Abstract
Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous disease whose prognosis is associated with clinical features, cell-of-origin and genetic aberrations. Recent integrative, multi-omic analyses had led to identifying overlapping genetic DLBCL subtypes. We used targeted massive sequencing to analyze 84 diagnostic samples from a multicenter cohort of patients with DLBCL treated with rituximab-containing therapies and a median follow-up of 6 years. The most frequently mutated genes were IGLL5 (43%), KMT2D (33.3%), CREBBP (28.6%), PIM1 (26.2%), and CARD11 (22.6%). Mutations in CD79B were associated with a higher risk of relapse after treatment, whereas patients with mutations in CD79B, ETS1, and CD58 had a significantly shorter survival. Based on the new genetic DLBCL classifications, we tested and validated a simplified method to classify samples in five genetic subtypes analyzing the mutational status of 26 genes and BCL2 and BCL6 translocations. We propose a two-step genetic DLBCL classifier (2-S), integrating the most significant features from previous algorithms, to classify the samples as N12-S, EZB2-S, MCD2-S, BN22-S, and ST22-S groups. We determined its sensitivity and specificity, compared with the other established algorithms, and evaluated its clinical impact. The results showed that ST22-S is the group with the best clinical outcome and N12-S, the more aggressive one. EZB2-S identified a subgroup with a worse prognosis among GCB-DLBLC cases.
Subject terms: Lymphoma, Molecular medicine, Next-generation sequencing
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin lymphoma, with 3.13 and 5.6 new cases diagnosed in Europe1 and the USA2 per 100,000 habitants per year, respectively. DLBCL is an aggressive and heterogeneous disease with a variable clinical outcome; it can arise de novo or after histological transformation from other low-grade lymphomas, typically from follicular lymphoma. Most DLBCL patients can be cured by standard immunochemotherapy with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone)3. However, a substantial percentage of them (30–40%) are refractory to treatment or relapse (R/R) after an initial response, and it is not possible to accurately predict which patients will benefit from rituximab-based therapy4. Currently, the International Prognostic Index (IPI), based on clinical and analytical characteristics, is the most significant predictive factor, in which higher scores are associated with unfavorable outcomes5–7.
The most recent revision of the WHO classification of lymphoid neoplasms recognizes the high-grade B-cell double-/triple-hit lymphoma (DH/TH), with MYC and BCL2 and/or BCL6 rearrangements, as a new provisional entity associated with an inferior outcome. DLBCLs co-expressing MYC and BCL2 (double-expressor lymphomas) also have a worse prognosis than other DLBCL-NOS (not otherwise specified), but their behavior is not as aggressive as that of DH/TH lymphomas8.
Gene-expression profiling (GEP) allows distinguishes three subtypes based on cell of origin (COO): germinal center B-cell (GCB)-like, activated B-cell (ABC)-like and unclassified subtypes9,10. This classification has been shown to be of prognostic value, with ABC-DLBCL being associated with poorer outcome9–11, but it does not fully explain the high DLBCL heterogeneity, or accurately predict the response to standard therapy. The truth is that all patients are treated identically, independently of their COO subtype. Therefore, we need to identify the genetic alterations of DLBCL associated with refractoriness and develop alternative treatments or novel pharmacological strategies to overcome this resistance.
In the last few years, deep-sequencing studies have allowed a better understanding of the DLBCL genomic landscape and provided further evidence of their molecular heterogeneity. Several recent studies have proposed new genetic subtypes based on the DLBCL genomic profile. Although they are somewhat different, the newly defined genetic subtypes share several characteristics. Schmitz and colleagues identified four genetic subgroups, which they referred to as MCD (characterized by the co-occurrence of MYD88L265P and CD79B mutations), BN2 (with BCL6 fusions and NOTCH2 mutations), N1 (with NOTCH1 mutations) and EZB (characterized by EZH2 mutations and BCL2 translocations). MCD and N1 are associated with poorer outcomes than the other subtypes12. Most recently, the same group developed the LymphGen algorithm, which allows a more precise genetic classification, adding the A53 (characterized by TP53 mutations and deletions) and ST2 (SGK1 and TET2 mutated) subtypes, to the previous ones13. Chapuy et al.14 distinguished five subsets of DLBCL, including two ABC-DLBCL groups, one with low risk and a possible marginal zone origin (C1), and the other a high-risk group (C5) enriched in cases with mutations in MYD88, CD79B, and PIM1; they also described two subsets of GCB-DLBCLs with favorable (C4) and poor (C3) outcomes, and an ABC/GCB-independent group (C2) with biallelic inactivation of TP53, CDKN2A loss, and associated genomic instability. Finally, in an attempt to bring these genetic classifications together, Lacy et al.15 characterized five molecular subtypes, NOTCH2, MYD88, BCL2, TET2/SGK1, and SOCS1/SGK1, according to the mutated genes that are most highly enriched in each. The genetic classifications partially overlap, suggesting the existence of accurate molecular subtypes that might have predictive and prognostic capability and help to select the most appropriate therapy for each DLBCL patient. However, for these genetic classifications to be genuinely useful, a feasible, consensus classification based on selected genetic alterations must be validated to contribute to a sensitive and specific classification that is of clinical value.
We propose a simplified classification based on the previous ones, validated using data obtained by targeted deep-sequencing analysis in a set of diagnostic samples from DLBCL patients treated with R-CHOP or similar regimens, 40% of whom (34/84) did not respond, or relapsed after treatment.
Results
Mutational profile and association with clinical outcome
Patient and tumor characteristics of the "Puerta de Hierro" (PdH) cohort (n = 84) are shown in Table 1 and Supplementary Table S1. FISH analysis and COO classification were performed on samples with available material. Two of the 44 samples studied by FISH (4.5%) were classified as DH. Regarding COO, Lymph2Cx-based assay determined that 53.7% (29/54) of the cases were GCB, 25.9% ABC (14/54) and 20.4% (11/54) unclassified, while the Hans algorithm classified 50.8% (33/65) as GCB and 49.2% (32/65) as non-GCB.
Table 1.
Summary of clinical data of the Puerta de Hierro (PdH) cohort.
| Clinical variable | N | Categories | N | Percentage |
|---|---|---|---|---|
| Sex | 84 | Male | 46 | 54.8 |
| Female | 38 | 45.2 | ||
| Age | 83 | ≥ 60 | 47 | 56.7 |
| < 60 | 36 | 43.4 | ||
| ECOG | 78 | 0 | 28 | 36.8 |
| 1 | 29 | 38.2 | ||
| 2 | 18 | 23.7 | ||
| 3 | 3 | 3.9 | ||
| 4 | 0 | 0 | ||
| 5 | 0 | 0 | ||
| Stage | 81 | I | 6 | 7.8 |
| II | 23 | 29.9 | ||
| III | 31 | 40.3 | ||
| IV | 31 | 40.3 | ||
| IPI | 83 | Low | 28 | 33.7 |
| Low-Intermediate | 23 | 27.7 | ||
| High-Intermediate | 18 | 21.7 | ||
| High | 14 | 16.9 | ||
| Treatment | 84 | R-CHOP | 76 | 90.5 |
| R-like | 8 | 9.5 | ||
| Response to treatment | 83 | Complete response | 66 | 79.5 |
| Partial response | 6 | 7.2 | ||
| No response | 11 | 13.3 | ||
| Refractory/Relapse | 84 | Refractory/Relapse | 34 | 40.5 |
| No relapse | 50 | 59.5 | ||
| Status | 84 | Exitus | 33 | 39.3 |
| Alive | 51 | 60.7 | ||
| COO (Lymph2Cx) | 54 | GCB | 29 | 53.7 |
| ABC | 14 | 25.9 | ||
| Unclassified | 11 | 20.4 | ||
| COO (Hans) | 65 | GCB | 33 | 50.8 |
| Non-GCB | 32 | 49.2 | ||
| FISH | BLC2 | 11/50 | 22.0 | |
| BCL6 | 18/45 | 40.0 | ||
| MYC | 5/49 | 10.2 | ||
| Double-HIT | 2/44 | 4.5 |
International Prognostic Index (IPI), rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), rituximab-like (R-like), cell of origin (COO), activated B-cell (ABC), germinal center B-cell (GCB), fluorescence in situ hybridization (FISH).
