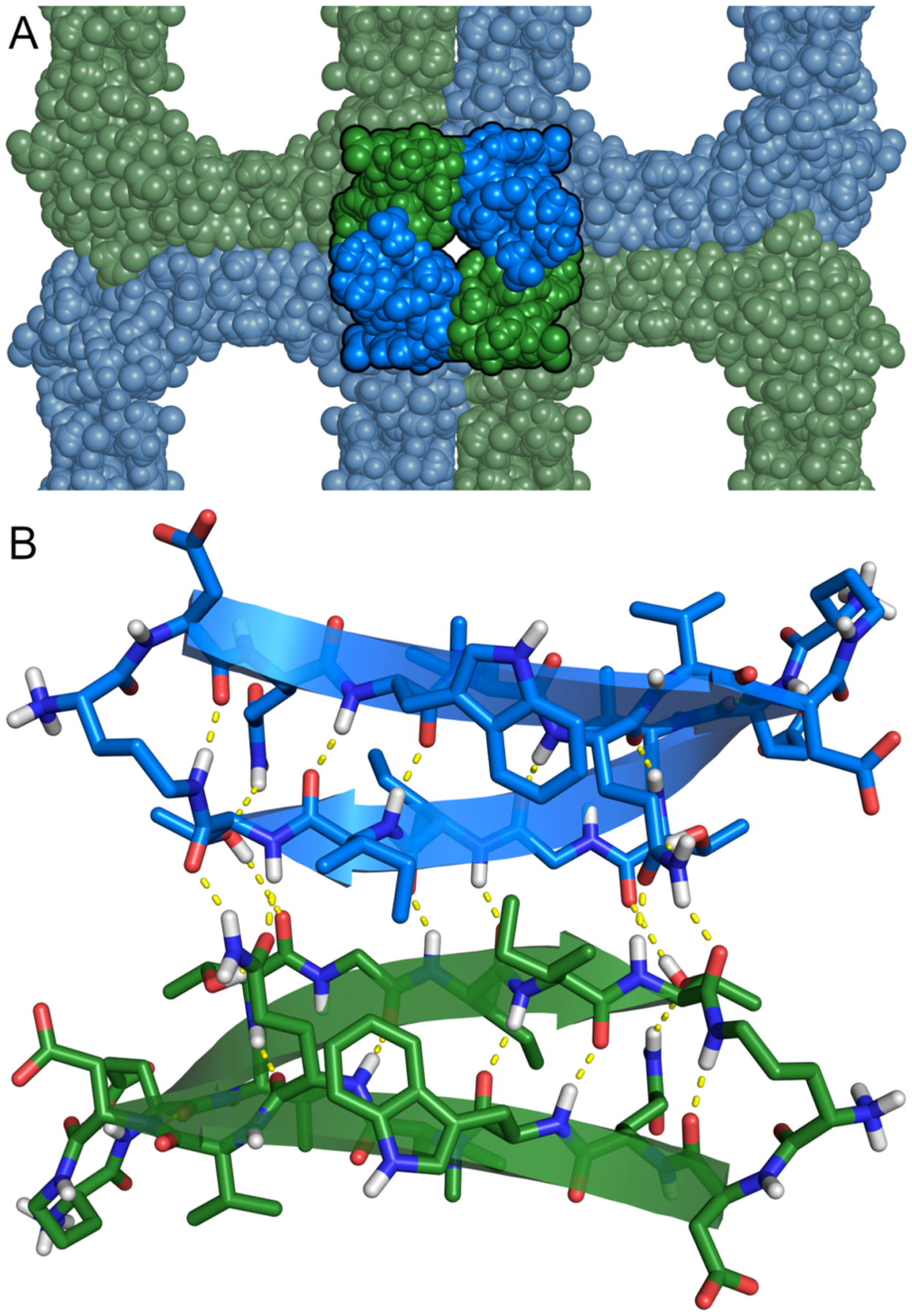
(A) The location of a tetramer in within the cubic assembly of peptide 1. (B) Hydrogen-bonded dimer within the tetramer.
