Summary
Although recent evidence suggests the involvement of iron accumulation in the pathogenesis of nonalcoholic steatohepatitis (NASH), the underlying mechanisms remain poorly understood. Previously, we reported a unique histological structure termed “crown-like structure (CLS),” where liver-resident macrophages (Kupffer cells) surround dead hepatocytes, scavenge their debris, and induce inflammation and fibrosis in NASH. In this study, using magnetic column separation, we show that iron-rich Kupffer cells exhibit proinflammatory and profibrotic phenotypic changes during the development of NASH, at least partly, through activation of MiT/TFE transcription factors. Activation of MiT/TFE transcription factors is observed in Kupffer cells forming CLSs in murine and human NASH. Iron chelation effectively attenuates liver fibrosis in a murine NASH model. This study provides insight into the pathophysiologic role of iron in NASH. Our data also shed light on a unique macrophage subset rich in iron that contributes to CLS formation and serves as a driver of liver fibrosis.
Subject areas: Biological Sciences, Biochemistry, Cell Biology
Graphical Abstract

Highlights
-
•
Kupffer cells are magnetically divided into Fe-hi and Fe-lo fractions in a NASH model
-
•
Fe-hi Kupffer cells exert proinflammatory and profibrotic properties in NASH
-
•
MiT/TFE transcription factors mediate iron-induced Kupffer cells' phenotypic changes
-
•
MiT/TFE transcription factors are activated in Kupffer cells in murine and human NASH
Biological Sciences; Biochemistry; Cell Biology
Introduction
Nonalcoholic fatty liver disease (NAFLD) is commonly observed in patients with the metabolic syndrome and is one of the most prevalent chronic liver diseases in the world. The term NAFLD refers to a spectrum of chronic liver disease ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), of which the latter predisposes patients to cirrhosis and hepatocellular carcinoma (Tilg and Moschen, 2010). As proposed by the multiple parallel hits hypothesis, the pathogenesis of NASH involves a multistep process; in this context, metabolic stresses such as lipid accumulation and insulin resistance and inflammatory stimuli such as proinflammatory cytokines and oxidative stress have been intensively investigated (Tilg and Moschen, 2010). Among various etiologies, aberrant iron metabolism is also considered to be a “hit” in the pathogenesis of NASH (Nelson et al., 2012). For instance, 10%–50% patients with NAFLD exhibit hepatic iron accumulation and/or hyperferritinemia (Valenti et al., 2006; Manousou et al., 2011; Nelson et al., 2012; Ryan et al., 2018). High-fat-diet feeding decreases duodenal expression of oxidoreductases in mice, resulting in suppression of iron absorption (Sonnweber et al., 2012). Moreover, phlebotomy effectively ameliorates liver damage (Sumida et al., 2006; Valenti et al., 2011). Because iron increases oxidative stress through the Fenton reaction, iron overload may be involved in the pathogenesis of various chronic diseases including NAFLD (Galaris and Pantopoulos, 2008). However, the pathophysiologic role of iron in NASH remains controversial (O'Brien and Powell, 2012).
Liver comprises parenchymal hepatocytes and a variety of stromal cells including macrophages and hepatic stellate cells, all of which interact with each other to remodel tissue structure during the development of NASH. We previously demonstrated that hepatocyte death, a prominent feature of NASH, triggers a phenotypic change of resident macrophages in the liver (Kupffer cells), leading to liver fibrosis (Itoh et al., 2013, 2017). Indeed, in murine and human NASH, there is a unique histological structure termed “crown-like structure (CLS),” in which dead hepatocytes with large lipid droplets are surrounded by CD11c-positive Kupffer cells; in contrast, CD11c-negative Kupffer cells are scattered throughout the liver (Itoh et al., 2013, 2017). During the maturation of CLSs, Kupffer cells become positive for CD11c through cross talk with dead hepatocytes and acquire proinflammatory and profibrotic properties (Itoh et al., 2017). Thus, CD11c-positive Kupffer cells may be a novel macrophage subset crucial for liver fibrosis in NASH (Itoh et al., 2017). However, the molecular mechanism underlying the phenotypic change of Kupffer cells remains to be elucidated.
There is considerable evidence that iron metabolism plays a critical role in macrophages at steady state and in response to infections (Soares and Hamza, 2016). Because iron is an essential micronutrient for both microbial pathogens and their mammalian hosts, iron deficiency in macrophages is thought to predispose the host to bacterial infections (Schaible and Kaufmann, 2004). With regard to the potential underlying mechanism, low intracellular iron selectively impairs the Toll-like receptor 4 signaling in macrophages (Wang et al., 2009). Moreover, increasing evidence suggests a role for iron metabolism in non-infectious diseases such as atherosclerosis, cancer, and neurological disorders (da Silva et al., 2017; Gillen et al., 2018; Guo et al., 2018). Iron overloading skews macrophages toward a proinflammatory phenotype in a mouse model of skin wound healing (Sindrilaru et al., 2011), suggesting a role for iron metabolism in macrophage function and polarization in sterile inflammation in vivo. In addition, a recent study revealed a positive association between hepatic iron deposition in reticuloendothelial system cells (presumably Kupffer cells) and the severity of NASH (Maliken et al., 2013). These observations led us to hypothesize that aberrant iron metabolism in Kupffer cells is crucial for chronic inflammation in NASH.
Here, we show that iron-rich Kupffer cells exhibit proinflammatory and profibrotic phenotypic changes during the development of NASH. Using magnetic column separation, we divided Kupffer cells prepared from a murine NASH model into two fractions: ferromagnetic Fe-hi and remaining Fe-lo. Fe-hi Kupffer cells were responsible for upregulation of proinflammatory and profibrotic genes. With regard to the molecular mechanism, MiT/TFE transcription factors were involved in iron accumulation-mediated phenotypic changes of Kupffer cells. Furthermore, we confirmed activation of MiT/TFE transcription factors in Kupffer cells in murine and human NASH. This study provides insight into the pathophysiologic role of iron in NASH and sheds light on a unique macrophage subset rich in iron.
