Introduction
Dramatic improvements in clinical outcomes have been seen for children and adolescents with cancer over the past five decades.1 The improvement in survival is attributed primarily to risk-stratification of therapies and treatment intensification with cytotoxic chemotherapy and multi-modal approaches. However, accelerating the progress of pediatric oncology requires both therapeutic advances and attention to diminishing the late effects of standard cytotoxic therapies. The ultimate goal of precision medicine in pediatric oncology is to develop more effective and less toxic therapies in children, adolescents, and young adults with cancer. With the advancement in diagnostic and molecular profiling technologies, precision medicine trials utilizing clinical molecular testing are becoming more common for adult malignancies. Similarly, there is an interest in how these technologies can be applied to tumors in children and adolescents to expand our understanding of the biology of pediatric cancers and evaluate the clinical implications of genomic testing for these patients with the ultimate goal of improving survival for pediatric malignancies. This article reviews the early studies in pediatric oncology showing the feasibility of this approach, describe the future plans to evaluate the clinical implications in multi-center clinical trials, and identify the challenges of applying genomics in the patient population.
Feasibility of precision medicine in pediatric oncology
Biomarker-driven directed therapies have been utilized in pediatric oncology, however, combining this treatment approach with individualized genomic analysis is in its nascent phase.2–4 Over the last 5 years, several pediatric oncology studies have explored the feasibility and use of genomics-driven precision medicine and provided the foundation for pursuing this approach. These pilot studies have explored different features of precision medicine and used various study designs, including patient population, timing of specimen acquisition, and inclusion of routine germline analysis. Of note, none of the published studies included prospective treatment arms as part of the study, although several studies include clinical follow-up to assess therapy response and outcomes to genomics-based recommendations.
The Baylor College of Medicine Advancing Sequencing in Childhood Cancer Care (BASIC3) study completed enrollment of a primary cohort of 287 newly diagnosed and previously untreated patients with solid, including CNS, tumors.5 Whole-exome sequencing (WES) was performed both on tumor samples and peripheral blood. In the report of the first 150 patients (<18 years of age) of which 121 tumors were sequenced, 33 patients (27%) were found to have somatic mutations of established or potential clinical use. An additional 24 patients (20%) were found to have mutations in consensus cancer genes that were not classified as targetable. Diagnostic germline findings related to patient phenotype (either cancer or other diseases or both) were discovered in 15 (10%) of 150 cases including 13 (8.6%) with pathogenic or likely pathogenic mutations in known cancer susceptibility genes. Treatment decisions or recommendations were not part of this study.
The University of Michigan Pediatric Michigan Oncology Sequencing (PEDS-MIONCOSEQ) study is modeled after the sequencing experience in adults with cancer.6 The preliminary results of a cohort of 102 pediatric and young adult participants (25 years of age and younger) with refractory or relapsed cancer and newly diagnosed patients with high-risk or rare cancer types have been published.6 Patients with both hematopoietic malignancies and solid tumors were included. A total of 91 patients underwent genomic analyses with WES of tumor and germline DNA as well as RNA sequencing (RNA-seq) of tumor. A multidisciplinary tumor board provided clinical recommendations. They identified 42 patients (46%) with potentially actionable findings that were not identified by standard diagnostic tests, which did not include sequencing. Nine of the patients had germline findings, 10 patients had somatic actionable gene fusions found through RNA-seq, and 2 patients had their diagnosis changed because of the analyses. A total of 23 patients had an individualized care decision made based on either tumor or germline sequencing results. Fourteen of the patients had a change in therapy, 9 patients underwent genetic counseling, and 1 patient required both. Of the 14 patients, 9 had a clinical response to the change in therapy lasting more than 6 months. The median turnaround time for return of the results was 54 days.
