Abstract
Background
Recently, microRNAs have been demonstrated to be potential non-invasive biomarkers for diagnosis, prognosis assessment or prediction of response to treatment in cancer. In this study, we evaluate the potential of miR-30b-5p as a biomarker for early diagnosis of breast cancer (BC) in tissue and plasma.
Methods
Expression of miR-30b-5p was determined in a series of 112 BC and 40 normal breast tissues. Circulating miR-30b-5p levels in plasma samples were determined in a discovery cohort of 38 BC patients and 40 healthy donors and in a validation cohort of 83 BC patients and 83 healthy volunteers. miR-30b-5p expression was measured by quantitative real-time PCR and receiver operating characteristics curve analysis was carried out.
Results
The miR-30b-5p expression was significantly lower in BC tissue than in healthy breast samples. In contrast, circulating miR-30b-5p levels were significantly higher in BC patients compared with healthy donors. Furthermore, circulating miR-30b-5p levels were significantly higher in patients with positive axillary lymph node and de novo metastatic patients. Receiver operating characteristics curve analysis demonstrated a good diagnostic potential of miR-30b-5p to detect BC even at an early stage of the disease.
Conclusion
Thus, we highlight the potential of miR-30b-5p as a non-invasive, fast, reproducible and cost-effective diagnostic biomarker of BC.
Key words: microRNA, breast cancer, plasma, diagnosis
Highlights
Our results show the following:
-
•
Low miR-30b-5p expression was detected in breast tumor tissue compared with healthy breast tissue.
-
•
High circulating miR-30b-5p levels in plasma were associated to breast cancer.
-
•
miR-30b-5p levels in plasma identify breast cancer of all subtypes and stages of the disease.
-
•
Circulating miR-30b-5p levels relate with patients' tumor burden.
-
•
Circulating miR-30b-5p is a potential non-invasive and cost-effective diagnostic BC biomarker.
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer around the world and accounts for 30% of all new cases of cancer diagnosis in women. It is the second cause of cancer-related death in the general population and the first among women.1,2 Nonetheless, BC has one of the highest 5-year survival rates (90%) due to the improvement of detection practices and treatments.2 Pathological diagnosis including histological subclassification based on estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2) status and Ki67 level allows classification of tumors in four main subtypes [luminal A, luminal B, HER2-positive (HER2+) and triple-negative (TN)], which help clinicians in treatment decisions, prognosis and prediction.3
Despite the improvement in diagnosis and treatment, approximately 20% of BC patients will develop metastasis. This setting is considered an incurable disease that often occurs due to either resistance to therapy or diagnosis at advanced stages.4 In this scenario, although 44% of the patients are diagnosed at stage I (AJCC cancer staging) with 5-year survival rates of 100%, 5% of patients are diagnosed at stage IV with 5-year survival rates of approximately 26%.5 The small percentage of women diagnosed at advanced stage of the disease gives evidence that mammography represents the best available screening option for BC.5,6 However, more accurate and meaningful early-diagnosis methods together with image techniques would improve BC survival rates.
In the past few years, microRNAs (miRNAs) were established as relevant molecular components of cells in normal or malignant processes.7 Particularly, miRNAs have been demonstrated to have an important role in cancer biology through post-transcriptional editing of target messenger RNAs (mRNAs) expression involved in tumor growth, invasion, metastasis or immune escape.8 In addition, several tumor-associated miRNA profiles have been proposed and investigated as biomarkers for diagnosis, survival, response to treatment or tumor subclassification.9, 10, 11, 12, 13, 14, 15 The development of early diagnostic tools is of most interest to the clinics since early diagnosis is associated with better prognosis. In this context, miRNAs have been demonstrated to be good early diagnostic biomarkers in several types of cancer including BC, among others.16, 17, 18, 19
miRNAs have been shown to be present in several types of body fluids, including blood, where those can be found as cell-free miRNAs, or in exosomes.20,21 One of the main advantages of circulating miRNAs is their high stability in body fluids,22 which is the main reason for them to be used in cancer diagnosis or prognosis. Moreover, the assessment of circulating miRNAs can be carried out with simple, low-cost and quick assays. These characteristics highlight the value of miRNAs as non-invasive biomarkers. Indeed, several miRNAs have been found to be differentially expressed in blood, plasma or serum from healthy donors compared with BC patients, supporting their use as non-invasive, early-stage diagnosis biomarkers.23, 24, 25, 26, 27, 28
Particularly, miR-30b-5p has been studied in tissue and fluid samples as a biomarker for several types of cancer, including lung,29 pancreas,30 colorectal31 and melanoma.32 Although the value of miR-30b-5p as a BC biomarker has been evaluated in several studies, the available data are rather controversial. Some authors found miR-30b-5p overexpression to be related to poor prognosis33,34 and oncogenic functions.35 By contrast, other studies associated high miR-30b expression with better response to endocrine therapy,36 decreased metastasis37 and tumor-suppressor functions.38
In this study, we evaluated miR-30b-5p expression in tissue and plasma samples from healthy donors and BC patients to determine its potential as a novel non-invasive biomarker for BC diagnosis.
Materials and methods
Cell lines
BT474, AU565, SKBR3, MCF-7, MDA-MB-231 and MDA-MB-436 BC cell lines and MCF-12A non-tumorigenic epithelial breast cell line were obtained from American Type Culture Collection (ATCC). All cell lines were cultured at 37°C and 5% CO2 in a humidified atmosphere in the recommended culture media.
Study cohort and sample collection
We collected 112 nonconsecutive samples from patients with primary BC (cohort #1) from 1988 to 2018 at Hospital Clinico Universitario of Valencia/Biomedical Research Institute INCLIVA (HCUV/INCLIVA, Valencia, Spain). Forty healthy breast tissue samples from reduction mammoplasties from HCUV/INCLIVA and Portuguese Oncology Institute of Porto (IPO-Porto, Porto, Portugal) were also selected. To be included, samples were required to have a formalin-fixed paraffin-embedded (FFPE) or optimal cutting temperature (OCT) compound embedded biopsies.
All samples had been analyzed by an expert pathologist. Hormonal receptors status was evaluated by immunohistochemistry (IHC) (estrogen- and progesterone receptor-positive were defined as ≥1% positively stained nuclei), and HER2 was assessed by an IHC score of 3+ (whereby 0 or 1+ indicated HER2-negative, 3+ indicated HER2+, and 2+ was considered borderline; borderline cases were also tested by FISH).
