Abstract
Objective.
Cancer patient-derived organoids (PDOs) grow as three dimensional (3D) structures in the presence of extracellular matrix and have been found to represent the original tumor’s genetic complexity. In addition, PDOs can be grown and subjected to drug sensitivity testing in a shorter time course and with lesser expense than patient-derived xenograft models. Many patients with recurrent ovarian cancer develop malignant effusions that become refractory to chemotherapy. Since these same patients often present for palliative aspiration of ascites or pleural effusions, there is a potential opportunity to obtain tumor specimens in the form of multicellular spheroids (MCS) present in malignant effusion fluids. Our objective was to develop a short duration culture of MCS from ovarian cancer malignant effusions in conditions selected to support organoid growth and use them as a platform for empirical drug sensitivity testing.
Methods.
In this study, malignant effusion specimens were collected from patients with high-grade serous ovarian carcinoma (HGSOC). MCS were recovered and subjected to culture conditions designed to support organoid growth. In a subset of specimens, RNA-sequencing was performed at two time points during the short-term culture to determine changes in transcriptome in response to culture conditions. Organoid induction was also characterized in these specimens using Ki67 staining and histologic analysis. Drug sensitivity testing was performed on all specimens.
Results.
Our model describes organoids formed within days of primary culture, which can recapitulate the histological features of malignant ascites fluid and can be expanded for at least 6 days. RNA-seq analysis of four patient specimens showed that within 6 days of culture, there was significant up-regulation of genes related to cellular proliferation, epithelial-mesenchymal transition, and KRAS signaling pathways. Drug sensitivity testing identified several agents with therapeutic potential.
Conclusions.
Short duration organoid culture of MCS from HGSOC malignant effusions can be used as a platform for empiric drug sensitivity testing. These ex vivo models may be helpful in screening new or existing therapeutic agents prior to individualized treatment options.
Keywords: Ovarian cancer, Patient-derived organoid models, Multicellular spheroids, Targeted experimental therapeutics
1. Introduction
Ovarian cancer is the deadliest gynecologic cancer and often is associated with malignant effusions of the abdomen or pleural space that becomes refractory to treatment. High-grade serous carcinoma of the ovary, fallopian tube and peritoneum is the predominant histology [1]. Second-line chemotherapy for platinum-resistant recurrent ovarian cancer has objective response rates b30%, and the five-year mortality rate between 70 and 90% has not changed significantly in the past ten years [1,2]. Ovarian cancer genomic analysis reveals that extensive allelic copy number changes rather than common driver mutations are characteristic of the disease [3], making genomically-targeted treatment more difficult. The lack of reliable and validated in vitro models for empiric testing of potential drug sensitivities has also hindered progress. Two dimensional (2D) in vitro cultures have been used as ovarian cancer models and more recently, patient-derived xenografts (PDX) have been successfully developed [4]. A major disadvantage of 2D culture is that genetic heterogeneity found in the original tumors is poorly represented [5]. PDX models, while maintaining genomic heterogeneity, are costly, time-consuming and not always successful. Some of these disadvantages can be overcome by using PDO cultures. PDOs grow as 3D structures within the extracellular matrix and may develop from tumor-initiating cells that recapitulate the original tumor’s genetic complexity [6]. PDOs have been grown with a high success rate over a short duration in serum-free media with defined supplements designed to support the stem cell-like niche [7]. Accumulating evidence suggests that this stem cell-like niche in malignant tumors is a repository of treatment resistance and tumor recurrence, and consequently has high clinical relevance [8–11]. Tumors from the colon, prostate, breast, pancreas, endometrium and other solid tumors have been propagated at a high rate of success using PDO techniques. The resulting PDOs closely resemble the original patient tumor in morphology, mutation profile and gene expression patterns [12–14]. PDOs provide a fast, effective model for use in functional assays of individual patient tumors, and early clinical trials are underway to correlate drug sensitivity in PDO cultures with response to targeted agents in patients with gastrointestinal cancers [13,15]. Prominent genomic changes associated with HGSOC include allelic copy number changes and TP53 mutations, as opposed to a group of mutated driver genes associated with other cancer types. Accordingly, the general lack of oncogene mutations in HGSOC affords few opportunities for molecularly targeted therapy. Empirical testing of drugs using ex vivo organoid technology may provide rapid screening of active drugs in these patients.
Multicellular spheroids (MCS) are metastatic units that can adhere to the mesothelium and invade the extracellular matrix to facilitate peritoneal dissemination, and they could be considered the driving force in tumor metastasis in ovarian cancer [16]. Consequently, ex vivo models using MCS that recapitulate the early stages of metastasis could be valuable in testing experimental therapeutic agents. The current study presents evidence that MCS from ovarian cancer malignant effusions subjected to short duration culture under conditions known to support organoid growth show proliferative characteristics, EMT gene expression signature, KRAS pathway activation, and variable response to experimental and conventional therapeutics. These ex vivo models can be used as a platform for empiric drug sensitivity testing and advancing treatment options for ovarian cancer.
2. Methods
2.1. Recovery of MCS from ovarian cancer effusions
High-grade serous ovarian cancer specimens from ascites or pleural effusion fluid were collected for the study. Only effusion fluids not needed for pathologic diagnosis and considered “leftover” were collected. Specimens were provided by the UC Davis Pathology Biorepository which is jointly funded by the UC Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute and the Department of Pathology at UC Davis. The study was approved through the UC Davis Institutional Review Board. All samples were coded with a study number and all personal health information was removed. Patient demographics, tumor characteristics, and clinical information were provided in a de-identified manner. Effusion fluids were centrifuged (365 ×g, 15 min) and cell pellets were resuspended in Dulbecco’s Phosphate Buffered Saline (DPBS, Gibco #14040–133). Cold ammonium chloride solution (Stemcell Technologies #07800) was added in a 4:1 dilution for red blood cell lysis. After 10 min on ice, the cells were centrifuged and resuspended in DPBS. The specimen was then passed through a 100-μm sieve to remove large aggregates and debris. The flow-through was passed through a 38-μm sieve to remove most mononuclear cells. MCS structures between 38- to 100-μm were recovered by backflushing, washed and cryopreserved (mFreSR, Stemcell Technologies #05855).
2.2. Initial culture of MCS and organoid formation
For initial organoid culture, previously cryopreserved 38–100 μm MCS were rapidly thawed, washed with cold DMEM/F12 (Gibco #11320–033) and resuspended in cold Cultrex® Reduced Growth Factor Basement Membrane Extract, Type 2 (BME) (F # 343301001). MCS were then deposited in 10 μL droplets in 6-well tissue culture plates. Approximately 0.05–0.1 mL of packed MCS material was used per 6-well tissue culture plate. A test droplet (10 μL) droplet was plated and visualized under the microscope. MCS were counted and we aimed to have approximately 100 per droplet. Material was resuspended if not in this range. The droplets were allowed to solidify for 20 min at 37 °C. Each well was filled with 2 mL “complete medium” (CM, similar to that described by Sachs et al. [7] with some modifications), as follows: DMEM/F12, 10% R-spondin1 conditioned medium, 2% B27 supplement (Gibco #17504–044), 10 mM HEPES (Gibco #15630080), 1% Glutamax (Gibco #35050), 1.25 mM N-acetyl cysteine (Sigma #S7250), 100 μg/mL Primocin (InvivoGen #ant-pm), 1% Antibiotic-Antimycotic (Gibco #15240–062), 1mM nicotinamide (Sigma #N0636), 0.5 μM A 83–01 (Sigma #SML0788), 5 nM Neuregulin 1 (Abcam #ab73753), 5 ng/mL FGF-7 (Peprotech #10019), 20 ng/mL FGF-10 (PeproTech #10026), 100 ng/mL Noggin (PeproTech #12010C), 5 ng/mL EGF (Gibco #PHG0314), 0.5 μM SB 202190 (Sigma #S70767), and 5 μM Y-27632 (Stemcell technologies #72304). For RSPO1 conditioned medium, the recombinant cell line HA-R-Spondin-Fc 293T was purchased from Trevigen (#3710–001–01) and serum-free conditioned CD 293 medium (Gibco #11913–019) was produced as described by the supplier’s instructions. The cultures were placed at 37 °C with 5% CO2.