The monomeric β-hairpins that constitute the building blocks of the crystal lattice have an unexpected folding pattern. Peptide 1 was designed to contain two hydrogen-bonded β-strands — DQWLQVD (top strand) and EVTGIIT (bottom strand) — linked by two δOrn turn units. In the X-ray crystallographic structure, the top and bottom strands do indeed form a hydrogen-bonded β-sheet, but not precisely as designed (Figure 4). Residues T33 and G34 in the bottom strand form a β-bulge,34 and six of the seven residues of the top strand (Q20 through D25) participate in β-sheet formation. The β-bulge causes a shift in the registration of the strands, which brings D19 into an extended loop with δOrn and causes T37 to pair with Q20 instead of D19 (Figure 4B).
Figure 4.
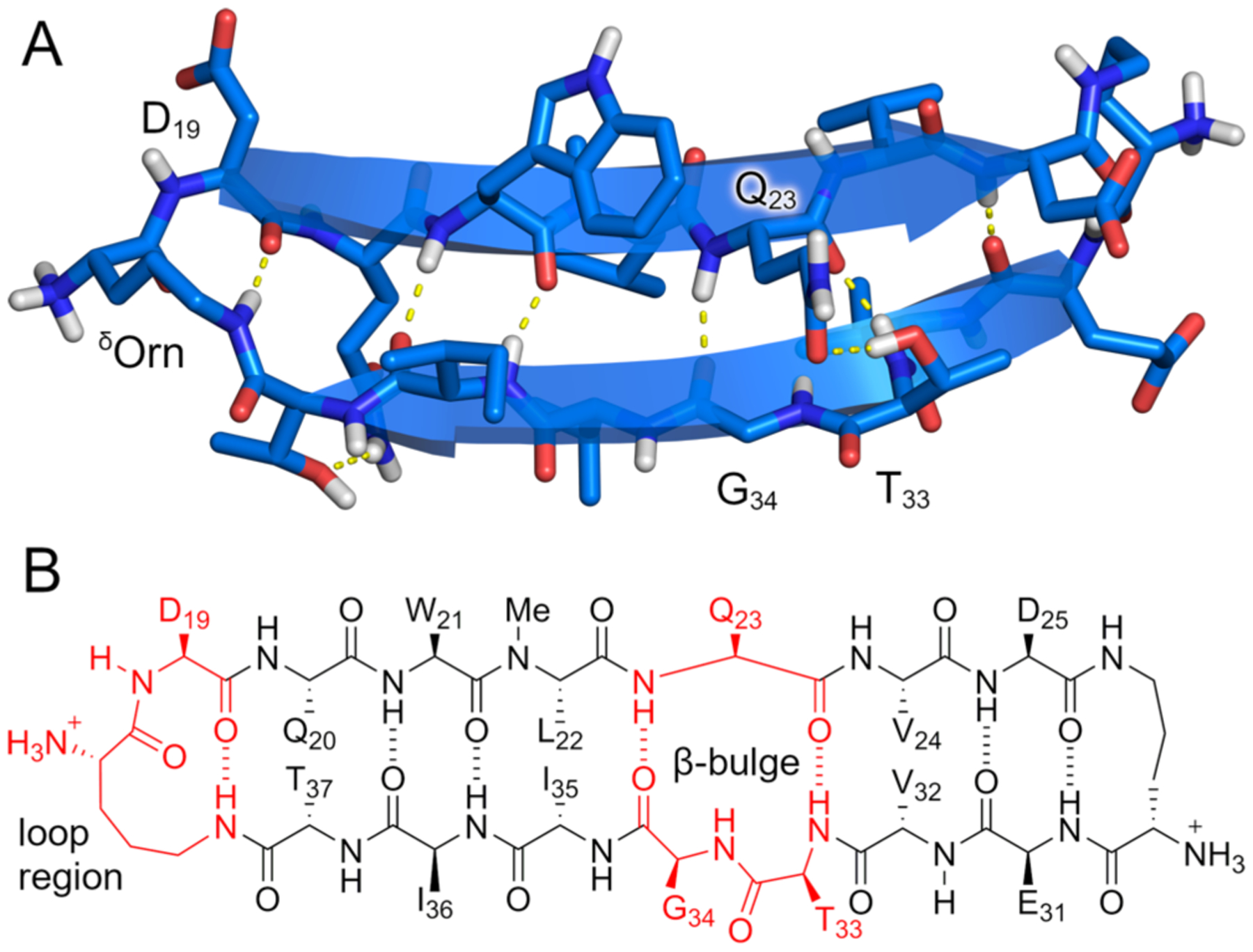
β-Bulge in the X-ray crystallographic structure of peptide 1. (A) X-ray crystallographic structure of the monomeric β-hairpin subunit of the crystal lattice. (B) Hydrogen bonding pattern observed in the X-ray structure.