Results
Iron loading accelerates hepatic proinflammatory and profibrotic changes in a murine NASH model
In this study, we employed genetically obese MC4R-KO (knockout) mice fed Western diet (WD) as a murine NASH model. The mice developed simple steatosis after 4 weeks of WD feeding, NASH-like liver phenotypes (micro- and macrovesicular steatosis, ballooning degeneration, massive infiltration of inflammatory cells, and pericellular fibrosis) at 16 weeks and multiple liver tumors at 1 year (Figure S1) (Itoh et al., 2011; Wang et al., 2016; Hasenour et al., 2020). To investigate the effect of iron loading on NASH, MC4R-KO mice fed standard diet (SD) (normal liver) or WD for 8 weeks (simple steatosis) received a single intraperitoneal injection of iron-dextran and were analyzed on day 1 (Figures 1A–1C). Iron accumulation detected by Perls' staining was mostly observed in F4/80+ macrophages (or Kupffer cells) (Figure 1B), suggesting the uptake of iron-dextran through phagocytotic activity. Efficient iron loading was confirmed by elevated mRNA levels of iron metabolism-related genes such as Hepcidin and Bmp6 (Figure 1C). In this experimental setting, iron loading resulted in a significant increase in mRNA levels of proinflammatory and profibrotic genes such as Itgax (CD11c, a marker for Kupffer cells exhibiting phenotypic changes in NASH), Tnfa (tumor necrosis factor alpha), Ccl3 (chemokine (C–C motif) ligand 3), and Timp1 (tissue inhibitor of metalloproteinase 1) in the livers of MC4R-KO mice fed WD, versus only a modest increase in MC4R-KO mice fed SD. Notably, Kupffer cells isolated from iron-treated MC4R-KO mice fed WD showed higher mRNA levels of proinflammatory and profibrotic genes than those from vehicle-treated mice (Figure 1D). Moreover, macrophage depletion using clodronate liposomes abolished the iron loading-induced increase in these mRNA levels in the livers of MC4R-KO mice fed WD (Figure S2). Collectively, these results indicate that Kupffer cells are responsible for iron-mediated proinflammatory and profibrotic changes in iron-treated steatotic livers.
Figure 1.
Iron loading accelerates proinflammatory and profibrotic changes in the liver in a murine model of NASH
(A–D) Effects of a single injection of iron-dextran on proinflammatory and profibrotic changes in the livers of MC4R-KO mice. (A) Experimental protocol. MC4R-KO mice fed standard diet (SD) or Western diet (WD) for 8 weeks (normal liver and simple steatosis, respectively) received a single intraperitoneal injection of iron-dextran (0.1 g/kg body weight) and were analyzed on day 1. (B) Co-staining of iron deposition (Perls' staining, blue) and macrophages (F4/80 immunostaining, brown) of the liver 1 day after iron-dextran injection. Insets: higher magnification of Kupffer cells. Scale bars, 50 μm. (C) Hepatic mRNA expression of genes related to iron metabolism, inflammation, and fibrosis. Data represent the mean ± SEM. n = 4. ∗p < 0.05 versus the respective SD, #p < 0.05 (Tukey-Kramer test). (D) mRNA expression of genes related to inflammation and fibrosis in Kupffer cells (CD11b+ F4/80hi) isolated from MC4R-KO mice fed WD for 10 weeks (simple steatosis) receiving a single injection of iron-dextran. Data represent the mean ± SEM. n = 5. ∗p < 0.05 versus vehicle (two-tailed unpaired Student's t test).
(E–H) Effects of repeated injection of iron-dextran in MC4R-KO mice during NASH development. (E) Experimental protocol. MC4R-KO mice were fed WD for 16 weeks, and the mice received once-weekly injection of iron-dextran (3 mg/week) during the last 11 weeks. (F) Hepatic mRNA expression of genes related to inflammation and fibrosis. Data represent the mean ± SEM. n = 5. ∗p < 0.05 versus WT/SD, #p < 0.05 (Tukey-Kramer test). (G) F4/80 immunostaining of the livers. Arrows indicate crown-like structures (CLSs). Scale bars, 50 μm. Data represent the mean ± SEM. n = 5. ∗p < 0.05 versus WT/SD (Tukey-Kramer test). (H) Type III collagen immunostaining of the livers. Scale bars, 50 μm. Data represent the mean ± SEM. n = 5. ∗p < 0.05 versus vehicle (two-tailed unpaired Student's t test).
See also Figures S2 and S3.
Moreover, we examined the long-term effect of iron loading on the development of NASH (Figures 1E–1H). MC4R-KO mice were fed WD for 16 weeks and received once-weekly injection of iron-dextran during the last 11 week (Figure 1E). Repeated injection of iron-dextran did not change body weight, liver weight, or blood parameters (serum ALT and AST, and blood glucose), but significantly decreased hepatic triglyceride contents and expression of the lipogenic gene Fasn (Figure S3). Notably, iron loading significantly increased mRNA levels of proinflammatory and profibrotic genes, together with type III collagen-positive areas, whereas it did not affect CLS formation (Figures 1F–1H). These findings indicate that iron loading accelerates proinflammatory and profibrotic changes in the liver during the development of NASH in MC4R-KO mice fed WD.
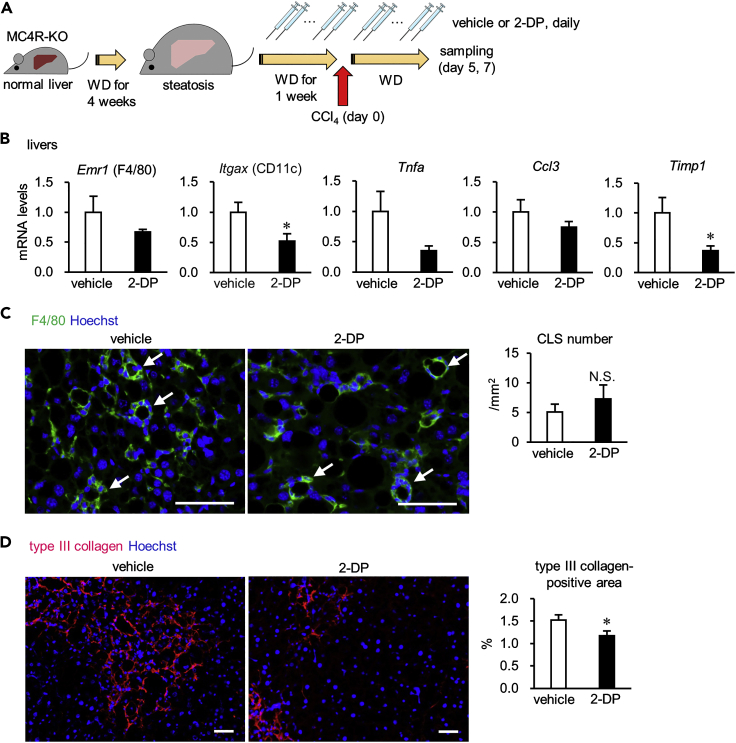
Iron chelation attenuates hepatic proinflammatory and profibrotic changes in an inducible NASH model
To further elucidate the role of iron metabolism in the pathogenesis of NASH, we examined the effects of the iron chelator 2,2′-dipyridyl (2-DP) on liver fibrosis in an inducible NASH model (Figure 2A), in which a single injection of carbon tetrachloride (CCl4, a potent hepatotoxic chemical) into MC4R-KO mice fed WD for 5 weeks (simple steatosis) recapitulates NASH-like liver phenotypes in the short term (Itoh et al., 2017). Using this model, we demonstrated that hepatocyte death triggers a phenotypic change in Kupffer cells, inducing CLS formation and fibrogenesis in the liver (Itoh et al., 2017). In this study, daily injection of 2-DP effectively decreased mRNA levels of proinflammatory and profibrotic genes and liver fibrosis, whereas it did not affect the number of CLSs, body weight, liver weight, hepatic triglyceride contents, or blood parameters (Figures 2B–2D and S4). These results confirm the role of iron metabolism in liver fibrosis in the NASH model.