The Individualized Cancer Therapy (iCAT) Study is a multicenter study led by investigators at Dana-Farber Cancer Institute/Boston Children’s Hospital to assess the feasibility of identifying actionable alterations and making individualized cancer therapy recommendations in pediatric and young adult patients (30 years of age and younger) with relapsed, refractory or high-risk extracranial solid tumors.7 A multidisciplinary expert panel reviewed the profiling results and iCAT treatment recommendations were made if an actionable alteration was present, and an appropriate drug was available. Of the 100 participants, 31 had tumor submitted only from diagnosis, whereas the rest had tumor submitted from recurrence or local control or multiple specimens from recurrence and diagnosis. Tumor profiling was successful in specimens from 89 patients. Overall, 31 (31%) patients received an iCAT treatment recommendation and 3 received matched therapy. There were no objective responses. Three patients had a change in their diagnosis based on the tumor profiling. Six patients had an actionable alteration, but an appropriate drug was not available through a clinical trial or as a Food and Drug Administration (FDA)-approved therapy with an age-appropriate dose and formulation, and so an iCAT recommendation could not be made. Lastly, 43 (43%) participants had results with potential clinical significance but not resulting in iCAT treatment recommendations were identified, including mutations indicating the possible presence of a cancer predisposition syndrome (if also found in the germline).
The Precision in Pediatric Sequencing (PIPseq) Program at Columbia University Medical Center instituted a prospective clinical NGS for high-risk pediatric cancer and hematologic disorders.8 WES and RNA-seq were performed on tumor and normal tissue from 101 high-risk pediatric patients. Results were initially reviewed by a molecular pathologist and subsequently by a multi-disciplinary molecular tumor board. Potentially actionable alterations were identified in 38% of patients, of which 16% subsequently received matched therapy. In an additional 38% of patients, the genomic data provided clinically relevant information of diagnostic, prognostic, or pharmacogenomic significance. RNA-seq was clinically impactful in 37/65 patients (57%) providing diagnostic and/or prognostic information for 17 patients (26%) and identified therapeutic targets in 15 patients (23%). Known or likely pathogenic germline alterations were discovered in 18/90 patients (20%) with 14% having germline alternations in cancer predisposition genes. American College of Medical Genetics (ACMG) secondary findings were identified in six patients.
The Individualized Therapy for Relapsed Malignancies in Childhood (INFORM) project is a nationwide German program for children and young adults with refractory, relapsed cancers that aim to identify therapeutic targets on an individualized basis.9 In the report of the pilot phase, 57 patients aged 1-40 years with hematopoietic and solid malignancies were enrolled. Seven patients for whom no standard therapy was available were enrolled at the time of primary diagnosis. Tumor specimens were analyzed by WES, low-coverage whole-genome, and RNA-seq along with methylation and expression microarray analyses. A customized 7-step scoring algorithm was utilized to prioritize molecular targets and reviewed by an interdisciplinary molecular tumor board before returning the results to the treating physician. Germline DNA was screened on each patient for damaging alterations in a predefined list of known cancer predisposition genes. Turnaround time was 28 days. Of 52 patients, 26 (50%) with next-generation sequencing (NGS) data on their tumors harbored a potentially actionable alteration with a prioritization score of intermediate or higher. Ten patients received targeted therapy based on these results with responses seen in some of the previously treated patients, although systematic follow-up was not an objective of this study. Underlying cancer predisposition was detected in 2 patients (4%). Comparative primary tumor-relapsed tumor analysis revealed substantial tumor evolution in addition to the detection in one case of an unsuspected secondary malignancy.
The Institut Curie reported their 1-year experience of genetic analysis and molecular biology tumor board discussions for targeted therapies in pediatric solid tumors.10 Tumor tissue from 60 pediatric patients (up to age 21.5 years) with poor prognosis or relapsed or refractory solid, including CNS, tumors were analyzed with panel-based NGS and array comparative genomic hybridization. The most recently available tumor tissue was analyzed but in the case in which there was inadequate material, a new biopsy was not requested and the initial diagnostic biopsy specimen was used. Recommendations from the molecular biology tumor board were given to the treating physicians. The mean turnaround time from the patient referral to the molecular biology tumor board and release of results was 42 ± 16 days. Of the 58 patients in whom molecular profiling was feasible, 23 (40%) had a potentially actionable finding with high-grade gliomas having the highest number of targetable alterations. Of the 23 patients, 6 received a matched targeted therapy with 5 being enrolled in a clinical trial and 1 by compassionate use. Two patients had a partial response. Despite having a targetable lesion, 4 patients could not receive therapy owing to lack of available clinical trials with the agents. The remaining 13 patients did not receive targeted therapy because of pursuit of conventional chemotherapy or change in health status. The investigators concluded that this approach is feasible, but only a small proportion of patients were able to receive the targeted therapy.
