Abstract
Horizontal transmission between distantly related species has been used to explain how Wolbachia infect multiple species at astonishing rates despite the selection for resistance. Recently, a terrestrial isopod species was found to be infected by an unusual strain of supergroup F Wolbachia. However, only Wolbachia of supergroup B is typically found in isopods. One possibility is that these isopods acquired the infection because of their recurrent contact with termites—a group with strong evidence of infection by supergroup F Wolbachia. Thus, our goals were: (1) check if the infection was an isolated case in isopods, or if it revealed a broader pattern; (2) search for Wolbachia infection in the termites within Brazil; and (3) look for evidence consistent with horizontal transmission between isopods and termites. We collected Neotroponiscus terrestrial isopods and termites along the Brazilian coastal Atlantic forest. We sequenced and identified the Wolbachia strains found in these groups using coxA, dnaA, and fpbA genes. We constructed phylogenies for both bacteria and host taxa and tested for coevolution. We found the supergroup F Wolbachia in other species and populations of Neotroponiscus, and also in Nasutitermes and Procornitermes termites. The phylogenies showed that, despite the phylogenetic distance between isopods and termites, the Wolbachia strains clustered together. Furthermore, cophylogenetic analyses showed significant jumps of Wolbachia between terrestrial isopods and termites. Thus, our study suggests that the horizontal transmission of supergroup F Wolbachia between termites and terrestrial isopods is likely. Our study also helps understanding the success and worldwide distribution of this symbiont.
Supplementary information
The online version of this article (10.1007/s10682-021-10101-4) contains supplementary material, which is available to authorized users.
Keywords: Horizontal transmission, Isoptera, Isopoda, Neotropical fauna, Symbiosis
Introduction
Wolbachia is a bacterial genus that includes obligate-intracellular bacteria that occur in a high proportion of arthropod species and many filarial nematodes (Werren et al. 2008). This evolutionary success that elevated Wolbachia to pandemic status was made possible by the efficient vertical transmission and the reproductive manipulation of hosts (Gerth et al. 2014). Despite the seeming success, several host lineages show a ‘loss’ of Wolbachia infection probably due to a selection for resistance (Zug and Hammerstein 2012). In fact, according to estimates, 0.14 loss events and 0.11 acquisition events are expected per million years (Bailly-Bechet et al. 2017). Nevertheless, Wolbachia maintain their infection prevalence at very high rates, particularly in arthropods, and this does not seem constrained by host phylogenetics (Zug and Hammerstein 2012). One possibility for the high prevalence rates might thus lie in the diversity of Wolbachia bacteria. To assess Wolbachia diversity, researchers used phylogenetic approaches that lead to their classification in ‘supergroups’ (Werren et al. 1995; Bandi et al. 1998; Casiraghi et al. 2005; Baldo et al. 2006b). To some extent, each Wolbachia ‘supergroup’ seems to associate with particular host groups; and the host groups differ among the supergroups. For example, supergroup A and B strains are the most abundant ones, being found in ∼ 40% of all terrestrial arthropods’ species (Zug and Hammerstein 2012). Wolbachia of supergroups C and D are found exclusively in co-evolutionary associations consistent with their mutualistic dependency with some filarial nematodes (Taylor et al. 2013). Supergroup F, on the other hand, can infect both arthropods and filarial hosts (Casiraghi et al. 2005). Thus, supergroup F is relatively atypical because of its ability to infect groups that are not phylogenetically close; hence, understanding what makes supergroup F atypical can allow us to understand the evolution of bacterial infection.
The mechanisms proposed to explain the phylogenetic widespread pattern of Wolbachia rely on horizontal transmission between distant species with close interactions. These close interactions approximate distant related species physically, creating an opportunity for horizontal transmission, as can be seen in interactions such as parasitoids and phoretic vectors (Covacin and Barker 2006; Gehrer and Vorburger 2012; Ahmed et al. 2015; Qi et al. 2019), use of common food sources (Sintupachee et al. 2006; Oliver et al. 2010; Stahlhut et al. 2010; Li et al. 2017), and predation and cannibalism (Yun et al. 2011; Le Clec’h et al. 2013; Brown and Lloyd 2015; Faria et al. 2016). As Wolbachia appears to transmit easily through hemolymph or the gut, the simplest route of transmission seems to be contacting the uninfected host through excretions or injuries (Rigaud and Juchault 1995). After entering the new host, Wolbachia can use cell-to-cell transfer to establish both somatic and germline infection (Frydman et al. 2006). Alternatively, Wolbachia could infect new hosts without cell-to-cell transfer. By using the host phagocytic and endocytic machinery, they can be transferred successfully to new hosts (White et al. 2017). Thus, one might expect that host groups infected with supergroup F can be infected by and infect any species in which they are in close contact with.
Terrestrial isopods are an interesting case of Wolbachia infection because, until recently, infections were almost exclusively from supergroup B (Rousset et al. 1992; Juchault et al. 1994; Bouchon et al. 1998, 2008; Nyirõ; et al. 2002; Ben Afia Hatira et al. 2008; Wiwatanaratanabutr et al. 2009; Almerão et al. 2012; Cordaux et al. 2012; Zimmermann et al. 2015). However, the Neotropical isopod Neotroponiscus littoralis from Northeast Brazil was infected with Wolbachia of supergroup F, a lineage that was never detected in crustaceans before (Zimmermann et al. 2015). Neotroponiscus is composed of ten small-sized and poorly studied species, endemic of South American Atlantic coastal forests (Cardoso et al. 2017). Contrary to most species of terrestrial isopods that live in leaf litter (Richardson and Araujo 2015), Neotroponiscus are generally found on banana and bromeliad leaves, decaying wood, and—most importantly—in termite nests (Lemos de Castro 1970a, b; Lenko 1971; Lisboa et al. 2013).
Termite nests are ideal places for horizontal transmission of bacteria because they provide shelter and food for many species (Costa et al. 2009). Unfortunately, we know very little about Wolbachia infection on Neotropical termite fauna. We know that Cavitermes tuberosus, a termite species from Northern Brazil, is infected by Wolbachia belonging to supergroup F (Hellemans et al. 2019) and another species, Serritermes serrifer, from Central Brazil, is infected by an unidentified Wolbachia supergroup (Lo and Evans 2007). Wolbachia of supergroup F are already found in symbiosis with ~ 20 species of termites belonging to three different families and distributed worldwide (Lo and Evans 2007; Roy and Harry 2007; Salunke et al. 2010; Hellemans et al. 2019; Yashiro and Lo 2019). Besides, supergroup F Wolbachia are generally rare, with phylogenetic analyses suggesting a recent emergence of this supergroup through frequent horizontal transmission (Panaram and Marshall 2007; Lefoulon et al. 2012, 2016). Given the proximity between Neotroponiscus isopods and termites that can be infected by supergroup F, we hypothesize that the supergroup F found on Neotroponiscus littoralis was acquired by horizontal transmission from the termite nests in which they inhabit.