Additionally, plasma samples were collected from two independent institutions. A discovery cohort (cohort #2) included 40 healthy donors and 38 BC patients with plasma samples available at Biobank from the Department of Pathology of IPO-Porto from 2015 to 2018. For the validation cohort (cohort #3), 83 BC patients from HCUV/INCLIVA from 2011 to 2019 and 83 healthy donors from the same institution and Valencian Biobanking Network were selected. All plasma samples were collected before any treatment. Peripheral blood was collected into EDTA tubes and centrifuged at 1600 g for 10 min at 4°C to obtain plasma. Plasma was stored at −80°C until further use. All relevant clinical data were obtained from medical records.
All patients provided signed informed consent for experimental analysis of samples. The study is compliant with all relevant ethical regulations regarding research involving human participants and received ethical approval from the Hospital Clínico/INCLIVA Research Ethics Committee (2019/196) and Institutional Ethical Committees of IPO-Porto (CES-IPOFG-120/015). Sample collection was carried out in accordance with the Declaration of Helsinki.
Total RNA extraction
RNA from cell lines was extracted using TRIzol® Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations.
Total RNA from tissue was isolated from eight 10 μm FFPE or OCT slides using the RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific, Waltham, MA) or mirVana Isolation Kit (Thermo Fisher Scientific), respectively, following the manufacturer's protocol. RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). miRNA extraction from 200 μl of plasma samples was carried out using the miRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.
Retrotranscription to cDNA
A total of 1000 ng of RNA from cells and tissue samples and 9.16 μl of RNA from plasma samples were retrotranscribed to cDNA using a High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific), following the manufacturer's protocol. Retrotranscription reaction was carried out at 16°C for 30 min, at 42°C for 30 min and at 85°C for 5 min. cDNA was stored at −20°C.
miRNA expression analysis
miRNA expression levels were evaluated by quantitative real-time PCR. Quantitative real-time PCR was carried out using specific Taqman miRNA assays for miR-30b-5p (Assay ID 000602, Thermo Fisher Scientific) and Taqman Universal MasterMix II no UnG (Thermo Fisher Scientific) for tissue miRNA expression or Xpert Fast Probe 2x MasterMix (GRiSP, Porto, Portugal) for plasma, following the manufacturer's protocol. A final volume of 10 μl was used for quantitative real-time PCR reaction and incubated at 98°C for 3 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s and 37°C for 30 s. Expression levels were detected using a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). Results were normalized according to the expression of housekeeping miR-16 and RNU-38B miRNAs (assay ID 000391 and ID 001004, Thermo Fisher Scientific). The threshold cycle value (CT) was determined for each measurement and miRNA expression was calculated relative to the control using the comparative critical threshold (2−ΔCT) method where: ΔCT = CTmiRNA − CThousekeeping control. Triplicates were carried out for each sample.
The Cancer Genome Atlas data analysis
The expression of miR-30b-5p in tissue from BC patients and healthy donors from available data from The Cancer Genome Atlas (TCGA) was analyzed. miRNA expression data were downloaded from the OncoMir Cancer Database (OMCD) (https://www.oncomir.umn.edu/omcd/basic_search.php).
Pathway analysis
To investigate the pathways targeted by miR-30b-5p, we carried out an in silico analysis using a web-based tool DIANA miRPath-v3.0 (http://www.microrna.gr/miRPathv3).39 We analyzed the experimentally validated miRNA-gene interactions (DIANA Tarbase v7.0). A threshold score of 0.8 was used.
Statistical analysis
The non-parametric Mann–Whitney U test was used to ascertain statistical significance of differences in miRNA expression levels between two groups. Receiver operating characteristics (ROC) curves were constructed and area under curve (AUC), specificity, sensitivity and accuracy were calculated as biomarker performance parameters. The best cut-off value was established based on the highest value obtained in ROC curve analysis according to Youden's J index.40,41 Statistical analysis was carried out using GraphPad Prism software (version 6.01; GraphPad Software, Inc., La Jolla, CA). Results were expressed as median with interquartile range. A P value <0.05 was considered significant (∗P value < 0.05, ∗∗P value < 0.01, ∗∗∗P value < 0.001, ∗∗∗∗P value < 0.0001).
Results
miR-30b-5p differential expression in BC cell lines and non-tumorigenic breast cell line
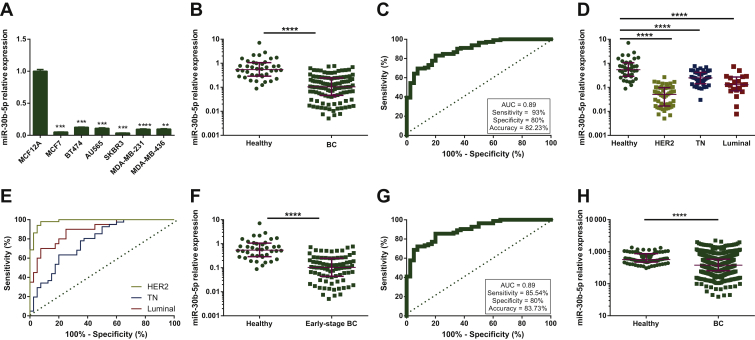
miR-30b-5p expression levels were determined in a set of BC cell lines including three subtypes: MCF-7 as Luminal; BT474, AU565 and SKBR3 as HER2+; MDA-MB-231 and MDA-MB-436 as TN BC and a healthy epithelial breast cell line MCF-12A. Herein, the MCF-12A cell line displayed significantly higher miR-30b-5p expression than all the tested BC cell lines (Figure 1A), thus implicating miR-30b-5p deregulation in BC development.
Figure 1.