Discussion
Coordination of Ca2+ by carboxylates in the peptide 1 structure is reminiscent of calcium binding in structures ranging from EF-hand proteins to metal-organic frameworks (MOFs). The D25-calcium binding site of peptide 1 contains three aspartate carboxylates and three water molecules in an octahedral arrangement (Figure 5A), while the D19-calcium binding contains three aspartate carboxylates and one water molecule (Figure 5B). EF-hands are a class of calcium-binding proteins featuring a helix-loop-helix motif, containing multiple carboxylates and other ligands that bind Ca2+. Calmodulin is a prominent example of this class of calcium-binding proteins, employing three aspartates to bind Ca2+, in a fashion similar to the binding mode within the peptide 1 cubes (binding of Figure 5C).35 Only a handful of calcium-based metal-organic frameworks have been reported, including MOFs suitable for removal of heavy metals from water36 and slow release of pesticides,37 far fewer than the number of MOFs containing transition metals.38 Calcium-based materials, such as alginate gels, are of interest for applications in the foodstuffs and medicines, where only non-toxic metals are permissible.39,40 We anticipate that calcium-based MOFs may also have potential applications in these areas.
Figure 5.
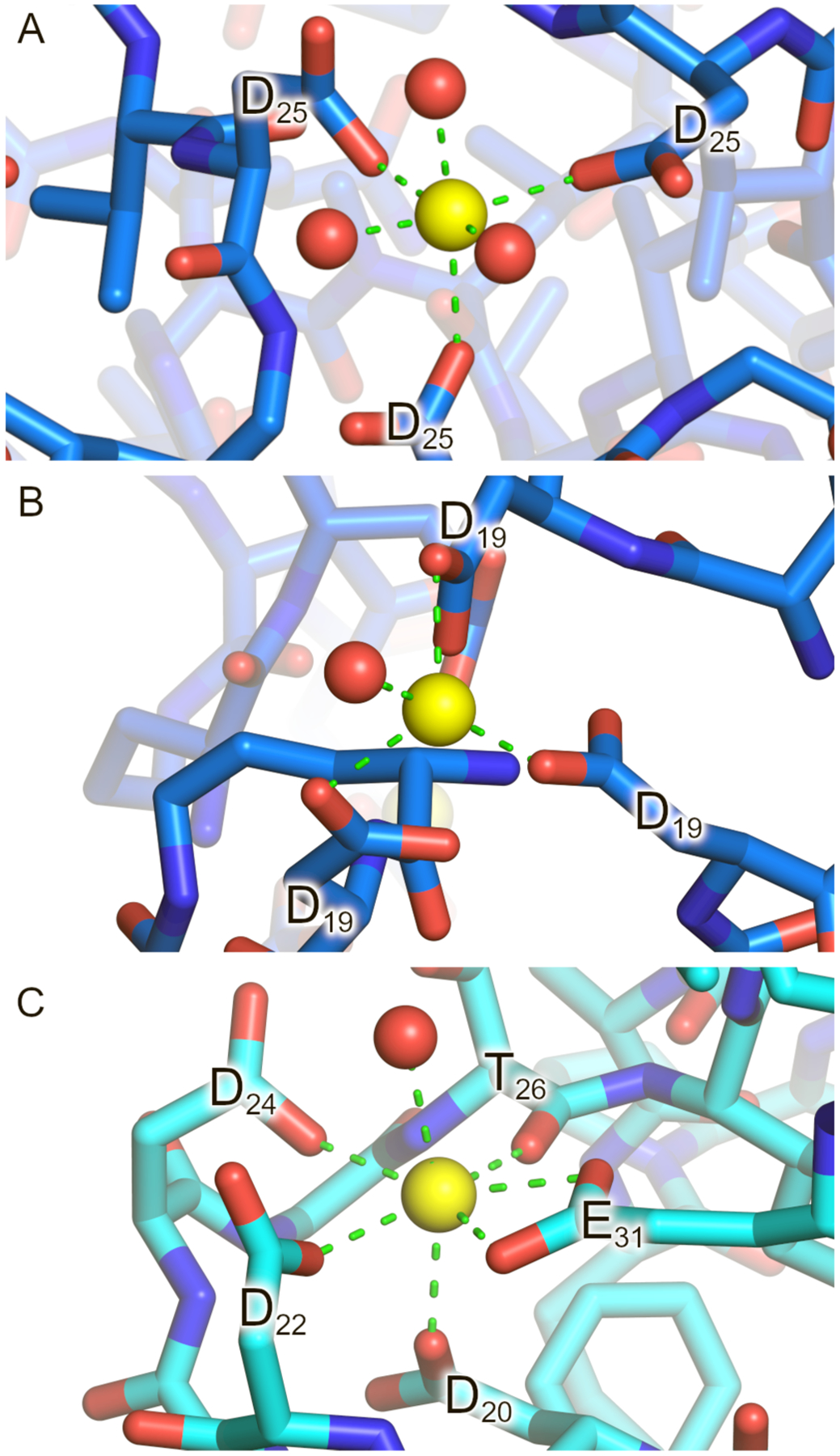
(A and B) Calcium binding geometry observed in the X-ray crystallographic structure of peptide 1. (C) Calcium binding site in the calmodulin protein (PDB 1CLL). Calcium ions are shown as yellow spheres and water molecules as red spheres.
The non-covalent interactions in the crystal structure of peptide 1 are diverse: metal coordination, hydrogen bonding, and hydrophobic packing. Although it is not possible to assess the relative contributions of each of these interactions in stabilizing the assembly, metal coordination is critical. We have not observed growth of any stable crystals in the absence of divalent metal ions (Ca2+, Ba2+, or Co2+). The interpenetrating cubes formed by peptide 1 and Ca2+ can thus be considered a calcium-based MOF with peptide ligands, and not merely an artificial calcium receptor.
