Abstract
The Protein synthesis Using Recombinant Elements (PURE) system enables transcription and translation of a DNA template from purified components. Therefore, the PURE system-catalyzed generation of RNAs and proteins constituting the PURE system itself represents a major challenge toward a self-replicating minimal cell. In this work, we show that all translation factors (except elongation factor Tu) and 20 aminoacyl-tRNA synthetases can be expressed in the PURE system from a single plasmid encoding 32 proteins in 30 cistrons. Cell-free synthesis of all 32 proteins is confirmed by quantitative mass spectrometry-based proteomic analysis using isotopically labeled amino acids. We find that a significant fraction of the gene products consists of proteins missing their C-terminal ends. The per-codon processivity loss that we measure lies between 1.3 × 10–3 and 13.2 × 10–3, depending on the expression conditions, the version of the PURE system, and the coding sequence. These values are 5 to 50 times higher than those measured in vivo in E. coli. With such an impaired processivity, a considerable fraction of the biosynthesis capacity of the PURE system is wasted, posing an unforeseen challenge toward the development of a self-regenerating PURE system.
Subject terms: Proteomics, Synthetic biology, Biosynthesis, Translation
Introduction
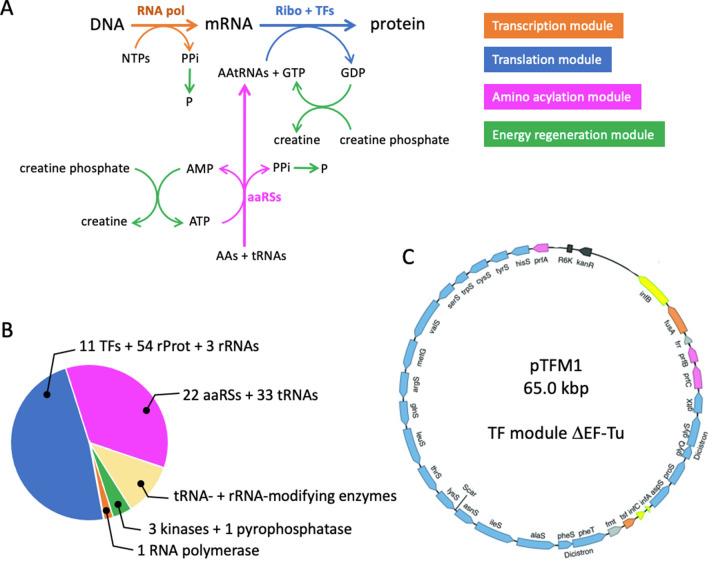
The creation of a man-made cellular system capable of autonomous replication is a grand challenge in synthetic biology1–4. It will be recognized as a milestone toward the bottom up construction of a minimal cell, and may shed light on the elementary constituents and processes that led to the emergence of early cells. Several cellular models that respond to the basic criteria of self-replication have been proposed and their in vitro construction has been experimentally challenged. The putative ‘RNA cell’ relies on two catalytic RNA molecules, called ribozymes, encapsulated in a vesicle1. Despite the apparent simplicity of this scenario, ribozymes that are able to self-replicate or catalyze the formation of membrane constituents from precursors do not exist yet. Alternatively, a ‘ribosome cell’ based on the extant biology might be more amenable to practical realization, although it is composed of many more components than an RNA cell. Translation of genetic information into proteins by the ribosome is universal to all living organisms, including to reduced bacterial cells. A major achievement within the conceptual framework of a ‘ribosome cell’ was the reconstitution of the E. coli translation machinery from purified factors5,6, a technology known as the PURE (Protein synthesis Using Recombinant Elements) system. Essential components of the PURE system are the T7 RNA polymerase for transcription, the E. coli ribosome, tRNAs and 31 translation factors, including the 20 aminoacyl-tRNA-ligases (aaRSs), as well as the translation initiation, elongation and release factors (Fig. 1A,B). Hence, regenerating the PURE system components from a minimal genomic DNA represents a major challenge towards a self-reproducing ‘ribosome cell’.
Figure 1.
Regenerating the PURE system from genes. (A) Schematic of the basic biochemical reactions in the PURE system. The set of reactions is broken down in four main modules. The fuel molecule is creatine phosphate. Hydrolysis of pyrophosphate (PPi) into phosphate (P) is catalyzed by a pyrophosphatase. (B) Macromolecules constituting the PURE system, plus the tRNA- and rRNA-modifying enzymes that will eventually have to be synthesized in vitro for complete PURE system biogenesis. Note that 22 proteins constitute the 20 aaRSs. (C) Schematic map of the pTFM1 plasmid encoding all aminoacyl-tRNA synthetases (aaRSs) and translation factors (TFs), except EF-Tu. Adapted from Ref.13. Other abbreviations: AAs amino acids, AAtRNAs aminoacyl-tRNAs, aaRSs aminoacyl-tRNA synthetases, RNA pol RNA polymerase, Ribo ribosome, rProt ribosomal protein.