(A) miR-30b-5p relative expression of BC cell lines compared with non-tumorigenic cell line MCF-12A. (B) miR-30b-5p relative expression in 112 BC tissues and 40 healthy breast tissues from cohort 1. (C) Receiver operating characteristics (ROC) curve analysis for miR-30b-5p expression in BC and healthy breast tissue from cohort 1. (D) miR-30b-5p relative expression in 51 HER2+, 41 TN and 20 Luminal BC tissues and 40 healthy breast tissues from cohort 1. (E) ROC curve analysis for miR-30b-5p expression in BC tissue with specific subtype and healthy breast tissue from cohort 1. (F) miR-30b-5p relative expression in 83 early stage (stage I and II) BC patients' tissues and 40 healthy breast tissues from cohort 1. (G) ROC curve analysis for miR-30b-5p expression in early-stage BC tissue and healthy breast tissue from cohort 1. (H) miR-30b-5p relative expression in 769 BC tissues and 74 healthy breast tissues from The Cancer Genome Atlas. Expression represented as reads per million microRNAs (miRNAs) mapped. Burgundy lines represent median with interquartile range. ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
AUC, area under curve; BC, breast cancer; HER2, human epidermal growth factor receptor 2; TN, triple-negative.
miR-30b-5p differential expression in BC and normal tissue (cohort #1)
Next, we investigated the expression of miR-30b-5p in a set of primary tumor tissue samples from 112 BC patients and 40 normal breast tissue samples (cohort #1) (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2020.100039). Clinico-pathological data of BC patients from cohort #1 are detailed in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100039. No statistically significant differences were found between the median age of patients and healthy donors (50 and 51 years old, respectively).
miR-30b-5p expression levels were significantly lower in BC tissues than in healthy breast tissues (P < 0.0001) (Figure 1B) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2020.100039). ROC curve analysis showed that miR-30b-5p expression in tissue could discriminate between healthy and BC tissues with AUC = 0.89 [95% confidence interval (CI) 0.83-0.94], P < 0.0001 (Figure 1C). When the best cut-off value was selected, miR-30b-5p expression identified BC with 83% sensitivity, 80% specificity and 82.23% accuracy.
Interestingly, all BC subtypes presented significantly different miR-30b-5p expression levels than normal breast tissue. Thereby, miR-30b-5p expression was significantly lower in HER2+, Luminal and TN compared with healthy controls (P < 0.0001) (Figure 1D). ROC curve analysis also revealed that miR-30b-5p expression levels may discriminate between normal tissue and each BC subtype (Figure 1E) with AUC = 0.98 (95% CI 0.96-1.00; P < 0.0001) in HER2+, AUC = 0.77 (95% CI 0.67-0.87; P < 0.0001) in TN and AUC = 0.88 (95% CI 0.79-0.97; P < 0.0001) in Luminal BC. The best cut-off value was selected for each subtype cohort and biomarker performance parameters were calculated (HER2+: 98% sensitivity, 92.50% specificity, 93.40% accuracy; TN: 63.40% sensitivity, 80% specificity, 74% accuracy; Luminal BC: 90% sensitivity, 75% specificity, 80% accuracy).
To explore the potential of miR-30b-5p as a BC biomarker, we next evaluated if miRNA expression might discriminate healthy tissue from BC even at the earliest stages of the disease. Notably, miR-30b-5p expression was significantly lower in early stages (stage I and II) BC than in healthy breast tissue; P < 0.0001 (Figure 1F). ROC analysis presented AUC = 0.89 (95% CI 0.84-0.95; P < 0.0001) with 85.54% sensitivity, 80% specificity and 83.73% accuracy when the best cut-off value was applied (Figure 1G).
Moreover, despite the small number of BC samples available at stage I, miR-30b-5p levels were also significantly lower in this stage than in healthy tissue; P < 0.0001 (Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2020.100039). ROC curve analysis showed an AUC = 0.87 (95% CI 0.78-0.97; P < 0.0001) and demonstrated that miR-30b-5p levels were able to discriminate stage I from healthy tissue with 73.70% sensitivity, 92.50% specificity and 84.44% accuracy when the best cut-off value was selected (Supplementary Figure S2B, available at https://doi.org/10.1016/j.esmoop.2020.100039).
To validate our results, an in silico analysis of miR-30b-5p expression in TCGA was carried out. Data from 769 BC patients and 74 healthy donors were available. miR-30b-5p expression was significantly lower in BC than in healthy tissue (P < 0.0001) (Figure 1H) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2020.100039).
miR-30b-5p differential expression in plasma samples from BC patients and healthy donors (cohort #2)
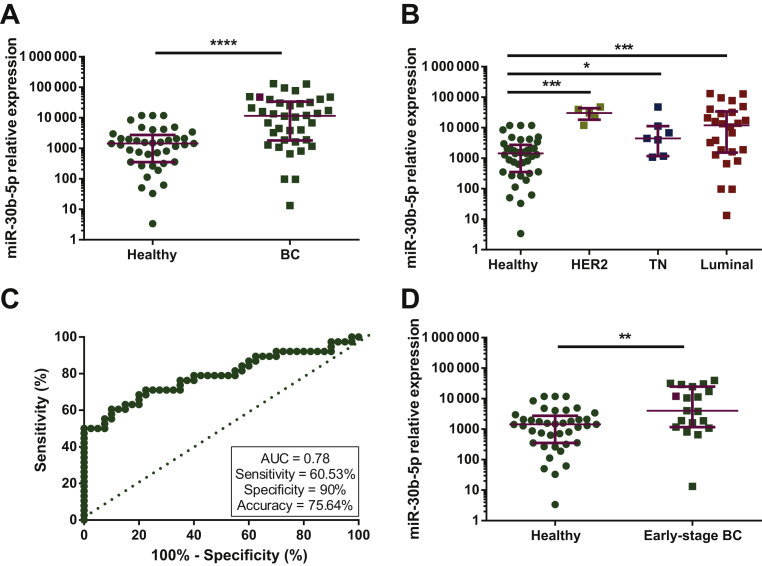
Given the value of miR-30b-5p as a diagnostic biomarker in breast tissue samples, we next investigated its potential as a non-invasive biomarker in liquid biopsy. We first evaluated circulating miR-30b-5p levels in a discovery cohort (cohort #2) of 38 plasma samples from BC patients and 40 plasma samples from healthy volunteers (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2020.100039). Clinico-pathological characteristics of patients from cohort #2 are described in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100039. No significant differences were observed in median age between BC and healthy donors (52 and 50 years old, respectively).