Considering this scenario, our goal is: (i) to investigate the presence of Wolbachia in other species of Neotroponiscus and test if the initial finding was an isolated event; (ii) to test for Wolbachia infection in the termites found in the Brazilian coastal forests; (iii) to test the phylogenetic and cophylogenetic relationships between the Wolbachia strains found in both taxa to ascertain if horizontal transmission is possible.
Materials and methods
Sample collection, DNA extraction and Wolbachia identification
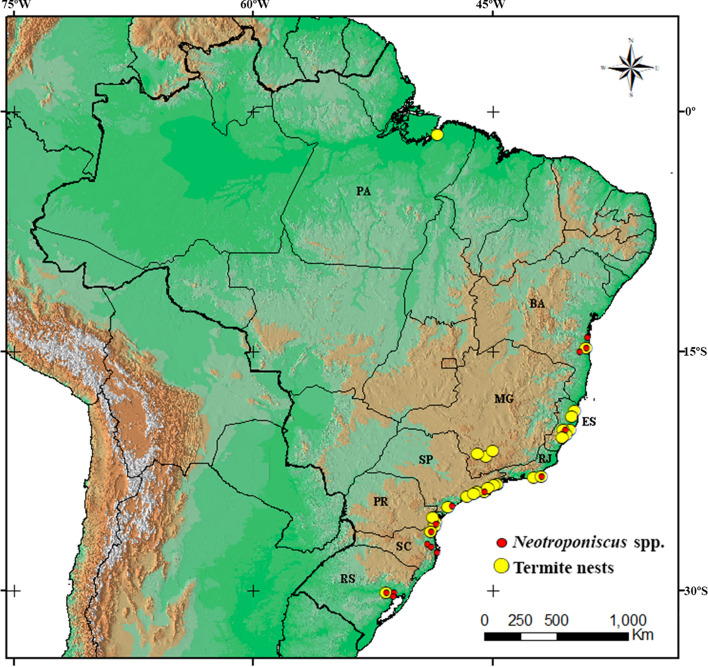
We sampled terrestrial isopod populations and termite nests in eight states along the Brazilian Atlantic Forest between the years 2013–2014 (Fig. 1) and stored in 100% ethanol at − 20 °C. We identified Neotroponiscus adults based on morphological criteria (Lemos de Castro 1970a, b). We also sampled the termites (soldiers and workers) and sent to an expert for identification. The number of sampled individuals of each species and the sampling sites are listed in Table 1.
Fig. 1.
Collection sites of Neotroponiscus isopods and termite nests examined for Wolbachia infection in Brazil
Table 1.
Terrestrial isopods and termites tested for Wolbachia infection and their collection sites
| Species | State | Municipality | Soldiers tested (infected) | Workers tested (infected) | Latitude | Longitude |
|---|---|---|---|---|---|---|
| Termites | ||||||
| Coptotermes sp. | Espirito Santo | Santa Tereza | 4 | 0 | − 19,9416 | − 40,5824 |
| Diversitermes castaniceps | São Paulo | Maresias | 1 | 6 | − 23,8247 | − 45,5286 |
| Diversitermes castaniceps | Espirito Santo | Sooterama | 5 | 2 | − 19,1164 | − 40,0733 |
| Diversitermes cf. castaniceps | São Paulo | Ubatuba | 14 | 0 | − 23,3542 | − 44,7652 |
| Microcerotermes sp. | Rio de Janeiro | Trindade | 0 | 7 | − 23,3299 | − 44,7057 |
| Nasutitermes cf. ephratae | Espirito Santo | Aracruz | 11 (5) | 3 | − 19,9434 | − 40,1628 |
| Nasutitermes cf. itapocuensis | Rio grande do Sul | Eldorado do Sul | 8 (3) | 6 (2) | − 30,1004 | − 51,6946 |
| Nasutitermes cf. jaraguae | São Paulo | Guarujá | 3 | 11 (1) | − 23,9066 | − 46,1639 |
| Nasutitermes corniger | Espirito Santo | São Mateus | 7 | 0 | − 18,7483 | − 39,8648 |
| Nasutitermes corniger | Bahia | Ilhéus | 10 | 6 | − 14,7977 | − 39,1727 |
| Nasutitermes gaigei | Pará | Belém | 3 | 11 | − 1,4537 | − 48,4768 |
| Nasutitermes jaraguae | Paraná | Matinhos | 0 | 7 | − 25,7060 | − 48,5720 |
| Nasutitermes jaraguae | São Paulo | Itamambuca | 8 | 6 | − 23,3822 | − 44,9660 |
| Nasutitermes jaraguae | São Paulo | Caraguatatuba | 0 | 14 | − 23,5702 | − 45,2971 |
| Nasutitermes jaraguae | Minas Gerais | Varginha | 7 | 7 | − 21,6226 | − 45,4305 |
| Nasutitermes jaraguae | Santa Catarina | Joinvile | 10 | 4 | − 26,3219 | − 48,8636 |
| Nasutitermes jaraguae | São Paulo | Bertioga | 14 (1) | 0 | − 23,7849 | − 46,0196 |
| Nasutitermes macrocephalus | Rio de Janeiro | Saquarema | 14 | 0 | − 22,9246 | − 42,4402 |
| Nasutitermes morphotype A | São Paulo | Iguapé | 9 | 5 | − 24,7636 | − 47,7859 |
| Nasutitermes morphotype A | Santa Catarina | Itapoá | 8 | 6 | − 25,9690 | − 48,6392 |
| Nasutitermes morphotype A | Paraná | Morretes | 10 | 4 | − 25,4135 | − 48,7714 |
| Nasutitermes morphotype A | São Paulo | Mongaguá | 3 | 4 | − 24,0844 | − 46,6103 |
| Nasutitermes sp. 1 | Espirito Santo | Cariacica | 9 | 5 | − 20,2415 | − 40,4173 |
| Nasutitermes sp. 2 | Rio de Janeiro | Cabo Frio | 3 (3) | 4 (3) | − 22,8671 | − 41,9850 |
| Neocapritermes opacus | Espirito Santo | Domingos Martins | 4 | 10 | − 20,3803 | − 40,6302 |
| Procornitermes araujoi | Minas Gerais | Alfenas | 4 (3) | 6 (5) | − 21,4250 | − 45,9460 |
| Procornitermes lespesii | Minas Gerais | Lavras | 7 | 7 (3) | − 21,2306 | − 44,9801 |
| Species | State | Municipality | Male tested (infected) | Female tested (infected) | Latitude | Longitude |
|---|---|---|---|---|---|---|
| Terrestrial isopods | ||||||
| Neotroponsicus argentinus | Bahia | Maraiúvis | 0 | 1 | − 14,1672 | − 39,0936 |
| Neotroponsicus argentinus | Paraná | Matinhos | 0 | 1 | − 25,8356 | − 48,5707 |
| Neotroponsicus argentinus | Santa Catarina | Joinville | 0 | 1 | − 26,3219 | − 48,8636 |
| Neotroponsicus argentinus | Santa Catarina | Florianópolis | 1 | 1 | − 27,6107 | − 48,5057 |
| Neotroponsicus argentinus | São Paulo | Iguape | 0 | 1 | − 24,7051 | − 47,5465 |
| Neotroponsicus carolii | Bahia | Itajú | 1 (1) | 0 | − 15,0328 | − 39,6042 |
| Neotroponsicus daguerri | Rio Grande do Sul | Eldorado do Sul | 2 | 9 | − 30,1004 | − 51,6946 |
| Neotroponsicus daguerri | Rio Grande do Sul | Porto Alegre | 1 | 1 | − 30,0967 | − 51,1860 |
| Neotroponsicus daguerri | Rio Grande do Sul | Barra do Ribeiro | 0 | 1 | − 30,3472 | − 51,2431 |
| Neotroponsicus lenkoi | São Paulo | Maresias | 0 | 1 | − 23,8247 | − 45,5286 |
| Neotroponsicus littoralis | Rio de Janeiro | Cabo Frio | 2 (2) | 3 (3) | − 22,8671 | − 41,9850 |
| Neotroponsicus littoralis | Bahia | Itabuna | 2 | 0 | − 14,7989 | − 39,1722 |
| Neotroponsicus perlatus | Espírito Santo | Santa Tereza | 1 | 1 | − 19,9347 | − 40,4705 |
| Neotroponsicus plaumani | Santa Catarina | Blumenau | 1 | 1 | − 27,0575 | − 49,0861 |
| Neotroponsicus plaumani | Santa Catarina | São João Batista | 0 | 1 | − 27,2644 | − 48,8533 |
Infected species are in bold
From these samples, total DNA was extracted with PureLink Genomic DNA Kit (Invitrogen/K1820-01). For detection of Wolbachia, three protein-coding genes were used: dnaA, coxA and fbpA (as previously described by Baldo et al. 2006a, b). We decided to use these genetic markers because they showed the best amplification results based on the study of Zimmerman et al. (2015), thus providing clear and unambiguous sequences. DNA from isopods and termites was also amplified with universal primers of the mitochondrial gene Cytochrome Oxidase I (COI) (Folmer et al. 1994). Polymerase chain reaction (PCR) products were electrophoresed on a 1.0% agarose gel and sequences were obtained using BigDye technology by Macrogen Inc., South Korea.