Several constituents of the PURE system have already been produced in the PURE system itself starting from genes. Awai et al. showed that 19 of the 20 aaRS enzymes could be synthesized in a soluble and active form7. In this study, the 20 aaRSs were expressed individually in a PURE system that contained lowered input concentration of the aaRS that was expressed, so that the activity of the de novo synthesized protein could be detected above the background level of activity stemming from the originally supplied aaRS. Attempts to reconstruct the E. coli ribosome were carried out in the PURE system by expressing the ribosomal proteins8. While all 54 proteins could be detected when produced separately, and all of the 21 proteins of the small subunit when co-expressed, only 29 of the 33 large ribosomal subunit proteins were detected in co-expression reactions. To differentiate newly synthesized components from the PURE system background, the reaction mixture was supplied with isotopically heavy arginine and lysine residues, and the translation products were detected by mass spectrometry similar to the well-established method of stable isotope labeling with amino acids in cell culture (SILAC)9. Hence, this strategy allows for quantification of the synthesized protein relative to the originally supplied protein in the PURE system. Efforts to synthesize the three ribosomal RNAs (rRNAs) are challenged by the numerous chemical modifications underwent by the 16S and 23S rRNAs to harbor their full activity spectrum10. To bypass the reconstitution of the enzymatic rRNA modification pathway in the PURE system, in vitro evolution has been applied to generate a 16S rRNA mutant that is active in the absence of post-transcriptional modifications11. Furthermore, 48 E. coli tRNAs have been synthesized in vitro from separate DNA templates using the T7 RNA polymerase, and most of them showed functionality in an E. coli cell-free translation system12. Actually, it has been proposed that only 33 E. coli-based tRNAs would be sufficient to decode all 20 amino acids2. The other PURE system elements that have to be regenerated as well are: the methionyl-tRNA-formyltransferase, T7 RNA polymerase, pyrophosphatase, and the enzymes of the energy recycling module, creatine kinase, myokinase and nucleoside-diphosphate kinase.
Herein, we show that all translation factors (TFs) (except elongation factor Tu, EF-Tu) and aaRSs can be expressed from a single plasmid in the PURE system. In the following, aaRSs will also be referred as TFs. We used the pTFM1 plasmid encoding 32 proteins in 30 cistrons: 20 aaRSs, three translation initiation factors, three release factors, ribosome recycling factor, two elongation factors, and the methionyl-tRNA-formyltransferase (Fig. 1C)13. All genes contain the same T7 promoter-lacO-RBS (RBS, ribosome binding site) block at the 5′ end of the coding sequence and T7 phi terminator at the 3′ end. Only the two aaRSs consisting of two subunits, namely the glycine tRNA-ligase and phenylalanine tRNA-ligase, are encoded as two-gene cistrons. We were able to detect the synthesis of all 32 proteins from a single plasmid. Moreover, we discovered that many truncated proteins were also generated, an issue that remains under-appreciated when interpreting the outcome of cell-free gene expression reactions.
Results
LC–MS detection of 32 proteins expressed in PURE system from a 30-cistron TF module plasmid
To detect proteins expressed with the PURE system from pTFM1 we used heavy isotope labeling and liquid chromatography-coupled mass spectrometry (LC–MS)8. We employed two commercially available versions of the PURE system, PUREfrex2.0 and PURExpress, in combination with a buffer containing 15N-labeled amino acids (Supplementary Table 1), so that all newly synthesized proteins contain 15N-amino acids and the concentration ratio of in situ expressed to originally supplied proteins can be determined by mass spectrometry (Fig. 2A). We assayed the detection efficiency of different candidate tryptic peptides by LC–MS/MS using Skyline14 and reaction monitoring guided by the E. coli MG1655 spectral library (The Global Proteome Machine, https://www.thegpm.org/). We finally selected a set of 64 peptides, each of the 32 proteins encoded by pTFM1 plus EF-Tu being covered by one to four peptides (Fig. 2D, Supplementary Tables 2 and 3) which could be measured in two LC–MS/MS runs with multiple reaction monitoring (MRM). We optimized trypsin reaction conditions to achieve complete digestion (Supplementary Fig. 1). As shown in Fig. 2B,C, all 32 proteins could be detected for both PUREfrex2.0 and PURExpress. The ratios of the peak areas of the 15N-labeled peptides to 14N-labeled peptides is a measure of the protein expression levels relative to the amount of the respective protein originally present in the PURE system. We found that the relative protein expression levels varied between 2 and 3000% and were generally higher for PUREfrex2.0 than for PURExpress.
Figure 2.
In vitro production of 32 translation factor proteins from a single plasmid. (A) Reaction scheme for pTFM1 expression in the PURE system. AAs amino acids, TFs translation factors, FMT methionyl-tRNA-formyltransferase, IF initiation factor, EF elongation factor, RF release factor, RRF ribosome recycling factor. (B,C) Relative expression levels of proteins expressed from pTFM1 with PUREfrex2.0 (B) and PURExpress (C) after 5 h incubation at 37 °C. Ratios of 15N-labeled peptides (newly synthesized proteins) to 14N peptides (original proteins) are displayed for all measured peptides; for proteins with multiple measured peptides, the peptides are ordered from N- to C-terminus (top to bottom). Error bars denote ± 1 standard deviation from triplicate experiments. Results from a different batch of PUREfrex2.0 are shown in Supplementary Fig. 2. (D) Specific peptides measured in (B) and (C).