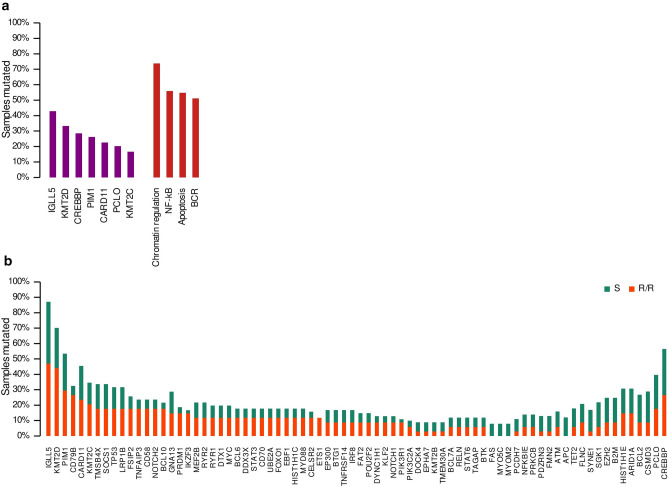
We performed targeted massive parallel sequencing in the 84 DLBCL samples, and were able to identify at least one somatic mutation in all the samples (Suppl. Table S2). All the genes included in the panel (125; see “Methods” section) were mutated at least once, except KRAS and CD22. A total of 1030 somatic mutations (SNVs and indels) were detected, considering missense, non-sense, and splicing mutations. The samples harbored a median of 12.3 mutations (range: 2–45), 13.4 mutations in R/R cases (range: 2–45), and 11.5 mutations in sensitive (S) cases (range: 3–30). The most recurrently mutated genes were IGLL5 (42.9%, 36/84 cases), KMT2D (33.3%, 28/84), CREBBP (28.6%, 24/84), PIM1 (26.2%, 22/84), CARD11 (22.6%, 19/84), PCLO (20.2%, 17/84), and KMT2C (19.4%, 13/84) (Fig. 1A; Suppl. Table S3).
Figure 1.
Mutational prevalence in genes and pathways. (A) Most frequently mutated genes and pathways in the whole cohort. (B) Mutational prevalence for refractory/relapsed (R/R) and sensitive (S) cases for genes with more than four samples mutated.
We also explored the DLBCL mutational landscape in predefined signaling pathways or lymphomagenesis-related gene sets. Genes included in every gene set, based on the previously published B-cell NHL gene signatures16, are summarized in Supplementary Table S4. Overall, the samples had a higher incidence of mutations in genes involved in chromatin regulation (73.8%, 62/84), NFκB (58.3%, 47/84), apoptosis (54.8%, 46/84), and BCR pathways (51.2%, 43/84) (Fig. 1A).
We assessed the prognostic value of IPI and the COO classification in our series. The IPI predicted shorter overall survival (OS) (Intermediate risk: p = 0.009, HR = 5.04; High risk: p = 0.003; HR = 7.9) and progression-free survival (PFS) (Intermediate risk: p = 0.002, HR = 6.72; High risk: p = 0.004; HR = 7.13) by Cox proportional-hazards model analysis for the intermediate- and high-risk groups. When we evaluated the prognostic value of the COO by the Kaplan–Meier survival method, GCB cases showed better clinical outcomes, but the magnitude of the differences was not great enough to be significant, given the small number of classified samples available (Fig. S1).
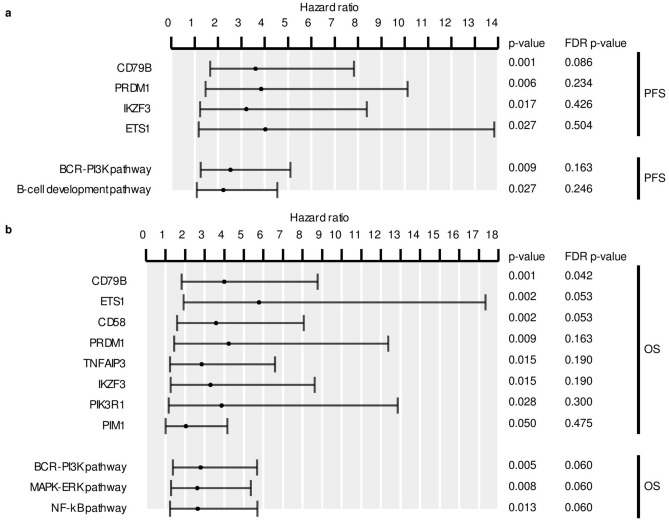
We then explored the clinical relevance of the gene/pathway mutations present in at least four samples. The analysis showed that CD79B mutations were associated with a higher risk of R/R (Figs. 1B, 2A). Similarly, patients with mutations in B-cell development and BCR-PI3K pathways were more prone to relapse after therapy. Although not significantly associated, several other genes and pathways were more frequently mutated in samples from R/R patients, such as PRDM1 (15% vs. 4%), ETS1 (12% vs. 0%), IKZF3 (15% vs. 2%), BCL10 (18% vs. 4%), NOTCH2 (18% vs. 6%), CD58 (16% vs. 6%), and MAPK-ERK (32% vs. 16%), and Toll pathways (18% vs. 8%) (Fig. 1B, Suppl. Tables S3 and S5). Furthermore, the multivariate analysis, combined with the IPI, showed that patients with mutations in CD79B had an independent association with shorter PFS (HR = 2.73, p = 0.01). The analysis also demonstrated that mutations in CD79B, ETS1, and CD58, were associated with shorter OS (Fig. 2B), as well as mutations in the BCR-PI3K, MAPK-ERK, and NFκB pathways (Fig. 2B). Mutations in CD79B (HR = 2.72, p = 0.02), and ETS1 (HR = 3.67, p = 0.03) were also associated with a worse OS, as indicated by the multivariate analysis with the IPI.
Figure 2.
Statistical analysis of genes and pathways in the PdH cohort. Mutated genes and pathways with significant p-values in the univariate Cox proportional-hazards model analysis for (A) progression-free (PFS) and (B) overall survival (OS). Error bars represent the 95% confidence intervals for the hazard ratios. FDR p-values for Benjamini–Hochberg correction are shown.