This single-institutional feasibility study (MOSCATO-01) at Gustave Roussy in France prospectively characterized genomic alterations in recurrent or refractory solid tumors of pediatric patients for selection of targeted therapy. Seventy-five patients underwent tumor biopsy or surgical resection of primary or metastatic tumor site on study. Tumor samples were analyzed by comparative genomic hybridization array NGS for 75 target genes, WES, and RNA-seq. Biological significance of the alterations and recommendation of targeted therapies available were discussed in a multidisciplinary tumor board. All patients were pretreated, 37% had CNS tumors, and 63% had an extra-cranial solid tumor. Successful molecular analysis in 69 patients detected in 61% of patients an actionable alteration in various oncogenic pathways and change in diagnosis was seen in three patients. Fourteen patients received 17 targeted therapies. This study demonstrated the feasibility of research biopsies in advanced pediatric malignancies for NGS and matching potential actionable mutations with targeted therapies.
These initial pilot studies demonstrated the feasibility of clinical sequencing for patients with childhood cancers and set the stage for subsequent precision medicine trials that prospectively assess the impact of molecularly targeted therapies in pediatric oncology. Our evolving understanding of the landscape of the cancer genome of pediatric cancers also necessitates the inclusion of unique genomic technologies, such as RNA analysis for fusion detection, and analyses of germline mutations for cancer susceptibility risk determination in forthcoming studies.
NCI-COG Pediatric MATCH: New era of precision medicine in pediatric oncology
In collaboration with and supported by the NCI, the Children’s Oncology Group (COG) is leveraging the information gained from earlier precision medicine studies a step further in their design of a histology agnostic trial in which eligibility to treatment arms is determined based on predefined lists of genomic aberration(s), or actionable mutation(s) of interest (aMOI). Pediatric MATCH is a national clinical trial under a single IND (NCT03155620; Figure 1). Relapsed tumor tissue from pediatric and young adult patients with recurrent or refractory tumors including CNS tumors, as well as lymphomas and histiocytic disorders is submitted for molecular profiling. In order to provide broader access to precision medicine trials for the adolescent and young adult oncology population, patients up to the age of 21 years are eligible to enroll on Pediatric MATCH.
Figure 1.
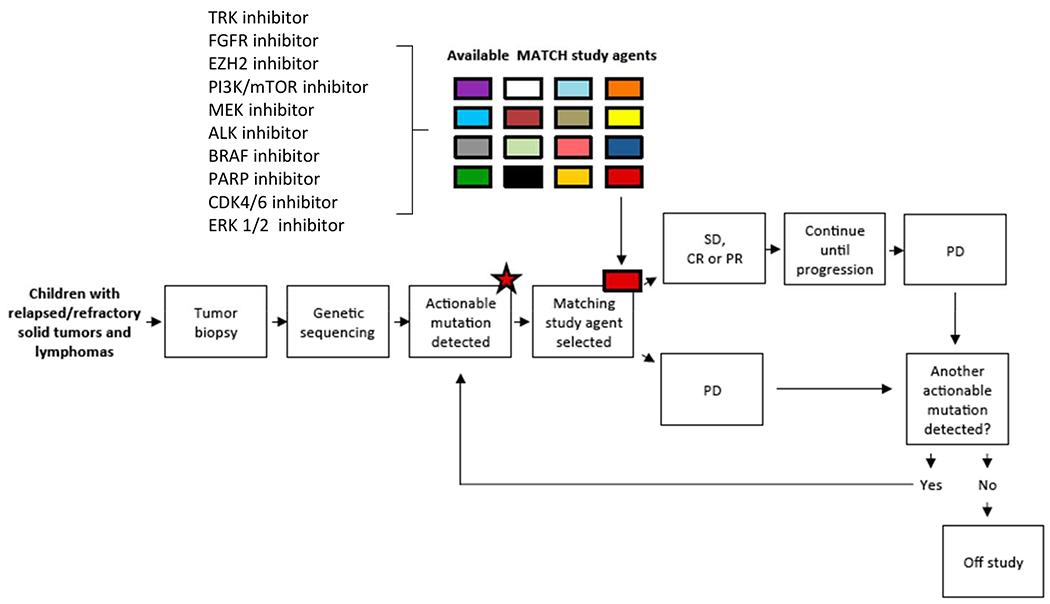
NCI-COG Pediatric Molecular Analysis for Therapeutic Choice (MATCH) Trial Schema. TRK, tyrosine receptor kinase; FGFR, fibroblast growth factor receptor; EZH2, enhancer of zeste homolog 2; PI3K/mTOR, phosphoinositide 3-kinase/mammalian target of rapamycin; ALK, anaplastic lymphoma kinase; PARP, poly-ADP ribose polymerase; ERK, extracellular signal-regulated kinase; SD, stable disease; CR, complete response; PR, partial response; PD, progressive disease. From Allen CE, Laetsch TW, Mody R, et al. Target and Agent Prioritization for the Children’s Oncology Group-National Cancer Institute Pediatric MATCH Trial. J Natl Cancer Inst. 2017;109(5); with permission.