Phylogenetic analysis
A BLAST search at the NCBI (http://www.ncbi.nlm.nih.gov) was used to identify the Wolbachia supergroup of the infected individuals in our study. Once we confirmed that all belonged to supergroup F, we included in our alignments other sequences from supergroup F available in GenBank (www.ncbi.nlm.nih.gov/genbank/) and Wolbachia MLST database (pubmlst.org/wolbachia/). Wolbachia sequences from supergroups A, B, C, D and E were used as out-group (Table S1). The dnaA, coxA and fbpA sequences were aligned using Muscle (Edgar 2004), implemented in MEGA version 7.0 (Kumar et al. 2016). Recombination analyses were conducted on single and concatenated alignments using the RDP3 package (Martin et al. 2010) with default settings. Sequences were considered linear and the highest acceptable P value cutoff was 0.05. The best-fit models of nucleotide substitution for each gene were selected with JModeltest 2.1.10 (Darriba et al. 2012). This approach suggested the following models: HKY + G for coxA, HKY + I + G for fbpA and HKY + I + G for dnaA. Phylogenetic reconstructions of Wolbachia were performed with Bayesian inference in BEAST version 1.8 (Drummond et al. 2012) using concatenated (coxA + fbpA and dnaA + coxA + fbpA) genes sequences. We used coxA + fbpA and dnaA + coxA + fbpA reconstructions to assess the robustness of the reconstructions because there are few dnaA sequences available on databases. Four simultaneous Markov chains were run for 50 million generations with a sampling frequency of 1,000 generations and an initial burn-in of 10%. Concatenated alignments were run with the alignments partitioned so that the specific model of evolution corresponded to each gene fragment. The runs were checked for convergence using Tracer version 1.6 (Rambaut et al., 2014). The results were visualized and checked with FigTree 1.4.2 (Rambaut 2014). All dnaA, coxA, fbpA and COI gene sequences generated in this study were deposited into GenBank under accession numbers KX024833 to KX024843 for coxA, KX036766 to KX036775 for fbpA, KX036776 to KX036786 for COI and KX036787 to KX036796 for dnaA.
Comparative phylogenetic analysis
For the cophylogenetic analysis, we generated a Wolbachia tree of representative strains from A, B, C, D, E and F supergroups using coxA, dnaA and fbpA sequences available in GenBank (Table S1) and the sequences from the present study. The closest relative taxa to Wolbachia, Anaplasma marginale and Ehrlichia canis, were used as outgroups. Similarly, host phylogeny was reconstructed using the corresponding COI mitochondrial sequences of host taxa (Table S2). To visualize host-bacteria associations, tanglegrams were generated in TreeMap 3.0, using the default settings (Charleston and Robertson 2002). Since we were interested in both the evolutionary history of the infection (i.e., who infected whom), and whether the phylogenies of hosts and Wolbachia were congruent, we used both event-based methods (for the reconstruction of phylogenies) and global fit (for congruency tests). Statistical congruence between host and Wolbachia phylogenies was performed using both the Procrustean Approach to Cophylogeny (PACo) program (Balbuena et al. 2013) and the ParaFit function (Legendre et al. 2002) from the ape R package (Paradis and Schliep 2019). The significance of the global goodness of fit of PACo and the Parafit global test was estimated using 10,000 permutations of the association matrix. The level of statistical significance of cospeciation was determined using reconciliation analyses implemented in the program JANE 4.0 (Conow et al. 2010). As recommended by Conow et al. (2010), the genetic algorithm parameters were set at the default values for selection strength and mutation rate, and number of generations. We thus performed analyses with 100 generations, population sizes of 100, and a default cost setting matrix of 0 for cospeciation, 1 for duplication of parasites, 2 for duplication and host switch, 1 for loss of parasite, and 1 for failure to diverge. In a conservative manner, we minimized the total cost results by maximizing the cospeciation events while allowing host switching.
Results
Infection detections
Thirty-six terrestrial isopods belonging to six species of Neotroponiscus were analyzed (Fig. 2). Besides the already known infected population of N. littoralis from Northeast Brazil, the presence of Wolbachia was also observed in individuals of this same species sampled in Southeast Brazil. We also detected Wolbachia infection in another species, N. carolii also sampled in Northeast Brazil (Table 1) representing the first Wolbachia infection record for this species.
Fig. 2.
Representatives of terrestrial isopods and termite species infected by Wolbachia strains of supergroup F. a Nasutitermes itapocuensis; b Procornitermes araujoi; c Neotroponiscus littoralis; d Neotroponicus carolii
Regarding the termites, 317 individuals belonging to 17 species were analyzed, which represents the largest survey of Wolbachia infection in Neotropical termites. Seven species from two genera, Nasutitermes and Procornitermes (Fig. 2), sampled in five Brazilian states were Wolbachia positive. The infection status of each species and the number of individuals screened are listed in Table 1. This is the first time that the infection is recorded in the termite species studied here.
