Abstract
Neuropsychiatric disorders, such as autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), schizophrenia (SCZ), bipolar disorder (BIP), and major depressive disorder (MDD) share common clinical presentations, suggesting etiologic overlap. A substantial proportion of SNP-based heritability for neuropsychiatric disorders is attributable to genetic components, and genome-wide association studies (GWASs) focusing on individual diseases have identified multiple genetic loci shared between these diseases. Here, we aimed at identifying novel genetic loci associated with individual neuropsychiatric diseases and genetic loci shared by neuropsychiatric diseases. We performed multi-trait joint analyses and meta-analysis across five neuropsychiatric disorders based on their summary statistics from the Psychiatric Genomics Consortium (PGC), and further carried out a replication study of ADHD among 2726 cases and 16299 controls in an independent pediatric cohort. In the multi-trait joint analyses, we found five novel genome-wide significant loci for ADHD, one novel locus for BIP, and ten novel loci for MDD. We further achieved modest replication in our independent pediatric dataset. We conducted fine-mapping and functional annotation through an integrative multi-omics approach and identified causal variants and potential target genes at each novel locus. Gene expression profile and gene-set enrichment analysis further suggested early developmental stage expression pattern and postsynaptic membrane compartment enrichment of candidate genes at the genome-wide significant loci of these neuropsychiatric disorders. Therefore, through a multi-omics approach, we identified novel genetic loci associated with the five neuropsychiatric disorders which may help to better understand the underlying molecular mechanism of neuropsychiatric diseases.
Subject terms: ADHD, Personalized medicine
Introduction
Neuropsychiatric disorders, such as autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), schizophrenia (SCZ), bipolar disorder (BIP), and major depressive disorder (MDD) are a spectrum of common, complex, polygenetic diseases1–4. Neuropsychiatric disorders have become a major public health concern. It is estimated that every year 38.2% of the European population (164.8 million persons) are suffering from a neuropsychiatric disorder5. Some neuropsychiatric disorders, such as the common childhood behavioral disorders ASD and ADHD, are characterized by the early age of onset6,7. For example, one in 69 children aged 8 suffer from ASD6. The average age at diagnosis for ADHD is 7 years old and symptoms can appear as early as 3 years old8.
It is often hard to define clear clinical boundaries between these diseases. MDD, SCZ, and BIP share frequent clinical symptoms, such as anxiety, psychosis, mood swings, and depression9. Accumulating evidence suggests a common genetic overlap between neuropsychiatric disorders. Familial co-segregation of SCZ and BIP was observed in a Swedish population study10, and the increased cross-disorder risk was found to be associated with a family history of SCZ, mood disorders, ASD, and ADHD11. These family based studies demonstrated the co-inheritance of neuropsychiatric disorders. Early onset MDD was found to have genetic overlap with SCZ and BIP based on polygenic risk score analysis12. Thus, it is important to understand the common and unique genetic and pathogenic features of these diseases, which may be of help to design efficient and effective treatments.
Each of these five neuropsychiatric disorders is highly heritable, with heritability estimates in twin studies ranging from 40 to 88%13–20, and a number of genome-wide significant (GWS) loci have been identified from large scale genome-wide association studies (GWASs), especially through the collective effort of the Psychiatric Genomics Consortium (PGC). Among the five neuropsychiatric disorders, SCZ has been best studied with ~200 GWS loci identified as summarized in the GWAS catalog21,22, whereas the number of GWS loci for MDD and BIP are somewhat less with 44 and 30 discovered loci, respectively, based on recent studies23,24. ADHD and ASD are disorders with the least number of GWS loci reported, though they are of similar prevalence and heritability as SCZ. GWS loci have been discovered in recent studies for ADHD and ASD by PGC25,26 with increased case numbers of over 15,000 for each disease. As common variants usually only confer a small effect on the disease risk, a large sample size is necessary for GWS locus identification. All the largest GWAS of each of these neuropsychiatric disorders to date were carried out by PGC ranging from 18,381 cases and 27,969 controls for ASD to 135,458 cases and 344,901 controls for MDD (including iPSYCH samples in the MDD cohort)21,23–26. Additional loci were found when trans-ancestry GWAS was conducted. A meta-analysis of SCZ in the largest combined cohort of 56,418 cases and 78,818 controls including both European ancestry and East Asian subjects generated 53 novel loci21. The single-nucleotide polymorphism (SNP)-based heritability (hG2) was estimated to be within the range of 8.7% (MDD) to 24% (SCZ) in these large-scale GWASs, still much lower than the heritability estimated from twin studies. Many genetic loci have been shown to be pleiotropic, associated with multiple related traits or disorders. For example, the well-known locus of gene GRIN2A was associated with SCZ21 and BIP23; the newly identified locus of SEMA6D was associated with both ADHD25 and MDD27. To identify additional loci associated with neuropsychiatry disorders with increased study power and to systematically examine pleiotropy of loci associated with neuropsychiatric disorders, cross-cohort meta-analyses have been conducted for the PGC cohorts. An early PGC study that meta-analyzed a cohort of 33,332 cases with the diagnosis of ADHD, ASD, BIP, MDD, or SCZ and 27,888 controls yielded the identification of 4 GWS loci with pleiotropic effects28. The recent meta-analysis of 8 neuropsychiatric disorders by PGC further reported 109 pleiotropic loci, including 23 associated with four or more disorders and 11 showing differential directions of effects between neuropsychiatric disorders with an increased sample size of 232,964 cases and 494,162 controls29. These studies have made significant advances in our understanding of the genetic architecture and molecular mechanism of neuropsychiatric disorders. To take advantage of recent improvements in technical and analytical methods, we conducted a meta-analysis of five correlated neuropsychiatric disorders of ADHD, ASD, BIP, MDD, or SCZ and a joint-analysis using the multi-trait analysis of GWAS (MTAG) method which has been shown to increase statistical power to identify genetic loci associated with each individual disease by incorporating information from other correlated traits. We further performed a replication study in an independent pediatric cohort for the novel loci of ADHD which usually presents as a childhood-onset neurodevelopmental disorder. Our study replicated multiple of the discoveries reported in the recent cross-cohort meta-analysis, using a complementary approach and further identified novel loci.
Methods
GWAS data of neuropsychiatric disorders
GWAS summary statistics were downloaded from the website of the PGC (https://www.med.unc.edu/pgc). All five largest cohorts of European ancestry for the five neuropsychiatric disorders of ADHD25, ASD26, BIP23, MDD30, and SCZ21 were selected, which are listed in Supplementary Table 1 and described in detail below.
ADHD|PMID: 30478444
This is a meta-analysis including 12 cohorts of European, North American, and Chinese ancestry collected by the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) and PGC, which contained 20,183 ADHD cases and 35,191 controls. The cases of iPSYCH were diagnosed by psychiatrists in accordance with ICD10 (F90.0) and genotyped using Illumina PsychChip while the cases of the remaining cohorts were diagnosed in hospitals or clinics by professional clinicians. Here we used the summary statistics of a meta-analysis of the European ancestry subjects composed of 19,099 cases and 34,194 controls.