2.3. Short-term organoid growth assay
Following initial culture of MCS in BME droplets, the developing organoids were recovered after 3–4 days of culture using Cell Recovery Reagent (Corning #354253), washed in cold DMEM/F12, suspended in CM + 2% BME and distributed, 50 μL per well containing approximately 100 organoids/well, into 96-well plates (low evaporation lid, Sigma #CLS3595), the wells of which had been previously coated with 15 μL of 7.5 mg/mL BME [17].
For cytotoxicity assays using targeted drugs or experimental therapeutic agents, a single drug concentration was selected that was at or near the pharmacokinetic maximum (Cmax) value associated with the therapeutic dosage in humans from published clinical trials. When pharmacokinetic data were not available, drugs were screened at a 1 micromolar concentration. Drugs tested and their sources are detailed in Table 1. After 24 h of culture (on culture day 1), 25 μL of each drug solution to be tested, dissolved in CM at 3 times the final target concentration, was added to each of 6 replicate wells. Except for carboplatin and abraxane which were dissolved in aqueous solutions, all drugs were prepared in DMSO. All wells, including controls, were adjusted to a DMSO concentration of 0.1%. Cultures were incubated until day 6, then assayed by CellTiter-Glo 3D Cell Viability Assay (Promega #G9681) as described by Francies et al. [17]. Identical frozen aliquots of 0.5 μM ATP solution (Pharmacia #27–2056–01), thawed and utilized on each day of the CellTiter-Glo (CTG) assay, were used to normalize results for growth comparisons. These control wells were assayed on day 0 (to check viability), day 1 and day 6. Representative images of control well organoids were obtained by video microscopy (Olympus IX81). Mean organoid colony areas were measured using ImageJ 1.52a software. For statistical comparison, the Student’s t-test was utilized. Fold-change in growth between day 1 and day 6 was determined by comparison of ATP-normalized CTG results for control wells. In samples performed in duplicate or triplicate, control well CTG results were averaged. For IC50 determination, 9 two-fold dilutions of each test drug were tested and IC50 values calculated with GraphPad Prism software (v8). Percent inhibition was calculated as follows: percent inhibition = 100–100 (D / C), where C = average luminosity among 6 control replicates, and D = average luminosity among 6 drug replicates. Percent inhibition of <50% in the drug screen was considered “resistant”.
Table 1.
Inhibitors used in drug assay.
| Drug | Mechanism of action | Published Cmax(μM) | Target concentration (μM) |
|---|---|---|---|
| Mocetinostat | HDAC inhibitor | 0.5 [39] | 0.5 |
| Trametinib | MEK1/2 inhibitor | 0.08 [40] | 0.1 |
| LY294002 | PI3Kα/δ/β inhibitor | 26 [41] | 26 |
| AZD5363 | AKT 1/2/3 inhibitor | 4.7 [42] | 5 |
| BBI503 | Stemness kinase inhibitor | – | 2.5 |
| MK-1775 | Wee-1 inhibitor | 2.66 [43] | 3 |
| Sorafenib | Raf-1, B-Raf, VEGFR-2 inhibitor | 12.5 [44] | 12.5 |
| APR-246 | p53 re-activation/apoptosis induction | 250 [34] | 200 |
| CB5083 | p97 inhibitor | – | 5 |
| Napabucasin | STAT3 inhibitor | 1.5 [45] | 1.5 |
2.4. Characterization of organoid induction from MCS to day 6 organoids
Organoid induction was studied over 6 days of culture by video microscopy, formalin-fixed paraffin-embedded H&E stain, Ki67 index, and RNA-Sequencing (RNA-seq) analysis. Uncultured day 0 MCS (thawed and washed in DMEM) were compared to matched day 6 organoids (recovered from BME droplet culture with Cell-Recovery solution) from four unique subjects. MCS and organoids were placed under coverslips, stained with trypan blue and representative images were obtained by video microscopy (Olympus IX81). Uncultured day 0 MCS and day 6 organoids were fixed and processed for histology and Ki67 staining as previously described [12].
2.5. Transcriptome analysis of HGSOC multi-cellular spheroids with RNA-sequencing (RNA-seq)
Day 0 MCS and Day 6 organoid samples were submitted to the UC Davis Comprehensive Cancer Center’s Genomics Shared Resource (GSR) for isolation of total cellular RNA and RNA-seq analysis. Total cellular RNA was isolated from snap-frozen spheroids using the TRIzol Reagent (Invitrogen) and a modified protocol that incorporates an additional extraction with phenol/chloroform/isoamyl alcohol (25,24,1, pH 4.3), followed by an additional clean-up with an RNeasy spin column (Qiagen). Stranded mRNA-seq libraries were prepared from 100 ng total RNA using the NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs, Ipswich, MA) according to the manufacturer’s standard protocol, and as previously described [18]. Subsequently, libraries were combined for multiplex sequencing on an Illumina HiSeq 4000 System (2 × 150 bp, paired-end, ≥20 × 106 reads per sample). A Salmon-tximport-DESeq2 workflow was utilized for primary and secondary analysis of the RNA-Seq data (FASTQ format). Sequence read mapping to the reference genome assembly (Dec. 2013, GRCh38/hg38) and transcript abundance estimation were performed with Salmon [19]. Transcript-level read counts were prepared with the R package tximport [20], annotated with GENCODE Human Release 29 (GRCh38.p12), and differential gene expression analysis was conducted with DESeq2 [21]. Principal component analysis (PCA), hierarchical clustering, and heatmap visualization were performed with the Strand NGS software package (Strand Life Sciences). Gene Set Enrichment Analysis (GSEA) was performed using the predefined hallmark gene set [22,23]. A ranked gene list was created. All values >1 were considered to have been upregulated in Day 6 vs Day 0 and all values <1, downregulated.
3. Results
Table 2 shows the demographic characteristics of 6 unique subjects from which 14 separate specimens were recovered for analysis. MCS from malignant effusions were structurally heterogeneous with a wide distribution of shapes and sizes. Fig. 1 shows images of a representative specimen before and after sieving. The amount of MCS material recovered (post sieving, 38–100 μm) per liter of malignant effusion varied with an average of 0.55 mL packed MCS material per liter of effusion (n = 14, range: 0.05–2.0). Seven additional specimens were collected (not reported in Table 2), however in 4 cases, chemotherapy was administered within 2 weeks of sampling. No material for further testing was obtained. In 3 cases, scant material was obtained which was insufficient to perform complete testing.
Table 2.
Clinicodemographics of participants.
| Participant | Specimen | Race/ethnicity | Age | Source | Stage/histologya | Prior treatment | Days from last infusion |
|---|---|---|---|---|---|---|---|
| 1 | OV469 | White/non-Hispanic | 56 | Ascites | IIIC HGS Peritoneal cancer |
Untreated | NA |
| 2 | OV778 | White/non-Hispanic | 74 | Ascites | IVB HGSOC | Untreated | NA |
| 3b | OV798 OV817 OV820 OV838 OV840 |
White/non-Hispanic | 61 | Pleural effusion | IIIC HGSOC | C1 paclitaxel/bevacizumab C4 paclitaxel/bevacizumab C5 paclitaxel/bevacizumab C6 paclitaxel/bevacizumab C1 gemcitabine |
21 14 7 14 2 |
| 4 | OV899 OV860 |
White/non-Hispanic | 74 | Ascites | IIIC HGSOC | Untreated C1 carboplatin/paclitaxel |
NA 4 |
| 5 | OV867 OV875 OV882 |
White/non-Hispanic | 62 | Ascites Ascites Ascites |
IVB HGSOC | C1 carboplatin/paclitaxel C1 carboplatin/doxorubicin C1 carboplatin/doxorubicin |
3 |
| 6c | OV870 OV888 |
White/non-Hispanic | 54 | Ascites Ascites |
IIIC HGSOC | See footnote | 46 |
C = Cycle; HGS = High grade serous; HGSOC = High grade serous ovarian cancer.