PURE system produces C-terminal truncated proteins over time
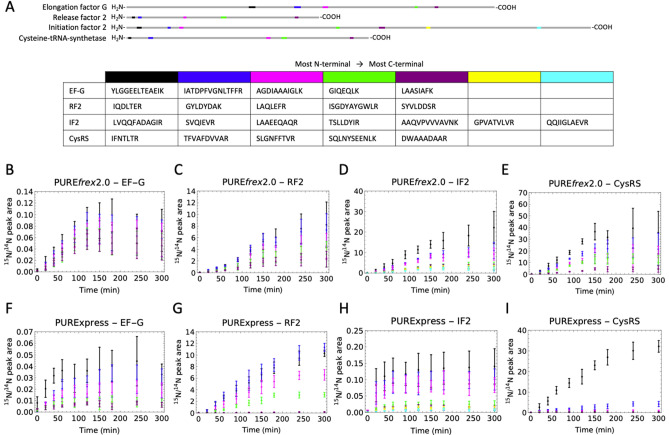
We noticed that for some proteins with multiple measured peptides the 15N-to-14N ratio tended to decrease from the more N-terminal peptides to the more C-terminal peptides, indicating synthesis of truncated products (Fig. 2B,C, Supplementary Fig. 2). This trend was noticeable for ~ 10 proteins (depending on the batch of the kit) out of 21 with PUREfrex2.0 and for ~ 16 proteins out of 21 with PURExpress. To investigate this effect in more detail we selected another set of peptides (Supplementary Table 4) covering four of the proteins (EF-G, RF2, IF2 and CysRS) from N- to C-terminus (Fig. 3A), and measured the 15N-to-14N ratio for these peptides over time (Fig. 3B–I). We chose these proteins because they each represent a different class of translation factor and because we identified a sufficient number of unique peptides that span the entire sequence of the corresponding protein. For all four proteins, a continuous decrease of the 15N-to-14N ratio towards the more C-terminal peptides was observed for all measured time points in both versions of the PURE system (Fig. 3B–I, note that the EF-G result with PUREfrex2.0 is more significant here than in Fig. 2B partly because of the inclusion of an additional peptide). This result confirms that the processivity of translation elongation is impaired for several proteins. Moreover, this behavior is not an artefact due to long expression times, when the system possibly runs out of nutrients or some of the protein machinery becomes inactive. Premature translation arrest also occurs at the start of the reaction, as indicated by the overabundance of N-terminal peptides within the first 40 min of expression (Fig. 3B–I). Therefore, this process does not result from detrimental effects that would become more prominent in the course of expression, like mRNA degradation. The average per-codon loss of processivity was calculated as the negative exponent of an exponential regression fit to the 15N-to-14N ratio as a function of the peptide position (Supplementary Figs. 4, 5). Values range between 1.3 × 10–3 and 4.5 × 10–3 for PUREfrex2.0, and between 4.0 × 10–3 and 13.2 × 10–3 for PURExpress, with CysRS and RF2 having the highest per-codon loss values in both systems, followed by IF2 and EF-G (Fig. 4). No major differences were observed after 20 min and 300 min expression durations (Fig. 4). These values are an order of magnitude higher than the processivity error measured in E. coli15,16. Interestingly, the expression lifetime, defined here as the time at which production of the most N-terminal peptide ceases, is different for the four proteins, but also between the two different PURE system versions (Supplementary Fig. 6). For instance, the expression lifetime of IF2 is ~ 415 min in PUREfrex2.0 and ~ 39 min in PURExpress, whereas the expression lifetime of CysRS is ~ 151 min in PUREfrex2.0 and ~ 339 min in PURExpress. This result shows that both the coding sequence of the DNA template and the nature of the cell-free gene expression system influence the kinetics of protein synthesis.
Figure 3.
Time-course of 15N-to-14N ratio of selected peptides for four proteins encoded in pTFM1. (A) Schematic of the full-length proteins displaying the positions of selected proteolytic peptides. For each cell-free synthesized protein, the set of signature peptides is chosen to span the entire polypeptide chain from the N- to C-termini. Color coding from N- to C-terminal peptides: black, blue, pink, green, magenta, yellow, cyan. The set of specific peptides measured for all four proteins is also indicated. EF-G elongation factor G, RF2 release factor 2, IF2 initiation factor 2, CysRS cysteine-tRNA-ligase. (B–I) Relative expression levels of the indicated peptides for the four annotated proteins with PUREfrex2.0 (B–E) and with PURExpress (F–I). Color coding of the peptides is similar as in (A). The five kinetics plots in (B) are displayed in separate graphs in Supplementary Fig. 3. All reactions were performed at 37 °C. Error bars denote ± 1 standard deviation from triplicate experiments.