Similar to the NCI MATCH study for adults, if an aberration is identified that has been defined as a driver mutation for a Pediatric MATCH study drug targeting the identified aberration, then the patient will have the opportunity to enroll onto the relevant single agent treatment arm. Consequently, the trial is providing access to the study agent(s) for each patient in addition to tumor genomic analysis and treatment assignment. This trial will screen over 1000 patients for multiple targets and evaluate investigational targeted therapies for clinical activity in patients carrying specific mutations that can inform future trials. This is the largest pediatric oncology trial for all solid tumors to identify the molecular aberration(s) in the tumor and provide the investigational agent for the treatment of the identified molecular aberrations within the same trial. Similar to the NCI MATCH study for adults, Pediatric MATCH employs an analytically validated NGS targeted assay of more than 4000 different mutations (single nucleotide variants, indels, copy number alterations, and gene fusions) across more than 140 genes.11 This type of basket or umbrella hybrid trial uses a rules-based treatment assignment, based on available preclinical and clinical data, which has not been used in the other pediatric trials.12 This offers the advantage of predefining treatment based on the presence of a molecular aberration, ensures availability of agents within the context of a trial, and negates assignment bias because all patients with a predefined aMOI are assigned a given treatment.
The primary aims of the study are to determine the objective response rate in pediatric patients with advanced solid tumors and lymphomas harboring a priori specified genomic alterations treated with pathway-targeting agents, and to determine the proportion of pediatric patients whose tumors have pathway alterations that can be targeted by existing anti-cancer drugs. A total of 20 patients will be enrolled per treatment arm (or stratum within the arm) depending on the aMOIs, and the agent will be considered of interest for further development if 3 or more patients of 20 show a response. The arms may be expanded in the trial to enroll additional patients if activity is seen for a particular agent. The study will have the flexibility to open and close arms. A patient’s tumor that progresses while on treatment will be eligible to go on another treatment arm if the tumor has additional genetic aberrations that are being targeted with another Pediatric MATCH agent.
The molecular targets and study drugs selected for the trial were identified and prioritized by the Pediatric MATCH Target and Agent Prioritization (TAP) Committee consisting of representatives from COG disease committees from 10 children’s hospitals, the NCI, and the FDA. The TAP committee systematically reviewed target and agent pairs for inclusion in Pediatric MATCH trial. Criteria used to prioritize the target and agent pairs included the frequency of the alterations in the target in pediatric malignancies, strength of the evidence linking the target to activity of the agent, whether the target can be detected with the testing platform, clinical and preclinical evidence for the specific agent, and other ongoing or planned biomarker defined clinical studies. Details of this process have been described previously.13 Two criteria established for identifying specific agents to be considered for inclusion in Pediatric MATCH were demonstrated activity against tumors with a particular genomic alteration, and the establishment of the an adult recommended phase 2 dose. The same levels of evidence for drug selection used in NCI MATCH were applied for Pediatric MATCH.11 Neither a completed pediatric phase 1 study nor a pediatric formulation was required to be considered for Pediatric MATCH trial. However, in case of an oral agent, appropriately sized capsules or tablets were required to dose pediatric patients. Currently, Pediatric MATCH trial has opened 10 treatment arms with the goal of investigating a total of 15-20 single agents based on ongoing review of new data and as agents become available based on the identified targets.