ASD|PMID: 30804558
Data from a Danish population-based cohort and five family based cohorts of trio samples of European ancestry were utilized in the meta-analysis of this publication. The cases with ASD were diagnosed according to the expert clinical consensus or ICD-10. Summary statistics from meta-analysis of all the samples, including 18,381 cases and 27,969 controls, were included in our analysis.
BIP|PMID: 31043756
The meta-analysis is composed of 32 cohorts of European descent, including 20,352 cases and 31,358 controls. Diagnoses were confirmed with the international consensus criteria of DSM-IV, ICD-9, or ICD-10. The meta-analysis summary statistics from all subjects were utilized in our study.
MDD|PMID: 30718901
This meta-analysis combined three previous studies of depression from UK Biobank, 23andme, and PGC, including a total of 246,363 cases and 561,190 controls. As reported in the original meta-analysis, the determination of case or control status of the subjects in the UK Biobank dataset was mostly self-reported, and the case status of samples in the PGC dataset was determined by structured clinical interviews or medical records. Due to the unavailability of full summary statistics of 23andme data, we only used the summary level data from PGC and UK Biobank with 170,756 cases and 329,443 controls.
SCZ|PMID: 31740837
The study is a meta-analysis of East Asian subjects (22,778 cases and 35,362 controls) and European descent subjects (33,640 cases and 43,456 controls). The summary statistics of European descent subjects were derived from Ripke et al.’s work31 and updated to 1000 genomes phase 3. We only used the summary statistics of European descent subjects in our study.
Multi-trait joint analysis
Joint analysis of the GWAS summary statistics was carried out using software MTAG32, which takes disease heterogeneity into account. The alternative allele frequency information of each SNP was taken from 1000 G data of European ancestry33.
Meta-analysis
A meta-analysis based on the summary statistics of the above five neuropsychiatric diseases was conducted with the P value-based method implemented in the software METAL with sample overlap correction34.
Defining independent genomic loci
PLINK was used to clump the GWS SNPs into linkage disequilibrium (LD)-independent genomic regions (–clump-r2 = 0.4, –clump-kb = 500, –clump-p1 = 5e-08, –clump-p2 = 5e-02)35. LD information from the 1000 G data of European ancestry was applied in clumping. Genomic loci with the distance between index SNPs less than 500 kb were merged. The MHC region (chr6:25–35 MB) was regarded as one locus owing to the extensive LD.
Determination of novel loci
The novel loci were determined in the following four steps by excluding known loci. (a) The results obtained for each neuropsychiatric disorder through MTAG analysis were clumped with the reported associations of each corresponding disease in GWAS Catalog36 using PLINK, and the results from the METAL analysis were clumped with the reported associations of all five diseases in GWAS Catalog. The parameters and LD information used here were the same as those used in the step of defining independent genomic loci. All GWS SNPs located in the same clump with the significant SNPs reported in the GWAS catalog were considered as known SNPs, and the loci containing these SNPs were considered as known loci. (b) All the significant loci reported in the original GWAS article of each neuropsychiatric disorder and the recent meta-analysis of eight neuropsychiatric disorders by PGC were considered as known loci. (c) GWS SNPs from our analyses located within a 1 MB range of a reported significant locus from the above two resources were considered to be within known loci. (d) If a locus containing GWS SNPs in our analyses was merged by stepwise conditional analysis with a known locus using GCTA-COJO37, the locus was regarded as known. The GWS loci left after the above exclusion steps were defined as novel loci.
Fine mapping of causal variants
A stepwise conditional analysis was further performed using software package GCTA-COJO to identify the independent associations at each locus, based on LD information from the 1000 G EUR reference panel. Fine mapping of causal variants at each novel locus was conducted using the software FINEMAP38 with the setting of five as the maximum number of allowed causal SNPs. The required input of LD file was also generated from PLINK with a 1000 G EUR reference panel. For each novel locus, SNPs in a 95% credible set with posterior probabilities of 1, in LD with the index SNP (r2 > 0.6) and with MTAG association P value < 1 × 10−4 were selected as the potential causal variants.
Estimation of SNP-based heritability and genetic correlation
The SNP-based heritability on the liability scale and genetic correlation of these neuropsychiatric disorders were estimated using the software LD score (LDSC)39 based on the MTAG analysis results. Precomputed LDSC from 1000 G European ancestry samples were used in both analyses with population prevalence for each trait being consistent with that reported in the original publication of each cohort listed in Supplementary Table 1. Besides, partitioning of the SNP-based heritability by functional category using the baseline model was conducted for 53 functional overlapping annotations40 with the MTAG summary statistics. The baseline model LDSC, regression weights, and allele frequencies computed from 1000 G phrase 3 data were downloaded from the website http://data.broadinstitute.org/alkesgroup/LDSCORE/.
Functional annotation
Functional annotation of MTAG results was conducted through web portal FUMA41. The mapping of SNPs to corresponding candidate genes took into account the distance between SNPs and genes, expression quantitative trait loci (eQTLs), and the long-range chromatin interactions (Hi-C) between SNPs and genes. The eQTL data sets used in our analysis were PsychENCODE42, BRAINEAC43, and GTEx v844 Brain; and Hi-C data specifically from brain tissues and cell types were used in our analysis, including data from PsychENCODE, adult and fetal brain, as well as neural progenitor cell42,45.
Histone modification site analysis
Histone modification analysis was conducted via web portal Haploreg and the overlap between histone modification regions and potential causal variants was evaluated based on epigenome data in the brain from epigenomics databases of ROADMAP project46,47.
Gene set enrichment and gene expression profile analyses
The gene set enrichment analysis and gene expression profile analysis among all the significant loci in MTAG results for each disease were performed via MAGMA48 implemented in FUMA. The resources of gene sets were from predefined MSigDB49 Gene Ontology gene sets with 5497 curated gene sets and 9986 GO terms. The significant enrichment of gene sets was based on hypergeometric tests and the significance threshold was set as P value < 0.05 after multiple-testing adjustment. The gene expression profile analysis was also carried out through FUMA with the Brainspan dataset which contains reads per kilobase of transcript per million mapped reads (RPKM) values of 21,001 genes for brain samples of 29 different ages and 19,601 genes for 11 general developmental stages50.
GWAS replication study
Study subjects
The replication study was conducted at the Center for Applied Genomics (CAG), the Children’s Hospital of Philadelphia (CHOP). The ICD codes used to retrieve the ADHD cases are listed in Supplementary Table 2. For controls, we included subjects without ADHD diagnosis or any other relevant neuropsychiatric conditions. The age range for subjects in our study is from 3 to 21 years old with an average age of 6.