Figure 4.
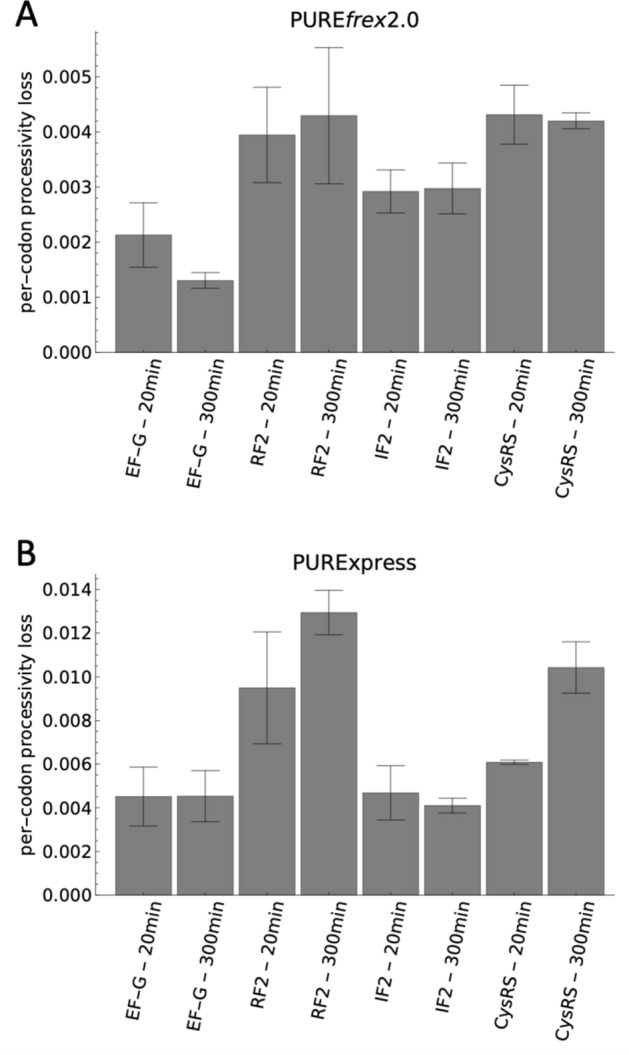
Quantification of processivity loss with pTFM1 as a template. Average per-codon processivity loss of the indicated proteins expressed in PUREfrex2.0 (A) or in PURExpress (B) for 20 min or 300 min. Bar heights are slope values obtained from weighted fitting of data shown in Supplementary Fig. 4 and error bars are weighted fit uncertainties.
To rule out the possibility that processivity errors originate either from an artefact of the homemade buffer that we used instead of the commercially supplied buffers (in order to substitute the unlabeled amino acids with 15N-labeled ones), or from the co-expression of 32 proteins, we performed control experiments in which a single protein, the bacterial tubulin homolog FtsZ, was expressed in both PURE system versions either with the buffer provided with the commercial kits or with our homemade buffer. In the latter case, the amino acid mix consisted of either 14N-labeled amino acids (each in equimolar amounts), or the same 15N-labeled amino acid mix as used for pTFM1 expression. Purified FtsZ protein, either unlabeled or 15N-labeled, was used as an internal standard to quantify the absolute concentrations of the synthesized peptides. We selected a set of seven FtsZ peptides that could be monitored in a single MRM experiment (Fig. 5A, Supplementary Table 5). Expression of this single protein resulted also in a significant decrease in concentration from N-terminal to more C-terminal peptides for all conditions and time points (Fig. 5B–G). Yield of synthesized FtsZ peptides was significantly higher with the commercial buffers (the most C-terminal peptide reached a concentration of ~ 5 µM) than with the homemade ones, while yields between the unlabeled amino acid mix and the 15N-labeled one were negligible in both versions of PURE system. For PUREfrex2.0, the processivity was similar for all tested buffers, while for PURExpress the processivity was lower in the homemade buffers compared to the commercial ones (Fig. 5H–J). This result suggests that, also in the case of expression from pTFM1, lower processivity in PURExpress might be an effect of the buffer rather than an intrinsically lower performance of PURExpress as compared to PUREfrex2.0.
Figure 5.
Impaired ribosome processivity occurs also in single-gene expression reactions. (A) Schematic of the full-length FtsZ protein displaying the positions of selected peptides. The specific peptides measured are appended. Color coding from N- to C-terminal: black, blue, pink, green, magenta, yellow, cyan. (B–G) Time-course of the concentration of the indicated peptides synthesized with PUREfrex2.0 (B–D) and with PURExpress (E–G). All reactions were performed at 37 °C. Error bars denote ± 1 standard deviation from triplicate experiments. Color coding of the peptides is similar as in (A). (H,I) End-point (5 h) FtsZ peptide concentration as a function of peptide position for reactions with the commercial buffer (dark blue, solid lines), with the homemade buffer containing unlabeled amino acids (blue, dashed lines) or with 15N-labeled amino acids (light blue, dotted lines). Straight lines are exponential regression fits. (J) Average per-codon processivity loss derived from the slope computed in (H,I). Error bars are weighted fit uncertainties.