Proposal of a combined two-step method for DLBLC genetic classification
Several attempts have been made to improve COO classification by considering tumor genetics in order to facilitate the use of precision-medicine approaches in DLBLC patients. This has led to new DLBCL genetic classifications, as described earlier. One of our aims was to look for a method to unify them and simplify the classification algorithms in such a way as to facilitate its routine use in pathology laboratories eventually to enable prospective sample classification. For this purpose, we developed a classification method based mainly on the Wright13 and Lacy15 subtypes characteristics, integrating the most significant features from both studies. To refine and evaluate our method, we used the United Kingdom population-based Haematological Malignancy Research Network (HMRN) cohort15, restricting our analysis to DLBLC-NOS patients treated with R-CHOP (n = 580).
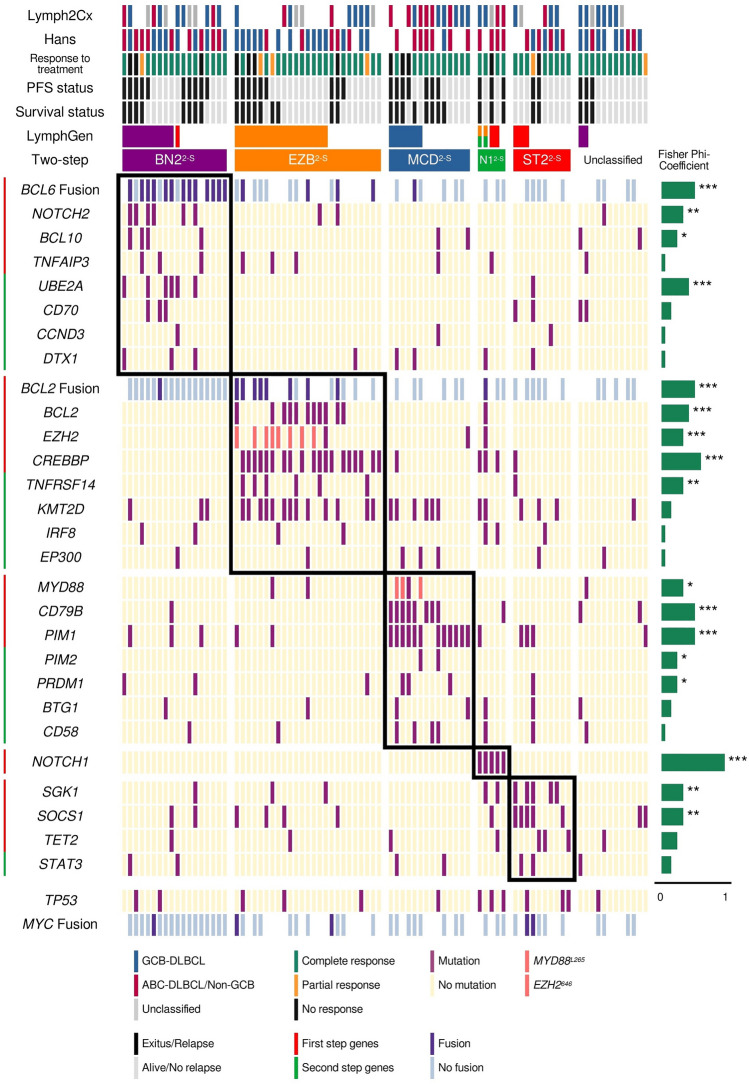
Several tests were carried out to select the best combination of genes, taking into account the sensitivity and specificity compared with the PdH and HMRN cohorts using the LymphGen algorithm13 (see “Methods” section). We selected the combination of genes to be used in the two-step (2-S) classifier, and then applied the approach to our cohort. Five cases were classified as N12-S, 18 as BN22-S, 25 as EZB2-S, 14 as MCD2-S, and ten as ST22-S. Twelve cases (14.3%) remained unclassified (Fig. 3).
Figure 3.
Genetic subtypes and association with overall survival (OS) and progression-free survival (PFS). Genetic classification in the five defined genetic subtypes of DLBCL. Clustering was performed using alterations in genes (rows) from 84 DLBCL samples (columns). The OS and event-free survival status, and the ABC/GCB classification based on Lymph2cx and Hans are represented at the top. The phi coefficient and Fisher's exact test significance are represented on the right of the figure (*p < 0.05; **p < 0.01; ***p < 0.001).
When we classified the PdH cohort using LymphGen, 11 cases were assigned as BN2, 16 as EZB, six as MCD, and six as ST2. Although we have five cases with NOTCH1 mutations, only two were assigned as EZB/N1 complex genotype by LymphGen. Forty-three cases (53%) remained unclassified. Compared with LymphGen, the two-step classifier showed a 100% sensitivity for the MCD2-S and EZB2-S, 81.8% for the BN22-S, and 50% for the ST22-S subtypes, taking the LymphGen classification as reference (Table 2; Fig. 3). The specificity values achieved were 89.7% for MCD2-S, 88% for BN22-S, 86.8% for EZB2-S, and 91.4% for ST22-S (Table 2; Fig. 3).
Table 2.
Sensitivity and specificity analysis comparing the three classifiers in the Puerta de Hierro (PdH) and HMRN cohorts.
| Sensitivity/specificity (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MCD2-S | BN22-S | EZB2-S | ST22-S | N12-S | ||||||
| PdH cohort | ||||||||||
| Two-step versus LymphGen | 100 | 89.7 | 81.8 | 88.0 | 100 | 86.8 | 50 | 91.4 | – | 94.1 |
| Two-step versus AIC cluster | 97.5 | 92.0 | 41.2 | 97.2 | 88.5 | 90.6 | 56.1 | 95.8 | – | – |
| HMRN cohort | ||||||||||
| Two-step versus LymphGen | 98.5 | 90.1 | 64.0 | 95.8 | 89.9 | 92.8 | 81.3 | 92.7 | 100 | 99.8 |
| LymphGen versus AIC cluster | 81.3 | 99.6 | 39.2 | 97.8 | 84.9 | 93.5 | 50.4 | 99.0 | – | – |
Due to the small size of our cohort, to validate the method, we applied the two-step as well as the LymphGen classifiers to the HMRN cohort, and compared them to the Akaike Information Criterion (AIC) clustering method employed in the original study15. The sensitivity values for the two-step classifier compared with the AIC cluster were 97.5% for MCD2-S, 41.2% for BN22-S, 88.5% for EZB2-S, and 56.1% for ST22-S. Regarding specificity, all the subtypes achieved values greater than 90% (Table 2; Fig. 4). No sensitivity and specificity values were obtained for N12-S because the AIC cluster did not include this group. The values comparing the two-step classification to LymphGen were similar, except for the BN22-S/BN2 and ST22-S/ST2, with 64.0 and 81.3% sensitivity, respectively (Table 2; Fig. 4). Seventeen samples were classified as N12-S by our classifier, 16 of them also ranked as N1 by LymphGen. BN2/NOTCH2 and ST2/SGK1-TET2-SOCS1 also showed differences between with sensitivity values of 39.2 and 50.4%, respectively (Table 2; Fig. 4).
Figure 4.