In a report of the first 422 patients enrolled from 93 COG sites on the Pediatric MATCH screening protocol, the median age is 13 years (range 1-21).14 A tumor sample was submitted for 390 patients, sequencing was attempted for 370 patients (95%), and results were confirmed for 357 patients (92%). The median turn-around time for the tumor genomic results was 15 days. An aMOI for at least one of the 10 current treatment arms was identified in approximately 25% of patients with tumor submitted for the Pediatric MATCH screening protocol. These patients are assigned to a treatment arm and must meet the eligibility criteria to enroll on therapy.
There are some notable differences for Pediatric MATCH as compared to NCI MATCH or similar precision medicine protocols. The number of molecular aberrations seen in pediatric tumors (~10%) is predicted to be much less than identified in adult malignancies.15,16 These projections are based primarily on actionable mutation frequencies in newly diagnosed tumors. However, the Pediatric MATCH aMOI detection rate is currently higher than predicted.14 In comparison, this Pediatric MATCH aMOI detection rate is higher than the match rate of for NCI-MATCH for the first 10 treatment arms (approximately 9%).11 One reason for this may be that patients with known targetable mutations from prior molecular testing are enrolling on the screening study at a higher rate on Pediatric MATCH. In addition, the Pediatric MATCH study is optimizing the chances of finding a targetable aberration in a patient’s tumor by requiring the submission of tumor specimen from a biopsy done after recurrence or progression and as close to the time of genomic analysis, as tumors are likely to acquire more mutations during over time.17–20 In fact, in some studies in adults, it is recommended that a metastatic (as opposed to primary) lesion is biopsied.21,22 To provide access for as many children and adolescents as possible and because the risks associated with biopsies in children differ from adults, there is more flexibility with the timing of the biopsy (need not be obtained just before study enrollment as long as it is from a recurrence) or in the case of brain stem gliomas from the time of diagnosis in the Pediatric MATCH study. Lastly, although tumors occurring in adults may have a larger number of mutations (on average), many of those mutations are passenger mutations that have been acquired over time that may have little relevance to the biology (or treatment) of the tumor. Thus, the number of targetable mutations might be more similar in children and adults than initially projected based on total number of mutations.
Pediatric cancers harbor a different spectrum and frequency of mutations compared to adult cancers. For example, frequent targetable kinase alterations seen in lung cancer and breast cancer such as EGFR and HER2, respectively, rarely occur in pediatric tumors. Therefore, such agents would not meet the selection criteria as treatment arms in Pediatric MATCH. Drug availability is another challenge, as agents are not yet available to target many of the recurrent aberrations identified in pediatric tumors. Based on the small number of pediatric cancers and even smaller subgroups of molecular aberrations identified in the tumors, agents have not been developed to target some of the detectable molecular aberrations, such as epigenetic alterations.
Similar to several of the pediatric studies described earlier, germline DNA is collected and analyzed from all patients enrolled on Pediatric MATCH. In contrast, NCI MATCH does not evaluate germline molecular aberrations. By including germline analysis using the same panel as for tumor sequencing, it is possible to determine which mutations of interest and actionable mutations of interest represent germline variants in cancer susceptibility genes. Clinical genomics laboratories interpret the germline findings and provide a report back to the treating oncologist identifying whether any of genomic aberrations included in the tumor sequencing report represent pathogenic or likely pathogenic germline variants in cancer susceptibility genes. The results of the germline analysis are not used for treatment assignment and are not meant to provide a comprehensive cancer susceptibility evaluation. However based on the results the treating pediatric oncologist may recommend formal genetic testing and counseling for the patient/family.
Other precision medicine trials in pediatric oncology
There is a similar precision medicine initiative for the conduct of a genomically-driven basket study for children and adolescents with relapsed or refractory cancers in Europe. The European Proof-of-Concept Therapeutic Stratification Trial of Molecular Anomalies in Relapsed or Refractory Tumors in Children (AcSé-ESMART) utilizes genomic data derived from multiple panels from several ongoing sequencing studies in Europe.23 In contrast to Pediatric MATCH, which utilizes predefined levels of evidence linking variants to targeted therapies, sequencing data are reviewed at a multidisciplinary molecular tumor board to determine whether an actionable variant is present and whether the variant is a match for one of the ESMART treatment arms or other targeted agent trials. To date, ESMART has seven treatment arms for five genomic targets/pathways. Each of the treatment arms are conducted as individual clinical trials, with a phase 1 dose escalation phase and a phase 2 expansion phase. In contrast to Pediatric MATCH, which thus far includes only single agents, many of the treatment arms in ESMART combines targeted agents with chemotherapy.