Ethics statement
The study was approved by the Institutional Review Board at CHOP and all participants provided written informed consent. The replication cohort is independent of the PGC cohort.
Genotyping
Genomic DNA of the samples in the replication study was extracted from whole blood or saliva samples following the phenol–chloroform protocol and genotyped on Illumina HumanHap550-V1/V3, HumanHap610-Quad, or Infinium Global Screening Array-24 (GSA) DNA Bead Chips at CAG.
Quality control (QC) filtering
QC steps were performed with PLINK (software release v1.9)35. Sample QC includes the following criteria. Samples with call rate <98%, or with ambiguous sex detected by PLINK as well as heterozygosity outliers were excluded. Duplicated and cryptically related samples were identified as PI_HAT ≥ 0.1875 in PLINK identity-by-state analysis, and one from each pair was excluded. SNP based QC was conducted according to the following measurements. SNPs with genotype rate <95%, minor allele frequency <0.01, or Hardy–Weinberg equilibrium P values < 10−5 were removed from further analysis.
Principal component analysis (PCA)
PCA was conducted with PLINK and samples that were not of European ancestry were excluded based on PCA plots. Afterward, PCA was performed again to derive the principal components which were used as covariates to control for population stratification.
A total of 5194 ADHD cases and 33,127 controls were genotyped, among which, 2726 ADHD cases and 16,299 healthy controls of European ancestry, with 495,535 variants shared on the V1/V3Quad arrays and 497203 variants on the GSA arrays passing QC.
Genome-wide imputation
For all samples genotyped on different platforms, unphased GWAS genotype data were converted to VCF files and uploaded to Michigan Imputation Server (https://imputationserver.sph.umich.edu/) for phasing using Eagle251 and imputation using Minimac352 with the Haplotype Reference Consortium reference panel. Afterward, variants with imputation score R2 < 0.4 were filtered out. Further, after SNP QC described above, 7,617,389 variants shared across genotyping platforms were included in the association testing.
Association analysis
After imputation and post-imputation quality control filtering, logistic regression was conducted to evaluate the association between SNP genotype and neuropsychiatric disease status, including sex and the first ten principal components as covariates.
Sign test
Summary statistics of overlapping SNPs in both the ADHD MTAG analysis and the pediatric replication cohort were extracted. PLINK clumping and GCTA-COJO step-wise conditional analyses were similarly performed to identify independent associations as described above, with the European ancestry population of 1000 Genomes as reference. The sign test was performed to compare the direction of effect at each GWS association between the ADHD MTAG analysis results and our pediatric replication cohort. For all the GWS associations (P value < 5 × 10−8) in the ADHD MTAG analysis, the proportion of concordant direction of effect (π) in the replication cohort was tested against a null hypothesis of π = 0.50 based on the one-sample test of the proportion with Yates’ continuity correction.
Test for replication of effect size
Adjustment for winner’s curse was first performed on the ADHD MTAG results via the method of FDR Inverse Quantile Transformation (FIQT)53. Then for all the GWS associations in the ADHD MTAG analysis, the effect size in the replication cohort was regressed on the adjusted effect size from the ADHD MTAG analysis. The slope of the regression was tested against the null hypothesis of the expected slope being zero or one respectively.
Test for genetic correlation of the replication cohort
The genetic correlation between the ADHD MTAG analysis and our replication cohort was similarly computed using LDSC regression as described above for the genetic correlation analyses between neuropsychiatric disorders based on genome-wide data.
Results
Joint analysis of GWAS summary statistics
To identify novel genetic factors associated with neuropsychiatric diseases, we conducted a multi-trait joint analysis based on summary statistics of the five neuropsychiatric GWASs of ADHD, ASD, BIP, MDD, and SCZ (Supplementary Table 1), taking phenotype correlation into account. The standardized genomic inflation factor for MTAG analysis of each neuropsychiatric disease was close to one, suggesting no evidence of inflation due to confounding (Supplementary Fig. 1). A number of loci reached the genome-wide significance (GWS) threshold for each disease, including 20 for ADHD, 9 for ASD, 54 for BIP, 74 for MDD, and 90 for SCZ, respectively (Supplementary Table 3). We identified 16 novel GWS loci, including 5 loci for ADHD, 1 for BIP, and 10 for MDD, which have not previously been reported to be associated with the corresponding neuropsychiatric disorder at the GWS level (Table 1, Supplementary Fig. 2, Supplementary Table 4). We also found SNPs at other loci in each of the MTAG results overlapped with the GWS loci reported in the recent meta-analysis of eight neuropsychiatric disorders29 (9 for ADHD, 7 for ASD, 49 for BIP, 25 for MDD, and 77 for SCZ) (Supplementary Table 5). While this reduced the total number of novel GWS loci found in our MTAG analyses, it demonstrated the reliability of our analyses, being consistent and complementary to the meta-analysis with the approach of association analysis based on SubSETs (ASSET). All of the novel loci identified showed evidence of association (P < 0.05) with two or more neuropsychiatric disorders and the direction of effects were consistent across the diseases (Supplementary Fig. 3, Supplementary Table 4). Of note, eleven of the 16 novel GWS loci exhibited consistent genome-wide marginal significance (P < 5 × 10−5) between ADHD and MDD. The locus at 18q12.3 reached GWS for the first time for both ADHD and MDD. Such concordance between ADHD and MDD is in line with recent evidence showing a strong genetic correlation between ADHD and MDD25, as well as the high level of comorbidity between ADHD and MDD. We noticed that genetic variants, including SNPs and CNVs, at several of the novel loci have been found to be associated with other neuropsychiatric or neurodegeneration diseases, including variants in gene IP6K1 which are associated with general cognitive ability54, educational attainment55, and intelligence56; variants at the TRAF3 locus which are associated with brain imaging in SCZ at genome-wide marginal significance level57; and mutations in SPTBN2 that cause spinocerebellar ataxia with neurodegenerative feature58.
Table 1.