Figure 2.
Iron chelation attenuates proinflammatory and profibrotic changes in the liver in an inducible NASH model
Effects of repeated injection of the iron chelator 2-DP on NASH-like liver phenotypes in the inducible NASH model.
(A) Experimental protocol. MC4R-KO mice were fed WD for 5 weeks (simple steatosis), received a single intraperitoneal injection of CCl4 at a dose of 0.15 mL/kg for induction of the NASH model, and then were analyzed on days 5 and 7. During the last 2 weeks of the experimental period, the mice received daily intraperitoneal injection of 2-DP (20 mg/kg body weight/day).
(B) Hepatic mRNA expression of genes related to inflammation and fibrosis on day 5. Data represent the mean ± SEM. n = 9. ∗p < 0.05 versus vehicle (two-tailed unpaired Student's t test).
(C and D) Immunostaining for F4/80 (C) and type III collagen (D) of the livers on day 7. Arrows indicate CLS. Scale bars, 50 μm. Data represent the mean ± SEM. n = 13–14. N.S., not significant (two-tailed unpaired Student's t test).
See also Figure S4.
Iron accumulation is involved in proinflammatory and profibrotic responses in NPCs during the development of NASH
As liver is composed of hepatocytes and non-parenchymal cells (NPCs) such as Kupffer cells, hepatic stellate cells, and sinusoidal cells, we next sought to determine which cell types are responsible for iron accumulation-mediated proinflammatory and profibrotic responses. First, primary NPCs prepared from the livers of MC4R-KO mice fed WD for 5 weeks (simple steatosis) were treated with FeCl3 for 48 h in culture (Figure 3A). Iron loading significantly increased mRNA levels of proinflammatory and profibrotic genes (Figure 3B). Conversely, primary NPCs prepared from the livers of MC4R-KO mice fed WD for 14 weeks (NASH) were treated with 2-DP for 12 h in culture (Figure 3C). Iron chelation significantly suppressed mRNA expression of these genes (Figure 3D). These observations suggest that iron metabolism in NPCs is involved in proinflammatory and profibrotic responses in the NASH model.
Figure 3.
Iron accumulation is involved in proinflammatory and profibrotic responses in NPCs during NASH pathogenesis
(A and B) Effects of FeCl3 on primary non-parenchymal cells (NPCs). (A) Experimental protocol. NPCs prepared from the livers of MC4R-KO mice fed WD for 5 weeks (simple steatosis) were cultured with or without FeCl3 for 48 h. (B) mRNA levels of genes related to inflammation and fibrosis in primary NPCs. Data represent the mean ± SEM. n = 4. ∗p < 0.05 versus vehicle (Tukey-Kramer test).
(C and D) Effects of the iron chelator 2-DP on primary NPCs. (C) Experimental protocol. NPCs prepared from the livers of MC4R-KO mice fed WD for 14 weeks (NASH) were cultured with or without 2-DP for 12 h. (D) mRNA levels of genes related to inflammation and fibrosis in primary NPCs. Data represent the mean ± SEM. n = 4. ∗p < 0.05 versus vehicle (Tukey-Kramer test).
Iron-rich Kupffer cells exhibit proinflammatory and profibrotic properties during the development of NASH
We have already demonstrated that there is a small subset of Kupffer cells that become positive for CD11c through interactions with dead hepatocytes during the development of NASH, thereby inducing sustained inflammation and fibrosis in the liver (Itoh et al., 2013, 2017). Because Kupffer cells are a major cellular source of iron accumulation in the liver (Theurl et al., 2016), we narrowed down the target cell type to Kupffer cells. Iron is the most predominant cellular metal with ferromagnetic features. Thus, using magnetic column separation, we divided NPCs into two fractions: ferromagnetic Fe-hi and remaining Fe-lo (Figure 4A). Depending on the cellular iron contents, Fe-hi cells were enriched on the magnetic column, whereas Fe-lo cells were in the flow through (Orr et al., 2014; da Silva et al., 2017; Hubler et al., 2018). Each fraction was subjected to fluorescence-activated cell sorting (FACS) analysis, and Fe-hi and Fe-lo Kupffer cells (CD11b+ F4/80hi) were purified (Figure 4B). Fe-hi Kupffer cells displayed higher cellular iron content than Fe-lo ones (Figure 4C). The number of Fe-hi and Fe-lo Kupffer cells was higher in MC4R-KO mice fed WD during the course of NASH development than in wild-type mice fed SD (Figure 4D). The ratio of Fe-hi to Fe-lo Kupffer cell number was significantly elevated in MC4R-KO mice fed WD for 16 weeks (NASH) (Figure 4E). In this experimental setting, mRNA expression of Itgax was markedly increased in Fe-hi Kupffer cells from MC4R-KO mice fed WD during the course of NASH development, whereas there was only a modest increase in Fe-lo cells (Figure 4F). Fe-hi Kupffer cells also expressed higher levels of mRNAs encoding proinflammatory and profibrotic genes than Fe-lo Kupffer cells. These findings strongly suggest that Fe-hi Kupffer cells expressing CD11c are responsible for inflammation and fibrosis in the livers of MC4R-KO mice fed WD.
Figure 4.
Iron-rich Kupffer cells exhibit proinflammatory and profibrotic properties during the development of NASH
(A) Experimental protocol for preparation of iron-rich (Fe-hi) and remaining (Fe-lo) tissue-resident macrophages or Kupffer cells (F4/80hi CD11b+). Hepatic nonparenchymal cells (NPCs) were prepared from the livers of MC4R-KO mice fed WD for 4, 8, and 16 weeks (simple steatosis, simple steatosis, and NASH, respectively) and wild-type mice fed SD for 4 weeks (normal liver). The NPCs were divided into Fe-hi and Fe-lo fractions by magnetic bead column separation. Then, Fe-hi and Fe-lo Kupffer cells were sorted by the flow cytometer.
(B) FACS gating to identify Kupffer cells in NPCs. F4/80hi CD11b+ cells in the CD45+ fraction were defined as Kupffer cells.