Wolbachia identification
All strains found both in isopods and termites belong to the supergroup F. Two strains were found in the terrestrial isopods, one on each Neotroponiscus species. Both strains clustered together in the phylogenies (Figs. 3 and 4). In termites, two strains of the supergroup F were found in Procornitermes (one in P. araujoi and one in P. lespesii). On the other hand, six strains were found in Nasutitermes: two in N. cf. ephratae and one on each of the following taxa: N. cf. itapocuensis, N. jaraguae, N. cf. jaraguae and Nasutitermes sp. As in the case of terrestrial isopods, Procornitermes and Nasutitermes sequences were very similar despite the large geographical distance among the sample locations (Figs. 3 and 4).
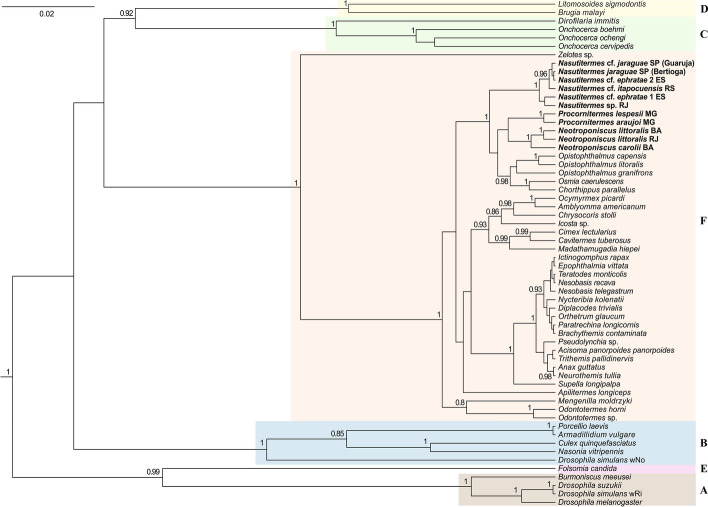
Fig. 3.
Unrooted phylogeny of Wolbachia based on concatenated datasets of coxA and fbpA sequences. Numbers above branches represent Bayesian posterior probabilities based on one run of 50 million generations. Posterior probabilities < 0.75 are not shown. Sequences were obtained from GenBank and MLST Wolbachia databases except those from species in bold. Letters represent Wolbachia supergroups A, B, C, D, E and F
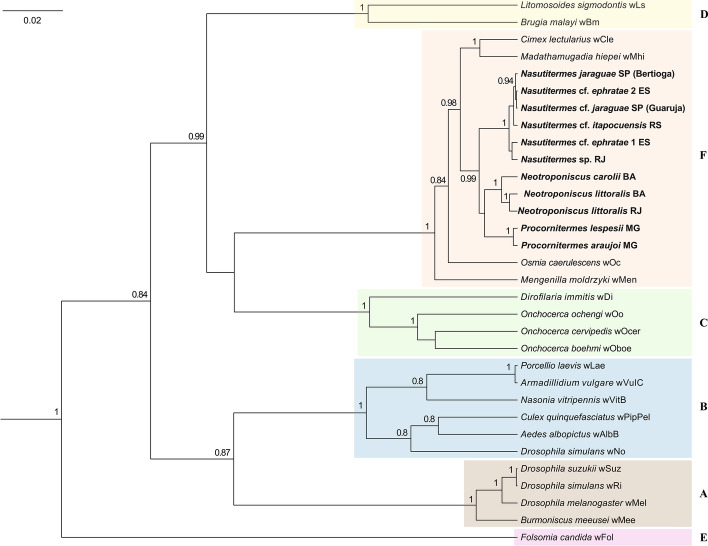
Fig. 4.
Unrooted phylogeny of Wolbachia based on concatenated datasets of coxA, dnaA and fbpA sequences. Numbers above branches represent Bayesian posterior probabilities based on one run of 50 million generations. Posterior probabilities < 0.75 are not shown. Sequences were obtained from GenBank and MLST Wolbachia databases except those from species in bold. Letters represent Wolbachia supergroups A, B, C, D, E and F
Phylogenetic correlation between isopods and termites
The Wolbachia strains found in terrestrial isopods are closely related to those found in termites, regardless of how the phylogeny was generated. Wolbachia sequences found in termites and terrestrial isopods were always grouped in the same clade regardless of the number of host species and sequences used to build the tree (i.e., fbpA + coxA or dnaA + fbpA + coxA sequences; Figs. 3 and 4), which denotes the robustness of the reconstructed phylogenies.
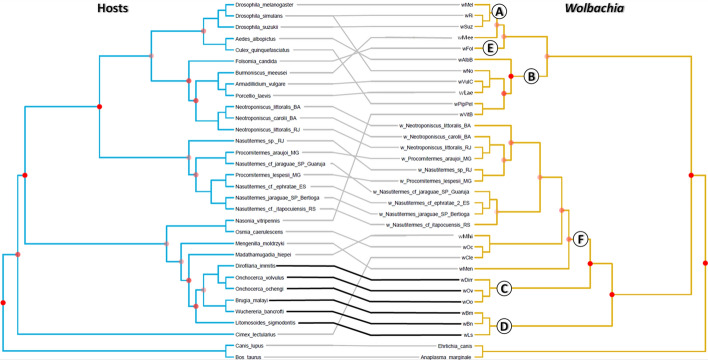
Moreover, the phylogenetic comparison of Wolbachia and their hosts showed no congruence in a substantial part of the tanglegram (Fig. 5). Overall, both ParaFit and PACo analyses provided no evidence for significant coevolution between Wolbachia and hosts (ParaFitGlobal = 76.06896, P = 0.3425657; m2 global value = 187.3399, P = 0.1905; 10,000 permutations). Apart from the outgroups, only six of the 33 individual host-bacteria links are significant (based on ParaFit1 or Parafit2, P ≤ 0.05). These six links are between Wolbachia supergroups C and D and their long-lasting associations with filarial nematodes. As can be seen in Fig. S1, these links contributed with most of the cophylogenetic structure (i.e., high squared residuals; Fig. S1). On the other hand, the less contributing host-Wolbachia associations to the cophylogenetic structure are those belonging to supergroups A, B and F. Indeed, terrestrial isopods and termites with their respective Wolbachia strains were among the associations that contributed least to the phylogenetic congruence between hosts and bacteria.
Fig. 5.