The interpenetrating cubes cannot form separately, but rather must form simultaneously from tetramers, dimers, single molecules, or other assemblies of peptide 1 during the crystallization process. For this reason, the lattice may also be interpreted as a result of the assembly of tetramers, with the tetramers acting as ligands bearing eight carboxylate groups. Regardless of the process by which the lattice assembles, it can ultimately be viewed as an assembly of cubes with Ca2+ at the vertices, because of the crucial role metal cations play in the assembly.
Intrinsic chirality and biocompatibility have allowed peptides to act as ligands in a number of MOFs possessing remarkable properties. These structures include MOFs exhibiting selective binding of chiral molecules,41 reversible conformational changes upon binding,42,43 and high mechanical stability.44 The relative flexibility of peptide chains, which imparts these interesting properties, is also a major limiting factor. In longer peptide chains, it is almost impossible to predict and control secondary structure. A notable example is a pentapeptide-cadmium complex reported by Yaghi et al. that exhibits no porosity.45 In the case of the framework structure formed by peptide 1, the greatly reduced conformational flexibility of the cyclic β-sheet peptide is sufficient to allow the formation of a well-defined assembly. The preorganized spatial arrangement of the two aspartate side chains allows the peptide to behave like a rigid ligand. Thus, a design principle emerges for the construction of other peptide-based MOFs, whereby a preference for a particular secondary structure allows preorganization of metal-binding ligands (e.g., Glu, Asp, or His).