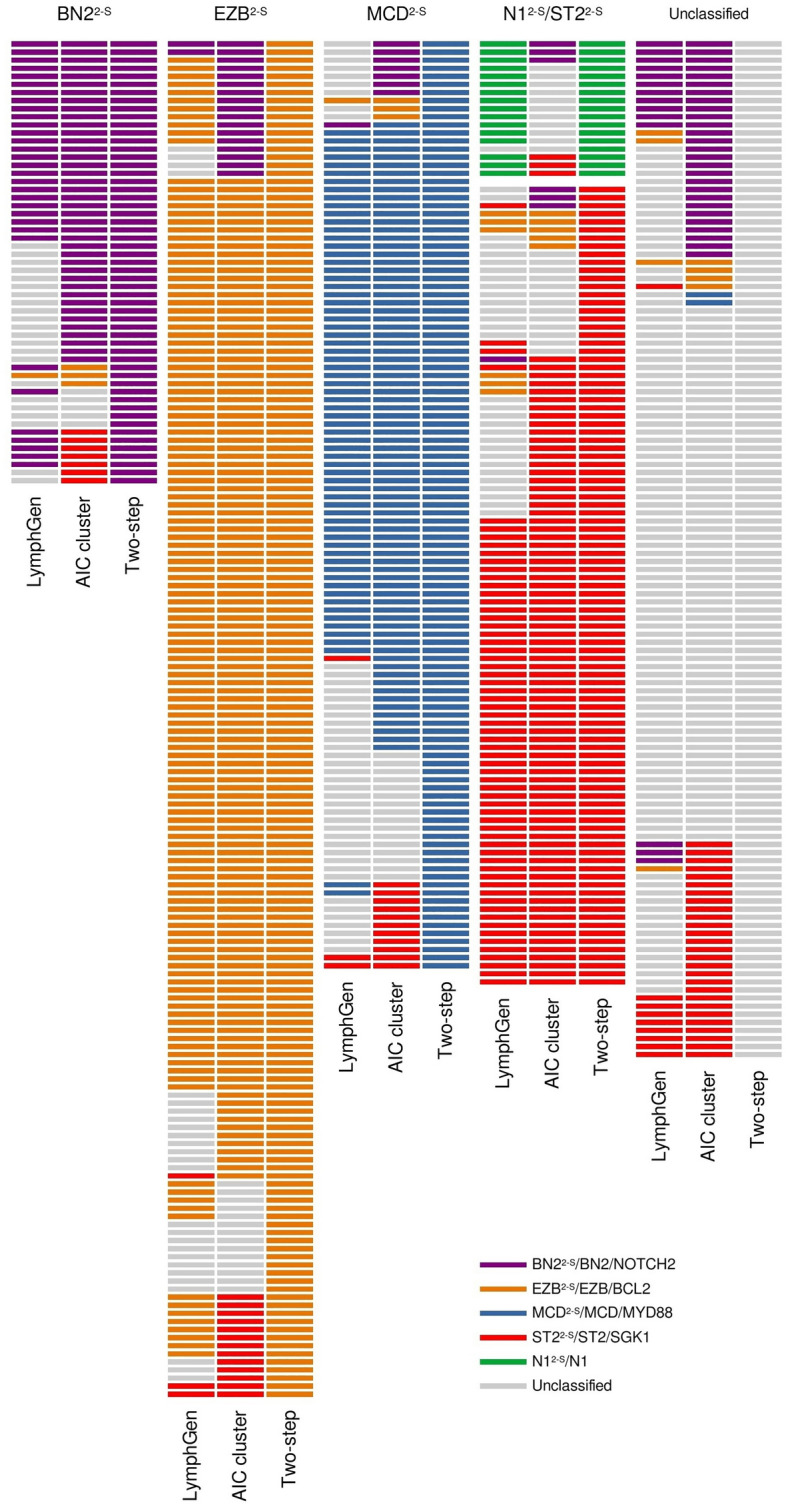
Comparison of the classifiers. Subtypes assigned to the HMRN cohort by LymphGen, AIC cluster and the two-step classifier.
The low sensitivity of the BN22-S subtype prompted us to analyze the samples included in this cluster more thoroughly. Twenty-eight percent of NOTCH2 cases in the HMRN cohort remained unclassified by our two-step classifier. Eighteen percent had more mutations in genes for the EZB2-S subtype and were, therefore, classified as such. Similar low sensitivity was obtained when compared with the LymphGen classification, in which 28% of the BN2 cases in the HMRN cohort were not assigned to any subtype by the two-step method. When we compared the AIC cluster with the LymphGen, 43% of the NOTCH2 cases were unclassified by LymphGen, and 13% were assigned to EZB.
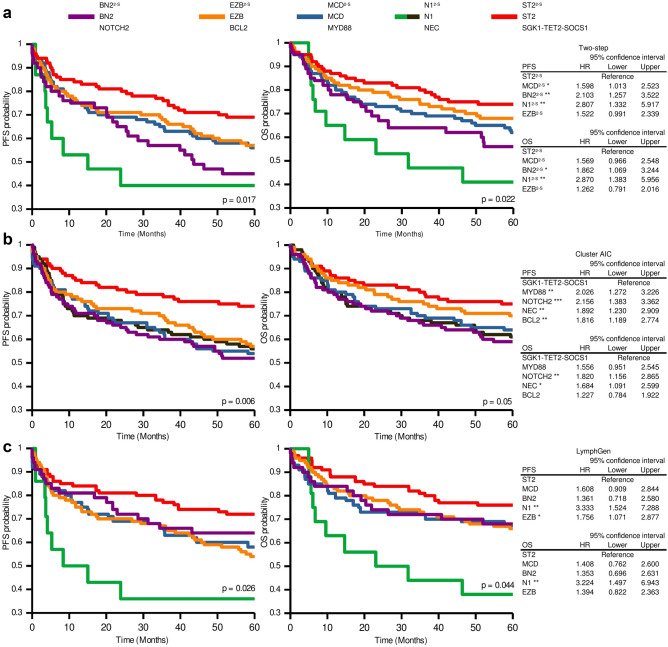
When we examined the new two-step classifier's potential clinical value, the survival analyses produced no significant results in the PhD cohort, although they did reveal a trend towards shorter OS and higher risk of relapse for MCD2-S and BN22-S cases compared with EZB2-S and ST22-S (Fig. S2A). MCD was the subtype with the worst OS and PFS prognosis for the LymphGen classification, although, again, the differences were not substantial enough to be statistically significant, given the small number of classified samples by both methods (67 and 39, respectively) (Fig. S2B).
Therefore, we evaluated the two-step classifier's clinical relevance using data from the DLBLC-NOS patients treated with R-CHOP included in the HMRN cohort. The analysis assigned a higher risk of relapse to the N12-S, BN22-S, and MCD2-S cases than to the ST22-S cases (Fig. 5A). N12-S and BN22-S also had shorter OS. We compared these results with those obtained by the original AIC cluster and LymphGen classifiers, and we found that the groups defined by the three methods had grosso modo similar associations with PFS and OS, as indicated by Kaplan–Meier and Cox proportional-hazards analysis (Fig. 5A–C). ST22-S/ST2/SGK1-TET2-SOCS1 is the group with the best clinical outcome. N12-S and N1 showed the shortest OS (Fig. 5A,C).