Stage at time of original diagnosis.
Prior to first specimen, patient underwent treatment with AZD2014, bevacizumab, carboplatin, cisplatin, doxorubicin, olaparib, paclitaxel, topotecan, in combination or as a single agent.
Prior to first specimen, patient underwent treatment with ABT888 (PARP inhibitor), bevacizumab, carboplatin, pegylated liposomal doxorubicin, paclitaxel, topotecan. Prior to collection of OV888, patient initiated therapy with anastrazole.
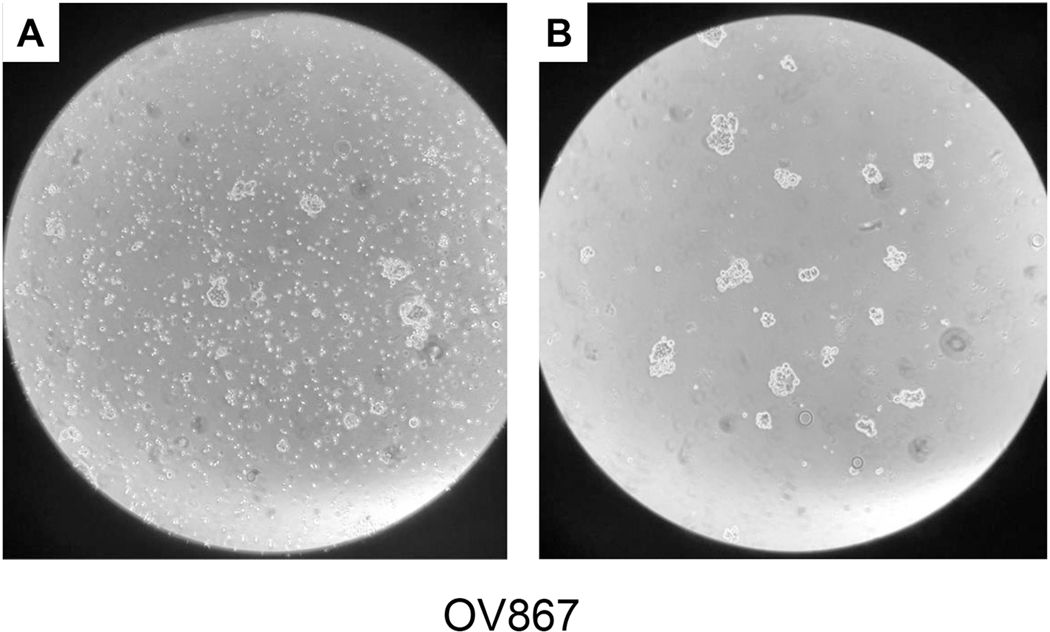
Fig. 1.
Representative specimen before and after sieving (38–100 μm). Malignant effusion specimens were centrifuged, treated with cold ammonium chloride and suspended in DPBS. This solution was then sieved in order to recover 38–100 μm material. Panel A represents heterogeneous pre-sieved material and panel B represents post-sieved MCS (38–100 μm) (100×).
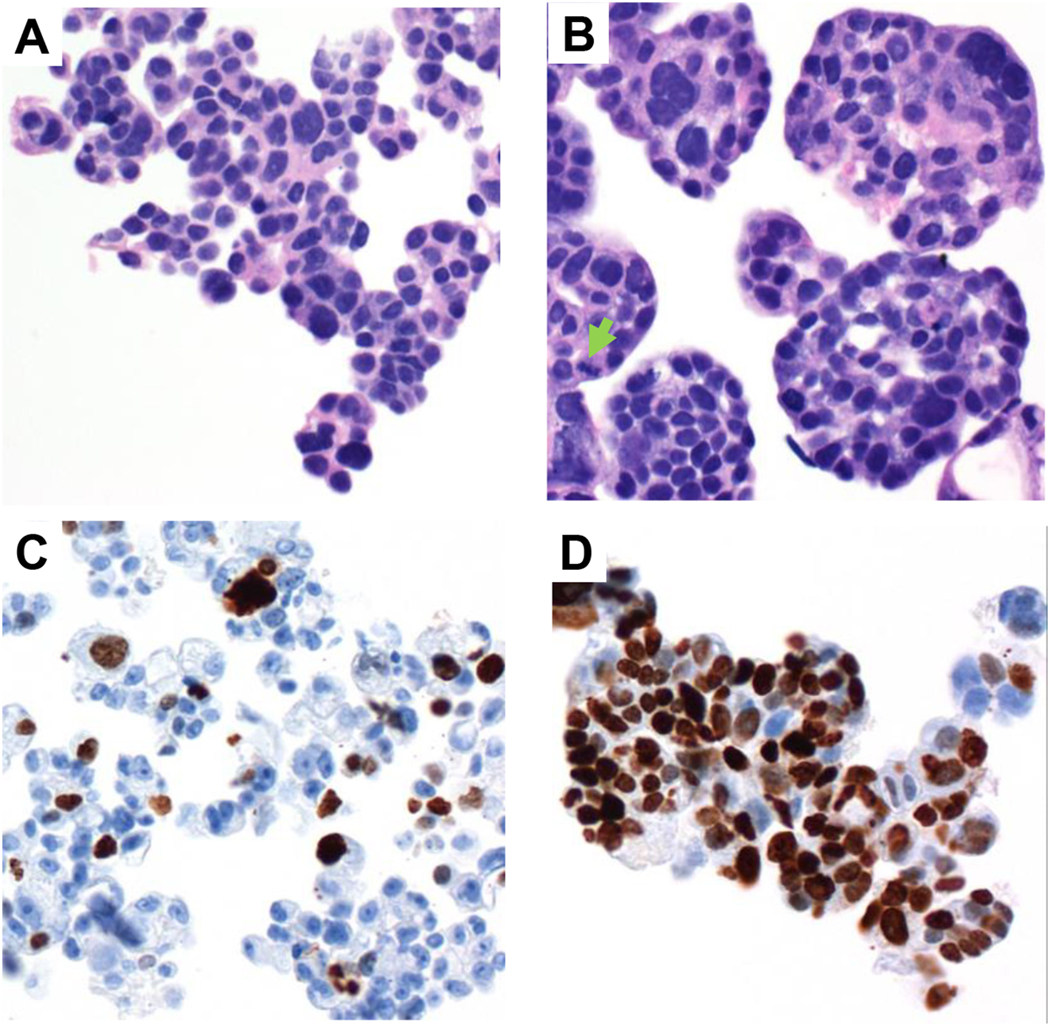
Fig. 2 shows representative photomicrographs of paraformaldehyde-fixed, paraffin-embedded material stained with hematoxylin/eosin (H&E). Variable numbers of non-viable appearing cells were seen in both MCS and day 6 specimens, but day 6 specimens showed larger cell groupings with conspicuous mitotic figures. Comparing MCS to day 6 organoids, Ki67 index increased on average by 3.5-fold. Individual Ki67 indices for each of the 4 specimens are shown in Table S1. Day 6 organoids also showed evidence of cell death with more trypan blue-stained cells in organoids recovered from BME droplets, and more eosinophilic debris along with fragmented nuclei (data not shown).
Fig. 2.
Characterization of organoid by H&E (A–B) and Ki67 (C–D). A represents MCS (day 0) while B depicts organoids on day 6. Variable numbers of non-viable appearing cells were seen in both MCS and day 6 specimens, but day 6 specimens showed larger cell groupings with frequently-seen mitotic figures (arrow). C represents Ki67 staining for MCS (day 0) while D represents organoids on day 6. Ki67 index increased on average by 3.5-fold. A–B: 400×, C–D: 200×.