Tanglegram of cophylogenetic relationships between hosts (left) and Wolbachia strains (right). Maximum likelihood phylogenies for Wolbachia bacteria (yellow) and their hosts (blue) rooted with A. marginale and E. canis as outgroups. All host-symbiont associations are shown in the tanglegram as grey and black connecting lines. Black lines indicate significant individual cospeciation links between Wolbachia and the host as indicated by ParaFit (P ≤ 0.01), while gray lines represent non-significant links. Significance of the congruence of the nodes to their counterpart subtree in the other tree as indicated by the Treemap z-scores are indicated by red dots. The opacity of the dot is related to the significance of the test: opaque dot for P < 0.001; 50% opacity for P ≤ 0.01, and 25% opacity for P ≤ 0.05. A, B, C, D, E and F Wolbachia supergroups are indicated by circled letters. The following codes represent the Wolbachia strains according to the host species: wAlbB = Aedes albopictus, wBm = Brugia malayi, wBn = Wuchereria bancrofti, wCle = Cimex lectularius, wDim = Dirofilaria immitis, wFol = Folsomia candida, wLae = Porcellio laevis, wLs = Litomosoides sigmodontis, wMee = Burmoniscus meeusei, wMel = Drosophila melanogaster, wMen = Mengenilla moldrzyki, wMhi = Madathamugadia hiepei, wNo = Drosophila simulans, wOc = Osmia caerulescens, wOo = Onchocerca ochengi, wOv = Onchocerca volvulus, wPipPel = Culex quinquefasciatus, wRi = Drosophila simulans, wSuz = Drosophila suzukii, wVitB = Nasonia vitripennis, wVulC = Armadillidium vulgare
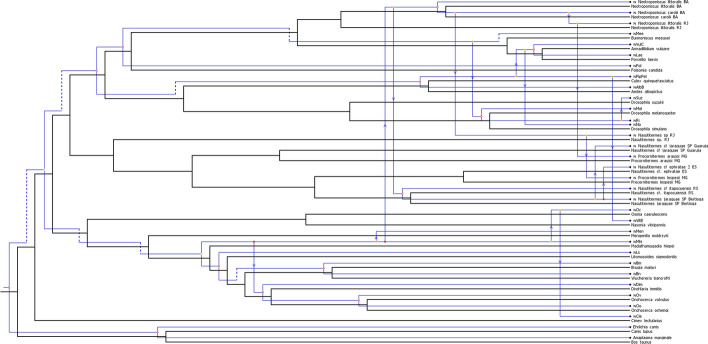
The events-based method suggested significant horizontal transfers between termites and isopods when reconstructing the evolutionary history of the phylogeny. In the Treemap analysis, 22 optimal reconstructions were found. The optimal solutions postulated 22 cospeciations, 24 losses, 18 host switches, and 25 duplication events. However, the lower cost solutions were found in the JANE 4.0 analysis with 115 isomorphic solutions for a total cost value of 42. The optimal solutions resulted in 15 cospeciations, 8 losses, 17 host switches, and no duplication events (Fig. 6). Accordingly, most of the host switches (Wolbachia horizontal transmissions) occurred in the F supergroup. At the highest phylogenetic level, we found host switches between isopods and termites: between N. carolii and the termite Nasutitermes sp. RJ, and between N. littoralis and P. araujoi. At a lower phylogenetic level, we found also several transfers within termites and within isopods as well (Fig. 6).
Fig. 6.
Optimal cophylogeny reconstruction of hosts and Wolbachia strains produced by JANE 4.0. One of 115 isomorphic solutions with 15 cospeciations, 17 host switches, and eight losses (total cost = 42). Blue and black lines indicate the phylogenies of Wolbachia and hosts, respectively. Empty circles represent cospeciations; arrows represent host switches; and dashed lines represent loss events. See Fig. 5 legend for abbreviation of Wolbachia strains
Discussion
Horizontal transmission events are supposed to be recurrent in Wolbachia. However, the probability of successful likely is highest between closely related hosts and decreases with increasing phylogenetic distance (Zug and Hammerstein 2012). Thus, horizontal transmission of Wolbachia between distantly related arthropods is relatively rare. This study presents evidence for the horizontal transmission between unrelated hosts involving strains of supergroup F, an atypical supergroup of Wolbachia that infects a large variety of arthropods hosts and nematodes (Casiraghi et al. 2005). In this study, we corroborate our hypothesis that the unexpected discovery of supergroup F Wolbachia in Neotropical terrestrial isopods could be the result of horizontal transmission from termites. Interestingly, Wolbachia strains found in Neotroponiscus isopods and termite species clustered together in phylogenies regardless of the relatedness of the host taxa. We also detected horizontal transfers between isopods and termites when reconstructing the evolutionary history of Wolbachia, confirming that the supergroup F Wolbachia and host taxa phylogenies did not have an underlying structure resembling coevolution. Thus, our results suggest that a horizontal transmission of supergroup F Wolbachia endosymbionts between termites and terrestrial isopods is likely.
The Wolbachia strains found in the termites Nasutitermes and Procornitermes are phylogenetically close to each other. Nasutitermes is the most common and abundant of all termite genera in Brazil. They usually build arboreal nests and are abundant in the same areas where Neotroponiscus occurs (Thorne et al. 1996; Constantino and Acioli 2006; Souza et al. 2012). Procornitermes, on the other hand, is a poorly studied genus composed of species that build subterranean and epigean nests (Cancello 1986; Moreira et al. 2018). However, Procornitermes can occur with Nasutitermes spp. in the soil and under wood material over the soil (Amaral-Castro et al. 2004). When we add the close phylogenetic relationship between the termites, it is not surprising that the Wolbachia strains are closely related. Indeed, Wolbachia is pre-adapted to infect closely related hosts due to their similar physiologies (Russell et al. 2009).
The horizontal transmission reported here likely occurred from micro-habitat sharing. Horizontal transmission of Wolbachia involves species living in close association, such as being connected via shared physical space or belonging to similar trophic levels (Stahlhut et al. 2010). These two prerequisites are met by termites and isopods and can explain how Wolbachia jumped hosts. First, Neotroponiscus species are frequently found in decaying wood and tree barks, where termites build arboreal nests, especially Nasutitermes (Boulogne et al. 2017). In fact, N. carolii isopods were already found occupying abandoned arboreal nests of Nasutitermes spp. termites (Lisboa et al. 2013). Such sites would be ideal for terrestrial isopods as they are rich in organic matter and provide a moist and shaded shelter (Costa et al. 2009). Therefore, it is relatively easy for the isopods to find the termite nests in these environments.
Second, terrestrial isopods and termites occupy the same ecological guild. Terrestrial isopods feed on litter, decomposing tree trunks, and nearly all kind of detritus which makes them keystone decomposers in many terrestrial ecosystems (Hornung 2011). It has been recently demonstrated a close cooperation between microbiota and terrestrial isopods for the lignocellulose degradation (Bredon et al. 2018, 2019, 2020). Similarly, due to the symbiotic organisms in the termites’ hindgut, termites are also important cellulose decomposers wherever they inhabit (Hongoh 2010; Brune 2014). Thus, both groups are decomposers of plant-derived organic matter. Being of the same guild (and having relatively similar roles) might thus account for the high bacterial diversity found in these animals, including the presence of Wolbachia (Dittmer et al. 2016; Diouf et al. 2018). Additionally, occupying similar ecological guilds can also make the gut environment relatively similar, facilitating the horizontal transmission of Wolbachia.