Figure 5.
Progression-free survival (PFS) and overall survival (OS) according to the genetic subtypes of the two-step, AIC cluster and LymphGen classifiers in the HMRN cohort. Kaplan–Meier analysis of genetic subtypes from (A) two-step, (B) AIC cluster and (C) LymphGen methods. Tables on the right of the figure show the hazard ratio (HR) values from the Cox proportional-hazards model of genetic subtypes for PFS and OS status. Error bars represent the 95% confidence intervals. Significance: *p < 0.05; **p < 0.01.
The three classifiers showed similar PFS and OS for MCD2-S/MCD/MYD88, EZB2-S/EZB/BCL2, and ST22-S/ST2/SGK1-TET2-SOCS1 subtypes, as demonstrated by the superimposed curves from the Kaplan–Meier analysis (Fig. S3). BN22-S/BN2/NOTCH2 is the group that differed the most depending on the classifier used, showing shorter OS and PFS with the two-step classification and the AIC cluster compared with LymphGen (Fig. S3A).
Among the GCB-DLBLC cases, EZB2-S cases showed worse clinical outcomes, although not statistically significant (Fig. 6). The 36% of the non-EZB2-S GCB-DLBCL cases were classified as ST22-S, 12% as MCD2-S, 11% as BN22-S; 40% of samples were unclassified. No differences were found between subtypes in the ABC-DLBLC cases.
Figure 6.
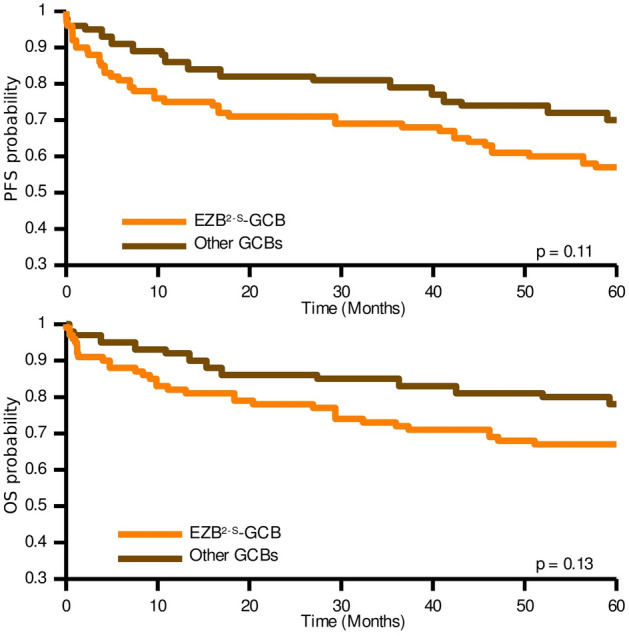
Kaplan–Meier analysis of progression-free survival (PFS) and overall survival (OS) for EZB2-S-GCB and other GCB cases in the HMRN cohort when applying the two-step classifier.
Finally, the predictive value of the subtypes in combination with the IPI was analyzed. We performed a multivariate Cox proportional-hazards analysis with the IPI and the two-step classifier-defined genetic subtypes in the HMRN cohort (Table S6), and subsequently validated the results in the PdH cohort. This model assigned progression and survival risk scores for each sample in the PdH cohort (Table S1), from which we generated three risk groups with different clinical outcomes, as indicated by the Kaplan–Meier analysis (p = 0.038 for OS; p = 0.005 for PFS) (Fig. S4).
Discussion
Our study confirmed the current landscape of genetic alterations in DLBCL and explored their involvement in clinical outcome and response to treatment. We also propose a modified genetic classification based on those of the Wright et al.13 and Lacy et al.15, validated its accuracy, and analyzed its ability to predict response to therapy and clinical outcome in R-CHOP-treated DLBCL patients.
Mutational data confirmed the well-known high degree of heterogeneity in DLBCLs. We found that the genes most frequently mutated in this cohort were IGLL5, KMT2D, CREBBP, PIM1, CARD11, PCLO, and KMT2C, similar to what has been noted in other DLBCL series12,14,17–21. When we integrated the genes in predefined signaling pathways or gene functions, chromatin remodeling, BCR, NFκB, and apoptosis pathways were the most frequently mutated gene sets (Fig. 1A).
The main problems for DLBCL patients are the high percentage of them (30–40%) that are refractory to treatment or relapse, and the lack of accurate predictive markers. We found that mutations in CD79B were associated with relapse, independent of the IPI. We also tested whether any alteration was related to primary refractoriness, but none was found to be enriched in samples from R/R patients (data not shown). Patients with mutations in CD79B, ETS1, and CD58 had significantly lower survival rates. CD79B and CD58 mutations had previously shown adverse prognostic value, and CD79B was already known to be one of the most frequently mutated genes at relapse22–25. Mutations in ETS1 were found more frequently in Burkitt lymphoma, but only sporadically in DLBLC. ETS1 regulates B-cell-relevant pathways, such as B-cell differentiation. ETS1 overexpression has been associated with gain of the 11q24.3 region. This region, characteristic of a Burkitt-like lymphoma subtype (BLL-11q), has also been found to be gained in up to a quarter of DLBCL cases26,27.
Several recent studies have undertaken integrative, multiplatform analysis of gene mutations, structural aberrations, and gene-expression profiling12,13,15,28,29 that have led to the proposal of new genetic subtypes determined by distinct genetic backgrounds and clinical behaviors. Although these classifications share several characteristics and genetic features, their different uses lead to some inconsistencies in determining definite clusters. For example, Schmitz12 and Wright13 and their colleagues used BCL2 and BCL6 fusions, essential drivers of DLBCL pathogenesis, as classifier alterations, whereas Lacy et al.15 did not use them to classify their samples; on the other hand, Chapuy et al.14 used CNVs to define their clusters, which are especially relevant for the C2 subtype. The algorithms' complexity and the large number of genes and structural alterations used to define the genetic subtypes made it challenging to use them as part of the clinical routine. Moreover, some classifications, like LymphGen13, produce a high percentage of unclassified tumors. Others, such as that of Chapuy14 and Lacy15 and their coworkers, could not be used for individual samples. Here, we propose an easier classifier, using a panel with a small number of genes (26) and BCL2 and BCL6 translocations detected by routine FISH technique, to define the genetic subtypes. This two-step classifier is based mainly on the approaches of Wright13 and Lacy15, and uses the most frequently mutated genes shared by the two studies, maintaining the names given by Schmitz12 and Wright13. We define five groups and classify the samples as N12-S, MCD2-S, BN22-S, EZB2-S, or ST22-S.
Given the limitations of this analysis arising from the low number of patients, we applied it in the external HMRN series, restricting the analyses to DLBCL-NOS patients treated with R-CHOP.
Despite the limitations of the study, the comparison of the two-step classifier specificity and sensitivity with that of LymphGen and the AIC cluster, and assessing its prognostic and predictive value in a large real-world series (HMRN), showed our method to be robust and demonstrated that the clinical outcomes of the subgroups did indeed differ.
N12-S was defined by the presence of NOTCH1 mutations regardless of all other alterations detected in the tumors. Although N1 was not included in the Lacy classification, we considered it an essential subtype because it is the most aggressive (Fig. 5), as previously found by Schmitz12 and Wright13, and demonstrated in the Lacy series15.