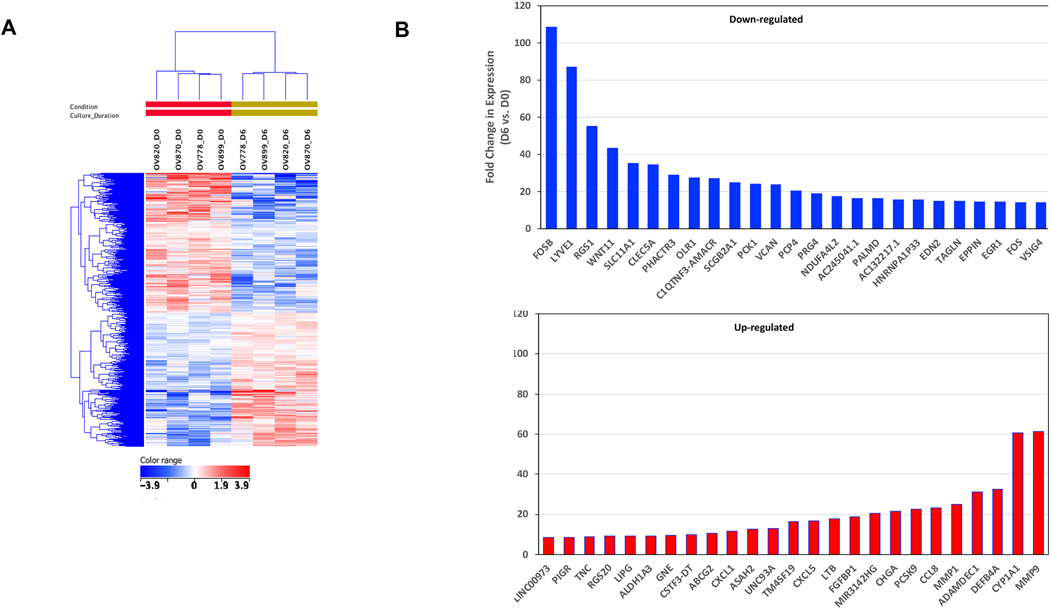
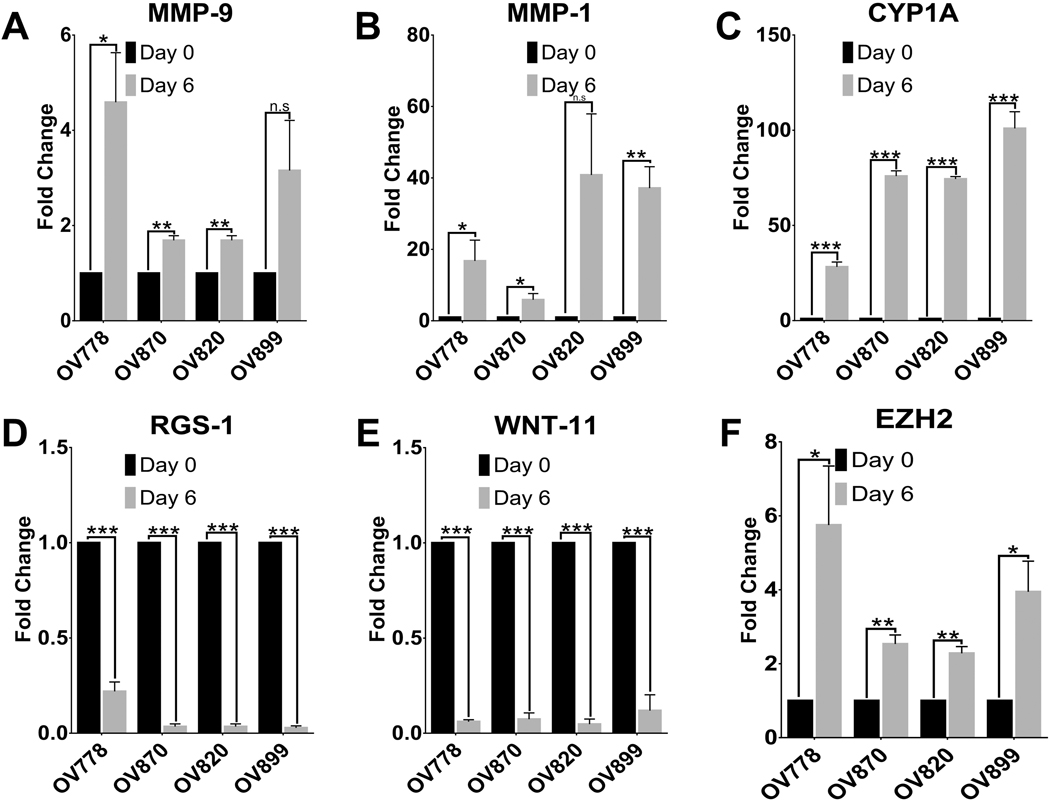
Molecular characterization of the same four pairs of Day 0 (D0) MCS and Day 6 (D6) organoids was conducted by RNA-Seq analysis. Organoid induction (culture D6 vs. uncultured D0 MCS) for all four specimens produced 1584 differentially-expressed genes (DEG; paired t-test, p < 0.05), and hierarchical clustering of the 775 up-regulated and 805 down-regulated genes resulted in organization of the D0 MCS and D6 organoids into two distinct sub-trees (Fig. 3A). The gene expression changes suggested a shift to an actively proliferating population (up-regulation of PCNA 1.7-fold and MKI67 3.3-fold), consistent with the Ki67 IHC. Also observed in all four specimens was a moderate downregulation of differentiation markers MUC1 (1.7-fold) and MUC16 (2.4-fold). Epithelial differentiation marker E-cadherin (CDH1) was slightly down-regulated (1.4-fold). Among the most highly up-regulated genes were several metalloproteinases involved in extracellular matrix remodeling and cell adhesion (MMP9 61-fold, MMP1 25-fold, and ADAMDEC1 31-fold) and a member of the P450 cytochrome oxidase family CYP1A1 (60-fold). Quantitative RT-PCR (qRT-PCR) analysis indicates MMP-9, MMP-1 and CYP1A are consistently upregulated upon organoid induction (D6 vs D0) (Fig. 4). Among the most markedly down-regulated genes were FOSB (108-fold) - a subunit of the AP-1 transcription factor; RGS1 (55-fold) - a regulator of G protein signaling; and WNT11 (43-fold) - a secreted signaling factor (Fig. 3B). The qRT-PCR analysis confirmed that RGS-1 and WNT11 are consistently downregulated upon organoid induction in these samples (Fig. 4). Except for ABCG2 and EZH2, none of the genes purported to be associated with cancer stemness potential (including CD44, CD133, ALDH1A1, NOTCH1, and STAT5B) showed consistent up-regulation during organoid induction in the four specimens examined. qRT-PCR showed upregulation of EZH2 upon organoid induction in all four specimens (Fig. 4). ABCG2 (an ATP-binding cassette transporter) was consistently upregulated, average 10.6-fold, among the four specimens but presented as a very low abundance transcript in all specimens. CD44 was upregulated in 3 of 4 specimens (average 3.0-fold) and STAT5B was slightly down-regulated in all four. GSEA indicated that organoid induction in these four specimens was strongly associated with up-regulation in Hallmark gene sets for epithelial-mesenchymal transition (p < 0.001), KRAS signaling (p < 0.001), IL6/JAK/STAT pathway signaling (p = 0.003), and other pathways commonly up-regulated in malignant tissues (e.g. MTORC signaling (p < 0.001) and E2F target signaling (p < 0.001). There were no significantly downregulated gene sets in this analysis. Fig. S1 illustrates these gene sets. These observations suggest that organoids induced under these conditions show transcriptional programs consistent with the proliferative phenotype.
Fig. 3.
Molecular characterization of organoid culture. A: Hierarchical clustering and heatmap of 1584 statistically significant differentially-expressed genes (DEG) in day 6 organoids compared to MCS (day 0); p < 0.05, >1× fold change. B: Fifty most highly DEGs (25 increased and decreased) in OVCA organoids after 6 days in organoid culture (MCS vs. day 6, paired t-test, p < 0.05). Expression changes are depicted as red or blue bars, respectively.