Although terrestrial isopods are primarily herbivores, they are occasionally found eating non-plant items which suggest a possible route of transmission. Although rarely, terrestrial isopods can act as predators, preying on several kinds of small living invertebrates (Edney et al. 1974), including eggs and their own young (Warburg 1994). The isopods that prey become prone to be infected by the Wolbachia bacteria found on their food items (Le Clec’h et al. 2013). It is unlikely that the small-sized Neotroponiscus prey on soldiers or worker termites, but they could prey on termite eggs or larvae. In fact, Wolbachia is the most abundant bacterial group in the early stages of life—particularly the eggs—of Nasutitermes arborum termites (Diouf et al. 2018), which could increase the likelihood of horizontal transmission. If we add that termites occur in high densities (e.g., nests of the arboreal termite N. corniger contain up to 800,000 individuals Thorne and Noirot 1982), it is thus possible that isopods consume anything left by the termites when abandoning the nest, being of decaying eggs or larvae, which increases the likelihood of horizontal transmission.
Supergroup F strains are known for being geographically and phylogenetically widespread among host taxa (Ros et al. 2009). However, they are also a relatively uncommon strain of Wolbachia. Thus, finding that the first supergroup F strain in isopods was not an isolated event already suggested that the evolutionary history of Wolbachia supergroup F is more complex. Indeed, supergroup F was also found in several species of termites, including other species of Nasutitermes termites that occur in distant regions, such as Japan, Malaysia, Australia, and Venezuela (Lo and Evans 2007). It was thus rather unsurprising when we found that Wolbachia strains found in scorpions, bees, and locusts nested within the monophyletic group of termites and isopods Wolbachia (Fig. 3). The horizontal transmission of Wolbachia might thus be more widespread in arthropods than we initially thought. Unfortunately, tests on how the supergroup F can transfer horizontally are still scarce (not to mention inexistent). Our study was the first to assess if horizontal transmission between isopods and termites was possible, but we still lack proper investigation. We are unsure why the scorpions, bees, and locusts that do not inhabit South America were included within the monophyletic group of South American strains. But, but the same pattern has been found in other studies, in which strains of supergroup F from hosts occurring in distinct continents grouped together (Baldo et al. 2007; Lefoulon et al. 2012). It thus seems a promising venue for further research.
The evolution of endosymbiotic bacteria such as Wolbachia is shaped by the ecology and evolution of many arthropods (Hellemans et al. 2019). Despite the importance of horizontal transmission for Wolbachia, the routes of transmission are not yet fully known (Gerth et al. 2013). Our study is one of the few that provides evidence for horizontal transmission based on in situ studies—especially on such distant related taxa. Our results suggest that the horizontal transmission of Wolbachia strains of supergroup F between termites and terrestrial isopods occurred. We hope more studies will be carried out to clarify the mechanisms governing horizontal transmission of Wolbachia and allow the understanding of the success and worldwide distribution of this symbiont.
Supplementary information
Below is the link to the supplementary information.
Acknowledgements
We are sincerely grateful to Dr. Reginaldo Constantino from Universidade de Brasília for the esteemed support in the identification of termites. CAPES for the scholarships granted to G.M.C. (PDSE 201713/2014-4) and to B.L.Z. CNPq for productivity fellowship Granted to P.B.A. (305900/2014-5) and to Laboratory of Molecular Biology, Departamento de Zoologia, UFRGS (Process Number 479412/2008-1). License Number: 40181-1.
Author contributions
BLZ, GMC and PBA conceived the idea and designed methodology. GMC collected the samples. BLZ and PHP generated the molecular data. BLZ and DB analyzed the molecular data. BLZ and AVP led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Data availability
All gene sequences were submitted to NCBI database (https://www.ncbi.nlm.nih.gov/genbank/) under Accession Numbers KX024833 to KX024843, KX036766 to KX036775, KX036776 to KX036786 and KX036787 to KX036796.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmed MZ, Li S-J, Xue X, Yin XJ, Ren SX, Jiggins FM, Greef JM, Qiu BL. The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLOS Pathog. 2015;11:e1004672. doi: 10.1371/journal.ppat.1004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almerão MP, Fagundes NJR, Araújo PB, Verne S, Grandjean F, Bouchon D, Araújo AM. First record of Wolbachia in South American terrestrial isopods: prevalence and diversity in two species of Balloniscus (Crustacea, Oniscidea) Genet Mol Biol. 2012;35:980–989. doi: 10.1590/S1415-47572012000600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Castro NR, Zanetti R, Moraes JC, Zanuncio JC, Freitas GD, Santos MS. Species of soil inhabiting termites (Insecta: Isoptera) collected in Eucalyptus plantations in the state of Minas Gerais, Brazil. Sociobiology. 2004;44:717–726. [Google Scholar]
- Bailly-Bechet M, Martins-Simões P, Szöllősi GJ, Mialdea G, Sagot MF, Charlat S (2017) How long does Wolbachia remain on board?. Mol Biol Evol 34:1183–1193 https://academic.oup.com/mbe/article/34/5/1183/2992913 [DOI] [PubMed]
- Balbuena JA, Míguez-Lozano R, Blasco-Costa I. PACo: a novel procrustes application to cophylogenetic analysis. PLoS ONE. 2013;8:e61048. doi: 10.1371/journal.pone.0061048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Prendini L, Corthals A, Werren JH. Wolbachia are present in southern african scorpions and cluster with supergroup F. Curr Microbiol. 2007;55:367–373. doi: 10.1007/s00284-007-9009-4. [DOI] [PubMed] [Google Scholar]
- Bandi C, Anderson TJC, Genchi C, Blaxter ML. Phylogeny of Wolbachia in filarial nematodes. Proc R Soc London Ser B Biol Sci. 1998;265:2407–2413. doi: 10.1098/rspb.1998.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Afia Hatira H, Charfi-Cheikhrouha F, Bouchon D (2008) Wolbachia in terrestrial isopods in Tunisia. In: Zimmer M, Charfi-Cheikhrouha FST (ed) Proceedings of the international symposium on terrestrial isopod biology. Shaker Verlag, Aachen, pp 117–24
- Bouchon D, Rigaud T, Juchault P. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc R Soc London Ser B Biol Sci. 1998;265:1081–1090. doi: 10.1098/rspb.1998.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon D, Cordaux R, Grève P (2008) Feminizing Wolbachia and the evolution of sex determination in isopods. In: Bourtzis K, Miller T (eds) Insect symbiosis, vol 3. Taylor and Francis Group, Boca Raton, pp 273–294. 10.1201/9781420064117.ch12