The BN22-S group had the worst clinical outcome of our series, with results similar to those of Lacy15, but unlike those of Chapuy14 and of Schmitz–Wright12,13 (Fig. S2). The associations of the BN2 and C1 subtypes with a more favorable outcome were explained as being a possible transformation from occult MZL. The BN2-NOTCH2-C1 subtype was more weakly defined than the others, probably due to the disagreement regarding the use of BCL6 translocations and the selection of genes defining it. They are based on several unequally shared characteristics, such as NOTCH2 (shared by Schmitz–Wright12,13 and Lacy15), BCL10 (Schmitz–Wright12,13 and Chapuy14) or BCL6 translocations (Lacy15 and Chapuy14). These differences may explain the smaller overlap and the different clinical behavior in the three series. We decided to include BCL2 and BCL6 translocations due to their importance in DLBCL pathogenesis, the availability of the data for most DLBCL samples, the strong relationship between BCL2 mutations and BCL2 translocation, and the fact that, in the Lacy series15, the NOTCH2 subtype was enriched in samples with BCL6 fusions.
ST22-S/ST2/SGK1-TET2-SOCS1 was the subtype with the best clinical outcome, as previously reported13,15, and confirmed here with the three classifiers. MCD2-S/MCD/ MYD88 and EZB2-S/EZB/BCL2 are the most consistent subtypes, as shown by the high sensitivity and specificity values when comparing the three classifiers and the overlap of the Kaplan–Meier curves (Fig. 5 and Fig. S3). Comparing EZB2-S with other GCB cases (Fig. 6) revealed the former to have a slightly worse clinical outcome.
Finally, given the limitations of analyzing CNVs in standard pathology laboratories, we decided not to include A53 (Wright13)/C2 (Chapuy14) in our classifier as they were identified by TP53 mutations and deletions together with widespread CNVs.
The original classifiers are of limited use in clinical practice because they are based on so many genetic alterations and such complex algorithms. Here we propose a genetic DLBCL classifier that integrates the findings of Schmitz–Wright12,13 and Lacy15 and that is based on an optimized panel with a minimal set of markers (26 genes and the BCL2 and BCL6 translocations) that can classify samples individually for the purpose of routine patient management. The validations carried out in this study demonstrated the close correlation between the two-step classifier and the other classifiers, as the specificity and sensitivity analysis showed it to be adequate for achieving a successful classification.
Despite the well-known heterogeneous clinical behavior of DLBCL patients, the diagnostic work-up and treatment are identical for every patient. Although COO classification provided significant prognostic information, and some targeted therapies have shown different efficacy depending on the COO, this has not led to any changes in the use of R-CHOP as the standard treatment for DLBLC patients. We hope that this classifier would make it possible to assign patients to a genetic subtype on a tumor-by-tumor basis, thereby guiding the clinical management of individual patients.
The genetic alterations defining each subtype, the association of functionally altered pathways, and previously performed functional studies suggested the appropriate therapeutic approach for each of them. N1 is the most interesting due to its poor response to standard immunochemotherapy. NOTCH1 aberrations frequently occur in hematological malignancies, mainly chronic lymphocytic leukemia and T-cell leukemia, making this gene a candidate target in the design of tailored therapy for this subtype. In this context, γ-secretase inhibitors (GSIs) are the most widely studied anti-NOTCH1 molecules in cancer. The clinically relevant GSI PF-03084014, combined with fludarabine, has antitumor effects in primary NOTCH1-mutated CLL cells30 and, when tested in phase I clinical trials, showed good tolerance and antitumor activity in solid and hematological malignancies31,32. Therefore, the combination of GSIs with chemotherapy is a promising strategy for treating N1 DLBCL patients. Several approaches have already been suggested for the other subtypes, such as BTK inhibitors for MCD cases, EZH inhibitors, and BCL2 inhibitors, such as venetoclax or navitoclax, for the EZB subtype13.
In conclusion, we propose a feasible classifier built with mutation information from 26 genes and BCL2/BCL6 translocations, based on the recently identified genetic subgroups12–15. This genetic classifier, combined with clinical data and other molecular characteristics, should eventually help researchers develop improved risk models for DLBCL patients and, more importantly, for guiding precision therapy.
Methods
Patients and samples
The study population (PdH cohort) consisted of 84 DLBCL patients, diagnosed in Spanish medical institutions between 2002 and 2016. Five of the patients (PDLB63-PDLB67) had a diagnosis of DLBCL after a histological transformation of follicular lymphoma without treatment prior to transformation33. Formalin-fixed, paraffin-embedded tissue (FFPET) sections from diagnostic biopsies were collected. The research project was approved by the Ethics Committee of Hospital Universitario Puerta de Hierro-Majadahonda (reference PI67-14) and conducted in compliance with the Declaration of Helsinki. All participants signed an informed consent form. Samples were collected and clinical data managed following protocols guaranteeing the confidentiality of donor data. Material and data from other centers were anonymously transferred to our laboratory after obtaining approval from the corresponding ethics committees, under the relevant Spanish legislation (Ley 14/2007 de Investigación Biomédica and Real Decreto 1716/2011). All samples were reviewed upon arrival by qualified hematopathologists to confirm their diagnosis.
Patients were treated with R-CHOP (77 patients) or R-CHOP-like (7 patients) regimens. The retrospective collection included FFPET sections from samples at diagnosis from 50 patients (59.5%) who responded to treatment, referred to hereafter as sensitive (S), and 34 samples (40.5%) from refractory (11 patients) and relapsed (23) (R/R) patients. The median follow-up was 6 years. For the IPI analysis, we reclassified the patients into three risk categories: low (0–1), intermediate (2–3) and high (4–5). Patient characteristics are summarized in Table 1 and Suppl. Table S1.
Targeted sequencing
Genomic DNA was extracted from FFPET using a truXTRAC FFPE DNA Kit (Covaris, Woburn, MA, USA) following the manufacturer's instructions.
Two SureSelect target enrichment custom panels were designed using the SureDesign (Agilent Technologies, Santa Clara, CA, USA) web-based tool (earray.chem.agilent.com/suredesign/). The genes included are involved in lymphomagenesis-relevant pathways and were selected based on previous studies17,18,34–37 and our earlier findings33. The designs covered the coding exons of the selected genes. The targeted regions (according to Human Assembly GRCh37/hg19) were captured using a SureSelectXT Target Enrichment System and SureSelectXT Low Input (Agilent), as described in the manufacturer's instructions. One hundred twenty-five genes were common to the two designs and are referred to as the common gene set (CGS) (Suppl. Table S7). Captured libraries were diluted to 8 pM for Illumina clustering, and paired-end sequencing was performed on MiSeq and NextSeq sequencers (Illumina Inc., San Diego, CA, USA).
Three independent analyses were carried out for each sample. We first used the tools available in the Variant Reporter instrument (Illumina). A second variant calling was done with VarScan 2.3.9 to detect the mutations of the files extracted from two sources: (1) BWA Enrichment of Illumina Base Space, and (2) the Burrows-Wheeler Aligner (BWA), Picard and Indel Realignment-Base Recalibration from the Genome Analysis Toolkit 3.8.1.0 (GATK).