It is important to note that larger peptides with well-defined secondary structures have previously been used with metals to construct various nanostructures. Notable examples include α-helix bundles46,47,48,49 and collagen triple helices.50 Many of these structures rely upon the incorporation of non-natural ligands — most often various pyridine derivatives — into the peptide sequences. The three-dimensional framework formed by peptide 1 is distinctive, because the metal binding involves side chains of unmodified aspartic acid residues. We envision that it will be possible to construct other large calcium-containing MOFs from peptides with well-defined secondary structures that display Glu or Asp side chains. We further envision that this design principle might be extended to other metals and amino acid side chains such as His or Cys.
Conclusion
The assembly formed by peptide 1 in the presence of divalent metal ions constitutes a rare example of a metal-organic framework in which a large polypeptide acts as a ligand. The MOF formed by peptide 1 is among the largest peptide-based MOFs. While the structure exhibits interpenetration, and thus does not contain solvent-accessible pores, it demonstrates important rules for the construction of peptide-based MOFs. The peptide’s cyclic β-sheet design exemplifies one such emergent principle, in which a preference for a particular secondary structure and supramolecular assembly allows the preorganization of metal-binding sites. We envision that different peptides with well-defined secondary structures, bearing multiple metal-binding residues may — in conjunction with metal ions — result in MOFs. Amyloidogenic peptides are particularly suited for this purpose, since they are predisposed toward supramolecular assembly. Amyloidogenic peptides other than medin might also serve as ligands for MOF structures if they contain, or are altered to contain, suitable metal-binding residues.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (GM097562) and the National Science Foundation (CHE 1808096). M. W. acknowledges the support from the Ministry of Science and Higher Education, Republic of Poland (Mobility Plus grant no. 1647/MOB/V/2017/0). Beamline 8.2.2 of the Advanced Light Source, a U.S. DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01. We thank Dr. Milan Gembicky for allowing us access to the Crystallography Facility of the University of California, San Diego and Dr. Jake Bailey for performing data collection. We also thank Dr. Bonnie Cuthbert for her assistance in the refinement of the crystal structure.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Burkoth TS; Benzinger TLS; Urban V; Morgan DM; Gregory DM; Thiyagarajan P; Botto RE; Meredith SC; Lynn DG Structure of the β-Amyloid(10–35) Fibril. J. Am. Chem. Soc 2000, 122, 7883–7889. [Google Scholar]
- 2.Benzinger TLS; Gregory DM; Burkoth TS; Miller-Auer H; Lynn DG; Botto RE; Meredith SC Two-Dimensional Structure of β-Amyloid(10–35) Fibrils. Biochemistry 2000, 39, 3491–3499. [DOI] [PubMed] [Google Scholar]
- 3.Benzinger TLS; Gregory DM; Burkoth TS; Miller-Auer H; Lynn DG; Botto RE; Meredith SC Propagating structure of Alzheimer’s β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu K; Jacob J; Thiyagarajan P; Conticello VP; Lynn DG Exploiting Amyloid Fibril Lamination for Nanotube Self-Assembly. J. Am. Chem. Soc 2003, 125, 6391–6393. [DOI] [PubMed] [Google Scholar]