Circulating miR-30b-5p levels were significantly increased in plasma from BC patients compared with healthy donors; P < 0.0001 (Figure 2A) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2020.100039). This upregulation was observed across all BC subtypes (HER2+, P < 0.0001; TN, P = 0.0304; Luminal, P = 0.0004) compared with healthy donors (Figure 2B). ROC curve analysis showed an AUC = 0.78 (95% CI 0.68-0.89; P < 0.0001). When the best cut-off value was selected, circulating miR-30b-5p levels identified BC with 60.53% sensitivity, 90% specificity and 75.64% accuracy (Figure 2C).
Figure 2.
Circulating miR-30b-5p levels in plasma from all 38 BC patients (A) or five HER2+, seven TN and 26 Luminal BC patients (B) and 40 healthy volunteers from cohort 2. (C) Receiver operating characteristics (ROC) curve analysis for miR-30b-5p expression in plasma from BC patients and healthy donors from cohort 2. (D) Circulating miR-30b-5p levels in plasma from 19 early-stage (stage I and II) BC patients and 40 healthy volunteers from cohort 2. Burgundy lines represent median with interquartile range.
∗P< 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
BC, breast cancer; HER2, human epidermal growth factor receptor 2; TN, triple-negative.
A similar analysis was carried out within the different BC stages. Circulating miRNA levels were significantly higher in earliest stages (stage I and II) compared with healthy donors (P = 0.004) (Figure 2D). In addition, if considering only stage I BC as the very initial stage of BC, miR-30b-5p was still significantly higher in patients than in controls (P < 0.0001) (Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2020.100039). These results suggest circulating miR-30b-5p as a potential diagnostic BC biomarker.
Validation of miR-30b-5p as a BC diagnostic biomarker in plasma samples (cohort #3)
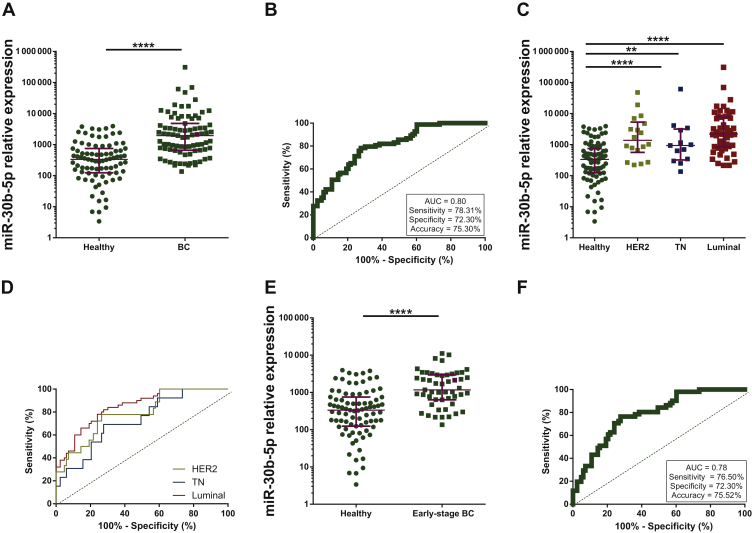
The previous results were further validated in an independent and larger set of 83 plasma samples from BC patients and 83 plasma samples from age-matched healthy donors (cohort #3) (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2020.100039). Clinico-pathological data of BC patients of cohort #3 are described in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100039.
In line with the results observed in cohort #2, circulating miR-30b-5p levels were significantly higher in plasma samples from BC patients compared with plasma from healthy donors (P < 0.0001) (Figure 3A) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2020.100039). The ROC curve analysis showed that mir-30b-5p was able to discriminate BC patients from healthy individuals with an AUC = 0.80 (95% CI 0.74-0.87) P < 0.0001 (Figure 3B). Biomarker performance parameters were evaluated applying best cut-off, and sensitivity, specificity and accuracy were 78.31%, 72.30% and 75.30%, respectively.
Figure 3.
(A) Circulating miR-30b-5p levels in plasma from 83 BC patients and 83 healthy volunteers from cohort 3. (B) Receiver operating characteristics (ROC) curve analysis for circulating miR-30b-5p in plasma from BC and healthy individuals from cohort 3. (C) Circulating miR-30b-5p levels in plasma from 18 HER2+, 13 TN and 50 Luminal BC patients and 83 healthy volunteers from cohort 3. (D) ROC curve analysis for circulating miR-30b-5p in plasma from BC patients with specific subtype and healthy donors from cohort 3. (E) Circulating miR-30b-5p levels in plasma from 51 early-stage (stage I and II) BC patients and 83 healthy volunteers from cohort 3. (F) ROC curve analysis for circulating miR-30b-5p in plasma from early stage BC patients and healthy donors from cohort 3. Burgundy lines represent median with interquartile range.
∗∗P < 0.01; ∗∗∗∗P < 0.0001.
AUC, area under curve; BC, breast cancer; HER2, human epidermal growth factor receptor 2; TN, triple-negative.
Similarly, miR-30b-5p expression levels were significantly lower in healthy controls than in all BC subtypes (HER2+, P < 0.0001; TN, P = 0.0079; Luminal, P < 0.0001) (Figure 3C). Specific ROC curve analysis for each subtype was consistent with previous findings demonstrating the potential of circulating miR-30b-5p as a diagnostic biomarker for HER2+ (AUC = 0.78, 95% CI 0.67-0.90, P = 0.0001; 77.78% sensitivity, 73.50% specificity, 74.25% accuracy), TN (AUC = 0.72, 95% CI 0.58-0.86, P = 0.0088; 69.23% sensitivity, 72.30% specificity, 71.80% accuracy) and Luminal BC (AUC = 0.84, 95% CI 0.77-0.91, P < 0.0001; 82% sensitivity, 72.30% specificity, 75.90% accuracy) (Figure 3D).
Importantly, circulating miR-30b-5p levels also effectively identified early BC stages in this cohort. miR-30b-5p expression levels were significantly higher in earliest stages of BC than in healthy controls (P < 0.0001) (Figure 3E). As expected, circulating miR-30b-5p was able to distinguish stage I and II BC from healthy samples with an AUC = 0.78 (95% 0.70-0.85), P < 0.0001 (Figure 3F). Biomarker performance parameters were also evaluated showing 76.50% sensitivity, 72.30% specificity and 75.52% accuracy. Similar results were obtained when stage I was evaluated separately. Circulating miR-30b-5p of stage I BC was significantly higher than in healthy controls (P < 0.0001). ROC analysis showed AUC = 0.79 (95% CI 0.70-0.89), P < 0.0001 and presented 80.90% sensitivity, 73.50% specificity and 69.20% accuracy (Supplementary Figure S4A and B, available at https://doi.org/10.1016/j.esmoop.2020.100039).