(C) Iron levels in Fe-hi and Fe-lo Kupffer cells prepared from the livers of MC4R-KO mice fed WD for 13 weeks (NASH). Data represent the mean ± SEM. n = 3–7. ∗p < 0.05 versus Fe-lo (two-tailed unpaired Student's t test).
(D) The number of Fe-hi and Fe-lo Kupffer cells in the livers. Data represent the mean ± SEM. n = 8. ∗p < 0.05 versus WT/SD (p < 0.05, Tukey-Kramer test).
(E) The ratio of Fe-hi to Fe-lo Kupffer cells in the livers. Data represent the mean ± SEM. n = 8. ∗p < 0.05 versus WT/SD (Tukey-Kramer test).
(F) mRNA levels of genes related to inflammation and fibrosis in Fe-lo and Fe-hi Kupffer cells. Data represent the mean ± SEM. n = 3–5. ∗p < 0.05 versus the respective WT/SD (Fe-lo or Fe-hi) (Dunnett's test), #p < 0.05 (two-tailed unpaired Student's t test).
Iron accumulation induces lysosomal dysfunction in macrophages
Next, we compared gene expression profiles between NASH liver-derived CD11c-positive Kupffer cells and iron-treated cultured macrophages to clarify how iron metabolism is involved in the phenotypic change of CD11c-positive Kupffer cells with proinflammatory and profibrotic properties (Figure 5A). We found that 305 genes were commonly upregulated in CD11c-positive Kupffer cells from NASH livers (versus CD11c-negative Kupffer cells from normal livers) and iron-treated RAW264 macrophages (versus untreated RAW264 macrophages). As expected, genes related to “cytokine-cytokine receptor interaction” and “chemokine signaling pathway” were enriched in these 305 genes, indicative of the proinflammatory and profibrotic phenotypes. Moreover, pathway analysis revealed that the two most enriched pathways were “phagosome” and “lysosome” (Figure 5B). These results are consistent with our previous findings that CD11c-positive Kupffer cells aggregate around dead hepatocytes to scavenge accumulated lipids and cell debris within CLSs (Itoh et al., 2013, 2017). It is also known that cellular iron accumulation triggers lysosomal dysfunction through increased production of reactive oxygen species (Boya and Kroemer, 2008). Moreover, ChIP Atlas enrichment analysis identified TFE3 as a potential transcription factor enriched in the enhancer regions of these 305 genes (Table S1).
Figure 5.
MiT/TFE transcription factors are activated in CD11c+ Kupffer cells and iron-treated cultured macrophages
(A–C) Transcriptome analysis of CD11c+ Kupffer cells in NASH and FeCl3-treated RAW264 cells. (A) Venn diagram showing the commonly upregulated (≥1.5-fold) 305 genes. CD11c+ and control Kupffer cells were prepared from MC4R-KO mice fed WD (NASH) and wild-type mice fed SD (normal liver) for 20 weeks, respectively. RAW264 cells were treated with or without FeCl3 (500 μM) for 24 h. (B) Pathway analysis of these 305 genes. The overlap included pathways related to phagocytosis and lysosome. (C) Heatmap of the overlapped 305 genes in RAW264 cells under indicated conditions. RAW264 cells, transfected with siRNA targeting GFP (si-GFP) or MiT/TFE transcription factors (si-Tfe3/Tfeb), were treated with or without FeCl3 (500 μM) for 24 h. Arrows indicate genes annotated with “Phagosome” or “Lysosome.”
(D) mRNA expression of genes annotated as “Phagosome” and “Lysosome.” Data represent the mean ± SEM. n = 3. ∗p < 0.05 and #p < 0.05 versus the respective si-GFP (vehicle or FeCl3) (Dunnett's test).
(E) mRNA levels of Tfe3 and Tfeb. Data represent the mean ± SEM. n = 3. ∗p < 0.05 versus si-GFP (Dunnett's test).
(F) Increased lysosomal stress in iron-treated RAW264 cells. RAW264 cells were treated with (black bar) or without (white bar) FeCl3 (500 μM) for 24 h, stained with acridine orange (AO) and then subjected to flow cytometry analysis. AO emits red fluorescence within acidic lysosomes, indicative of intact lysosomal membranes, whereas green fluorescence reflects the release of AO from lysosomes into the cytosol, indicating stressed lysosomes. Data represent the mean ± SEM. n = 5. ∗p < 0.05 versus vehicle (two-tailed unpaired Student's t test).
(G) Immunostaining of TFE3 in primary hepatic macrophages. Hepatic macrophages prepared from MC4R-KO mice fed WD for 6 weeks (simple steatosis) were treated with or without FeCl3 (200 μM) for 6 h. Scale bars, 25 μm.
(H–J) Hepatic mRNA levels of Atp6v0d2. Data represent the mean ± SEM. (H and I) MC4R-KO mice receiving a single (H) or repeated (I) injection of iron-dextran (experimental protocols are shown in Figures 1A and 1D, respectively). (H) n = 4. ∗p < 0.05 versus the respective SD (Tukey-Kramer test). (I) n = 5. ∗p < 0.05 versus WT/SD, #p < 0.05 (Tukey-Kramer test). (J) The inducible NASH model receiving 2-DP injection (experimental protocol, Figure 2A). n = 9. ∗p < 0.05 (two-tailed unpaired Student's t test).
(K) mRNA levels of Atp6v0d2 in Fe-lo and Fe-hi Kupffer cells prepared from MC4R-KO mice fed WD for 8 weeks (simple steatosis) and WT mice fed SD (normal liver) (experimental protocol, in Figure 4A). Data represent the mean ± SEM. n = 3–5. ∗p < 0.05 versus the respective WT/SD (Fe-lo or Fe-hi), #p < 0.05 (Tukey-Kramer test).
TFE3 is a member of the MiT/TFE transcription factors, which regulate lysosomal biogenesis and functions in response to lysosomal dysfunction (Puertollano et al., 2018). In this study, knockdown of two MiT/TFE transcription factors, Tfe3 (Transcription factor E3) and Tfeb (Transcription factor EB), effectively suppressed iron treatment-induced upregulation of phagosome- and lysosome-related genes such as Atp6v0d2 (ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2), Cd63, and Ctsk (Cathepsin K) (Figures 5C and 5D). We validated these small interfering RNAs (siRNAs) by examining the effect of each of them on Tfe3 and Tfeb mRNA expression (Figure 5E). Moreover, using acridine orange staining, we observed that iron treatment induced lysosomal dysfunction in RAW264 macrophages (Figure 5F). In addition, iron treatment increased nuclear localization of TFE3 in primary hepatic macrophages isolated from steatotic livers, whereas it did not increase mRNA levels of Tfe3 and Tfeb (Figures 5G and S5). Furthermore, Atp6v0d2 expression was significantly elevated in the livers of MC4R-KO mice receiving a single injection (Figure 5H) or repeated injection (Figure 5I) of iron-dextran (related data are shown in Figures 1A–1C and 1E–1H, respectively). 2-DP treatment significantly suppressed Atp6v0d2 expression in the inducible NASH model (Figure 5J; related data were shown in Figure 2). Finally, Atp6v0d2 expression was markedly upregulated in Fe-hi Kupffer cells prepared from MC4R-KO mice fed WD for 8 weeks (Figure 5K; related data are shown in Figure 4). These findings, taken together, indicate activation of MiT/TFE transcription factors in CD11c-positive Kupffer cells, probably through lysosomal dysfunction induced by iron accumulation.