Novel genome-wide significant loci from multi-trait joint-analyses of neuropsychiatric disorders.
| Disease | Locus | Chr | POS(hg19) | Lead SNP | Cytoband | Gene | A1 | A2 | Freq | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADHD | 1 | 3 | 49809841 | rs9870755 | 3p21.31 | IP6K1 | T | C | 0.16 | 0.090 | 0.016 | 6.14E−09 |
| 2 | 5 | 92995013 | rs71639293 | 5q15 | FAM172A | A | G | 0.81 | 0.090 | 0.016 | 2.53E−08 | |
| 3 | 11 | 49356806 | rs10839264 | 11p11.12 | FOLH1 | T | C | 0.08 | 0.118 | 0.021 | 2.27E−08 | |
| 4 | 18 | 39300226 | rs8083506 | 18q12.3 | PIK3C3,KC6 | T | C | 0.21 | −0.089 | 0.015 | 6.19E−09 | |
| 5 | 20 | 44730245 | rs6032660 | 20q13.12 | NCOA5 | A | G | 0.74 | −0.083 | 0.014 | 2.77E−09 | |
| BIP | 6 | 1 | 95578207 | rs12563424 | 1p21.3 | TMEM56 | T | C | 0.64 | −0.064 | 0.011 | 1.29E−08 |
| MDD | 7 | 1 | 191027948 | rs10920885 | 1q31.2 | RP11-309H21.3, LOC440704 | T | G | 0.81 | −0.032 | 0.005 | 2.26E−09 |
| 8 | 3 | 29756378 | rs9834021 | 3p24.1 | RBMS3 | T | C | 0.49 | −0.023 | 0.004 | 3.18E−08 | |
| 9 | 7 | 114059156 | rs2894699 | 7q31.1 | AC073626.2, FOXP2 | T | C | 0.43 | −0.025 | 0.004 | 4.15E−09 | |
| 10 | 11 | 28642653 | rs4636654 | 11p14.1 | RP11-960D24.1, METTL15 | A | G | 0.40 | −0.027 | 0.004 | 2.83E−10 | |
| 11 | 11 | 66456387 | rs2276138 | 11q13.2 | SPTBN2 | T | C | 0.48 | −0.023 | 0.004 | 2.18E−08 | |
| 12 | 13 | 97208898 | rs1414622 | 13q32.1 | HS6ST3 | T | C | 0.92 | 0.043 | 0.008 | 3.03E−08 | |
| 13 | 14 | 103301072 | rs7146581 | 14q32.32 | TRAF3 | T | C | 0.22 | 0.027 | 0.005 | 2.3E−08 | |
| 14 | 15 | 66529936 | rs4776768 | 15q22.31 | MEGF11 | T | C | 0.28 | −0.025 | 0.005 | 3.71E−08 | |
| 15 | 18 | 39296488 | rs8099746 | 18q12.3 | PIK3C3,KC6 | T | C | 0.80 | 0.030 | 0.005 | 1.27E−08 | |
| 16 | 22 | 46457723 | rs62228096 | 22q13.31 | hsa-mir-4763, LOC150381 | A | G | 0.35 | −0.026 | 0.004 | 4.84E−09 |
Disease the name of the neuropsychiatric disorders, ADHD attention deficit hyperactivity disorder, BIP bipolar disorder, MDD major depressive disorder, CHR chromosome, POS the position of each SNP on the human genome build hg19, SNP the most significantly associated SNP at each locus, Gene closest gene, A1 the effect allele, A2 the non-effect allele, Beta the association coefficient, Freq the frequency of the effect allele, SE the standard error of the effect size estimate; P P value in MTAG analysis.
A meta-analysis of GWAS summary statistics of five neuropsychiatric disorders
We further conducted a meta-analysis based on PGC GWAS summary statistics of the five neuropsychiatric disorders using the software METAL, which yielded 3095 GWS SNPs (P value < 5 × 10−8) located at 43 loci, including one novel locus. This novel locus on chr22 has also reached GWS in the MTAG analysis of MDD (Table 2, Fig. 1, Supplementary Figure 4, Supplementary Table 6).
Table 2.
Novel locus from a meta-analysis of five neuropsychiatric disorders.
| Chr | POS | SNP | A1 | A2 | META_P | Direction | Gene | Disease | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 46457723 | rs62228096 | A | G | 4.48E-08 | ––––– | hsa-mir-4763, LOC150381 | ADHD | −0.022 | 0.014 | 3.42E−04 |
| ASD | −0.019 | 0.014 | 2.47E−03 | ||||||||
| BIP | −0.005 | 0.014 | 4.22E−01 | ||||||||
| MDD | −0.023 | 0.005 | 6.38E−07 | ||||||||
| SCZ | −0.018 | 0.012 | 2.82E−04 |
Chr chromosome, POS the position of each SNP on the human genome build hg19, SNP the most significantly associated SNP at each locus, A1 the effect allele, A2 the reference allele, META_P P value in the meta-analysis, Direction summary of effect direction for each study, with one “+” or “−” per the study, and “+” indicates A1 allele confers risk effect while “−” indicates A1 allele confers protective effect, Gene closest gene; disease the name of five neuropsychiatric disorders, ADHD attention deficit hyperactivity disorder, ASD autism spectrum disorder, BIP bipolar disorder, MDD major depression disorder, SCZ schizophrenia, Beta the association coefficient in the original summary statistics of each disease-specific cohort, SE the standard error of the effect size estimate in original summary statistics of each disease-specific cohort, P P value in the original summary statistics of each disease-specific cohort.
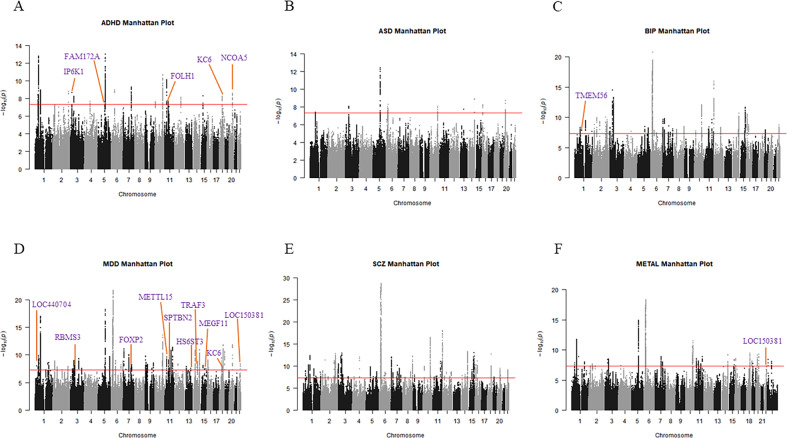
Fig. 1. Manhattan plots showing the association statistics for multi-trait joint analysis and meta-analysis of five neuropsychiatric diseases.
Manhattan plots of the susceptibility loci of five neuropsychiatric diseases: ADHD (A), ASD (B), BIP (C), MDD (D), SCZ (E), METAL (F). SNP locations are plotted on the x-axis according to their chromosomal position. The y-axis shows the −log10 of P values per SNP derived from the meta-analysis. The horizontal red line represents the genome-wide significance threshold of P value < 5 × 10−8. The purple text shows the closest gene to the most prominent SNPs at each novel locus.
Fine mapping and biological annotation of causal variants at novel GWS loci
To understand how the novel loci of ADHD, BIP, and MDD contribute to the development of neuropsychiatric disorders, we conducted fine-mapping to find the likely causal variants at each locus. We found multiple potential causal variants at each locus with the marginal posterior inclusion probabilities of one and the association P value in meta-analysis less than 5 × 10−5. The results showed that the index SNP at each locus serves as one of the potential causal variants. The detailed information for each locus is shown in Supplementary Table 7.