Association of circulating miR-30b-5p to locally advanced and metastatic BC
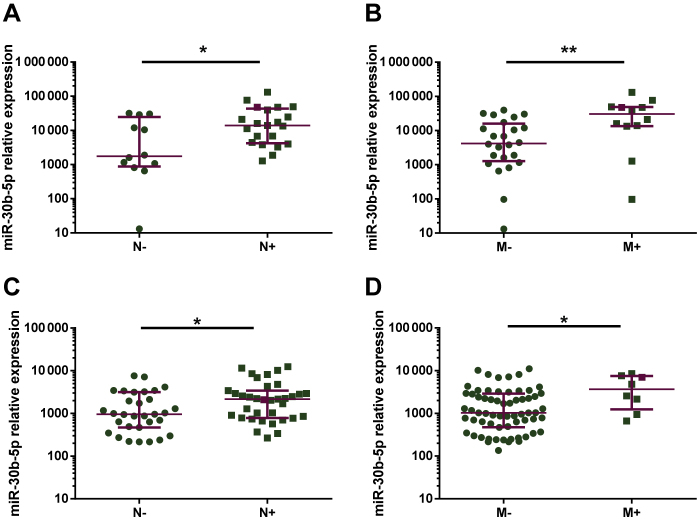
In cohort #2, patients with positive lymph nodes displayed significantly higher circulating miR-30b-5p levels than patients with negative lymph nodes (P = 0.0216) (Figure 4A). Moreover, circulating miRNA levels were significantly higher in de novo metastatic BC patients than in non-metastatic patients (P = 0.0056) (Figure 4B).
Figure 4.
Circulating miR-30b-5p levels in plasma from: 12 breast cancer (BC) patients with negative axillary lymph nodes and 21 BC patients with positive axillary lymph nodes from cohort 2 (A), 24 non-metastatic and 12 de novo metastatic BC patients from cohort 2 (B), 31 BC patients with negative axillary lymph nodes and 35 BC patients with positive axillary lymph nodes from cohort 3 (C) and 64 non-metastatic and eight de novo metastatic BC patients from cohort 3 (D). Burgundy lines represent median with interquartile range.
∗∗P < 0.01; ∗P < 0.05.
M-, non-metastatic; M+, de novo metastatic; N-, negative axillary lymph node; N+, positive axillary lymph node.
In agreement, in cohort #3, patients with positive axillary lymph nodes and metastatic disease also presented higher circulating miR-30b-5p levels (P = 0.0378 and P = 0.0275, respectively) (Figure 4C and D). Therefore, circulating miRNA levels might associate with patients' tumor burden.
Discussion
Screening mammography is the most commonly used technique worldwide for the detection of early BC in asymptomatic women and significantly reduces mortality.1,2 However, 10%-20% of patients are diagnosed with advanced stage of BC which is still characterized by poor prognosis. Thereby, more accurate and effective early-diagnosis methods are needed, as well as research on risk-based screening strategies.4, 5, 6
miRNAs were first reported to be related to cancer biology in 2002,42 and since then, a wide variety of studies support their impact in the pathogenesis of cancer.7,8 The potential of miRNAs as detection biomarkers in different types of cancer has also been demonstrated by several authors.16, 17, 18, 19 Moreover, miRNAs stability in body fluids makes them one of the best options for non-invasive detection techniques. Mitchell et al.22 demonstrated in 2008 that detection of circulating miRNAs in plasma can serve as diagnostic biomarkers for common human cancer types. Several studies have proposed plasma circulating miRNAs as early cancer detection biomarkers. Namely, miR-182-5p and miR-375-3p were identified in prostate cancer,43 miR-448, miR-506, miR-4316, miR-4478 and miR-31 in lung cancer,44,45 miR-24, miR-320a and miR-423-5p in colorectal cancer46 and miR-214, miR-96-5p, miR-21-5p, miR-505, miR-195 and miR-99a in BC.23,26,47,48
In this scenario, we assessed miR-30b-5p expression in BC tissue and plasma and tested its diagnostic performance. In cohort #1 miR-30b-5p expression levels were significantly lower in BC tissue than in normal breast tissue. When ROC curve analysis was carried out, miR-30b-5p expression in tissue showed 83% sensitivity, 80% specificity and 82.23% accuracy, thus supporting its potential as a diagnostic biomarker for BC. Moreover, miR-30b-5p expression identified all BC subtypes and early stage of the disease. These results are concordant with those of Hafez et al.49 in a cohort of Egyptian BC patients.
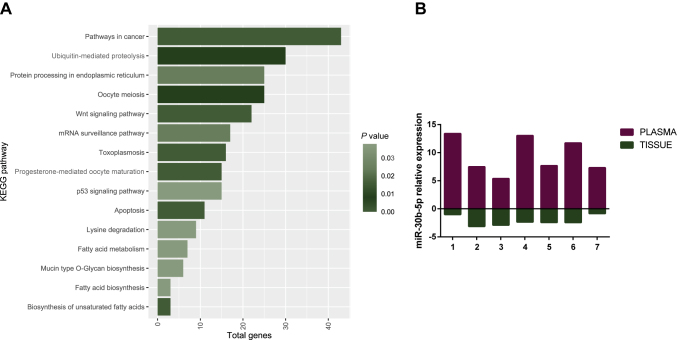
The impact of the miR-30 family in tumorigenesis, metastasis and drug resistance in cancer has been widely investigated. Moreover, Kyoto Encyclopedia for Genes and Genomes (KEGG) pathway analysis for miR-30b-5p showed significant enrichment in cancer pathways including the Wnt signaling pathway, the p53 signaling pathway and the apoptosis pathway (Figure 5A).
Figure 5.
KEGG pathway enrichment analysis for miR-30b-5p using DIANA Tools MirPath v3.0 (A). miR-30b-5p shows opposite regulation between tumor tissue and plasma matched samples from seven BC patients from cohorts 1 and 3. Relative expression represented by log two scale fold change (B).