MiT/TFE transcription factors are involved in iron accumulation-mediated macrophage phenotypic changes
Next, we investigated how MiT/TFE transcription factors influence proinflammatory and profibrotic properties in macrophages. Chloroquine and bafilomycin A1, which potently induce lysosomal dysfunction, markedly upregulated expression of Itgax, Tnfa, Ccl3, and Timp1 in RAW264 macrophages (Figure 6A). Similarly, FeCl3 treatment dose-dependently increased the expression of these genes, except for Timp1 (Figure 6B). Similar results were observed in primary hepatic macrophages prepared from steatotic livers (Figure 6C). Moreover, iron-induced upregulation of these genes was significantly inhibited by knockdown of Tfe3 and Tfeb in RAW264 macrophages (Figure 6D). As addressed above, Itgax (CD11c) is a marker for Kupffer cells constituting CLSs in NASH. Therefore, our observations indicate that iron accumulation in Kupffer cells is involved in lysosomal dysfunction and subsequent activation of MiT/TFE transcription factors, which contributes to phenotypic changes and sustained inflammation during the development of NASH.
Figure 6.
MiT/TFE transcription factors are involved in iron accumulation-induced macrophage phenotypic changes
(A) Effects of lysosomal stress on mRNA levels of genes related to inflammation and fibrosis in RAW264 cells. RAW264 cells were treated with chloroquine (50 μM) or bafilomycin A1 (100 nM) for the indicated time. Data represent the mean ± SEM. n = 4. ∗p < 0.05 versus vehicle (two-tailed unpaired Student's t test).
(B and C) Effects of iron loading on mRNA levels of genes related to inflammation and fibrosis in RAW264 cells (B) and primary hepatic macrophages (C). RAW264 cells were treated with FeCl3 for 24 h. Hepatic macrophages isolated from MC4R-KO mice fed WD for 6 weeks (simple steatosis) were treated with or without FeCl3 (200 μM) for 6 or 12 h. Data represent the mean ± SEM. n = 3–6. ∗p < 0.05 versus vehicle (Tukey-Kramer test (B) or two-tailed unpaired Student's t test (C)).
(D) Role of MiT/TFE transcription factors in iron loading-induced upregulation of inflammation and fibrosis-related genes in RAW264 cells. RAW264 cells were transfected with siRNA targeting GFP (si-GFP) or MiT/TFE transcription factors (si-Tfe3/Tfeb). Forty-eight hours after transfection, the cells were treated with or without FeCl3 (500 μM) for additional 24 h. Data represent the mean ± SEM. n = 3. ∗p < 0.05 and #p < 0.05 versus the respective si-GFP (vehicle or FeCl3) (Dunnett's test).
MiT/TFE transcription factors are activated in CD11c+ Kupffer cells constituting CLSs in NASH in mice and humans
To prove this notion in vivo, we performed immunohistochemical analysis of TFE and TFEB. Nuclear immunostaining of TFE3, indicating activation of TFE3, was observed in macrophages forming CLSs in the livers of MC4R-KO mice fed WD for 16 weeks (NASH), whereas there was only faint staining of TFE3, probably in hepatocytes, in wild-type mice fed SD (normal liver) (Figures 7A and 7B). We confirmed that CD11c-positive Kupffer cells in CLSs were the main cell type expressing TFE3 in the NASH model (Figure 7C). Notably, most CLSs were positive for TFE3 immunostaining (Figures 7B and 7C). Similarly, more than half of CLSs were immunostained with TFEB (Figure 7D). A single injection of iron-dextran to lean mice (normal liver) increased immunostaining of TFE3 in Kupffer cells (Figure 7E). Moreover, TFE3 immunostaining was observed in macrophages forming CLSs in a different experimental model of NASH induced by deficiency of methionine and choline (Figures 7F and 7G). As shown in Figure 7H, deficiency of methionine and choline in wild-type mice exhibited histological features characteristic of NASH, along with CLS formation and upregulation of Itgax and Atp6v0d2. Finally, we examined TFE3 expression using liver biopsy specimens from patients with NAFLD/NASH (Table S2). In humans, CLSs can be detected by CD68 immunostaining, as reported previously (Itoh et al., 2013). Immunostaining using serial sections revealed that approximately 40% of CLSs were immunostained with TFE3 (Figure 7I). These observations, taken together, suggest that MiT/TFE transcription factors are activated in CD11c-positive Kupffer cells constituting CLSs in murine and human NASH, thereby contributing to the pathogenesis of NASH.
Figure 7.
MiT/TFE transcription factors are activated in CD11c+ Kupffer cells constituting CLSs in murine and human NASH
(A–D) Immunostaining of TFE3 and TFEB in CLSs in the NASH model using MC4R-KO mice. (A) Experimental protocol. (B–D) Immunostaining of F4/80 and TFE3 (B), CD11c and TFE3 (C), and F4/80 and TFEB (D) in the livers of MC4R-KO mice fed WD for 16 weeks (NASH) and WT mice fed SD (normal liver). Data represent the mean ± SEM. n = 4–5. Scale bars, 25 μm.
(E) Immunostaining of F4/80 and TFE3 in the livers of MC4R-KO mice fed SD (normal liver) receiving a single injection of iron-dextran. Scale bars, 25 μm.
(F–H) Effects of dietary deficiency of methionine and choline on hepatic mRNA levels of inflammation and fibrosis-related genes. (F) Experimental protocol. (G) Immunostaining of F4/80 and TFE3 in the livers of WT mice fed methionine and choline-deficient diet for 4 weeks. Scale bars, 25 μm. (H) Hepatic mRNA expression of genes related to inflammation and fibrosis, together with Atp6v0d2, a target gene of MiT/TFE transcription factors. Data represent the mean ± SEM. n = 3. ∗p < 0.05 versus SD (two-tailed unpaired Student's t test).
(I) Immunostaining of CD68, CD11c, and TFE3 in the livers of patients with NAFLD/NASH. Scale bars, 50 μm.
See also Table S2.