- Boulogne I, Constantino R, Amusant N, Falkowski M, Rodrigues AM. Houël E (2017) Ecology of termites from the genus Nasutitermes (Termitidae: Nasutitermitinae) and potential for science-based development of sustainable pest management programs. J Pest Sci. 2017;90:19–37. doi: 10.1007/s10340-016-0796-x. [DOI] [Google Scholar]
- Bredon M, Dittmer J, Noël C, Moumen B, Bouchon D. Lignocellulose degradation at the holobiont level: teamwork in a keystone soil invertebrate. Microbiome. 2018;6:162. doi: 10.1186/s40168-018-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredon M, Herran B, Lheraud B, Bertaux J, Grève P, Moumen B, Bouchon D. Lignocellulose degradation in isopods: new insights into the adaptation to terrestrial life. BMC Genom. 2019;20:462. doi: 10.1186/s12864-019-5825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredon M, Herran B, Bertaux J, Grève P, Moumen B, Bouchon D. Isopod holobionts as promising models for lignocellulose degradation. Biotechnol Biofuels. 2020;13:49. doi: 10.1186/s13068-020-01683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AN, Lloyd VK. Evidence for horizontal transfer of Wolbachia by a Drosophila mite. Exp Appl Acarol. 2015;66:301–311. doi: 10.1007/s10493-015-9918-z. [DOI] [PubMed] [Google Scholar]
- Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- Cancello EM. Revisão de Procornitermes Emerson (Isoptera, Termitidae, Nasutitermitinae) Papéis Avulsos Zool (São Paulo) 1986;36:189–236. [Google Scholar]
- Cardoso GM, Araujo PB, Bichuette ME. Two new species of Neotroponiscus Arcangeli, 1936 (Crustacea, Isopoda, Oniscidea) from Brazilian caves. Stud Neotrop Fauna Environ. 2017;52:122–130. doi: 10.1080/01650521.2017.1299440. [DOI] [Google Scholar]
- Casiraghi M, Bordenstein SR, Baldo L, Lo N, Beninati T, Wernegreen JJ, Werren JH, Bandi C. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology. 2005;151:4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- Charleston MA, Robertson DL. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst Biol. 2002;51:528–535. doi: 10.1080/10635150290069940. [DOI] [PubMed] [Google Scholar]
- Conow C, Fielder D, Ovadia Y, Libeskind-Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino R, Acioli ANS. Termite diversity in Brazil (Insecta: Isoptera) In: Moreira F, Siqueira JO, Brussaard L, editors. Soil biodiversity in Amazonian and other Brazilian ecosystems. Wallingford: CAB International; 2006. pp. 117–128. [Google Scholar]
- Cordaux R, Pichon S, Ben Afia Hatira H, Doublet V, Grève P, Marcadé I, Braquart-Varnier C, Souty-Grosset C, Charfi-Cheikhrouha F, Bouchon D. Widespread Wolbachia infection in terrestrial isopods and other crustaceans. Zookeys. 2012;176:123–131. doi: 10.3897/zookeys.176.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa DA, de Carvalho RA, de Lima Filho GF, Brandao D. Inquilines and invertebrate fauna associated with termite nests of Cornitermes cumulans (Isoptera, Termitidae) in the Emas National Park, Mineiros, Goiás. Brazil Sociobiology. 2009;53:443. [Google Scholar]
- Covacin C, Barker SC. Supergroup F Wolbachia bacteria parasitise lice (Insecta: Phthiraptera) Parasitol Res. 2006;100:479–485. doi: 10.1007/s00436-006-0309-6. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diouf M, Miambi E, Mora P, Frechault S, Robert A, Rouland-Lefèvre C, Hervé V. Variations in the relative abundance of Wolbachia in the gut of Nasutitermes arborum across life stages and castes. FEMS Microbiol Lett. 2018;365:fny046. doi: 10.1093/femsle/fny046. [DOI] [PubMed] [Google Scholar]
- Dittmer J, Lesobre J, Moumen B, Bouchon D. Host origin and tissue microhabitat shaping the microbiota of the terrestrial isopod Armadillidium vulgare. FEMS Microbiol Ecol. 2016;92:fiw063. doi: 10.1093/femsec/fiw063. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edney EB, Allen W, McFarlane J. Predation by terrestrial isopods. Ecology. 1974;55:428–433. doi: 10.2307/1935231. [DOI] [Google Scholar]
- Faria VG, Paulo TF, Sucena E. Testing cannibalism as a mechanism for horizontal transmission of Wolbachia in Drosophila. Symbiosis. 2016;68:79–85. doi: 10.1007/s13199-015-0354-y. [DOI] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Frydman HM, Li JM, Robson DN, Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature. 2006;441:509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- Gehrer L, Vorburger C. Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol Lett. 2012;8:613–615. doi: 10.1098/rsbl.2012.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth M, Röthe J, Bleidorn C. Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using multilocus sequence typing data and host phylogeny. Mol Ecol. 2013;22:6149–6162. doi: 10.1111/mec.12549. [DOI] [PubMed] [Google Scholar]
- Gerth M, Gansauge MT, Weigert A, Bleidorn C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat Commun. 2014;5:5117. doi: 10.1038/ncomms6117. [DOI] [PubMed] [Google Scholar]
- Hellemans S, Kaczmarek N, Marynowska M, Calusinska M, Roisin Y, Fournier D. Bacteriome-associated Wolbachia of the parthenogenetic termite Cavitermes tuberosus. FEMS Microbiol Ecol. 2019;95:fiy235. doi: 10.1093/femsec/fiy235. [DOI] [PubMed] [Google Scholar]
- Hongoh Y. Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci Biotechnol Biochem. 2010;74:1145–1151. doi: 10.1271/bbb.100094. [DOI] [PubMed] [Google Scholar]
- Hornung E. Evolutionary adaptation of oniscidean isopods to terrestrial life: structure, physiology and behavior. Terr Arthropod Rev. 2011;4:95–130. doi: 10.1163/187498311X576262. [DOI] [Google Scholar]
- Juchault P, Frelon M, Bouchon D, Rigaud T. New evidence for feminizing bacteria in terrestrial isopods: evolutionary implications. Comptes rendus l’Académie des Sci. 1994;317:225–230. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clec’h W, Chevalier FD, Genty L, Bertaux J, Bouchon D, Sicard M. Cannibalism and predation as paths for horizontal passage of Wolbachia between terrestrial isopods. PLoS ONE. 2013;8:e60232. doi: 10.1371/journal.pone.0060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefoulon E, Gavotte L, Junker K, Barbuto M, Uni S, Landmann F, Laaksonen S, Saari S, Nikander S, Lima SS, Casiraghi M, Bain O, Martin C. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int J Parasitol. 2012;42:1025–1036. doi: 10.1016/j.ijpara.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Lefoulon E, Bain O, Makepeace BL, d’Haese C, Uni S, Martin C, Gavotte L. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ. 2016;4:e1840. doi: 10.7717/peerj.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Desdevises Y, Bazin E. A statistical test for host-parasite coevolution. Syst Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- Lemos de Castro A. Quatro espécies novas de isópodos terrestres do gênero Neotroponiscus Arcangeli (Oniscidae, Bathytropinae) do Brasil. Bol do Mus Nac. 1970;275:1–15. [Google Scholar]
- Lemos de Castro A (1970a) Isópodos terrestres do gênero Neotroponiscus Arcangeli (Oniscidae: Bathytropinae). In: Anais da Academia Brasileira de Ciências, pp 89–95