KEGG, Kyoto Encyclopedia for Genes and Genomes.
miR-30b was suggested to function as a tumor suppressor in several types of cancer such as colorectal,50, 51, 52 esophageal,53 gastric,54,55 hepatocellular,56,57 thyroid,58,59 lymphoma,60 renal,61 lung62,63 and pancreatic cancer.64 In fact, in BC the low miR-30b expression in tumor tissue has been associated with poor relapse-free survival, being proposed to prevent tumor progression and metastasis development through targeting of CDH11, ITGA5 and ITGB3.37 Furthermore, low expression of the miR-30 family has been reported to maintain self-renewal of breast tumor initiating cells by targeting ITGB3 and Ubc9.38 Moreover, high miR-30b expression has been also associated with improved response to endocrine therapy in luminal BC,36 as well as with trastuzumab response.65
Nonetheless, the role of the miR-30 family in cancer biology remains controversial. Some studies present this miRNA family as tumor promoters in cancer-related processes. Specifically, a pro-metastatic role of miR-30b/30d has been proposed in melanoma.32 A plausible explanation of these different roles in cancer may be due to miR-30's ability to target different mRNAs in line with what has been described for other well-studied miRNAs.
Given the advantages of miRNAs as non-invasive biomarkers detectable in body fluids, we further tested circulating miR-30b-5p biomarker performance in two independent cohorts of patients: a discovery cohort (cohort #2) and cohort #3 in which results were validated in a blindly fashion. We demonstrated that circulating miR-30b-5p was significantly higher in BC plasma samples than healthy donors in both cohorts. Moreover, we proved the potential of circulating miR-30b-5p as a diagnostic biomarker with AUC values of 0.78 and 0.80 in the discovery and validation cohorts, respectively. In addition, we showed that the diagnostic performance was applicable to all subtypes of BC and early stage of the disease. Similar results on circulating miR-30b-5p levels were previously obtained by other research teams in whole blood and sera from a rather limited cohort of BC patients and healthy controls.34,66
Another interesting observation was the significant correlation between circulating miR-30b-5p levels and patients' tumor burden. Our results showed that circulating miR-30b-5p levels were positively correlated with positive axillary lymph nodes and distant metastasis, in accordance with previous studies.33 In fact, the overexpression of circulating miR-30b in different liquids as sera, extracellular vesicles, plasma or cyst fluid has been associated with cancer and poor prognosis in several tumors such as lung,29,67 colorectal,68 colangiocarcinoma69 and pancreatic cancer.30 The opposite trend of miR-30b-5p expression found in tissue and plasma samples was certainly unexpected. Nonetheless, several studies also found opposite levels for other circulating miRNAs versus breast tissue expression26,28,66,70,71 as well as other tumor types.72, 73, 74, 75 Different explanations have been anticipated for this apparent paradox. First, released miRNAs do not necessarily reflect the expression in the tissue of origin. Indeed, processes by which miRNAs are released into circulation and respective function remain a challenge. This mechanism might well be via inter-organ cell communication. It has been proposed that cells might have a mechanism to selectively release specific miRNAs and that extracellular miRNAs should be considered independent of cellular miRNA levels, when considering diagnostic markers.66,76 To check the feasibility of this hypothesis, we compared miR-30b-5p expression levels in available material for matched tissue and plasma from seven patients. Herein, miRNA expression levels in tissues were opposite to those in plasma (Figure 5B), thus supporting that mir-30b-5p is being selectively released and then its abundance is decreased in tumor cells.
A second hypothesis is that extracellular miRNAs might be the product of dead cells and tissue injury that persist in circulation due to their high stability.77
Additionally, it has been argued that other possible sources of circulating miRNAs might be tumor microenvironment cells. Given the importance of tumor microenvironment in cancer initiation and progression, its contribution to the circulating miRNA profile should not be neglectable.28
The last hypothesis proposes that these miRNAs may originate from blood cells,78 however this is a widely discussed, controversial issue.79
To our knowledge, this is the first study evaluating miR-30b-5p expression levels both in tissue and plasma sets simultaneously. Moreover, these are the largest cohorts of patients in which miR-30b-5p levels have been evaluated in tissue and plasma samples. Importantly, applicability of circulating miR-30b-5p in plasma as a diagnostic biomarker of BC would be cost-effective given that plasma acquisition would be easily accessible and tests could be carried out with several samples simultaneously. Moreover, we demonstrated the reproducibility of circulating miR-30b-5p levels in plasma as a BC diagnostic biomarker by blindly validating its potential in an independent cohort from a different institution. In addition, circulating miR-30b-5p fold change in BC compared with donors was not statistically different between the discovery and validation cohorts (cohort #2 and #3).
Conclusion
Taken together, the data presented in this study demonstrate the value of miR-30b-5p expression levels as an early diagnostic BC biomarker. Both low miRNA expression in tumor tissue and high circulating miR-30b-5p levels in plasma were associated with BC. miR-30b-5p levels were able to identify BC in three different patients' cohorts, independently of the subtype and stage of the disease. We also showed that circulating miR-30b-5p levels relate with patients' tumor burden. We highlight the potential of circulating miR-30b-5p as a non-invasive, fast, reproducible and cost-effective diagnostic BC biomarker.
Acknowledgements
The authors of this study thank the INCLIVA Biobank (PT17/0015/0049; B.000768 ISCIII) and the Valencian Biobanking Network integrated into the Spanish National Biobanks Network for its collaboration. We are also grateful to the nursing staff from IPO-Porto and INCLIVA. We are especially grateful to the patients and healthy volunteers, both from INCLIVA and IPO-Porto, that accepted to participate in this study and to Associations Amunt Contra el Cáncer and Ágora-Foia de Castalla.
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) and co-financed by FEDER funds [grant numbers PI18/01219 to PE and CB16/12/00481 to ALL]. AA-A and AL were funded by Asociación Española Contra el Cáncer (AECC). IG-C was funded by Generalitat Valenciana [grant number ACIF/2016/030]. VC was funded by Liga Portuguesa Contra o Cancro/Fundação PT. SS was funded by FCT Fundação para a Ciência e Tecnologia [grant number SFRH/BD/143717/2019]. JMC was funded by Sociedad Española de Oncología Médica (Río Hortega-SEOM).