Annotation was carried out with Annovar. All variants identified by the three complementary methods were visualized using an Integrative Genomics Viewer (Broad Institute and UC San Diego, San Diego, CA, USA). Data have been deposited in the Sequence Read Archive (SRA) (accession number PRJNA648645).
After annotation, the variants were subjected to additional, more stringent, and quality- and relevance-based filtering applying the following criteria: quality read depth of bases ≥ 50; depth of variant-supporting bases ≥ 5; localization (exonic, UTRs and splice site); variant effect (non-synonymous); variant allele frequency ≥ 5% and not listed as a single nucleotide polymorphism, or listed but with a MAF < 0.01% (The Exome Aggregation Consortium, 1000 Genomes Project of the International Genome Sample Resource (IGSR), Single Nucleotide Polymorphism Database (dbSNP) v138 of the National Center for Biotechnology Information (NCBI)). The median coverage was 764x (89–1908) (Suppl. Table S8).
Tissue microarray construction, immunohistochemistry and in situ hybridization analyses
Thirty-one DLBCL samples for which sufficient biopsy material was available were included in a tissue microarray (TMA). Known positive and negative controls (one tonsil, one lymphadenopathy, and a known triple-hit DLBLC case) were also included. A tissue arrayer (Beecher Instruments, Silver Spring, MD, USA) was used for TMA construction. Hematoxylin and eosin-stained sections from each biopsy were used to define the appropriate areas, and two representative cores (1 mm diameter) were used for the TMA.
TMA sections were stained with antibodies against CD10 (clone 56C6), BCL-6 (PG-B6p), MUM1 (MUM1p), BCL-2 (124), and p53 (D07), all of which were mouse monoclonal antibodies, obtained from Agilent-Dako (Santa Clara, CA, USA), and MYC (Y69, rabbit polyclonal; Agilent-Dako). The COO based on IHC was established by the Hans algorithm38. EBER in situ hybridization (EBER1 DNA probe, Roche-Ventana; Basel, Switzerland) was carried out to detect EBV.
Fluorescence in situ hybridization (FISH) was performed following the routine protocol at the Pathology Department of our Institution. ZytoLight Dual Color Break Apart probes (ZytoVision GmbH, Bremerhaven, Germany) were used to analyze BCL2 (18q21 region), BCL6 (3q27), and C-MYC (8q24).
IHC and FISH data from the samples not included in the TMA were collected from the original pathological reports.
Nanostring LST nCOUNTER gene-expression assay
We used the NanoString LST assay performed with the nCounter Dx Analysis System (NanoString Technologies, Seattle, WA, USA) to determine COO. Total RNA from 54 FFPET sections from diagnostic samples was isolated using a truXTRAC FFPE total NA kit (Covaris) following the manufacturer's instructions. RNA quality and quantity were assessed with an RNA 6000 Nano kit (Agilent) using the Agilent 2100 Bioanalyzer System (Agilent).
The previously reported protocol for the NanoString LST gene expression assay was used39,40. Briefly, the probes corresponding to the 20 LST genes41,42 and based on the Lymph2Cx gene expression assay were hybridized in a multiplexed reaction to 400 ng of the total RNA for 16 h at 65 °C. Automated removal of excess probe and immobilization of probe–transcript complexes on a streptavidin-coated cartridge were carried out in the nCounter Prep Station, and raw gene expression counts were collected on the Digital Analyzer. Data were analyzed using nSolver 4.0 software (NanoString Technologies) for COO classification.
Genetic classification
We performed a simplified genetic classification based on those proposed by Schmitz-Wright12,13 and Lacy15 and their research teams. We used the alterations of specific genes to develop a two-step classification method and classified the samples as N12-S, BN22-S, EZB2-S, MCD2-S, or ST22-S. The A53 LymphGen subtype was not included due to the lack of copy number variation data in our series and most published series. The genes from the Schmitz-Wright12,13 and Lacy15 studies were selected by calculating their power to classify determined by the Fisher's exact test, as featured in these studies (Suppl. Table S9). Several tests were then carried out to select the best combination of genes, taking into account the sensitivity and specificity in the PdH and HMRN cohorts according to the LymphGen and Lacy classification (AIC cluster). In the first step, at least one of the top genes should be mutated: NOTCH1, for N12-S (NOTCH1 mutated samples were classified as N12-S, regardless the presence of other alterations); MYD88, CD79B, and PIM1 for MCD2-S; BCL6 translocation, NOTCH2, BCL10, and TNFAIP3 mutations for BN22-S; BCL2, EZH2, and CREBBP for EZB2-S; and SGK1, TET2, and SOCS1 for ST22-S. Samples with the same score for two or more subtypes, or samples with no mutations, were classified in the second step, in which we added the following genes for each subtype: PRMD1, BTG1, PIM2, and CD58 for MCD2-S; UBE2A, CD70, CCND3, and DTX1 for BN22-S; TNFRSF14, KMT2D, IRF8, and EP300 for EZB2-S, and STAT3 for ST22-S. In this second step, at least two genes from any of the genes that define each subtype should be mutated to assign the sample to the corresponding subtype (Fig. S5 and Suppl. Table S9).
We selected the HMRN cohort15 as an external validation cohort for the two-step method, restricting the analysis to patients treated with R-CHOP (n = 580).
Sensitivity and specificity
The sensitivity and specificity of each subtype defined by the two-step method were determined using the LymphGen and AIC classifiers as references in the PdH and HMRN cohorts. Sensitivity was calculated as the percentage of true positives relative to the reference classifier:
Specificity was calculated as the percentage of true negatives relative to the reference classifier:
Statistical analysis
Statistical analyses were performed using R 3.6.1 (https://www.R-project.org, R Foundation for Statistical Computing, Vienna, Austria). Associations between factors were assessed with Fisher's exact test and the predictive value of the selected variants by Cox proportional-hazards models, adjusted using the Benjamini–Hochberg correction method.
OS was calculated as a binomial factor for survival or non-survival of each sample. Progression-free survival (PFS) was calculated as a binomial factor, assigning the event to the treated patients who did not respond to treatment or who relapsed. We performed univariate and multivariate logistic regression analyses of genes and pathways mutated in at least four samples to establish each variable's predictive value. Hazard ratios (HRs) with 95% confidence intervals were estimated for each variable. The survival function of genetic groups, as measured by OS and time to relapse (PFS), was also calculated by the Kaplan–Meier method.