- Lenko K. Subsídios para o conhecimento dos isópodos inquilinos de formigas no Brasil (Isopoda, Oniscoidea) Rev Bras Entomol. 1971;15:1–10. [Google Scholar]
- Li SJ, Ahmed MZ, Lv N, Shi PQ, Wang XM, Huang JL, Qiu BL. Plantmediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017;11:1019–1028. doi: 10.1038/ismej.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa JT, Couto ECG, Santos PP, Delabie JHC, Araujo PB. Terrestrial isopods (Crustacea: Isopoda: Oniscidea) in termite nests (Blattodea: Termitidae) in a cocoa plantation in Brazil. Biota Neotrop. 2013;13:393–397. doi: 10.1590/S1676-06032013000300039. [DOI] [Google Scholar]
- Lo N, Evans TA. Phylogenetic diversity of the intracellular symbiont Wolbachia in termites. Mol Phylogenet Evol. 2007;44:461–466. doi: 10.1016/j.ympev.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira EA, Alvarez TM, Persinoti GF, Paixão DAA, Menezes LR, Cairo JPF, Squina FM, Costa-Leonardo AM, Carrijo T, Arab A. Microbial Communities of the gut and nest of the humus- and litter-feeding termite Procornitermes araujoi (Syntermitinae) Curr Microbiol. 2018;75:1609–1618. doi: 10.1007/s00284-018-1567-0. [DOI] [PubMed] [Google Scholar]
- Nyirõ G, Oravecz O, Márialigeti K. Detection of Wolbachia pipientis infection in arthropods in Hungary. Eur J Soil Biol. 2002;38:63–66. doi: 10.1016/S1164-5563(01)01127-X. [DOI] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Panaram K, Marshall JL. F supergroup Wolbachia in bush crickets: what do patterns of sequence variation reveal about this supergroup and horizontal transfer between nematodes and arthropods? Genetica. 2007;130:53–60. doi: 10.1007/s10709-006-0020-7. [DOI] [PubMed] [Google Scholar]
- Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Qi LD, Sun JT, Hong XY, Li YX. Diversity and phylogenetic analyses reveal horizontal transmission of endosymbionts between whiteflies and their parasitoids. J Econ Entomol. 2019;112:894–905. doi: 10.1093/jee/toy367. [DOI] [PubMed] [Google Scholar]
- Rambaut A (2014) Figtree version 1.4.2. In: http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Drummond AJ, Suchard M (2014) Tracer v1. 6. In: http://beast.bio.ed.ac.uk
- Richardson A, Araujo PB (2015) Lifestyles of terrestrial crustaceans. In: Thiel M, Watling L (eds) The natural history of the Crustacea: lifestyles and feeding biology, vol 2. Oxford University Press, Oxford, pp 299–336
- Rigaud T, Juchault P. Success and failure of horizontal transfers of feminizing Wolbachia endosymbionts in woodlice. J Evol Biol. 1995;8:249–255. doi: 10.1046/j.1420-9101.1995.8020249.x. [DOI] [Google Scholar]
- Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse Is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia Supergroup recovered from spider mites (Acari: Tetranychidae) Appl Environ Microbiol. 2009;75:1036–1043. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M. Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc R Soc London Ser B Biol Sci. 1992;250:91–98. doi: 10.1098/rspb.1992.0135. [DOI] [PubMed] [Google Scholar]
- Roy V, Harry M. Diversity of Wolbachia isolated from the Cubitermes sp. affinis subarquatus complex of species (Termitidae), revealed by multigene phylogenies. FEMS Microbiol Lett. 2007;274:102–111. doi: 10.1111/j.1574-6968.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, Werren JH, Pierce NE. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution. 2009;63:624–640. doi: 10.1111/j.1558-5646.2008.00579.x. [DOI] [PubMed] [Google Scholar]
- Salunke BK, Salunkhe RC, Dhotre DP, Khandagale AB, Walujkar SA, Kirwale GS, Ghate HV, Patole MS, Shouche YS. Diversity of Wolbachia in Odontotermes spp. (Termitidae) and Coptotermes heimi (Rhinotermitidae) using the multigene approach. FEMS Microbiol Lett. 2010;307:55–64. doi: 10.1111/j.1574-6968.2010.01960.x. [DOI] [PubMed] [Google Scholar]
- Sintupachee S, Milne JR, Poonchaisri S, Baimai V, Kittayapong P. Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb Ecol. 2006;51:294–301. doi: 10.1007/s00248-006-9036-x. [DOI] [PubMed] [Google Scholar]
- Souza HBA, Alves WF, Vasconcellos A. Termite assemblages in five semideciduous Atlantic Forest fragments in the northern coastland limit of the biome. Rev Bras Entomol. 2012;56:67–72. doi: 10.1590/S0085-56262012005000013. [DOI] [Google Scholar]
- Stahlhut JK, Desjardin CA, Clark ME, Baldo L, Russell JA, Werren JH, Jaenike J. The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol Ecol. 2010;19:1940–1952. doi: 10.1111/j.1365-294X.2010.04572.x. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Voronin D, Johnston KL, Ford L. Wolbachia filarial interactions. Cell Microbiol. 2013;15:520–526. doi: 10.1111/cmi.12084. [DOI] [PubMed] [Google Scholar]
- Thorne BL, Noirot C. Ergatoid reproductives in Nasutitermes corniger (Motschulsky) (Isoptera: Termitidae) Int J Insect Morphol Embryol. 1982;11:213–226. doi: 10.1016/S0020-7322(82)80006-8. [DOI] [Google Scholar]
- Thorne BL, Collins MS, Bjorndal KA. Architecture and nutrient analysis of arboreal carton nests of two neotropical Nasutitermes species (Isoptera: Termitidae), with notes on embedded nodules. Florida Entomol. 1996;79:27–37. doi: 10.2307/3495751. [DOI] [Google Scholar]
- Warburg MR. Marsupial contents and losses due to putative intramarsupial cannibalism by the mancas in three oniscid isopod species. J Crustac Biol. 1994;14:560–567. doi: 10.1163/193724094X00128. [DOI] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc London Ser B Biol Sci. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- White PM, Pietri JE, Debec A, Russell S, Patel B, Sullivan W. Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl Environ Microbiol. 2017;83:e03425–e3516. doi: 10.1128/AEM.03425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwatanaratanabutr I, Kittayapong P, Caubet Y, Bouchon D. Molecular phylogeny of Wolbachia strains in arthropod hosts based on groE-homologous gene sequences. Zool Sci. 2009;26:171–177. doi: 10.2108/zsj.26.171. [DOI] [PubMed] [Google Scholar]
- Yashiro T, Lo N. Comparative screening of endosymbiotic bacteria associated with the asexual and sexual lineages of the termite Glyptotermes nakajimai. Commun Integr Biol. 2019;12:55–58. doi: 10.1080/19420889.2019.1592418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Peng Y, Liu FX, Lei C. Wolbachia screening in spiders and assessment of horizontal transmission between predator and prey. Neotrop Entomol. 2011;40:164–169. doi: 10.1590/S1519-566X2011000200002. [DOI] [PubMed] [Google Scholar]
- Zimmermann BL, Bouchon D, Almerão MP, Araujo PB. Wolbachia in Neotropical terrestrial isopods. FEMS Microbiol Ecol. 2015;91:fiv025. doi: 10.1093/femsec/fiv025. [DOI] [PubMed] [Google Scholar]
- Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene sequences were submitted to NCBI database (https://www.ncbi.nlm.nih.gov/genbank/) under Accession Numbers KX024833 to KX024843, KX036766 to KX036775, KX036776 to KX036786 and KX036787 to KX036796.