Fig. 4.
qRT-PCR analysis of candidate gene expression affected upon organoid induction. To validate the RNA-seq data, we performed quantitative RT-PCR assay on selected genes. Consistent with our RNA-seq data we observed a significant upregulation of genes MMP-9, MMP-1, CYP1A and EZH2 in the day 6 organoids in all samples in comparison to Day 0 (panels a, b, c and f). Similarly, significant downregulation observed in genes WNT-11 and RGS-1 were also consistent with our RNA seq data (panels d and e). These results indicate that genes that are significantly up-regulated and downregulated between the two groups of our organoid cultures can be validated and confirmed through q-RT-PCR analysis, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
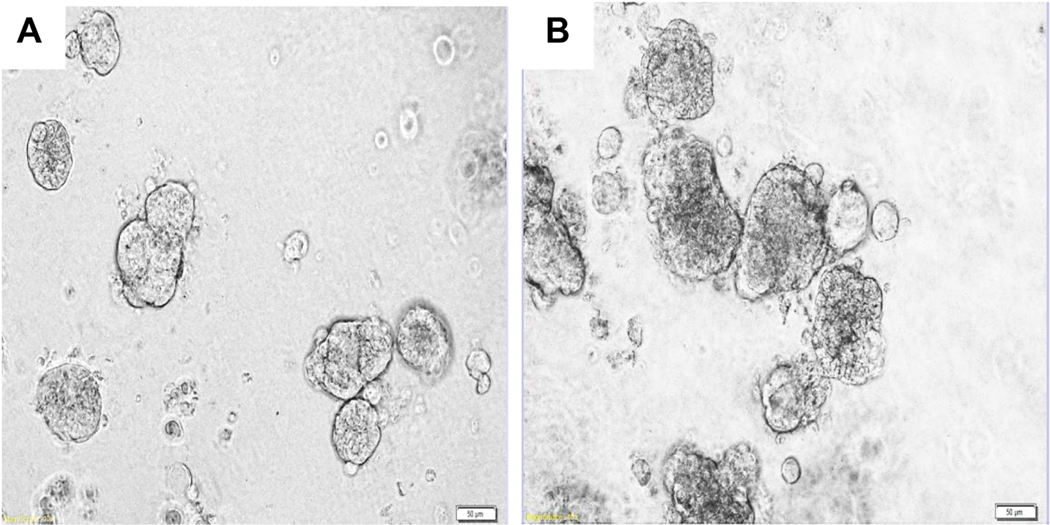
The short-term drug sensitivity assay utilized organoids cultured for 3–4 days in BME droplets, recovered, then distributed in 96 well-plates and cultured for an additional 6 days in the presence of drugs prior to CTG assay. During this phase of the assay (between days 1 and 6 of assay culture), growth was also demonstrated in the control wells. Organoid areas consistently increased (from a Day 1 average of 3500μm2 to a Day 6 average of 6500μm2 for an increase of 1.87-fold, n = 7). Fig. 5 depicts a representative appearance of day 1 and day 6 control well organoids. Fold increases in the ATP content (as measured by CellTiter Glo assay, adjusted with internal ATP standard) of untreated control wells between day 1 and day 6 of the assay were less marked (mean 1.035, n = 14, range (0.22–2.73). One value was excluded as the fold growth was 5.76 which was deemed to be an extreme outlier.
Fig. 5.
Organoid growth in drug assay. Organoid growth was studied via a number of methods. Representative appearance of OV820 Day 1 (A; 400×) and Day 6 (B; 400×) control well organoids in the drug assay are depicted here. Organoids appeared larger on day 6.
Experimental therapeutic agents (some in clinical trials and some still in the preclinical phase) were tested for activity in this organoid platform using a single screening concentration. Table 3 shows the results of screening testing for 10 drugs in 10 specimens from 6 unique subjects. Drugs that showed the most consistent activity (APR-246, CB-5083, MK-1775, and Sorafenib) were further assayed for IC50 values in duplicate or triplicate when there was sufficient material. IC50 assays were performed without screening plates in 4 specimens in order to conserve material. For these specimens, drugs were chosen based on screening testing in another specimen from the same patient. Carboplatin was also subjected to IC50 measurement. Fig. S3 shows representative triplicate IC50 curves. Table 4 shows IC50 values for a total of 118 assays mostly done in triplicate, on 13 different specimens (derived from 6 unique subjects) with 5 different drugs (carboplatin and 4 experimental therapeutic agents).
Table 3.
Drug screening assays
| Agent | % inhibition (log p value) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OV469 | OV778 | OV798a | OV817a | OV820a | OV840a | OV899b | OV860b | OV867 | OV870 | |
| 40.39 | 56.46 | 34.49 | 20.72 | 18.65 | 31.10 | 36.24 | 22.00 | 30.25 | 18.4 | |
| Olaparib | (−2.82) | (−1.73) | (−1.53) | (−4.67) | (−3.11) | (−1.58) | (−1.78) | (−1.62) | (−2.72) | (−3.15) |
| Paclitaxel | 46.29 | 17.69 | 66.95 | 61.61 | 62.39 | 81.89 | 0.28 | 22.95 | 29.88 | 21.91 |
| (−1.84) | (−0.39) | (−2.83) | (−6.38) | (−5.09) | (−4.35) | (−0.02) | (−1.33) | (−1.99) | (−2.59) | |
| 46.01 | −45.02 | 41.4 | 20.84 | 92.84 | 66.31 | 36.89 | 51.05 | 3.57 | 12.39 | |
| Mocetinostat | (−1.85) | (−1.09) | (−1.22) | (−3.39) | (−6.35) | (−2.88) | (−2.75) | (−3.32) | (−0.45) | (−3.35) |
| 37.81 | −91.96 | −66.94 | 9.18 | 44.87 | −18.14 | 19.31 | 17.06 | −39.75 | 10.34 | |
| Trametinib | (−2.74) | (−1.70) | (−2.53) | (−1.07) | (−3.64) | (−0.68) | (−0.52) | (−1.82) | (−1.61) | (−1.81) |
| 50.53 | −84.45 | 74.93 | 61.12 | 78.77 | 77.77 | −7.86 | 28.48 | 28.69 | 0.37 | |
| LY294002 | (−2.90) | (−2.46) | (−2.14) | (−6.28) | (−5.50) | (−3.59) | (−0.65) | (−1.89) | (−1.44) | (−0.4) |
| 79.70 | −90.78 | 58.90 | 38.65 | 80.16 | 69.81 | 24.57 | 39.56 | 39.63 | 29.30 | |
| AZD5363 | (−3.88) | (−3.35) | (−1.78) | (−4.65) | (−5.21) | (−3.53) | (−1.37) | (−2.20) | (−2.79) | (−3.67) |
| 40.28 | −94.71 | −8.59 | 30.46 | −98.36 | 37.60 | 7.19 | 8.69 | 4.68 | ||
| BBI503 | (−1.26) | (−1.43) | (−0.14) | – | (−2.46) | (−2.67) | (−2.66) | (−0.21) | (−0.67) | (−1.07) |
| 80.59 | 3.57 | 90.32 | 94.77 | 98.21 | 91.43 | 65.79 | 57.97 | 63.51 | 75.15 | |
| MK-1775 | (−3.81) | (−0.07) | (−2.42) | (−7.00) | (−5.78) | (−4.18) | (−4.29) | (−3.75) | (−3.90) | (−6.19) |
| 85.34 | 33.00 | −32.89 | 99.11 | 97.71 | −10.31 | 99.89 | 17.32 | 3.95 | 98.99 | |
| Sorafenib | (−4.31) | (−1.16) | (−0.62) | (−7.21) | (−5.78) | (−0.30) | (−5.16) | (−1.01) | (−0.20) | (−6.84) |
| 99.98 | 99.78 | 99.96 | 99.98 | 99.99 | 99.96 | 98.2 | 99.9 | 99.90 | 99.98 | |
| APR-246 | (−4.54) | (−3.65) | (−2.74) | (−7.21) | (−5.84) | (−4.26) | (−6.35) | (−5.14) | (−4.70) | (−6.82) |
| 99.97 | 99.38 | 99.94 | 99.98 | 99.96 | 99.91 | 95.39 | 92.57 | 97.95 | 99.79 | |
| CB-5083 | (−4.54) | (−3.66) | (−2.74) | (−7.21) | (−5.85) | (−4.27) | (−5.86) | (−5.04) | (−4.61) | (−6.83) |
| 93.29 | −60.6 | 0.73 | 30.34 | 13.02 | −99.69 | 14.06 | 75.78 | 65.93 | 25.80 | |
| Napabucasin | (−4.22) | (−1.02) | (−0.01) | (−4.39) | (−1.90) | (−1.85) | (−1.44) | (−3.86) | (−3.67) | (−4.44) |
Results are reported as % inhibition (log p value).