Absolute quantification of translation factors expressed from pTFM1 in PURE system
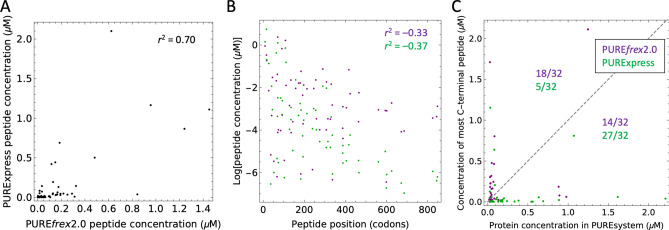
The ratios of 15N-to-14N peak intensities do not allow for a comparison between expression levels in reactions with PUREfrex2.0 and PURExpress, nor between different proteins within the same reactions. With the aim to provide absolute quantification of PURE system TFs, we designed a QconCAT (Quantification conCATemer), an artificial protein generated by concatenation of proteolytic peptides used as reference standards for quantification of the corresponding TF peptides. Our QconCAT is composed of one to four peptides from all proteins encoded on pTFM1, as well as two peptides from EF-Tu and two quantification peptides (Supplementary Figs. 7, 8). Although the QconCAT protein expressed well in lysogeny broth (LB) medium, we were unable to purify the protein from E. coli cells grown in isotope-labeled medium. Therefore, the DNA sequence was split into two halves and both halves were recloned into a pRSET-B vector harboring an N-terminal His-tag. After testing the purified, isotope-labeled QconCAT halves against the purified, unlabeled full-length QconCAT, we determined the concentrations of all corresponding proteins in PUREfrex2.0 and PURExpress. For proteins with multiple peptides, the concentration differences were within the error margin. From these measurements, we could calculate the absolute concentrations of the peptides expressed from pTFM1 in both PURE systems. Concentration values span a few orders of magnitude, with most peptides having a concentration below 0.2 µM and only a few reaching the micromolar range (Fig. 6). Absolute expression levels are significantly correlated between the two systems with a correlation coefficient of 0.70 (Fig. 6A). This correlation can in part be explained by the negative correlation of the peptide concentration with respect to its position within the coding sequence due to processivity errors (Fig. 6B). Moreover, significant disparity in expression levels between the different proteins was observed even when comparing peptides located at roughly the same position in the primary sequence. We then assessed the correlation of the measured peptide concentration against predictions from three different mRNA design tools: RBS calculator, RBS designer and UTR designer17–19, as well as an empirical 3-codon score20. The peptides belonging to the beta subunits of the glycine- and phenylalanine-tRNA ligases were excluded from the analysis because these proteins are expressed as second protein of an operon with the corresponding alpha subunits, which is expected to influence the expression level. No significant correlations were found for any of the tested predictive tools (Supplementary Fig. 9).
Figure 6.
Correlation analyses of absolute peptide concentrations for pTFM1 expression. Correlations of absolute peptide expression levels between PUREfrex2.0 and PURExpress (A), with the peptide position within the coding sequence (B), and with their initial amounts in the commercial kits (C). (A,B) Values of r2 denote Pearson correlation coefficients. (B,C) Data points for PUREfrex2.0 and PURExpress are colored in magenta and in green, respectively. (C) The most C-terminal peptide for each protein was used for analysis. The appended dashed line depicts the slope equal to 1. PUREfrex2.0 and PURExpress reactions were run in the homemade buffer containing 15N-labeled amino acids. Concentrations of synthesized proteins were derived from 15N-to-14N peak ratios, while the concentrations of PURE system components were determined in separate experiments using QconCAT as an internal standard. For each PURE system kit, the number of proteins whose concentration falls above ([protein]expressed > [protein]input) or below ([protein]expressed < [protein]input) the diagonal is indicated.
Finally, we compared the absolute concentration of all 32 synthesized proteins with respect to their original concentrations in PUREfrex2.0 and PURExpress (Fig. 6C). The most C-terminal peptide for each translation factor was used as it best estimates the amount of full-length protein. Under these expression conditions, PUREfrex2.0 is able to produce more proteins than initially contained in the commercial kit for > 50% of the TFs, versus ~ 15% for PURExpress. For proprietary reasons, we cannot specify which protein corresponds to which data point in Fig. 6C, as it would conflict with the policy of New England Biolabs and GeneFrontier Corporation to not reverse engineer their products. Whereas processivity loss is similar for both PURE systems in their respective commercial buffers, translation elongation is particularly affected in the homemade buffer with 15N-labeled amino acids for PURExpress compared to PUREfrex2.0 (Fig. 5J). Therefore, we expect that expression of pTFM1 in optimal reactions would lead to doubled (or more) concentrations for a higher fraction of TFs, and this effect would be more pronounced for PURExpress than for PUREfrex2.0.