Discussion
Iron accumulation has been proposed as a “hit” in the multiple parallel hits hypothesis (Nelson et al., 2012). However, the underlying mechanism(s) and the responsible cell type(s) are incompletely understood. This study provided novel evidence on the role of iron accumulation in Kupffer cells during the development of liver fibrosis in NASH. Our findings demonstrate that (1) proinflammatory and profibrotic CD11c-positive Kupffer cells in NASH comprise the Fe-hi fraction, (2) iron accumulation in Kupffer cells activates MiT/TFE transcription factors to increase proinflammatory and profibrotic properties, (3) activated MiT/TFE transcription factors are observed in Kupffer cells forming CLSs in mouse models of NASH and human NASH, and (4) iron chelation attenuated liver fibrosis in a murine NASH model (Figure S6). Thus, we defined Kupffer cells as a target cell type responsible for iron accumulation-mediated chronic inflammation in NASH. This is consistent with our observations that iron loading and chelation did not affect serum markers for hepatocyte injury and CLS formation. This study also provides insight into the role of iron in the multiple parallel hits hypothesis.
Among various cell types involved in the pathogenesis of NASH, macrophages are crucial for the progression of inflammation and fibrosis. Although substantial attention has hitherto been paid to the role of infiltrating macrophages, we recently revealed the critical role of a novel subset of CD11c-positive Kupffer cells in inflammation and fibrosis in NASH (Itoh et al., 2017). Although surface markers are still indispensable for evaluating macrophage subpopulation, this study proposes a novel categorization of Kupffer cells based on their cellular iron content, in which the Fe-hi fraction corresponds to the CD11c-positive Kupffer cells during the development of NASH. In this regard, Orr et al. previously reported that alternatively activated macrophages undergo an inflammatory shift, together with altered iron metabolism, in adipose tissue during the development of obesity (Orr et al., 2014). In addition to the nomenclature based on their origin (tissue resident versus bone marrow-derived) and polarization (proinflammatory versus anti-inflammatory) (Satoh et al., 2017), recent evidence suggests a pathogenesis-specific macrophage subset (Itoh et al., 2017). Collectively, our data link a macrophage subset responsible for liver fibrosis with the aberrant cellular metabolism of iron in the pathogenesis of NASH.
It is important to discuss how iron accumulates in a particular group of Kupffer cells during the development of NASH. We previously reported that Kupffer cells give rise to formation of CLSs, where they scavenge debris of dead hepatocytes and residual lipids, and thus gradually become positive for CD11c during CLS maturation, probably through interaction with dead cells (Itoh et al., 2017). Given that CD11c-positive Kupffer cells in CLSs comprise the Fe-hi fraction, iron may accumulate in these Kupffer cells in a phagocytosis-dependent manner. Indeed, Kupffer cells exhibit high phagocytic activity (Varol et al., 2015). It is therefore conceivable that phagocytosis of dead hepatocytes causes iron accumulation in Kupffer cells forming CLSs, thereby inducing phenotypic changes including elevated expression of CD11c. In addition, our preliminary data suggest that mRNA expression of Ferroportin, an iron exporter, is markedly reduced in Kupffer cells from NASH livers (Y.K., unpublished data). In the next step, it will be interesting to examine whether the Fe-hi Kupffer cells in NASH originate from the same subpopulation at steady state. Cell fate tracing experiments would help elucidate the causality of aberrant iron metabolism in Kupffer cell polarization in vivo during the development of NASH.
With regard to the molecular mechanism underlying iron accumulation-induced phenotypic changes of Kupffer cells, we demonstrated the involvement of lysosomal stress and subsequent activation of MiT/TFE transcription factors. MiT/TFE transcription factors regulate gene expression directly or coordinately with SMADs and FOXOs (Hua et al. 1999; Nakagawa et al., 2006). In addition to their previously known role in lysosomal biogenesis, this study showed that MiT/TFE transcription factors are capable of regulating expression of proinflammatory and profibrotic genes in macrophages. On the other hand, other signaling pathways downstream of lysosomal stress may contribute to this process, as the effect of knockdown of MiT/TFE transcription factors was only partial. Lysosomal stress triggers diverse cellular responses such as inflammasome activation, autophagy induction, and leakage of lysosomal cathepsins (Papadopoulos and Meyer, 2017). Notably, intracellular calcium release under lysosomal stress conditions activates signaling pathways of mitogen-activated protein kinases and nuclear factor-κB (Okada et al., 2014; Chen et al., 2018). Accordingly, further studies are needed to clarify the molecular mechanisms underlying iron accumulation-induced phenotypic changes of Kupffer cells during the development of NASH.
Although this study focused on Kupffer cells, recent evidence suggested a role for iron in hepatocytes in the pathogenesis of NASH. For instance, lipid peroxidation causes hepatocyte ferroptosis, an iron-dependent non-apoptotic form of cell death, in the pathogenesis of NASH (Yang and Stockwell, 2016). Several studies reported that inhibition of ferroptosis effectively prevents the progression of steatosis, hepatocyte death, inflammation, and fibrosis in certain types of murine NASH models (Tsurusaki et al., 2019; Li et al., 2020; Qi et al., 2020). Conversely, iron accumulation may affect lipid metabolism in the liver; i.e., dietary iron overload or iron-dextran injection suppresses steatosis and liver injury in diet-induced obese mice (Padda et al., 2015; Folgueras et al., 2018). Another study also showed that dietary iron deficiency upregulates hepatic lipogenic gene expression in rats (Davis et al., 2012). In line with these reports, we observed in this study that chronic iron-dextran challenge suppressed triglyceride accumulation and upregulation of Fasn mRNA in the liver; nevertheless, the treatment resulted in exacerbation of liver fibrosis. This is probably because MC4R-KO mice exhibit morbid obesity with severe hepatic steatosis at a level sufficient to provoke chronic inflammation in the liver even if the symptoms are attenuated to a certain extent. Moreover, MiT/TFE transcription factors exert anti-steatotic effects in hepatocytes from obese mice (Nakagawa et al., 2006; Settembre et al., 2013). Based on these observations, it will be intriguing to investigate whether MiT/TFE transcription factors and/or ferroptosis in hepatocytes trigger CLS formation and the subsequent phenotypic changes of Kupffer cells during the development of NASH.
In summary, we demonstrated that iron-rich Kupffer cells exhibit proinflammatory and profibrotic properties in a murine model of NASH. Our data suggest that iron accumulation induces lysosomal stress, resulting in activation of MiT/TFE transcription factors in CD11c-positive Kupffer cells forming CLSs, thereby contributing to inflammation and fibrosis in NASH. Although NASH results from various etiologies, activation of MiT/TFE transcription factors in CLSs was observed in murine NASH models with or without obesity, as well as human NASH. Therefore, this study provides insight into the molecular mechanism underlying the pathogenesis of NASH.