We further performed functional annotation on the likely causal variants at each novel locus identified, examining the involvement of these SNPs in chromatin interactions, histone modification, and eQTL effects in brain tissues. Among the likely causal variants, 20 overlap with histone marks at gene enhancer or promoter regions in brain tissues (Supplementary Table 8). A total of 22 causal SNPs are brain eQTL SNPs in different brain regions (Supplementary Table 9). In addition, 12 loci showed chromatin interactions with distant genes (Supplementary Fig. 5, Supplementary Table 10). Therefore, multiple lines of evidence support the role of the potential causal SNPs at these novel loci in regulating gene expression.
Gene expression profile and gene set enrichment analysis
Gene expression profile analysis of genes at all the novel loci demonstrated the enrichment of differentially expressed gene sets at early developmental stages (Supplementary Fig. 6).
We also conducted a gene-set analysis to gain insights into the pathways involving genes at GWS loci of neuropsychiatric disorders. We found that the GWS loci of each disorder were notably enriched in genes in GO cellular component of a postsynaptic membrane or postsynaptic density membrane, which was consistent with other reports on the involvement of postsynaptic components in the pathology of neuropsychiatric disorders59. In addition, GWS loci were also enriched in GO biological functions of synaptic signaling, membrane depolarization, neuron differentiation for particular neuropsychiatric disorders (Supplementary Table 11). Our results indicated the highly significant enrichment of GWS loci of neuropsychiatric disorders in various aspects of neuronal activity.
The SNP-based heritability and genetic correlation of neuropsychiatric disorders
To evaluate the contribution of common variants to the phenotypic variance of each neuropsychiatric disorder, we conducted a SNP-based heritability estimation based on MTAG results. The SNP-based heritability on the liability scale to each disease ranged from 0.109 to 0.382 (ADHD h2SNP = 0.339, ASD h2SNP = 0.217, BIP h2SNP = 0.382, MDD h2SNP = 0.109, SCZ h2SNP = 0.241), slightly increased when compared to what was reported in the cohort’s original publication, with consistent population prevalence (Supplementary Fig. 7); however, SNP-based heritability was still much less than that estimated from family based or population-based samples (Supplementary Table 12). Further analyses partitioning SNP-based heritability showed significant enrichment in the h2SNP from SNPs at the functional category of Conserved LindbladToh in all the five neuropsychiatric disorders, which implicated the eminent contribution of conserved regions to the SNP-based heritability of neuropsychiatric diseases. We also observed significant h2SNP partitioning to the categories of H3K9ac_peaks and H3K4me1 across five disorders, suggesting that these neuropsychiatric GWS loci may be involved in disease pathogenesis through transcriptional regulation (Supplementary Table 13).
We also assessed the relationship between these neuropsychiatric disorders by LDSC regression analyses and observed a highly significant genetic correlation between them (Supplementary Fig. 8), which is consistent with the reported estimates in previous studies by PGC29. Among them, SCZ and BIP showed the highest genetic correlation, followed by that between MDD and ADHD which was also reflected by the shared novel loci between them (Table 1, Supplementary Table 4).
The replication study
Among the neuropsychiatric disorders which yielded GWS signals in our MTAG analyses, ADHD is less studied than BIP and MDD. ADHD is an early onset childhood disease; it has been reported that almost all patients with clinical features of ADHD retrospectively recalled an onset before age 16. We are, therefore, particularly interested in understanding the genetic mechanism behind ADHD. We conducted a replication study on 2726 ADHD cases and 16,299 healthy controls of European ancestry in a pediatric cohort at CAG, CHOP, which was genotyped on three GWAS chip types. We conducted genome-wide imputation and association testing with no evidence of population stratification (λ = 1.013, Supplementary Fig. 9). Due to the limited number of ADHD cases in the replication cohort, we only observed modest associations. Among the five novel loci for ADHD in the MTAG analysis, the lead SNPs at two loci were of nominal association P value < 0.05 in the replication study, including rs10839264 at 11p11.12 (P value = 0.0487), rs8083506 at18q12.3 (P value = 0.0253); and rs6032660 at 20q13.12 is of marginal significance with P value = 0.0528 (Supplementary Table 14).
We further conducted a sign test on all the ADHD GWS associations and observed sign concordance significantly higher than expected by chance (π = 75% concordant; P = 6.27 × 10−3) between the pediatric replication cohort and the ADHD MTAG analysis results. We further tested the replication of the effect sizes of the ADHD GWS associations from MTAG analysis in the replication cohort by regressing the effect size in the replication cohort on the estimated effect size from the ADHD MTAG analysis adjusted for winner’s curse (Supplementary Fig. 10). The result demonstrated a significantly positive slope (slope = 0.645, P = 2.28 × 10−3) and it did not show a significant difference from 1 (P = 0.0735), which suggested that the effect size estimates in the pediatric replication cohort were significantly correlated with the adjusted effect size in the ADHD MTAG analysis. Therefore, though the index SNPs at the novel GWS associations from the ADHD MTAG analysis showed only modest association in the replication cohort due to the sample size limitation, the direction and magnitude of the effect size of all the primary ADHD GWS loci were significantly correlated between the two cohorts. We then examined the genetic correlation of genome-wide data between the ADHD MTAG analysis and the replication cohort and found a strong positive correlation (rg = 0.663, SE = 0.183, P = 3 × 10−4). Considering the average age-of-onset in adulthood for BIP and MDD, the effect size of the GWS loci, and the limited sample size of cases with either of these two disorders in our pediatric cohorts, they are not suitable for replication study of MTAG analysis results for BIP and MDD.
Discussion
In our study, we conducted an integrative analysis of five common and comorbid neuropsychiatric disorders. Through a joint analysis based on correlated traits and fixed-effect meta-analysis, we identified 16 novel GWS loci associated with one or more diseases. As the index SNPs may just be the tag SNPs for the causal variants, we performed fine-mapping to identify potential causal variants at each locus. Furthermore, the index SNPs and the causal variants are located in the non-coding region of the human genome, likely functioning by regulating target gene expression, therefore we identified potential target genes by positional mapping, regulatory and functional mapping. In cells, the protein products encoded by these target genes do not function alone, instead, they cooperate through protein-protein interaction, and by signaling transduction, so we further performed pathway enrichment and PPI network analysis and found the enrichment of synaptic signaling, membrane depolarization, neuron differentiation among the target genes. We then conducted a replication study with an independent pediatric cohort. The identification of pleiotropic GWS loci between neuropsychiatric disorders may help to understand the shared underlying molecular mechanisms for the design of a common therapeutic strategy and drug repositioning for neuropsychiatric disorders.
In comparison to the recently published cross-disorder meta-analysis of eight neuropsychiatric diseases, we found that many GWS loci obtained from the MTAG analysis overlapped with those reported in the recent meta-analysis29. While this reduced the total number of novel GWS loci found in our MTAG analyses, it demonstrated the reliability of our analyses, being consistent and complementary to the meta-analysis with the approach of association analysis based on SubSETs (ASSET). Similarly, the MTAG analysis on intelligence and education by Hill et al.56 and the ASSET analysis on cognitive ability, education, and SCZ by Lam et al.60 yielded consistent and complementary results, as compared and discussed in the latter study. In addition, our MTAG analysis results explicitly demonstrated the direction and strength of disease-specific association in the analyses with boosted power, compared to the ASSET analysis.