Discussion
Co-expression of 32 different proteins from a single 30-cistron plasmid was realized in the PURE system. Because the gene products are constituents of the PURE system itself, this work contributes to ongoing efforts to regenerate a minimal protein synthesis machinery from a DNA template7,8,11.
Detection of C-terminal truncated translation products reveals hampered ribosomal processivity in the PURE system. This process seems to be a general bottleneck as it affects the yield of synthesized full-length protein for many of the 33 genes expressed using two different PURE system variants, in single-gene as well as in 30-cistron expression reactions. The per-codon processivity loss is 5–50 times higher than that measured in E. coli15,16,21,22. Production of truncated products with the PURE system, but also with cell lysates, has been reported before, in particular with the expression of eukaryotic proteins23–26. Here, we show that processivity of translation elongation is significantly hampered with native E. coli sequences under various cell-free gene expression conditions.
Processivity issues were not mentioned in two recent studies, where ribosomal proteins8 or translation factors27 expressed in PURE system were also analyzed by LC–MS. In the latter study, TFs were expressed from three different plasmids in PURExpress27. We imported the MaxQuant ouput MS data from Ref.27 and plotted them as displayed in Supplementary Figs. 4, 5 to verify the occurrence of processivity errors (Supplementary Data file). A clear trend showing a decreased abundance of C-terminal peptides was observed for at least 18 out of the 32 proteins (Supplementary Data file). This finding emphasizes the need for systematic investigations of all gene products for unbiased monitoring and quantification of expressed proteins. Comparing the ratios of 15N-to-14N peak intensities for the different TFs obtained in the study of Libicher et al.27 and ours, we found a rather low correlation with Pearson correlation coefficients ~ 0.5 (Supplementary Fig. 10A,B). Besides, no correlation was found when comparing the protein concentrations quantitated in our study and in Fig. 2F of Shepherd et al.13 (Supplementary Fig. 10C,D). These differences may originate from the different DNA templates (single plasmid vs three plasmids in Refs.13,27), PURE compositions (PUREfrex2.0 vs modified PURExpress in Ref.27), expression conditions (in vitro vs in vivo in Ref.13) or quantification methods.
Possible causes for the observed processivity errors of translation elongation include ribosome stalling or destabilization, peptidyl-tRNA drop-off and premature termination. Supplementing the PURE system with ribosome rescue factors28–33, peptidyl-tRNA-hydrolase23,34, EF-G35–38, methylated RF1 and RF239, or the ribosomal protein bL3140,41 might therefore enhance processivity. Degradation of mRNA by nuclease contaminants is not substantial in the PURE system compared to cell extracts34,42,43. Given that proteins missing their C-terminal ends are already detected at short incubation times (Figs. 3, 4), stalling of translating ribosomes on mRNA truncated at the 3′ end is unlikely the main cause of impaired processivity. Moreover, identical results were obtained when the murine RNase inhibitor was supplied to PURE system reactions (data not shown). Overall, a complex set of side reactions may impede translation elongation in the PURE system. The fact that PURExpress is more susceptible to processivity errors than PUREfrex2.0 under the tested conditions indicates that optimization of the abundance and stoichiometric amount of the different components might help improve both the expression yield and translation processivity. In addition to optimizing the protein hardware of the PURE system, optimization of buffer components may also increase the system performance. This idea is supported by the observation that expression yield and processivity were improved in PURExpress when using the commercial buffer instead of the homemade one. In particular, magnesium ion and spermidine concentrations have a huge effect on many of the individual rates, in particular the EF-G-catalyzed translocation reaction, as well as peptidyl transfer. While translocation is faster at lower Mg2+ concentrations44, ternary complex binding and peptidyl transfer are however faster at higher Mg2+ concentrations, albeit coupled to a trade-off between rate and fidelity45. Systematic attempts to improve the PURE system by varying its composition have revealed complex interactions between different components46–49, further challenging the formulation of a high-fidelity, high-yielding gene expression system by rational design.
The biosynthesis capacity of the PURE system is one to two orders of magnitude lower than the yield required to reproduce the input proteins50. Such a suboptimal performance precludes the realization of a self-replicating PURE machinery. In fact, optimizing the PURE system composition and DNA sequence for better usage of resources and higher fidelity of translation would yield larger amounts of full-length products, without necessarily implying to increase the total mass of synthesized proteins. Furthermore, enhancing translation initiation would increase the fraction of translating ribosomes, hence the amount of output proteins34. Other important considerations include the proper folding of the polypeptide chains into functional proteins51, as well as the controlled co-expression of multiple proteins required for the reconstitution of complex biological functions. Although we could detect all 32 proteins encoded on pTFM1, controlling expression levels to yield functional feedback of the de novo synthesized translation factors and, hence, more sustainable expression, remains difficult. Absolute quantification of synthesized peptides has revealed that there exists no correlation of the expression levels with the tested predictive tools (Supplementary Fig. 9). Moreover, further investigations are needed to empirically correlate the amount of a large set of cell-free synthesized proteins with the initial coding sequence20. Nonetheless, we observed significantly lower yields for the GlyRS and PheRS beta subunits that were expressed from the second position of a cistron (the genes of their respective alpha subunits were in the first position) (Fig. 2B,C), as previously reported for similarly designed constructs with PURExpress52.