- 5.Mehta AK; Lu K; Childers WS; Liang Y; Dublin SN; Dong J; Snyder JP; Pingali SV; Thiyagarajan P; Lynn DG Facial Symmetry in Protein Self-Assembly. J. Am. Chem. Soc 2008, 130, 9829–9835. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y; Pingali SV; Jogalekar AS; Snyder JP; Thiyagarajan P; Lynn DG Cross-Strand Pairing and Amyloid Assembly. Biochemistry 2008, 47, 10018–10026. [DOI] [PubMed] [Google Scholar]
- 7.Chen KH; Corro KA; Le SP; Nowick JS X-ray Crystallographic Structure of a Giant Double-Walled Peptide Nanotube Formed by a Macrocyclic β-Sheet Containing Aβ16–22. J. Am. Chem. Soc 2017, 139, 8102–8105. [DOI] [PubMed] [Google Scholar]
- 8.Dai B; Li D; Xi W; Luo F; Zhang X; Zou M; Cao M; Hu J; Wang W; Wei G; Zhang Y; Liu C Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc. Natl. Acad. U. S. A 2015, 112, 2996–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krysmann MJ; Castelletto V; McKendrick JE; Clifton LA; Hamley IW Self-Assembly of Peptide Nanotubes in an Organic Solvent. Langmuir 2008, 24, 8158–8162. [DOI] [PubMed] [Google Scholar]
- 10.Castelletto V; Hamley IW; Harris PJF Self-assembly in aqueous solution of a modified amyloid beta peptide fragment. Biophys. Chem 2008, 138, 29–35. [DOI] [PubMed] [Google Scholar]
- 11.Castelletto V; Hamley IW; Hule RA; Pochan D Helical Ribbon Formation by a βAmino Acid Modified Amyloid β Peptide Fragment. Angew. Chem. Int. Ed 2009, 48, 2317–2320. [DOI] [PubMed] [Google Scholar]
- 12.Reches M; Gazit E Casting Metal Nanowires within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [DOI] [PubMed] [Google Scholar]
- 13.Reches M; Gazit E Self assembly of peptide nanotubes and amyloid like structures by charged termini capped diphenylalanine peptide analogues. Isr. J. Chem 2005, 45, 363–371. [Google Scholar]
- 14.Smith AM; Williams RJ; Tang C; Coppo P; Collins RF; Turner ML; Saiani A; Ulijn RV Fmoc Diphenylalanine Self Assembles to a Hydrogel via a Novel Architecture Based on π–π Interlocked β Sheets. Adv. Mater 2008, 20, 37–41. [Google Scholar]
- 15.Zhang S; Andreasen M; Nielsen JT; Liu L; Nielsen EH; Song J; Ji G; Sun F; Skrydstrup T; Besenbacher F; Nielsen NC; Otzen DE; Dong M Coexistence of ribbon and helical fibrils originating from hIAPP20–29 revealed by quantitative nanomechanical atomic force microscopy. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 2798–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elgersma RC; Meijneke T; Posthuma G; Rijkers DTS; Liskamp RMJ Self-Assembly of Amylin(20–29) Amide Bond Derivatives into Helical Ribbons and Peptide Nanotubes rather than Fibrils. Chem. Eur. J 2006, 12, 3714–3725. [DOI] [PubMed] [Google Scholar]
- 17.Wang S; Lin Y; Spencer RK; Thomas MR; Nguyen AI; Amdursky N; Pashuck ET; Skaalure SC; Song CY; Parmar PA; Morgan RM; Ercius P; Aloni S; Zuckermann RN; Stevens MM Sequence-Dependent Self-Assembly and Structural Diversity of Islet Amyloid Polypeptide-Derived β-Sheet Fibrils. ACS Nano 2017, 11, 8579–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenidis K; Waldner M; Bernhagen J; Fischle W; Bergmann M; Weber M; Merkle M; Voelter W; Brunner H; Kapurniotu A Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol 2000, 295, 1055–1071. [DOI] [PubMed] [Google Scholar]
- 19.Goux WJ; Kopplin L; Nguyen AD; Leak K; Rutkofsky M; Shanmuganandam VD; Sharma D; Inouye H; Kirschner DA The Formation of Straight and Twisted Filaments from Short Tau Peptides. J. Biol. Chem 2004, 279, 26868–26875. [DOI] [PubMed] [Google Scholar]