Disclosure
The authors have declared no conflicts of interest.
Contributor Information
P. Eroles, Email: pilar.eroles@uv.es.
J.M. Cejalvo, Email: jmcejalvo@incliva.es.
Supplementary data
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Senkus E., Kyriakides S., Penault-Llorca F. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(suppl 6):vi7–v23. doi: 10.1093/annonc/mdt284. [DOI] [PubMed] [Google Scholar]
- 4.Cardoso F., Senkus E., Costa A. 4th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 4) Ann Oncol. 2018;29(8):1634–1657. doi: 10.1093/annonc/mdy192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller K.D., Nogueira L., Mariotto A.B. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 6.Nattinger A.B., Mitchell J.L. Breast cancer screening and prevention. Ann Intern Med. 2016;164(11):ITC81–ITC94. doi: 10.7326/AITC201606070. [DOI] [PubMed] [Google Scholar]
- 7.Ebert M.S., Sharp P.A. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasinski A.L., Slack F.J. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J., Getz G., Miska E.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Dvinge H., Git A., Gräf S. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld N., Aharonov R., Meiri E. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 12.Andorfer C.A., Necela B.M., Thompson E.A., Perez E.A. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17(6):313–319. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Sebio A., Paré L., Páez D. The LCS6 polymorphism in the binding site of let-7 microRNA to the KRAS 3′-untranslated region: its role in the efficacy of anti-EGFR-based therapy in metastatic colorectal cancer patients. Pharmacogenet Genomics. 2013;23(3):142–147. doi: 10.1097/FPC.0b013e32835d9b0b. [DOI] [PubMed] [Google Scholar]
- 14.Wynendaele J., Böhnke A., Leucci E. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70(23):9641–9649. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 15.Tormo E., Ballester S., Adam-Artigues A. The miRNA-449 family mediates doxorubicin resistance in triple-negative breast cancer by regulating cell cycle factors. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-41472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S.B. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 18.Sochor M., Basova P., Pesta M. Early breast cancer in serum. BMC Cancer. 2014;14(1):1–7. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iorio M.V., Ferracin M., Liu C.G. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 20.Cortez M.A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A.K., Calin G.A. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arroyo J.D., Chevillet J.R., Kroh E.M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell P.S., Parkin R.K., Kroh E.M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzenbach H., Milde-Langosch K., Steinbach B., Müller V., Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat. 2012;134(3):933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
- 24.Hamam R., Ali A.M., Alsaleh K.A. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadaki C., Stratigos M., Markakis G. Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Res. 2018;20(1):1–17. doi: 10.1186/s13058-018-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matamala N., Vargas M.T., González-Cámpora R. Tumor MicroRNA expression profiling identifies circulating MicroRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 27.Madhavan D., Peng C., Wallwiener M. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37(5):461–470. doi: 10.1093/carcin/bgw008. [DOI] [PubMed] [Google Scholar]
- 28.Cuk K., Zucknick M., Heil J. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132(7):1602–1612. doi: 10.1002/ijc.27799. [DOI] [PubMed] [Google Scholar]
- 29.Li C., Qin F., Hu F. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8(1):1–21. doi: 10.1186/s13578-018-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Paris P.L., Chen J. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett. 2015;356(2):404–409. doi: 10.1016/j.canlet.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandimalla R., Gao F., Matsuyama T. Genome-wide discovery and identification of a novel miRNA signature for recurrence prediction in stage II and III colorectal cancer. Clin Cancer Res. 2018;24(16):3867–3877. doi: 10.1158/1078-0432.CCR-17-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaziel-Sovran A., Segura M.F., Di Micco R. MiR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20(1):104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Estevão-Pereira H., Lobo J., Salta S. Overexpression of circulating MiR-30b-5p identifies advanced breast cancer. J Transl Med. 2019;17(1):1–10. doi: 10.1186/s12967-019-02193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K., Wang Y.W., Wang Y.Y. Identification of microRNA biomarkers in the blood of breast cancer patients based on microRNA profiling. Gene. 2017;619:10–20. doi: 10.1016/j.gene.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Wu T., Song H., Xie D. Mir-30b-5p promotes proliferation, migration, and invasion of breast cancer cells via targeting ASPP2. Biomed Res Int. 2020;2020:7907269. doi: 10.1155/2020/7907269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amorim M., Lobo J., Fontes-Sousa M. Predictive and prognostic value of selected microRNAs in luminal breast cancer. Front Genet. 2019;10:815. doi: 10.3389/fgene.2019.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croset M., Pantano F., Kan C.W.S. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Cancer Res. 2018;78(18):5259–5273. doi: 10.1158/0008-5472.CAN-17-3058. [DOI] [PubMed] [Google Scholar]
- 38.Yu F., Deng H., Yao H., Liu Q., Su F., Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29(29):4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 39.Vlachos I.S., Zagganas K., Paraskevopoulou M.D. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Schisterman E.F., Perkins N.J., Liu A., Bondell H. Optimal cut-point and its corresponding Youden index to discriminate individuals using pooled blood samples. Epidemiology. 2005;16(1):73–81. doi: 10.1097/01.ede.0000147512.81966.ba. [DOI] [PubMed] [Google Scholar]
- 42.Calin G.A., Dumitru C.D., Shimizu M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bidarra D., Constâncio V., Barros-Silva D. Circulating microRNAs as biomarkers for prostate cancer detection and metastasis development prediction. Front Oncol. 2019;9:900. doi: 10.3389/fonc.2019.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powrózek T., Krawczyk P., Kowalski D.M. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumor Biol. 2016;37(2):2049–2055. doi: 10.1007/s13277-015-3971-4. [DOI] [PubMed] [Google Scholar]