All the novel loci identified in our study are compelling, among which a few loci from ADHD MTAG analysis achieved weak replication as results from an independent pediatric cohort showed moderate sign concordance and correlation of effect size for the GWS loci although none of the P-value surpassed the multiple-testing threshold. The index SNP and the causal variants at locus 11p11.12 are eQTL SNPs for genes GRM5 and FOLH1. GRM5 encodes metabotropic glutamate receptor 5 belonging to the G protein-coupled receptor family, which adopts a conformation change upon ligand binding, inducing downstream signaling transduction through guanine nucleotide-binding proteins and leading to the activation of down-stream effectors, such as phospholipase C. Accumulating evidence suggests an important function of GRM5 in regulating synaptic plasticity and neural network activity. The expression of GRM5 is highly enriched in different brain regions, especially the cerebral cortex. A deletion within GRM5 has been reported to be found in parents and their children with ADHD61 and variants in the GRM5 gene have been linked to other neuropsychiatric disorders, like SCZ62,63. Grm5 knockout mice displayed consistent defects in nerve system and abnormal neurological and behavior phenotypes at different genetic background64,65, and similar behavioral deficits have been observed in rats treated with specific negative allosteric modulators of mGluR566,67. GRM5 has been regarded as a promising drug target for neuropsychiatric and neurodegenerative disorders68,69. Another eQTL gene for SNPs at ADHD locus 11p11.12 is FOLH1, the protein product of which is a type II transmembrane glycoprotein with glutamate carboxypeptidase activity. Its substrates include the nutrient folate and the neuropeptide N-acetyl-l-aspartyl-l-glutamate (NAAG). Both immunohistochemistry and RNAseq data demonstrated an enhanced expression of FOLH1 in intestine and brain tissues, in both the central and peripheral nervous system. The role of FOLH1 in the brain involves hydrolysis of neuropeptide NAAG to generate glutamate and thus to regulate excitatory neurotransmission70. Coding variant H475Y in this gene results in reduced absorption of folates and consequently hyperhomocysteinemia which confers an increased risk for defects in the central nervous system and cognitive function71. The genetic variants of FOLH1 and other genes in the folate metabolism pathway have also been associated with the symptoms of SCZ72, in concordance with the epidemiological studies that inadequate maternal folate intake during pregnancy is a risk factor for SCZ73.
Another locus that reached GWS for ADHD and was replicated at nominal significance in our replication cohort is 18q12.3, of which the candidate gene is PIK3C3 (phosphatidylinositol 3-kinase catalytic subunit type 3), regulating degradative endocytic trafficking. Several studies have reported that rare CNVs overlapping with this gene are associated with SCZ74 and a variant at the promoter of the PIK3C3 gene is also associated with increased risk for SCZ and BIP among both European and African American ancestries75,76, as well as aberrant DNA methylation level in patients with ASD77. In a mouse model, knockout of Pik3c3 led to abnormal sensory neuron morphology, increased neuron apoptosis and axon degeneration, as well as some dysregulation of the immune system like increased T cell apoptosis, etc78.
Several SNPs in high LD with the index SNP rs6032660 at locus 20q13.12 are close to gene NCOA5 and overlap with the promoter or enhancer histone marks H3K4me1, H3K4me3, H3K27ac, and H3K9ac at various brain regions. Furthermore, they are eQTL SNPs for genes CD40 and SLC12A5. Both NCOA527,79 and SLC12A530,80 have been reported to be associated with depression. SNPs at the locus of CD40 are also associated with educational attainment at the GWS level81. The protein product of CD40 is a receptor on antigen-presenting cells, playing critical roles in immune and inflammatory responses. Concordantly, this locus is associated with diverse autoimmune diseases, such as rheumatoid arthritis82, multiple sclerosis83, and systemic lupus erythematosus (SLE)84. CD40 is therefore a pleiotropic locus for both psychiatric disorders and immune diseases, which supports the notion of a mechanistic link between the immune system and the neural system. In addition, in the early stage of Alzheimer’s disease, CD40 has been reported to play a unique role in the amyloid β-protein (Aβ)-induced microglial activation85. A CD40 knockout mouse model exhibits abnormal dendrite size and morphology86. Taken together, several loci that have been implicated to be underlying genetic loci for ADHD reached the stringent GWS threshold for the first time in our study by a multi-trait joint analysis approach which boosted study power by incorporating information for association with the other correlated traits.
Though we did not find an appropriate replication cohort for the loci identified in MTAG analysis of BIP and MDD, the biological relevance and the functional annotations of the candidate genes at these loci suggest that they are bona fide associations with these neuropsychiatric disorders. TMEM56, which was the nearest gene of SNP rs12563424 at the novel BIP locus 20q13.12 showed significant differential gene expression in some neurological disorders. TMEM56 was downregulated in the cerebellum with Atxn2 knockout87 and was upregulated in nucleus accumbens in the maternal immune activation model of neurodevelopmental disorders88. SNP rs12563424 was also reported to be significantly associated with BIP at a conditional false-discovery rate (FDR) in a recent study89. SPTBN2, a candidate gene at the locus of MDD, is involved in the regulation of the glutamate signaling pathway and mutations in this gene result in a class of spinocerebellar ataxia with the feature of neurodegeneration90.
Among the candidate genes at the newly identified loci, several functions in immune regulation, suggesting the contribution of these pathways to the pathogenesis of neuropsychiatric disorders. The impact of dysregulated immune function on the development of neuropsychiatric disorders has been implicated in previous studies. For example, Tiosano et al. reported that there is a positive association between SCZ and SLE, a prototypic autoimmune disease, across patients of different ages, gender, and socioeconomic status, which suggests an overlap in genetic basis and molecular mechanism between SLE and SCZ91. Epidemiological evidence and animal models implicate maternal infections and activation of the maternal immune system in neuropathology in the fetus which can further impair lifelong neuropathology and result in altered behavior in offspring, making maternal infection during pregnancy a risk factor for neuropsychiatric disorders92.
Both common SNPs and rare CNVs have been implicated in the pathogenesis of complex human diseases, especially neuropsychiatric disorders. CNV studies revealed CNVs in neurodevelopmental genes confer increased risk to various neuropsychiatric disorders93,94. As we discussed above, many common variants have been associated with these diseases though usually with a much smaller odds ratio compared to rare CNVs. Overlaps in genes between common variants and rare CNVs have been seen, such as the 11p13.12 locus containing the GRM5 gene. It would therefore be interesting to investigate the relationship between the rare CNVs and the common variants associated with the same disease to determine if the common variants or the haplotype defined by common variants are tagging for the rare CNVs, or if the SNPs confer synthetic effects mimicking the effects of the CNVs. It is also plausible that the common variants may serve as genetic modifiers for rare CNVs.