The initial results for the European pediatric precision medicine initiative have been reported.24 From 2016-2017, 174 patients with a median age of 13 years (range, 1-32) were included in the European molecular profiling trial (MAPPYACTS). Currently, the analysis for 104 patients has been completed. Seventy-six percent of patients had at least one “actionable” variant. Based on the detected alteration, 21 patients were included in the ESMART trial since it opened in August 2016, with two patients enrolling on two different treatment arms: CDK4/6 inhibitor ribociclib plus chemotherapy (1) or everolimus (5); DNA repair interfering combinations WEE1 inhibitor AZD1775 plus chemotherapy (4) and PARP inhibitor olaparib plus chemotherapy (1); dual mTOR inhibitor vistusertib alone (1) or with chemotherapy (5); or nivolumab and cyclophosphamide (6).24
Umbrella trials focusing on specific diseases or histologies in pediatric oncology are currently underway in patients with relapsed and refractory or newly diagnosed cancers. These studies require a good understanding of the gene variants to be encountered in a specific diagnosis and its activity to targeted agents. Moreover, the feasibility of conducting these studies requires that the frequency of variants is sufficient to justify a clinical trial within a disease group. Examples of these studies are shown in Table 1.
Table 1.
Examples of the range of precision medicine trials in pediatric oncology
| Study Design | Clinical Trial | Sponsor | NCT |
|---|---|---|---|
| Relapsed or Refractory Cancers | |||
| Basket studies across multiple histologies | Pediatric MATCH | NCI-COG | |
| AcSé-ESMART | Gustave Roussey | NCT02813135 | |
| Histology-specific umbrella studies | NEPENTHE (Neuroblastoma) | CHOP | NCT02780128 |
| Ruxolitinib or Dasatinib + chemotherapy in Ph-like ALL | MD Anderson | NCT02420717 | |
| RELPALL (ALL) | St. Jude | NCT03515200 | |
| Newly Diagnosed Cancers | |||
| Histology-specific umbrella studies | Dasatinib + chemotherapy for Ph-like ALL | COG | NCT02883049 |
| Ruxolitinib + chemotherapy for CRLF2/JAK/STAT mutations in ALL | COG | NCT02723994 | |
| Crizotinib + chemotherapy in ALK aberrant neuroblastoma | COG | NCT03126916 | |
| Total therapy XVII JAK/STAT mutations in ALL/lymphoma | St. Jude | NCT03117751 | |
| Clinical/Molecular Risk-Directed Therapy in Medulloblastoma | St. Jude | NCT01878617 | |
| BIOMEDE (DIPG) | Gustave Roussy | NCT02233049 | |
NCT, ClinicalTrials.gov Identifier/Number; COG, Children’s Oncology Group; CHOP, Children’s Hospital of Philadelphia; ALL, acute lymphoblastic leukemia; DIPG, diffuse intrinsic pontine glioma
A major challenge with these precision medicine clinical trials is that the treatment regimens are tailored to an increasingly smaller subset of genomically-defined patients. Clinical trials such as these are intended to be screening trials in order to detect a signal in a histology-agnostic cohort based on the genetic marker. Additional studies will need to be designed in order to assess the true activity of an agent in a pre-specified cohort. Likewise, future study designs and statistical methods will need to address these issues as we analyze clinical trial results and consider incorporating this information into standard of care therapies.
Summary
Molecular characterization has the potential to advance the management of pediatric cancer malignancies. The clinical integration of genome sequencing into standard clinical practice has been limited. Although there are still many obstacles remaining as precision medicine is applied to pediatric oncology, these studies represent the first step in exploring this application of genomic-directed treatment of patients with childhood cancer.
SYNOPSIS.