Clinically significant inhibition defined as >50% inhibition, displayed in bold.
Subject 3.
Subject 4.
Table 4.
IC50 results.
| Subject | Specimen | Carboplatin | APR-246 | CB-5083 | MK-1775 | Sorafenib |
|---|---|---|---|---|---|---|
| 1 | OV469 | 16.22 [2] | - | - | - | - |
| 2 | OV778 | 34.1 | 31.75 | 1.35 | ||
| (18.7–44.5) [3] | (23.6–39.9) [2] | (0.5–2.2) [2] | - | - | ||
| 3 | OV798 | - | 5.8 [1] | 1.0 [1] | 0.2 [1] | >12.5 [1] |
| OV820 | 36.9 | 38.2 | 1.17 | 0.6 | 2.7 | |
| 3 | (28.7–38.3) [3] | (15.8–77.8) [3] | (0.6–1.5) [3] | (0.2–1.1) [3] | [1] | |
| OV838 | 38.4 | 94.2 | 1.18 | 1.4 | - | |
| 3 | (34.8–41.4) [3] | (85.5–130.5) [2] | (0.9–1.4) [3] | (0.9–2.2) [3] | - | |
| 3 | OV840 | - | 7.4 [1] | 0.6 [1] | 0.4 [1] | >12.5 [1] |
| OV899 | 39.4 | 47.9 | 0.5 | 0.8 | 1.8 | |
| 4 | (28.0–54.5) [3] | (31.9–57) [3] | (0.4–0.5) [3] | (0.8–0.8) [2] | (1.8–5.7) [2] | |
| 4 | OV86O | 47.1 (42.2–55.9) [3] |
22.2 (16.5–27.9) [2] |
0.4 (0.4–0.4) [2] |
1.1 [1] | - |
| 5 | OV867 | 65.4 (45.3–98.8) [3] |
18.0 (7–38.8) [3] |
1.8 (0.8–2.4) [3] |
1.2 [1] | - |
| 5 | OV875 | 41.3 [1] | 29.1 [1] | 1.3 [1] | - | - |
| 5 | OV882 | 49.9 (19.7–70.6) [3] |
62.88 (31.1–90.9) [3] |
1.4 (1–1.9) [3] |
0.5 (0.3–0.8) [3] |
2.8 [1] |
| OV870 | 65.6 | 92.23 | 1.5 | 0.2 | 3.5 | |
| 6 | (50.3–87.2) [3] | (34.3–148.3) [3] | (0.8–2.9) [3] | (0.1–0.4) [3] | (3.2–4.8) [3] | |
| OV888 | 75.3 | 126.3 | 1.2 | 0.3 | 1.6 | |
| 6 | (61.9–94.2) [3] | (99.6–140.6) [3] | (0.5–1.9) [3] | (0.1–0.5) [3] | (1.2–2.0) [3] | |
Results are displayed as IC50 (μM) (range) [n].
The pattern for taxane inhibition in this assay was complex. Five specimens from 4 unique subjects were subjected to paclitaxel in IC50 assays. OV820 yielded an IC50 of 0.08 μM. IC50 assays in the remaining specimens (n= 4) with maximum paclitaxel concentration of 40 μM resulted in resistance. Because of solubility issues with paclitaxel, a higher concentration could not be achieved for testing. Nab-paclitaxel was used to achieve higher concentrations in the assay. OV820 and OV838 (participant 3) showed an inflection in the viability curve at low nab-paclitaxel concentrations (range of 0.01–0.26 μm). OV838 also showed more inhibition with higher concentrations (range of 5.91–54.97 μM, Fig. S2). Other specimens (OV870 and OV888) had a more traditional one phase inhibition with the IC50 achieved at a high concentration (range 20.8–51.7 μM). OV469 and OV899 were completely unaffected by taxanes, even at the highest concentration. In all, 23 IC50 assays were performed with taxanes (5 with paclitaxel and 18 with nab-paclitaxel).
4. Discussion
Under the culture conditions used here, organoids developed from MCS within days of initiating culture. Based on a detailed analysis of four specimens, organoid induction was characterized by the following: 1) an increase in the mean area of organoid structures; 2) more evidence of cell death; 3) increased cell proliferation; 4) an upregulation of genes related to cellular proliferation, epithelial-mesenchymal transition, and KRAS signaling pathways. Organoid induction under these conditions provides a model for the study of a highly proliferative cell population derived from a subset of original MCS, perhaps recapitulating the initiation of metastatic tumor growth following implantation of MCS onto the peritoneal surface. Such a model may be useful in empiric drug sensitivity testing.
This organoid induction model was adapted for drug sensitivity testing of 10 targeted agents as well as carboplatin and taxanes. Drug sensitivity results using a single targeted drug concentration on 10 specimens originating from 6 unique patients showed that four drugs (APR-246, CB-5083, MK-1775, and Sorafenib) had the most consistent inhibitory effects. These four targeted agents along with carboplatin, paclitaxel and/or nab-paclitaxel were subjected to IC50 testing. IC50 values for APR-246, MK1775 and Sorafenib were close to or lower than the published Cmax value associated with therapeutic dosage. Although there was some consistency, response to drugs in the screening assay was variable between subjects, highlighting the importance of an empiric assay for each patient and the need for individualized therapy.
IC50 values for carboplatin (range 18.7–98.8 μM) were generally lower than or approximately equal to published Cmax values. Although contemporary carboplatin doses are expressed as AUC values, prior pharmacokinetic studies [24] determined Cmax values for a range of carboplatin doses. Carboplatin delivered at 300 and 450 mg/m2 yielded Cmax values of 85 and 149 μM, respectively. IC50 values for carboplatin in the organoid assay described here (Table 4 and Supplemental Fig. S3) were mostly below these Cmax values suggesting that organoid viability should be substantially affected by the concentrations of carboplatin that can be achieved in vivo with standard dosing. The maximum concentration used in the assay described here was 314 μM. Therefore, had there been another inflection point approaching the published Cmax, it likely would have been detected. IC50 values for carboplatin suggests that this testing platform is probably not useful for determining clinically significant platinum sensitivity.
Paclitaxel was tested to a maximum of 40 μM showing that the IC50 was above this level (except for specimen OV820), however, solubility issues prevented testing higher concentrations. Consequently, nab-paclitaxel in aqueous solution was also tested and resulted in variable responses. Specimens from one unique subject (participant1) were particularly sensitive to taxanes and two inflection points were noted; one at a very low concentration and a second at a high concentration (Supplemental Fig. S2). In other specimens, IC50 value was achieved at a concentration above the published Cmax while still others were entirely resistant. This is consistent with the literature which suggests an approximately 20% response rate to single-agent Taxol in recurrent ovarian cancer [25,26] and with Jabs et al. who noted that paclitaxel resulted in inhibition in two out of nine patient cancer organoids [27].