Limitations of the study
In terms of the study limitation, our data are mainly based on the NASH model using MC4R-KO mice. Although we verified the activation of MiT/TFE transcription factors in Kupffer cells forming CLS in another NASH model using a methionine and choline-deficient diet and patients with NAFLD/NASH, other causes than iron accumulation may activate MiT/TFE transcription factors. Given that 10%–50% of patients with NAFLD exhibit hepatic iron accumulation and/or hyperferritinemia (Valenti et al., 2006; Manousou et al., 2011; Nelson et al., 2012; Ryan et al., 2018), further work would be required to fully understand the molecular mechanisms underlying activation of MiT/TFE transcription factors in Kupffer cells during the development of NASH.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Takayoshi Suganami (suganami@riem.nagoya-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The accession number for the microarray data reported in this article is NCBI's Gene Expression Omnibus: GSE157472.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Joel K. Elmquist (University of Texas Southwestern Medical Center) for his generous gift of MC4R-KO mice. We also thank Dr. Takahisa Nakamura (Cincinnati Children's Hospital) and the members of the Suganami laboratory for their helpful discussions, Dr. Tasuku Hirayama (Gifu Pharmaceutical University) and Dr. Yasumitsu Ogra (Chiba University) for technical advice, and Center for Animal Research and Education (CARE), Nagoya University for support on animal experiments. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20H03447, 20H05503, and 20H04944 to T.S.; 18K08508 to M.T.; and 19K16539 and 18J00710 to Y.K.) and Japan Agency for Medical Research and Development (CREST) (JP20gm1210009s0102 and 20fk0210082s0101 to T.S.). This study was also supported by research grants from The Hori Sciences and Arts Foundation, Smoking Research Foundation, Daiko Foundation (T.S.), Mochida Memorial Foundation for Medical and Pharmaceutical Research (Y.K.), and Takeda Science Foundation (T.S. and Y.K.).
Author contribution
Y.K., M.T., and T.S. designed the experiments. Y.K., M.T., M.I., and K.O. performed experiments. Y.K., M.T., M.I., and T.S. interpreted the data. I.H. and I.S. provided human samples. A.I. and Y.O. contributed to critical discussion. Y.K., M.T., and T.S. drafted and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2020.102032.
Contributor Information
Miyako Tanaka, Email: tanaka@riem.nagoya-u.ac.jp.
Takayoshi Suganami, Email: suganami@riem.nagoya-u.ac.jp.
Supplemental information
References
- Boya P., Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- Chen D., Xie J., Fiskesund R., Dong W., Liang X., Lv J., Jin X., Liu J., Mo S., Zhang T. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018;9:873. doi: 10.1038/s41467-018-03225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva M.C., Breckwoldt M.O., Vinchi F., Correia M.P., Stojanovic A., Thielmann C.M., Meister M., Muley T., Warth A., Platten M. Iron induces anti-tumor activity in tumor-associated macrophages. Front. Immunol. 2017;8:1479. doi: 10.3389/fimmu.2017.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.R., Rendina E., Peterson S.K., Lucas E.A., Smith B.J., Clarke S.L. Enhanced expression of lipogenic genes may contribute to hyperglycemia and alterations in plasma lipids in response to dietary iron deficiency. Genes. Nutr. 2012;7:415–425. doi: 10.1007/s12263-011-0278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueras A.R., Freitas-Rodríguez S., Ramsay A.J., Garabaya C., Rodríguez F., Velasco G., López-Otín C. Matriptase-2 deficiency protects from obesity by modulating iron homeostasis. Nat. Commun. 2018;9:1350. doi: 10.1038/s41467-018-03853-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaris D., Pantopoulos K. Oxidative stress and iron homeostasis: mechanistic and health aspects. Crit. Rev. Clin. Lab. Sci. 2008;45:1–23. doi: 10.1080/10408360701713104. [DOI] [PubMed] [Google Scholar]
- Gillen K.M., Mubarak M., Nguyen T.D., Pitt D. Significance and in vivo detection of iron-laden microglia in white matter multiple sclerosis lesions. Front. Immunol. 2018;9:255. doi: 10.3389/fimmu.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Akahori H., Harari E., Smith S.L., Polavarapu R., Karmali V., Otsuka F., Gannon R.L., Braumann R.E., Dickinson M.H. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J. Clin. Invest. 2018;128:1106–1124. doi: 10.1172/JCI93025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenour C.M., Kennedy A.J., Bednarski T., Trenary I.A., Eudy B.J., da Silva R.P., Boyd K.L., Young J.D. Vitamin E does not prevent Western diet-induced NASH progression and increases metabolic flux dysregulation in mice. J. Lipid Res. 2020;61:707–721. doi: 10.1194/jlr.RA119000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Miller Z.A., Wu G., Shi Y., Lodish H.F. Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor μE3, and Smad proteins. Proc. Natl. Acad. Sci. U S A. 1999;96:13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubler M.J., Erikson K.M., Kennedy A.J., Hasty A.H. MFehi adipose tissue macrophages compensate for tissue iron perturbations in mice. Am. J. Physiol. Cell Physiol. 2018;315:C319–C329. doi: 10.1152/ajpcell.00103.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Kato H., Suganami T., Konuma K., Marumoto Y., Terai S., Sakugawa H., Kanai S., Hamaguchi M., Fukaishi T. Hepatic crown-like structure: a unique histological feature in non-alcoholic steatohepatitis in mice and humans. PLoS One. 2013;8:e82163. doi: 10.1371/journal.pone.0082163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Suganami T., Kato H., Kanai S., Shirakawa I., Sakai T., Goto T., Asakawa M., Hidaka I., Sakugawa H. CD11c+ resident macrophages drive hepatocyte death-triggered liver fibrosis in a murine model of nonalcoholic steatohepatitis. JCI Insight. 2017;2:e92902. doi: 10.1172/jci.insight.92902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Suganami T., Nakagawa N., Tanaka M., Yamamoto Y., Kamei Y., Terai S., Sakaida I., Ogawa Y. Melanocortin 4 receptor-deficient mice as a novel mouse model of nonalcoholic steatohepatitis. Am. J. Pathol. 2011;179:2454–2463. doi: 10.1016/j.ajpath.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang T.X., Huang X., Li Y., Sun T., Zang S., Guan K.L., Xiong Y., Liu J., Yuan H.X. Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Int. 2020;40:1378–1394. doi: 10.1111/liv.14428. [DOI] [PubMed] [Google Scholar]