In our study, we increased study power by conducting joint analyses of correlated traits of neuropsychiatric disorders, and further performed a replication study of ADHD using a pediatric cohort, which provides the advantage of studying an early onset neuropsychiatric disease. However, the pediatric cohort also has two major limitations. It cannot be an ideal replication for certain other neuropsychiatric disorders with the age of onset above 20 years of age and with a different spectrum of clinical features, and the sample size of our replication cohort is small compared to the discovery PGC cohort. In our ADHD replication study, none of the five loci passed the multiple-testing adjusted threshold of P value < 0.01. For common variants with small effect sizes, the size of the replication cohort limits the ability to fully recapitulate the associations found in the discovery cohort. Failure to achieve corrected significance also reflects the heterogeneity between studies, which could arise from the patients’ characteristics or the different phenotyping and cohort ascertainment between cohorts. However, sign test and correlation test of all GWS associations, as well as the genetic correlation based on genome-wide SNP data, suggest the overall consistency between the ADHD MTAG analysis and our pediatric cohort which provides replication of nominal significance. The other novel loci found in our study are also highly biologically relevant and we anticipate that more of them will be replicated in future studies.
In conclusion, through meta-analysis and multi-trait joint analyses of GWAS data of five neuropsychiatric disorders and an independent replication analysis, multiple novel genetic loci were identified with a predisposition to neuropsychiatric disorders. Furthermore, the causal variants at these loci may regulate target gene expression at least in part by affecting histone modifications at various brain regions. Taken together, our research provides novel insight into the genetic basis of neuropsychiatric disorders which may uncover new therapeutic opportunities and provide a molecular basis for drug repositioning.
Supplementary information
Acknowledgements
We thank all the participants enrolled in the study at CAG, CHOP. This project was funded by the National Natural Science Foundation of China (No. 81771769), the Natural Science Foundation of Tianjin (No. 18JCYBJC42700), and the Institute Development Funds to the Center for Applied Genomics at CHOP.
Author contributions
J.L., Q.H.X., and H.H. conceived and designed the study. X.M.Y., J.G., J.Y.L., and X.H.Q. performed the analysis. X.M.Y. and X.Y.H. wrote the draft of the paper. J.L., Q.H.X., and H.H. reviewed and edited the paper critically. Other authors have partially participated in the study. All authors read and approved the paper.
Data availability
To have access to the CAG replication cohort, please contact the corresponding author Dr. Hakon Hakonarson.
Code availability
All analyses followed the standard pipeline of MTAG, METAL, and GWAS analyses and all the scripts are available from the authors upon request.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xueming Yao, Joseph T. Glessner, Junyi Li, Xiaohui Qi
Contributor Information
Qianghua Xia, Email: qhxia@tmu.edu.cn.
Hakon Hakonarson, hakonarson@email.chop.edu.
Jin Li, Email: jli01@tmu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-01195-5).
References
- 1.Jansen AG, et al. Psychiatric polygenic risk scores as predictor for attention deficit/hyperactivity disorder and autism spectrum disorder in a clinical child and adolescent sample. Behav. Genet. 2020;50:203–212. doi: 10.1007/s10519-019-09965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi, X. et al. Integrating genome-wide association study and methylation functional annotation data identified candidate genes and pathways for schizophrenia. Progr. Neuropsychopharmacol. Biol. Psychiatry10.1016/j.pnpbp.2019.109736 (2019). [DOI] [PubMed]
- 3.Johansson V, et al. A population-based heritability estimate of bipolar disorder—in a Swedish twin sample. Psychiatry Res. 2019;278:180–187. doi: 10.1016/j.psychres.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Otte C, et al. Major depressive disorder. Nat. Rev. Dis. Prim. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 5.Wittchen HU, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur. Neuropsychopharmacol. 2011;21:655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Baio J, et al. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill. Summ. 2018;67:1–23. doi: 10.15585/mmwr.ss6706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felt BT, et al. Diagnosis and management of ADHD in children. Am. Fam. Physician. 2014;90:456–464. [PubMed] [Google Scholar]
- 8.Kieling C, et al. The age at onset of attention deficit hyperactivity disorder. Am. J. Psychiatry. 2010;167:14–16. doi: 10.1176/appi.ajp.2009.09060796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpov B, et al. Anxiety symptoms in a major mood and schizophrenia spectrum disorders. Eur Psychiatry. 2016;37:1–7. doi: 10.1016/j.eurpsy.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Selzam S, et al. A polygenic p factor for major psychiatric disorders. Transl. Psychiatry. 2018;8:205. doi: 10.1038/s41398-018-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6:29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Power RA, et al. Genome-wide association for major depression through age at onset stratification: major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol. psychiatry. 2017;81:325–335. doi: 10.1016/j.biopsych.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry. 2019;24:562–575. doi: 10.1038/s41380-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corfield EC, Yang Y, Martin NG, Nyholt DR. A continuum of genetic liability for minor and major depression. Transl. Psychiatry. 2017;7:e1131. doi: 10.1038/tp.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandin S, et al. The heritability of autism spectrum disorder. J. Am. Med. Assoc. 2017;318:1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordsletten AE, et al. Evaluating the impact of nonrandom mating: psychiatric outcomes among the offspring of pairs diagnosed with schizophrenia and bipolar disorder. Biol. Psychiatry. 2020;87:253–262. doi: 10.1016/j.biopsych.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 18.McGuffin P, et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 19.Edvardsen J, et al. Heritability of bipolar spectrum disorders. Unity or heterogeneity? J. Affect. Disord. 2008;106:229–240. doi: 10.1016/j.jad.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 2000;97:12–17. doi: 10.1002/(SICI)1096-8628(200021)97:1<12::AID-AJMG3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Lam M, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardinas AF, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl EA, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard DM, et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 2018;9:1470. doi: 10.1038/s41467-018-03819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address p.m.h.e. and Cross-Disorder Group of the Psychiatric Genomics C. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482. e1411. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard DM, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turley P, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature526, 68 (2015). [DOI] [PMC free article] [PubMed]
- 34.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. s361–s363. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 362, 10.1126/science.aat8464 (2018). [DOI] [PMC free article] [PubMed]