It is interesting to note that among the experimental therapeutics, Wee1 inhibitor MK-1775 (AZD1775) and VCP (p97) inhibitor CB-5083 show consistent growth inhibitor effects in low micromolar range in all PDO models tested. Wee1 kinase regulates G2 checkpoint, and tumor cells with mutations in TP53 are susceptible to G2 checkpoint abrogation. Therefore, it is not surprising that PDO derived from HGSOC characterized by mutations in TP53 show sensitivity to MK-1775. Although overall survival data is immature, a recent phase II randomized clinical trial of MK-1775 (NCT0137161) shows a significant increase in the progression-free survival when MK-1775 was added to the carboplatin-paclitaxel combination therapy compared to the doublet [28]. Valosin-containing protein (VCP, also known as p97) is an AAA-ATPase with diverse functions in protein and organelle homeostasis. A recent genome-wide shRNA screen identifies VCP as a potential drug target in ovarian cancer because VCP knockdown causes lineage-specific dependencies in ovarian cancer [29]. A recently developed p97 inhibitor CB-5083 shows a broad spectrum of cytotoxicity in over 300 cancer cell lines as well as antitumor activities in several cancer cell line xenografts and patient-derived xenografts [30]. CB-5083 and other VCP inhibitors show cytotoxicity against established ovarian cancer cell lines in 2D culture [31]. Finally, it is important to note that some PDOs (OV798 and OV840) show enhanced sensitivity to APR-246, which reactivates specific p53 mutants [32,33]. Interestingly, four other PDOs (OV778, OV820, OV870, and OV899) are less sensitive to APR-246. It is not surprising that OV820 and OV870 are less sensitive to ARP-246 because they have truncation mutations (Fig. S4). Moreover, OV778 and OV899 harbor missense mutations (Fig. S4) but they are also less sensitive to APR-246. These results suggest that some missense mutants are not amenable to rescue by APR-246, and cells harboring these mutations are relatively insensitive to APR-246. Nonetheless, the IC50 for APR-246 in these models are in the high micromolar range that is clinically achievable based on the Cmax values obtained from clinical trials. Therefore, APR-246 may exert cytotoxic activity through additional targets, such as thioredoxin reductase [34,35]. Collectively, these experimental therapeutic agents hold promise for further evaluation of their clinical activities in patients with HGSOC.
The short-term organoid culture platform described here has limitations. Cellular material from palliative aspiration of refractory malignant effusions is a non-renewable resource, although additional material can sometimes be obtained if palliative aspiration is repeated. Some malignant effusions contain insufficient cellular material for detailed study. The assay employs a subpopulation of MCS structures separated by selective sieving; larger or smaller structures could theoretically display different results. Sieving to remove mononuclear cells may remove tumor-associated fibroblasts or immune cells important in the tumor microenvironment. However, mononuclear cells in the malignant effusions analyzed here usually represented more than half of the total cellular material. Consequently, to assess tumor cells’ response to treatment, the mononuclear cell fraction was excluded in these experiments.
Repeated measurement of IC50 values for some drugs tested here showed variability. Potential sources of variability include different viability of cryopreserved aliquots of the same specimen, variability induced by exposure to chemotherapy soon before specimen acquisition and variability of the distribution of organoids into each assay well. In situ methods for measuring the proportion of living vs. dead cells inside each organoid could increase the accuracy of this platform [27]. The medium described by Sachs et al. was selected since genomic analysis has established that at least one subtype of breast cancer shares the features of high copy number changes and TP53 mutations that characterize HGSOC [7,36]. Hill et al. recently reported the results of short-term ovarian cancer PDO cultures showing the potential value of organoid culture in assessing targetable defects, particularly those with DNA repair defects [37].
Though primary tumors can be removed surgically, metastatic tumors usually require drug therapy. Malignant effusion represents a form of metastatic disease that often becomes refractory to drug treatment. Palliative removal of recurrent effusions is a common and temporarily effective means of reducing symptoms of these conditions but also allows for access to individual patient’s tumor cells for empirical testing. The drug testing platform described here is feasible with this group of patients. Though the development of long-term organoid lines that can be continuously passaged and manipulated experimentally has many advantages, the time it takes to establish each line may preclude real-time clinical application for individual patients. Also, there is the possibility that genetic drift or subclone selection may occur during long-term culture such that the resulting organoid line is less representative of the original tumor. Short-term culture has the advantage of allowing for drug testing within one week of specimen acquisition, and though limited by the quantity of cellular material initially obtained, it opens the possibility of empirical testing of potential therapeutic agents in a time frame that could affect clinical outcomes. Because of the complexities of ovarian cancer genomic changes, the assay approach described here was developed to allow empirical testing of drugs that might not be considered as options for salvage therapy.
An important question for any study using PDOs is to relate the observations to clinical results. The goal of the present study was not to correlate clinical outcomes to assay results, however based on limited de-identified clinical information available, all patients did progress or recur after first line therapy (platinum and taxane). Interestingly, the majority of unique subjects PDOs were also resistant to taxane at standard doses. Two specimens (OV899 and OV778) are from participants who had not yet received chemotherapy at the time of effusion collection. These patients both progressed on platinum/taxane therapy and PDOs from both specimens were resistant to taxane. In the literature, initial reports comparing organoid drug sensitivity assay results and clinical outcomes are beginning to accumulate. Sachs et al. demonstrated that PDOs from breast cancer had responses to treatment that matched the patient’s outcomes [7]. Currently, the TUMOROID and SENSOR trials are using PDOs to allocate patients for treatment with targeted agents based on PDO drug sensitivities [38]. Similar clinical trials for HGSOC can be helpful in determining effective and personalized treatment for patients.
Finally, these PDOs may also be useful for dissecting the molecular mechanisms associated with treatment resistance. PDOs grown under 3D condition can be selected with chemotherapy in parallel to patient receiving treatment. Resistant cells that persist and expand after ex vivo drug exposure can be re-evaluated for genetic alterations to help explain ex vivo resistant mechanisms. In parallel, corresponding recurrent patient samples can be re-evaluated for genetic alterations and compared with genetic alterations identified from ex vivo selected PDOs. These comparative studies may provide new mechanistic insights into clinically relevant drug resistance mechanisms.
5. Conclusions
PDO cultures were successfully grown from MCS present in HGSOC in serum-free conditions. A short-duration PDO culture can be used to study drug susceptibilities for individual tumors. PDO-based in vitro drug sensitivity testing of MCS from ovarian cancer malignant effusions can provide quick information regarding targeted treatments that may be beneficial for a group of patients who often exhaust standard therapeutic options. Future clinical studies will be needed to correlate PDO assay results with patient outcomes.
Supplementary Material
HIGHLIGHTS.
Patient-derived organoids (PDOs) can be derived from malignant ascites and pleural effusions.
PDOs can be used for empirical drug testing of novel therapeutics.
RNA-sequencing analysis indicates gene signature associated with proliferation upon organoid induction.
PDOs represent an efficient model that recapitulates histological features of malignant ascites and pleural effusions.
Acknowledgments and grant support
The authors are grateful to Stephenie Y. Liu and Ryan R. Davis (Genomics Shared Resource, Department of Pathology and Laboratory Medicine, UC Davis School of Medicine) for their assistance in conducting the RNA-seq studies. The UC Davis Comprehensive Cancer Center Genomics Shared Resource is supported by Cancer Center Support Grant P30CA093373 (PI: Lara) from the National Cancer Institute. Additional funding was provided by the Department of Obstetrics and Gynecology at UC Davis School of Medicine and the Gynecologic Oncology Fellowship program at UC Davis School of Medicine.
Footnotes
Declaration of competing interest
The authors declare no conflict of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygyno.2020.03.026.