- 45.Yan H.J., Ma J.Y., Wang L., Gu W. Expression and significance of circulating microRNA-31 in lung cancer patients. Med Sci Monit. 2015;21:722–726. doi: 10.12659/MSM.893213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang Z., Tang J., Bai Y. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34(1):1–10. doi: 10.1186/s13046-015-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heneghan H.M., Miller N., Kelly R., Newell J., Kerin M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15(7):673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrido-Cano I., Constâncio V., Adam-Artigues A. Circulating mir-99a-5p expression in plasma: a potential biomarker for early diagnosis of breast cancer. Int J Mol Sci. 2020;21(19):1–14. doi: 10.3390/ijms21197427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hafez M.M., Hassan Z.K., Zekri A.R.N. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pacific J Cancer Prev. 2012;13(2):591–598. doi: 10.7314/apjcp.2012.13.2.591. [DOI] [PubMed] [Google Scholar]
- 50.Liao W.T., Ye Y.P., Zhang N.J. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232(4):415–427. doi: 10.1002/path.4309. [DOI] [PubMed] [Google Scholar]
- 51.Fan M., Ma X., Wang F. MicroRNA-30b-5p functions as a metastasis suppressor in colorectal cancer by targeting Rap1b. Cancer Lett. 2020;477:144–156. doi: 10.1016/j.canlet.2020.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H., Xu Z., Qin H., Gao Z., Gao L. MiR-30b regulates migration and invasion of human colorectal cancer via SIX1. Biochem J. 2014;460(1):117–125. doi: 10.1042/BJ20131535. [DOI] [PubMed] [Google Scholar]
- 53.Liu S.G., Qin X.G., Zhao B.S. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5(5):1639–1642. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu E.D., Li N., Li B.S. miR-30b, down-regulated in gastric cancer, promotes apoptosis and suppresses tumor growth by targeting plasminogen activator inhibitor-1. PLoS One. 2014;9(8):1–12. doi: 10.1371/journal.pone.0106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian S.B., Yu J.C., Liu Y.Q. MiR-30b suppresses tumor migration and invasion by targeting EIF5A2 in gastric cancer. World J Gastroenterol. 2015;21(31):9337–9347. doi: 10.3748/wjg.v21.i31.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao D., Zhou Z., Huang H. miR-30b-3p inhibits proliferation and invasion of hepatocellular carcinoma cells via suppressing PI3K/Akt pathway. Front Genet. 2019;10:1274. doi: 10.3389/fgene.2019.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin X., Chen J., Wu L., Liu Z. MiR-30b-5p acts as a tumor suppressor, repressing cell proliferation and cell cycle in human hepatocellular carcinoma. Biomed Pharmacother. 2017;89:742–750. doi: 10.1016/j.biopha.2017.02.062. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y., Liu X., Zhong L. The combined use of miRNAs and mRNAs as biomarkers for the diagnosis of papillary thyroid carcinoma. Int J Mol Med. 2015;36(4):1097–1103. doi: 10.3892/ijmm.2015.2305. [DOI] [PubMed] [Google Scholar]
- 59.Hu Y., Zhang X., Cui M. Verification of candidate microRNA markers for parathyroid carcinoma. Endocrine. 2018;60(2):246–254. doi: 10.1007/s12020-018-1551-2. [DOI] [PubMed] [Google Scholar]
- 60.Reddemann K., Gola D., Schillert A. Dysregulation of MicroRNAs in angioimmunoblastic T-cell lymphoma. Anticancer Res. 2015;35(4):2055–2061. [PubMed] [Google Scholar]
- 61.Liu W., Li H., Wang Y. MiR-30b-5p functions as a tumor suppressor in cell proliferation, metastasis and epithelial-to-mesenchymal transition by targeting G-protein subunit α-13 in renal cell carcinoma. Gene. 2017;626:275–281. doi: 10.1016/j.gene.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 62.Qi Z., Zhang B., Hu Q., Xu F., Chen B., Zhu C. MicroRNA-30b inhibits non-small cell lung cancer cell growth by targeting the epidermal growth factor receptor. Neoplasma. 2013;60(5):607–616. doi: 10.4149/neo_2018_170217N118. [DOI] [PubMed] [Google Scholar]
- 63.Cheng Y., Yang S., Shen B. Molecular characterization of lung cancer: a two-miRNA prognostic signature based on cancer stem-like cells related genes. J Cell Biochem. 2020;121(4):2889–2900. doi: 10.1002/jcb.29525. [DOI] [PubMed] [Google Scholar]
- 64.Xiong Y., Wang Y., Wang L. MicroRNA-30b targets Snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. J Cancer. 2018;9(12):2147–2159. doi: 10.7150/jca.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tormo E., Adam-Artigues A., Ballester S. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu J., Zheng Z., Wang J. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014;5:149. doi: 10.3389/fgene.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X., Zhang Q., Zhang M. Serum microRNA signature is capable of early diagnosis for non-small cell lung cancer. Int J Biol Sci. 2019;15(8):1712–1722. doi: 10.7150/ijbs.33986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ostenfeld M.S., Jensen S.G., Jeppesen D.K. miRNA profiling of circulating EpCAM+ extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles. 2016;5(1):1–14. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voigtländer T., Gupta S.K., Thum S. MicroRNAs in serum and bile of patients with primary sclerosing cholangitis and/or cholangiocarcinoma. PLoS One. 2015;10(10):1–14. doi: 10.1371/journal.pone.0139305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markou A., Zavridou M., Sourvinou I. Direct comparison of metastasis-related miRNAs expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem. 2016;62(7):1002–1011. doi: 10.1373/clinchem.2015.253716. [DOI] [PubMed] [Google Scholar]
- 71.Chan M., Liaw C.S., Ji S.M. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19(16):4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 72.Torres A., Torres K., Pesci A. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer. 2012;12:369. doi: 10.1186/1471-2407-12-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka M., Oikawa K., Takanashi M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5):1–5. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zedan A.H., Hansen T.F., Assenholt J., Pleckaitis M., Madsen J.S., Osther P.J.S. MicroRNA expression in tumour tissue and plasma in patients with newly diagnosed metastatic prostate cancer. Tumor Biol. 2018;40(5):1–11. doi: 10.1177/1010428318775864. [DOI] [PubMed] [Google Scholar]
- 75.Wang W., Yin Y., Shan X. The value of plasma-based microRNAs as diagnostic biomarkers for ovarian cancer. Am J Med Sci. 2019;358(4):256–267. doi: 10.1016/j.amjms.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 76.Pigati L., Yaddanapudi S.C.S., Iyengar R. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pritchard C.C., Kroh E., Wood B. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brase J.C., Wuttig D., Kuner R., Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:1–9. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.