- Maliken B.D., Nelson J.E., Klintworth H.M., Beauchamp M., Yeh M.M., Kowdley K.V. Hepatic reticuloendothelial system cell iron deposition is associated with increased apoptosis in nonalcoholic fatty liver disease. Hepatology. 2013;57:1806–1813. doi: 10.1002/hep.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousou P., Kalambokis G., Grillo F., Watkins J., Xirouchakis E., Pleguezuelo M., Leandro G., Arvaniti V., Germani G., Patch D. Serum ferritin is a discriminant marker for both fibrosis and inflammation in histologically proven non-alcoholic fatty liver disease patients. Liver Int. 2011;31:730–739. doi: 10.1111/j.1478-3231.2011.02488.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Shimano H., Yoshikawa T., Ide T., Tamura M., Furusawa M., Yamamoto T., Inoue N., Matsuzaka T., Takahashi A. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat. Med. 2006;12:107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- Nelson J.E., Klintworth H., Kowdley K.V. Iron metabolism in nonalcoholic fatty liver disease. Curr. Gastroenterol. Rep. 2012;14:8–16. doi: 10.1007/s11894-011-0234-4. [DOI] [PubMed] [Google Scholar]
- O’Brien J., Powell L.W. Non-alcoholic fatty liver disease: is iron relevant? Hepatol. Int. 2012;6:332–341. doi: 10.1007/s12072-011-9304-9. [DOI] [PubMed] [Google Scholar]
- Okada M., Matsuzawa A., Yoshimura A., Ichijo H. The lysosome rupture-activated TAK1-JNK pathway regulates NLRP3 inflammasome activation. J. Biol. Chem. 2014;289:32926–32936. doi: 10.1074/jbc.M114.579961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr J.S., Kennedy A., Anderson-Baucum E.K., Webb C.D., Fordahl S.C., Erikson K.M., Zhang Y., Etzerodt A., Moestrup S.K., Hasty A.H. Obesity alters adipose tissue macrophage iron content and tissue iron distribution. Diabetes. 2014;63:421–432. doi: 10.2337/db13-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padda R.S., Gkouvatsos K., Guido M., Mui J., Vali H., Pantopoulos K. A high-fat diet modulates iron metabolism but does not promote liver fibrosis in hemochromatotic hjv-/- mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G251–G261. doi: 10.1152/ajpgi.00137.2014. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C., Meyer H. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr. Biol. 2017;27:R1330–R1341. doi: 10.1016/j.cub.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Ferguson S.M., Brugarolas J., Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37:e98804. doi: 10.15252/embj.201798804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Kim J.W., Zhou Z., Lim C.W., Kim B. Ferroptosis affects the progression of nonalcoholic steatohepatitis via the modulation of lipid peroxidation–mediated cell death in mice. Am. J. Pathol. 2020;190:68–81. doi: 10.1016/j.ajpath.2019.09.011. [DOI] [PubMed] [Google Scholar]
- Ryan J.D., Armitage A.E., Cobbold J.F., Banerjee R., Borsani O., Dongiovanni P., Neubauer S., Morovat R., Wang L.M., Pasricha S.R. Hepatic iron is the major determinant of serum ferritin in NAFLD patients. Liver Int. 2018;38:164–173. doi: 10.1111/liv.13513. [DOI] [PubMed] [Google Scholar]
- Satoh T., Nakagawa K., Sugihara F., Kuwahara R., Ashihara M., Yamane F., Minowa Y., Fukushima K., Ebina I., Yoshioka Y. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature. 2017;541:96–101. doi: 10.1038/nature20611. [DOI] [PubMed] [Google Scholar]
- Schaible U.E., Kaufmann S.H.E. Iron and microbial infection. Nat. Rev. Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- Settembre C., de Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Jürgen Klisch T. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindrilaru A., Peters T., Wieschalka S., Baican C., Baican A., Peter H., Hainzl A., Schatz S., Qi Y., Schlecht A. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J. Clin. Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M.P., Hamza I. Macrophages and iron metabolism. Immunity. 2016;44:492–504. doi: 10.1016/j.immuni.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnweber T., Ress C., Nairz M., Theurl I., Schroll A., Murphy A.T., Wroblewski V., Witcher D.R., Moser P., Ebenbichler C.F. High-fat diet causes iron deficiency via hepcidin-independent reduction of duodenal iron absorption. J. Nutr. Biochem. 2012;23:1600–1608. doi: 10.1016/j.jnutbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Sumida Y., Kanemasa K., Fukumoto K., Yoshida N., Sakai K., Nakashima T., Okanoue T. Effect of iron reduction by phlebotomy in Japanese patients with nonalcoholic steatohepatitis: a pilot study. Hepatol. Res. 2006;36:315–321. doi: 10.1016/j.hepres.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Theurl I., Hilgendorf I., Nairz M., Tymoszuk P., Haschka D., Asshoff M., He S., Gerhardt L.M.S., Holderried T.A.W., Seifert M. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 2016;22:945–951. doi: 10.1038/nm.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H., Moschen A.R. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- Tsurusaki S., Tsuchiya Y., Koumura T., Nakasone M., Sakamoto T., Matsuoka M., Imai H., Yuet-Yin Kok C., Okochi H., Nakano H. Hepatic ferroptosis plays an important role as the trigger for initiating inflammation in nonalcoholic steatohepatitis. Cell. Death Dis. 2019;10:449. doi: 10.1038/s41419-019-1678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L., Moscatiello S., Vanni E., Fracanzani A.L., Bugianesi E., Fargion S., Marchesini G. Venesection for non-alcoholic fatty liver disease unresponsive to lifestyle counselling-a propensity score-adjusted observational study. QJM. 2011;104:141–149. doi: 10.1093/qjmed/hcq170. [DOI] [PubMed] [Google Scholar]
- Valenti L., Dongiovanni P., Piperno A., Fracanzani A.L., Maggioni M., Rametta R., Loria P., Casiraghi M.A., Suigo E., Ceriani R. α1-antitrypsin mutations in NAFLD: high prevalence and association with altered iron metabolism but not with liver damage. Hepatology. 2006;44:857–864. doi: 10.1002/hep.21329. [DOI] [PubMed] [Google Scholar]
- Varol C., Mildner A., Jung S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- Wang L., Harrington L., Trebicka E., Hai N.S., Kagan J.C., Hong C.C., Lin H.Y., Babitt J.L., Cherayil B.J. Selective modulation of TLR4-activated inflammatory responses by altered iron homeostasis in mice. J. Clin. Invest. 2009;119:3322–3328. doi: 10.1172/JCI39939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zheng Z., Caviglia J.M., Corey K.E., Herfel T.M., Cai B., Masia R., Chung R.T., Lefkowitch J.H., Schwabe R.F. Hepatocyte TAZ/WWTR1 promotes inflammation and fibrosis in nonalcoholic steatohepatitis. Cell Metab. 2016;24:848–862. doi: 10.1016/j.cmet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the microarray data reported in this article is NCBI's Gene Expression Omnibus: GSE157472.