- 20.Yoo S; Kreutzer AG; Truex NL; Nowick JS Square channels formed by a peptide derived from transthyretin. Chem. Sci 2016, 7, 6946–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laganowsky A; Liu C; Sawaya MR; Whitelegge JP; Park J; Zhao M; Pensalfini A; Soriaga AB; Landau M; Teng PK; Cascio D; Glabe C; Eisenberg D Atomic View of a Toxic Amyloid Small Oligomer. Science 2012, 335, 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mucchiano G; Cornwell III GG; Westermark P Senile Aortic Amyloid. Evidence for two Distinct Forms of Localized Deposits. Am. J. Pathol 1992, 140, 871–877. [PMC free article] [PubMed] [Google Scholar]
- 23.Häggqvist B; Näslund J; Sletten K; Westermark GT; Mucchiano G; Tjernberg LO; Nordstedt C; Engström E; Westermark P Medin: An integral fragment of aortic smooth muscle cell-produced lactadherin forms the most common human amyloid. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies HA; Rigden DJ; Phelan MM; Madine J Probing Medin Monomer Structure and its Amyloid Nucleation Using 13C-Direct Detection NMR in Combination with Structural Bioinformatics. Sci. Rep 2017, 7, 45224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson A; Södererg L; Westermark GT; Sletten K; Engström U; Tjernberg LO; Näslund J; Westermark P Unwinding fibril formation of medin, the peptide of the most common form of human amyloid. Biochem. Biophys. Res. Commun 2007, 361, 822–828. [DOI] [PubMed] [Google Scholar]
- 26.Reches M; Gazit E Amyloidogenic hexapeptide fragment of medin: homology to functional islet amyloid polypeptide fragments. Amyloid 2004, 11, 81–89. [DOI] [PubMed] [Google Scholar]
- 27.Madine J; Copland A; Serpell LC; Middleton DA Cross-β Spine Architecture of Fibrils Formed by the Amyloidogenic Segment NFGSVQFV of Medin from Solid-State NMR and X-ray Fiber Diffraction Measurements. Biochemistry 2009, 48, 3089–3099. [DOI] [PubMed] [Google Scholar]
- 28.Nowick JS; Brower JO A New Turn Structure for the Formation of β-Hairpins in Peptides. J. Am. Chem. Soc 2003, 125, 876–877. [DOI] [PubMed] [Google Scholar]
- 29.Spencer R; Chen KH; Manuel G; Nowick JS Recipe for β Sheets: Foldamers Containing Amyloidogenic Peptide Sequences. Eur. J. Org. Chem 2013, 2013, 3523–3528. [Google Scholar]
- 30.Hughes E; Burke RM; Doig AJ Inhibition of Toxicity in the β-Amyloid Peptide Fragment β-(25–35) Using N-Methylated Derivatives. A general strategy to prevent amyloid formation. J. Biol. Chem 2000, 275, 25109–25115. [DOI] [PubMed] [Google Scholar]
- 31.Kreutzer AG; Nowick JS Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer’s Disease and Other Amyloid Diseases. Acc. Chem. Res 2018, 51, 3, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.For a related structure noted by a reviewer see:; Sontz PA; Bailey JB; Ahn S; Tezcan FA A Metal Organic Framework with Spherical Protein Nodes: Rational Chemical Design of 3D Protein Crystals. J. Am. Chem. Soc 2015, 137, 36, 11598–11601. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y; O’Keeffe M; Treacy MMJ; Yaghi O The geometry of periodic knots, polycatenanes and weaving from a chemical perspective: a library for reticular chemistry. Chem. Soc. Rev 2018, 47, 4642–4664. [DOI] [PubMed] [Google Scholar]
- 34.Richardson JS; Getzoff ED; Richardson DC The β bulge: A common small unit of nonrepetitive protein structure. Proc. Natl. Acad. Sci. U. S. A 1978, 75, 2574–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyaya R; Meador WE; Means AR; Quiocho FA Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol 1992, 228, 1177–1192. [DOI] [PubMed] [Google Scholar]