Materials and methods
DNA constructs
pTFM1 was amplified and purified as previously described13. pET11a-ftsZ-his6 was constructed as follows. Gene fragments were PCR amplified from chromosomal E. coli BL21 DNA with primers 5′-TTAACTTTAAGAAGGAGATATACATATGTTTGAACCAATGGAACTTACC-3′ and 5′-TCCTTTCGGGCTTTGTTAGCAGCCGGATCCTTAATCAGCTTGCTTACGCAG-3′. These primers contain overhangs for the pET11-a plasmid. Next, the PCR products were digested with DpnI (New England BioLabs Inc.) and assembled to a linearized pET11-a plasmid (equimolar concentrations) via Gibson Assembly for 1 h at 50 °C. The assembly products were transformed into E. coli TOP10 competent cells via heat shock, the cells were centrifuged and resuspended in 50 µL of fresh liquid LB media and incubated at 250 rpm for 1 h at 37 °C. Cultures were plated on solid LB medium with 0.05 ng µL–1 ampicillin and were grown overnight at 37 °C. Selected colonies were cultured in 1 mL of liquid LB medium with 0.05 µg µL–1 ampicillin at 250 rpm for 6 h at 37 °C. Plasmid purification was carried out using PureYield Plasmid Miniprep System (column method, Promega). Production of linear DNA constructs from the above purified plasmids was performed by PCR using primers 5′-TAATACGACTCACTATAGGGGAATTGTGAGCGGATAACAATTCCCCT-3′ and 5′-CAAAAAACCCCTCAAGACCCGTTTAGAGG-3′. PCR products were analyzed on a standard DNA agarose gel (1%; EtBr or SYBR safe).
Protein purification
15N-labeled FtsZ was expressed from a pET11a-ftsZ-his6 vector in E. coli C41(DE3). Cells were grown to saturation in LB medium, diluted 1:100 into M9 medium with 15NH4Cl, grown over-night and again diluted 1:100 into fresh M9 medium containing 15NH4Cl. At OD600 = 0.5, cells were induced with 1 mM IPTG and harvested after 3 h at 37 °C. Unlabeled FtsZ was expressed in E. coli C41(DE3) cells in LB medium under the same induction conditions. FtsZ was purified as described previously53. Protein concentrations were determined by Bradford assay.
QconCAT purification
QconCAT halves were expressed in BL21(DE3) cells in M9 medium with 15NH4Cl and ampicillin54. A pre-culture was diluted 1:100 to a 50-mL expression culture. Protein expression was induced at OD600 = 0.5 with 1 mM IPTG and cells were grown for 3 h at 37 °C. Cells were harvested by centrifugation and the pellet was dissolved in 1 mL B-PER. 10 µL of 10 mg mL–1 lysozyme and 10 µL of DNaseI (ThermoScientific, 1 U µL–1) were added and the sample was incubated for 10 min at room temperature. The lysate was centrifuged for 20 min at 16,000g and the pellet was resuspended in 2 mL of a 1:10 dilution of B-PER in MilliQ water. The sample was twice again centrifuged and the pellet was resuspended in 2 mL of 1:10 diluted B-PER and centrifuged again. The pellet was resuspended in 600 µL of 10 mM Tris–HCl pH 8.0, 6 M guanidinium chloride and incubated at room temperature for 30 min. After spinning down the unsolubilized protein content the supernatant was loaded onto an equilibrated mini NiNTA spin column and the flow-through was reloaded twice to maximize protein binding. The column was washed twice with 600 µL of 10 mM Tris–HCl pH 6.3, 8 M urea and the QconCAT was eluted with 3 × 200 µL of 10 mM Tris–HCl pH 4.5, 8 M urea, and 400 mM imidazole. The eluate was dialyzed overnight and for additional 4 h against 10 mM Tris–HCl pH 8.0 and 100 mM KCl using a 10-kDa cut-off slide-a-lyzer cassette (ThermoScientific). Purification of the full-length QconCAT was carried out following the same protocol except for expression in LB medium.
tRNA deaminoacylation
50 µL of 15 mg mL–1 tRNA solution (Roche) was mixed with 300 µL of 1 M HCl, vortexed and incubated at room temperature for 15 min. A solution consisting of 300 µL of 1 M NaOH, 60 µL of 3 M sodium acetate, and 1.8 mL of ice-cold ethanol was added. After vortexing the solution was incubated at − 80 °C for 1 h and the tRNA pool was pelleted by centrifugation using a table-top centrifuge (5415R, Eppendorf) at maximum speed. The pellet was washed with ice-cold 75% ethanol, air-dried, and re-dissolved in MilliQ water.