References
- [1].Oronsky B, Ray CM, Spira AI, Trepel JB, Carter CA, Cottrill HM, A brief review of the management of platinum-resistant-platinum-refractory ovarian cancer, Med. Oncol 34 (2017) 103. [DOI] [PubMed] [Google Scholar]
- [2].Heintz AP, Odicino F, Maisonneuve P, Quinn MA, Benedet JL, Creasman WT, et al. Carcinoma of the ovary. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95 Suppl 1:S161–92. [DOI] [PubMed] [Google Scholar]
- [3].Cancer Genome Atlas Research N, Integrated genomic analyses of ovarian carcinoma, Nature. 474 (2011) 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cybulska P, Stewart JM, Sayad A, Virtanen C, Shaw PA, Clarke B, et al. , A genomically characterized collection of high-grade serous ovarian cancer xenografts for preclinical testing, Am. J. Pathol 188 (2018) 1120–1131. [DOI] [PubMed] [Google Scholar]
- [5].Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC, In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform, Front. Bioeng. Biotechnol 4 (2016) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. , Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 13308–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sachs N, Clevers H, Organoid cultures for the analysis of cancer phenotypes, Curr. Opin. Genet. Dev 24 (2014) 68–73. [DOI] [PubMed] [Google Scholar]
- [8].Blagosklonny MV, Cancer stem cell and cancer stemloids: from biology to therapy, Cancer Biol. Ther 6 (2007) 1684–1690. [DOI] [PubMed] [Google Scholar]
- [9].Reya T, Morrison SJ, Clarke MF, Weissman IL, Stem cells, cancer, and cancer stem cells, Nature. 414 (2001) 105–111. [DOI] [PubMed] [Google Scholar]
- [10].Visvader JE, Lindeman GJ, Cancer stem cells in solid tumours: accumulating evidence and unresolved questions, Nat. Rev. Cancer 8 (2008) 755–768. [DOI] [PubMed] [Google Scholar]
- [11].Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP The developing cancer stem-cell model: clinical challenges and opportunities, Lancet Oncol. 13 (2012) e83–e89. [DOI] [PubMed] [Google Scholar]
- [12].Girda E, Huang EC, Leiserowitz GS, Smith LH, The use of endometrial cancer patient-derived organoid culture for drug sensitivity testing is feasible, Int. J. Gynecol. Cancer 27 (2017) 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weeber F, Ooft SN, Dijkstra KK, Voest EE, Tumor organoids as a pre-clinical cancer model for drug discovery, Cell Chem. Biol 24 (2017) 1092–1100. [DOI] [PubMed] [Google Scholar]
- [14].Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. , Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium, Gastroenterology. 141 (2011) 1762–1772. [DOI] [PubMed] [Google Scholar]
- [15].Neal JT, Kuo CJ, Organoids as models for neoplastic transformation, Annu. Rev. Pathol 11 (2016) 199–220. [DOI] [PubMed] [Google Scholar]
- [16].Al Habyan S, Kalos C, Szymborski J, McCaffrey L, Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer, Oncogene. 37 (2018) 5127–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Francies HE, Barthorpe A, McLaren-Douglas A, Barendt WJ, Garnett MJ Drug sensitivity assays of human cancer organoid cultures . Methods Mol Biol, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chavez M, Silvestrini MT, Ingham ES, Fite BZ, Mahakian LM, Tam SM, et al. , Distinct immune signatures in directly treated and distant tumors result from TLR adjuvants and focal ablation, Theranostics. 8 (2018) 3611–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression, Nat. Methods 14 (2017) 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell Syst. 1 (2015) 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oguri S, Sakakibara T, Mase H, Shimizu T, Ishikawa K, Kimura K, et al. , Clinical pharmacokinetics of carboplatin, J. Clin. Pharmacol 28 (1988) 208–215. [DOI] [PubMed] [Google Scholar]
- [25].Trimble EL, Adams JD, Vena D, Hawkins MJ, Friedman MA, Fisherman JS, et al. , Paclitaxel for platinum-refractory ovarian cancer: results from the first 1,000 patients registered to National Cancer Institute Treatment Referral Center 9103, J. Clin. Oncol 11 (1993) 2405–2410. [DOI] [PubMed] [Google Scholar]
- [26].Einzig AI, Wiernik PH, Sasloff J, Runowicz CD, Goldberg GL, Phase II study and long-term follow-up of patients treated with taxol for advanced ovarian adenocarcinoma, J. Clin. Oncol 10 (1992) 1748–1753. [DOI] [PubMed] [Google Scholar]
- [27].Jabs J, Zickgraf FM, Park J, Wagner S, Jiang X, Jechow K, et al. , Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations, Mol. Syst. Biol 13 (2017) 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Oza AM, Weberpals JI, Provencher DM, Grischke E-M, Hall M, Uyar D, et al. An international, biomarker-directed, randomized, phase II trial of AZD1775 plus paclitaxel and carboplatin (P/C) for the treatment of women with platinum-sensitive, TP53-mutant ovarian cancer. Journal of Clinical Oncology. 2015;33:5506–. [DOI] [PubMed] [Google Scholar]
- [29].Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, et al. , Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 12372–12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, et al. , Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis, Cancer Cell 28 (2015) 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bastola P, Neums L, Schoenen FJ, Chien J, VCP inhibitors induce endoplasmic reticulum stress, cause cell cycle arrest, trigger caspase-mediated cell death and synergistically kill ovarian cancer cells in combination with Salubrinal, Mol. Oncol 10 (2016) 1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, et al. , Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound, Nat. Med 8 (2002) 282–288. [DOI] [PubMed] [Google Scholar]
- [33].Bykov VJ, Zache N, Stridh H, Westman J, Bergman J, Selivanova G, et al. , PRIMA-1 (MET) synergizes with cisplatin to induce tumor cell apoptosis, Oncogene. 24 (2005) 3484–3491. [DOI] [PubMed] [Google Scholar]
- [34].Bykov VJ, Zhang Q, Zhang M, Ceder S, Abrahmsen L, Wiman KG, Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient cancer therapy, Front. Oncol 6 (2016) 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Perdrix A, Najem A, Saussez S, Awada A, Journe F, Ghanem G, et al. PRIMA-1 and PRIMA-1(Met) (APR-246): From mutant/wild type p53 reactivation to unexpected mechanisms underlying their potent anti-tumor effect in combinatorial therapies. Cancers (Basel). 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr., et al. , Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids, Cancer Discov. 8 (2018) 1404–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Weeber F, Predicting Treatment Outcome by DNA and Organoids: Utrecht University, 2017. [Google Scholar]
- [39].Garcia-Manero G, Assouline S, Cortes J, Estrov Z, Kantarjian H, Yang H, et al. , Phase 1 study of the oral isotype specific histone deacetylase inhibitor MGCD0103 in leukemia, Blood. 112 (2008) 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Infante JR, Papadopoulos KP, Bendell JC, Patnaik A, Burris HA 3rd, Rasco D, et al. , A phase 1b study of trametinib, an oral mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours, Eur. J. Cancer 49 (2013) 2077–2085. [DOI] [PubMed] [Google Scholar]
- [41].Mahadevan D, Chiorean EG, Harris WB, Von Hoff DD, Stejskal-Barnett A, Qi W, et al. , Phase I pharmacokinetic and pharmacodynamic study of the pan-PI3K/mTORC vascular targeted pro-drug SF1126 in patients with advanced solid tumours and B-cell malignancies, Eur. J. Cancer 48 (2012) 3319–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tamura K, Hashimoto J, Tanabe Y, Kodaira M, Yonemori K, Seto T, et al. , Safety and tolerability of AZD5363 in Japanese patients with advanced solid tumors, Cancer Chemother. Pharmacol 77 (2016) 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Do K, Wilsker D, Ji J, Zlott J, Freshwater T, Kinders RJ, et al. , Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors, J. Clin. Oncol 33 (2015) 3409–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vaishampayan UN, Burger AM, Sausville EA, Heilbrun LK, Li J, Horiba MN, et al. , Safety, efficacy, pharmacokinetics, and pharmacodynamics of the combination of sorafenib and tanespimycin, Clin. Cancer Res 16 (2010) 3795–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hubbard JM, Grothey A, Napabucasin: an update on the first-in-class cancer stemness inhibitor, Drugs. 77 (2017) 1091–1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.