- 36.Pournara AD; Margariti A; Tarlas GD; Kourtelaris A; Petkov V; Kokkinos C; Economou A; Papaefstathiou GS; Manos MJ A Ca2+ MOF combining highly efficient sorption and capability for voltammetric determination of heavy metal ions in aqueous media. J. Mater. Chem 2019, 7, 15432–15443. [Google Scholar]
- 37.Yang J; Trickett CA; Alahmadi SB; Alshammari AS; Yaghi OM Calcium L Lactate Frameworks as Naturally Degradable Carriers for Pesticides. J. Am. Chem. Soc 2017, 139, 811–8121. [DOI] [PubMed] [Google Scholar]
- 38.Moghadam PZ; Li A; Wiggin SB; Tao A; Maloney AGP; Wood PA; Ward SC; Fairen-Jimenez D Development of a Cambridge Structural Database Subset: A Collection of Metal–Organic Frameworks for Past, Present, and Future. Chem. Mater 2017, 29, 2618–2625. [Google Scholar]
- 39.Smisrød O; Haug A The Effect of Divalent Metals on the Properties of Alginate Solutions. I. Calcium Ions. Acta Chem. Scand 1965, 19, 329–340 [Google Scholar]
- 40.Morris ER; Rees DA; Thom D Characterization of polysaccharide structure and interactions by circular dichroism: order–disorder transition in the calcium alginate system. J. Chem. Soc., Chem. Commun 1973, 245–246 [Google Scholar]
- 41.Navarro-Sánchez J; Argente-García AI; Moliner-Martínez Y; Roca-Sanjuán D; Antypov D; Campíns-Falcó P; Rosseinsky MJ; Martí-Gastaldo C Peptide Metal-Organic Frameworks for Enantioselective Separation of Chiral Drugs. J. Am. Chem. Soc 2017, 139, 4294–4297. [DOI] [PubMed] [Google Scholar]
- 42.Rabone J; Yue Y-F; Chong SY; Stylianou KC; Bacsa J; Bradshaw D; Darling GR; Berry NG; Khimyak YZ; Ganin AY; Wiper P; Claridge JB; Rosseinsky MJ An Adaptable Peptide-Based Porous Material. Science 2010, 329, 1053–1057. [DOI] [PubMed] [Google Scholar]
- 43.Katsoulidis AP; Antypov D; Whitehead GFS; Carrington EJ; Adams DJ; Berry NG; Darling GR; Dyer MS; Rosseinsky MJ Chemical Control of Structure and Guest Uptake by a Conformationally Mobile Porous Material. Nature 2019, 565, 213–217. [DOI] [PubMed] [Google Scholar]
- 44.Martí-Gastaldo C; Warren JE; Stylianou KC; Flack NLO; Rosseinsky MJ Enhanced Stability in Rigid Peptide-Based Porous Materials. Angew. Chem. Int. Ed 2012, 51, 11044–11048. [DOI] [PubMed] [Google Scholar]
- 45.Peri D; Ciston J; Gándara F; Zhao Y; Yaghi OM Crystalline Fibers of Metal–Peptide Double Ladders. Inorg. Chem 2013, 52, 13818–13820. [DOI] [PubMed] [Google Scholar]
- 46.Tavenor NA; Murnin MJ; Horne WS Supramolecular Metal-Coordination Polymers, Nets, and Frameworks from Synthetic Coiled-Coil Peptides. J. Am. Chem. Soc 2017, 139, 2212–2215. [DOI] [PubMed] [Google Scholar]
- 47.Nepal M; Sheedlo MJ; Das C; Chmielewski J Accessing Three-Dimensional Crystals with Incorporated Guests through Metal-Directed Coiled-Coil Peptide Assembly. J. Am. Chem. Soc 2016, 138, 11051–11057. [DOI] [PubMed] [Google Scholar]
- 48.Anzini P; Xu C; Hughes S; Magnotti E; Jiang T; Hemmingsen L; Demeler B; Conticello VP Controlling Self-Assembly of a Peptide-Based Material via Metal-Ion Induced Registry Shift. J. Am. Chem. Soc 2013, 135, 10278–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brodin JD; Ambroggio XI; Tang C; Parent KN; Baker TS; Tezcan FA Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem 2012, 4, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pires MM; Przybyla DE; Rubert Pérez CM; Chmielewski J Metal-Mediated Tandem Coassembly of Collagen Peptides into Banded Microstructures. J. Am. Chem. Soc 2011, 133, 14469–14471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