Supplementary information
Acknowledgements
We are indebted to the patients who contributed to this study and to the members of GOTEL (Grupo Oncológico para el Tratamiento y el Estudio de los Linfomas). We are especially thankful to E. Ramil of the Sequencing Unit of the Instituto de Investigación Sanitaria Puerta de Hierro-Segovia de Arana (IDIPHISA) and Á. Zaballos from the Genomics Unit of Instituto de Salud Carlos III (ISCIII). We acknowledge the Biobank of the Hospital Puerta de Hierro, the Biobank of the Sistema Sanitario Público de Andalucía and the Spanish Hospital Biobank Network, supported by the ISCIII, Complejo Hospitalario de Toledo and MD Anderson Cancer Center-Madrid, as well as the Pathology Departments of the centers collaborating in the study. We also thank L. Cereceda from the Instituto de Investigación Sanitaria Fundación Jimenez Díaz for her invaluable help, A. Royuela from IDIPHISA’s Clinical Biostatistics Unit and C. Parejo from IDIPHISA’s TIC Unit—Medical Oncology Department. This work was supported by the Spanish Ministry of Economy and Competence (MINECO) SAF2013-47416-R; ISCIII-MINECO AES-FEDER (Plan Estatal de I+D+I 2008-2011 and 2013-2016) (PI14/00221, DTS17/00039, PI17/00272, PI16/01294, CIBERONC CB16/12/00291); by GILEAD (GL18/00019) and Dirección General de Universidades e Investigación Consejería de Educación e Investigación de la Comunidad de Madrid (B2017/BMD-3778). LP and JGR are recipients of iPFIS predoctoral fellowships (IFI18/0004 and IFI14/0003, by ISCIII-MINECO AES-FEDER (Plan Estatal I+D+I 2017-2020), and Plan Estatal I+D+I 2013-2016, respectively), IFM is supported by B2017/BMD-3778, CT is recipient of a bioinformatician fellowship (CA18/00022), and NYC is supported by the Asociación Española Contra el Cáncer. MSB has been a beneficiary of a Miguel Servet II contract (CPII16/00024), by ISCIII-MINECO AES-FEDER (Plan Estatal I+D+I 2013-2016) and the Fundación de Investigación Biomédica Puerta de Hierro. The funders had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Author contributions
M.S.B., M.P., and D.P.C. designed the study. M.S.B. supervised the study. L.P., D.P.C., S.G., A.S., and M.R. performed the experimental work. C.Q., P.M.A., J.F.G., F.R.G.A., A.R., F.I.C., M.G.C., M.L., M.M., M.P.V., J.G.C., A.S., M.A.P., and M.P. collected clinical data and patient samples. L.P., I.F.M., D.P.C., J.G.R., and N.Y.C. analyzed and interpreted the data. I.F.M., C.T., and A.H. performed the statistical and computational analysis. L.P., I.F.M., D.P.C., and M.S.B. wrote the manuscript. All the authors critically reviewed and approved the manuscript. L.P., I.F.M., and D.P.C. contributed equally to this work.
Code availablity
Accession codes: SRA (PRJNA648645). The R code for the two-step classifier can be downloaded from https://github.com/Lymphoma-IDIPHISA/Two-step-classifier.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lucía Pedrosa, Ismael Fernández-Miranda and David Pérez-Callejo
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80376-0.
References
- 1.Sant M, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116:3724–3734. doi: 10.1182/blood-2010-05-282632. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, C. K. Diffuse Large B-Cell Lymphoma—Cancer Stat Facts. (2019).
- 3.Coiffier, B. et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood 116, 2040 (2010). [DOI] [PMC free article] [PubMed]
- 4.Pérez-Callejo D, González-Rincón J, Sánchez A, Provencio M, Sánchez-Beato M. Action and resistance of monoclonal CD20 antibodies therapy in B-cell Non-Hodgkin Lymphomas. Cancer Treat. Rev. 2015;41:680–689. doi: 10.1016/j.ctrv.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 5.International Non-Hodgkin’s Lymphoma Prognostic Factors Project A predictive model for aggressive non-Hodgkin’s lymphoma. N. Engl. J. Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 6.Sehn LH, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109:1857–1861. doi: 10.1182/blood-2006-08-038257. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, et al. An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood. 2014;123:837–842. doi: 10.1182/blood-2013-09-524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swerdlow SH, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 11.Wright G, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright GW, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37:551–568.e14. doi: 10.1016/j.ccell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapuy B, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacy SE, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020;135:1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal M, et al. Functional signatures identified in B-cell non-Hodgkin lymphoma profiles. Leukemia Lymphoma. 2009;50:1699–1708. doi: 10.1080/10428190903189035. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011;43:830. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Miranda NFCC, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124:2544–2553. doi: 10.1182/blood-2013-12-546309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:3879. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin RD, et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin. Cancer Res. 2016;22:2290–2300. doi: 10.1158/1078-0432.CCR-15-2123. [DOI] [PubMed] [Google Scholar]
- 23.Braggio E, et al. Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin. Cancer Res. 2015;21:3986–3994. doi: 10.1158/1078-0432.CCR-14-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, et al. Mutations or copy number losses of CD58 and TP53 genes in diffuse large B cell lymphoma are independent unfavorable prognostic factors. Oncotarget. 2016;7:83294–83307. doi: 10.18632/oncotarget.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mottok A, et al. Integrative genomic analysis identifies key pathogenic mechanisms in primary mediastinal large B-cell lymphoma. Blood. 2019;134:802–813. doi: 10.1182/blood.2019001126. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Farre B, et al. Burkitt-like lymphoma with 11q aberration: a germinal center-derived lymphoma genetically unrelated to Burkitt lymphoma. Haematologica. 2019;104:1822–1829. doi: 10.3324/haematol.2018.207928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonetti P, et al. Deregulation of ETS1 and FLI1 contributes to the pathogenesis of diffuse large B-cell lymphoma. Blood. 2013;122:2233–2241. doi: 10.1182/blood-2013-01-475772. [DOI] [PubMed] [Google Scholar]
- 28.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolen CR, et al. Prognostic impact of somatic mutations in diffuse large B-cell lymphoma and relationship to cell-of-origin: data from the phase III GOYA study. Haematologica. 2019 doi: 10.3324/haematol.2019.227892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Guerra M, et al. The γ-secretase inhibitor PF-03084014 combined with fludarabine antagonizes migration, invasion and angiogenesis in NOTCH1-mutated CLL cells. Leukemia. 2015;29:96–106. doi: 10.1038/leu.2014.143. [DOI] [PubMed] [Google Scholar]
- 31.Messersmith WA, et al. A Phase I, dose-finding study in patients with advanced solid malignancies of the oral γ-secretase inhibitor PF-03084014. Clin. Cancer Res. 2015;21:60–67. doi: 10.1158/1078-0432.CCR-14-0607. [DOI] [PubMed] [Google Scholar]
- 32.Papayannidis C, et al. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015;5:e350. doi: 10.1038/bcj.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Rincón J, et al. Unraveling transformation of follicular lymphoma to diffuse large B-cell lymphoma. PLoS ONE. 2019;14:e0212813. doi: 10.1371/journal.pone.0212813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karube K, et al. Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J. Pathol. 2014;234:423–430. doi: 10.1002/path.4428. [DOI] [PubMed] [Google Scholar]
- 36.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasqualucci L, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6:130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni, M. M. Digital multiplexed gene expression analysis using the NanoString nCounter system. Current protocols in molecular biology Chapter 25, Unit25B.10 (2011). [DOI] [PubMed]
- 40.Watatani Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–2883. doi: 10.1038/s41375-019-0473-1. [DOI] [PubMed] [Google Scholar]
- 41.Scott DW, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J. Clin. Oncol. 2015;33:2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott DW, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession codes: SRA (PRJNA648645). The R code for the two-step classifier can be downloaded from https://github.com/Lymphoma-IDIPHISA/Two-step-classifier.