Over the past decades, improvements in outcomes have been seen in children and adolescents with cancer. Nevertheless, challenges remain in trying to improve the outcomes for all children diagnosed with cancer and particularly in patients who present with metastatic disease or with cancers that are resistant or recur with standard treatment approaches. Precision medicine trials utilizing individualized tumor molecular profiling for selection of targeted therapies are ongoing in various adult malignancies. Similar approaches are being applied to children and adolescents with cancer and are currently under investigation in pediatric oncology. The purpose of this article is to describe how precision medicine is being applied to pediatric oncology and the unique challenges with these efforts.
KEY POINTS.
The ultimate goal of precision medicine in pediatric oncology is to develop more effective and less toxic therapies in children, adolescents and young adults with cancer.
Precision clinical trials designed to assess the impact of molecularly targeted therapies in pediatric oncology are ongoing in the United States and Europe.
Our understanding of the cancer genomic landscape, advancement in genomic technologies, and drug development in enhanced targeted therapies may lead to future opportunities for precision medicine in pediatric oncology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT
DWP is the co-inventor on current and pending patents related to cancer genes discovered through sequencing of several adult cancer types and participates in royalty sharing related to those patents. The other Authors have nothing to disclose.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2009;27(31):5175–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosse YP, Lim MS, Voss SD, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study. Lancet Oncol. 2013; 14(6):472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz KR, Carroll A, Heerema NA, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia. 2014;28(7):1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Roy A, Yang Y, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016;2(5):616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mody RJ, Wu YM, Lonigro RJ, et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA. 2015;314(9):913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris MH, DuBois SG, Glade Bender JL, et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. 2016;2(5):608–615. [DOI] [PubMed] [Google Scholar]
- 8.Oberg JA, Glade Bender JL, Sulis ML, et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med. 2016;8(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worst BC, van Tilburg CM, Balasubramanian GP, et al. Next-generation personalised medicine for high-risk paediatric cancer patients - The INFORM pilot study. Eur J Cancer. 2016;65:91–101. [DOI] [PubMed] [Google Scholar]
- 10.Pincez T, Clement N, Lapouble E, et al. Feasibility and clinical integration of molecular profiling for target identification in pediatric solid tumors. Pediatr Blood Cancer. 2017;64(6). [DOI] [PubMed] [Google Scholar]
- 11.Conley BA, Gray R, Chen A, et al. Abstract CT101: NCI-molecular analysis for therapy choice (NCI-MATCH) clinical trial: interim analysis. Cancer Research. 2016;76(14 Supplement):CT101–CT101. [Google Scholar]
- 12.Takebe N, Yap TA. Precision medicine in oncology. Curr Probl Cancer. 2017;41(3):163–165. [DOI] [PubMed] [Google Scholar]
- 13.Allen CE, Laetsch TW, Mody R, et al. Target and Agent Prioritization for the Children’s Oncology Group-National Cancer Institute Pediatric MATCH Trial. J Natl Cancer Inst. 2017;109(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, Janeway KA, Patton D, et al. Identification of targetable molecular alterations in the NCI-COG Pediatric MATCH trial. J Clin Oncol. 2019;37(suppl; abstr 10011). [Google Scholar]
- 15.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schleiermacher G, Javanmardi N, Bernard V, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol. 2014;32(25):2727–2734. [DOI] [PubMed] [Google Scholar]
- 18.Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47(8):864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schramm A, Koster J, Assenov Y, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet. 2015;47(8):872–877. [DOI] [PubMed] [Google Scholar]
- 20.Padovan-Merhar OM, Raman P, Ostrovnaya I, et al. Enrichment of Targetable Mutations in the Relapsed Neuroblastoma Genome. PLoS Genet. 2016;12(12):e1006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Tourneau C, Kamal M, Tsimberidou AM, et al. Treatment Algorithms Based on Tumor Molecular Profiling: The Essence of Precision Medicine Trials. J Natl Cancer Inst. 2016;108(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno L, Pearson ADJ, Paoletti X, et al. Early phase clinical trials of anticancer agents in children and adolescents - an ITCC perspective. Nat Rev Clin Oncol. 2017;14(8):497–507. [DOI] [PubMed] [Google Scholar]
- 24.Geoerger B, Schleiermacher G, Pierron G, et al. Abstract CT004: European pediatric precision medicine program in recurrent tumors: first results from MAPPYACTS molecular profiling trial towards AcSe-ESMART proof-of-concept study. Cancer Research. 2017;77(13 Supplement):CT004–CT004. [Google Scholar]