- 43.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt AD, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh PR, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bigdeli TB, et al. A simple yet accurate correction for winner’s curse can predict signals discovered in much larger genome scans. Bioinformatics. 2016;32:2598–2603. doi: 10.1093/bioinformatics/btw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies G, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hill WD, et al. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry. 2019;24:169–181. doi: 10.1038/s41380-017-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potkin SG, et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicita F, et al. Heterozygous missense variants of SPTBN2 are a frequent cause of congenital cerebellar ataxia. Clin. Genet. 2019;96:169–175. doi: 10.1111/cge.13562. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Christian KM, Song H, Ming GL. Synaptic dysfunction in complex psychiatric disorders: from genetics to mechanisms. Genome Med. 2018;10:9. doi: 10.1186/s13073-018-0518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam M, et al. Pleiotropic meta-analysis of cognition, education, and schizophrenia differentiates roles of early neurodevelopmental and adult synaptic pathways. Am. J. Hum. Genet. 2019;105:334–350. doi: 10.1016/j.ajhg.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elia J, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devon RS, et al. The genomic organisation of the metabotropic glutamate receptor subtype 5 gene, and its association with schizophrenia. Mol. Psychiatry. 2001;6:311–314. doi: 10.1038/sj.mp.4000848. [DOI] [PubMed] [Google Scholar]
- 63.Matosin N, Fernandez-Enright F, Lum JS, Newell KA. Shifting towards a model of mGluR5 dysregulation in schizophrenia: Consequences for future schizophrenia treatment. Neuropharmacology. 2017;115:73–91. doi: 10.1016/j.neuropharm.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Xu J, Zhu Y, Contractor A, Heinemann SF. mGluR5 has a critical role in inhibitory learning. J. Neurosci. 2009;29:3676–3684. doi: 10.1523/JNEUROSCI.5716-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu YM, et al. Mice lacking metabotropic glutamate receptor 5 show impaired learning and reduced CA1 long-term potentiation (LTP) but normal CA3 LTP. J. Neurosci. 1997;17:5196–5205. doi: 10.1523/JNEUROSCI.17-13-05196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zou D, Huang J, Wu X, Li L. Metabotropic glutamate subtype 5 receptors modulate fear-conditioning induced enhancement of prepulse inhibition in rats. Neuropharmacology. 2007;52:476–486. doi: 10.1016/j.neuropharm.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Vales K, et al. The difference in effect of mGlu2/3 and mGlu5 receptor agonists on cognitive impairment induced by MK-801. Eur. J. Pharmacol. 2010;639:91–98. doi: 10.1016/j.ejphar.2009.11.067. [DOI] [PubMed] [Google Scholar]
- 68.Akkus F, et al. Metabotropic glutamate receptor 5 neuroimaging in schizophrenia. Schizophr. Res. 2017;183:95–101. doi: 10.1016/j.schres.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 69.Kumar A, Dhull DK, Mishra PS. Therapeutic potential of mGluR5 targeting in Alzheimer’s disease. Front. Neurosci. 2015;9:215. doi: 10.3389/fnins.2015.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luthi-Carter R, Barczak AK, Speno H, Coyle JT. Hydrolysis of the neuropeptide N-acetylaspartylglutamate (NAAG) by cloned human glutamate carboxypeptidase II. Brain Res. 1998;795:341–348. doi: 10.1016/S0006-8993(98)00244-3. [DOI] [PubMed] [Google Scholar]
- 71.Devlin AM, et al. Glutamate carboxypeptidase II: a polymorphism associated with lower levels of serum folate and hyperhomocysteinemia. Hum. Mol. Genet. 2000;9:2837–2844. doi: 10.1093/hmg/9.19.2837. [DOI] [PubMed] [Google Scholar]
- 72.Roffman JL, et al. Genetic variation throughout the folate metabolic pathway influences negative symptom severity in schizophrenia. Schizophr. Bull. 2013;39:330–338. doi: 10.1093/schbul/sbr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirkbride JB, et al. Prenatal nutrition, epigenetics and schizophrenia risk: can we test causal effects? Epigenomics. 2012;4:303–315. doi: 10.2217/epi.12.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glessner JT, et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc. Natl Acad. Sci. USA. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stopkova P, et al. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol. Psychiatry. 2004;55:981–988. doi: 10.1016/j.biopsych.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 76.Saito T, Aghalar MR, Lachman HM. Analysis of PIK3C3 promoter variant in African-Americans with schizophrenia. Schizophr. Res. 2005;76:361–362. doi: 10.1016/j.schres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 77.Wong CC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry. 2014;19:495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bult CJ, et al. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagel M, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 2018;50:920–927. doi: 10.1038/s41588-018-0151-7. [DOI] [PubMed] [Google Scholar]
- 80.Baselmans BML, et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 2019;51:445–451. doi: 10.1038/s41588-018-0320-8. [DOI] [PubMed] [Google Scholar]
- 81.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen JM, et al. The association of CD40 polymorphisms with CD40 serum levels and risk of systemic lupus erythematosus. BMC Genet. 2015;16:121. doi: 10.1186/s12863-015-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan J, Town T, Mullan M. CD40-CD40L interaction in Alzheimer’s disease. Curr. Opin. Pharmacol. 2002;2:445–451. doi: 10.1016/S1471-4892(02)00180-7. [DOI] [PubMed] [Google Scholar]
- 86.Carriba P. & Davies A. M. CD40 is a major regulator of dendrite growth from developing excitatory and inhibitory neurons. eLife6, 10.7554/eLife.30442 (2017). [DOI] [PMC free article] [PubMed]
- 87.Halbach MV, et al. Atxn2 knockout and CAG42-knock-in cerebellum shows similarly dysregulated expression in calcium homeostasis pathway. Cerebellum. 2017;16:68–81. doi: 10.1007/s12311-016-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richetto J, et al. Genome-wide transcriptional profiling and structural magnetic resonance imaging in the maternal immune activation model of neurodevelopmental disorders. Cereb. Cortex. 2017;27:3397–3413. doi: 10.1093/cercor/bhw320. [DOI] [PubMed] [Google Scholar]
- 89.O’Connell, K. S. et al. Identification of genetic overlap and novel risk loci for attention-deficit/hyperactivity disorder and bipolar disorder. Molecular Psychiatry10.1038/s41380-019-0613-z (2019). [DOI] [PubMed]
- 90.Ikeda Y, et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 91.Tiosano S, et al. Schizophrenia among patients with systemic lupus erythematosus: population-based cross-sectional study. Epidemiol. Psychiatr. Sci. 2017;26:424–429. doi: 10.1017/S2045796016000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Estes ML, McAllister AK. Maternal immune activation: Implications for neuropsychiatric disorders. Science. 2016;353:772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kendall KM, et al. Association of rare copy number variants with risk of depression. Association of rare copy number variants with risk of depression. JAMA Psychiatry. 2019;76:818–825. doi: 10.1001/jamapsychiatry.2019.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Warland A, et al. Schizophrenia-associated genomic copy number variants and subcortical brain volumes in the UK Biobank. Mol. Psychiatry. 2020;25:854–862. doi: 10.1038/s41380-019-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To have access to the CAG replication cohort, please contact the corresponding author Dr. Hakon Hakonarson.
All analyses followed the standard pipeline of MTAG, METAL, and GWAS analyses and all the scripts are available from the authors upon request.