PURE system reactions
PURExpress was purchased from New England Biolabs and PUREfrex2.0 from GeneFrontier Corporation (Japan). Enzyme and ribosome solutions (PUREfrex2.0) or solution B (PURExpress) were mixed either with their respective commercial feeding solution (solution I for PUREfrex2.0, solution A for PURExpress) according to the supplier’s recommendations or with an equimolar volume of a homemade buffer consisting of 20 mM HEPES–KOH pH 7.6, 180 mM potassium glutamate, 14 mM magnesium acetate, 2 mM DTT, 2 mM spermidine, 100 mM creatine phosphate, 0.1 mg mL–1 10-formyl-tetrahydrofolate (prepared from 5-formyl-tetrahydrofolate according to the protocol described in Ref.55), 3 mM ATP, 3 mM GTP, 1 mM UTP, 1 mM CTP, 0.75 mg mL–1 deaminoacylated tRNA, 1.35 mg mL–1 amino acid mix (15N-labeled amino acid mix was from Cambridge Isotope Laboratories; 14N amino acid mix for control reactions contained equimolar amounts of all amino acids). Plasmid DNA was added to a final concentration of 5 ng µL–1.
Trypsin digest
Enzymatic digestion of proteins was performed as previously described54. Per LC–MS injection, 1.5 µL of PURE system reaction was mixed with 3 µL of 100 mM Tris–HCl pH8.0, 0.3 µL of 20 mM CaCl2, and 0.97 µL MilliQ water. Samples were incubated at 90 °C for 10 min and after cooling to room temperature 0.22 µL of 1 mg mL–1 trypsin (trypsin-ultra, MS-grade, New England Biolabs) was added. Samples were then incubated at 37 °C overnight. After addition of 0.6 µL 10% trifluoroacetic acid samples were centrifuged in a table-top centrifuge (5415R, Eppendorf) for 10 min at maximum speed. The supernatant was transferred to a glass vial with small-volume insert for LC–MS/MS analysis. For absolute quantitative proteomic analysis three different concentrations of PUREfrex2.0 and PURExpress samples were mixed with a fixed concentration of both QconCAT halves. Samples were digested with trypsin as described above and, before LC–MS/MS analysis, they were supplemented with 110 nM of 13C-Arg/Lys labeled SILs (Pepscan presto, Lelystad, The Netherlands) corresponding to the two quantification peptides on the QconCAT halves.
Proteomic analysis
LC–MS/MS analysis was performed on a 6460 Triple Quad LCMS system (Agilent Technologies, USA) using Skyline software14. 5.5 µL of sample was injected per run to an ACQUITY UPLC Peptide CSH C18 Column (Waters Corporation, USA). The peptides were separated in a gradient of buffer A (25 mM formic acid in MilliQ water) and buffer B (50 mM formic acid in acetonitrile) at a flow rate of 500 µL per minute and at a column temperature of 40 °C. The column was equilibrated with 98% A. After injection, the gradient was changed linearly over 20 min to 70% buffer A, over the next 4 min to 60% buffer A, and over the next 30 s to 20% buffer A. This ratio was held for another 30 s and the column was finally flushed with 98% buffer A to equilibrate for the next run. Selected peptides were measured by multiple reaction monitoring (MRM). For reactions with expression of pTFM1 measurements were split over three LC–MS/MS runs (Supplementary Tables 2, 3, 4). For reactions including 15N-labeled amino acids, transitions for peptides containing 15N-amino acids were monitored, except for glutamate because of the excess of the light glutamate contained in the buffer.
Kinetic model
Timeseries data were fitted to the equation , where t denotes time and describes the peptide concentration at time t. The expression timespan is calculated from the fitted parameters as .
Supplementary Information
Acknowledgements
We thank Josefine Liljeruhm for supplying pTFM1, and Tomoko Miyagi (GeneFrontier Corporation) and Isaac Meek (New England Biolabs) for reading the manuscript. This work was financially supported by the Netherlands Organization for Scientific Research (NWO/OCW) through the ‘BaSyC—Building a Synthetic Cell’ Gravitation grant (024.003.019). ACF was supported by the Swedish Research Council (NT project grants 2016-1 and 2017-04148).
Author contributions
A.D. designed and performed the experiments, and analysed the data. C.D. conceived and supervised the research. A.D. and C.D. wrote the manuscript. D.F. contributed preliminary mass spectrometry measurements. A.C.F. supervised the preparation of pTFM1, reviewed and edited the manuscript.
Data availability
All data reported in the current study are available from the corresponding author upon reasonable request. This also includes the original .nd file (created in Mathematica version 11.3, Wolfram Research) used to generate the data displayed in the Supplementary Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80827-8.
References
- 1.Szostak JW, Bartel DP, Luisi PG. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 2.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl. Acad. Sci. U. S. A. 2011;108(9):3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwille P, Spatz J, Landfester K, Bodenschatz E, Herminghaus S, Sourjik V, et al. MaxSynBio: Avenues towards creating cells from the bottom up. Angew. Chem. Int. Ed. 2018;57(41):13382–13392. doi: 10.1002/anie.201802288. [DOI] [PubMed] [Google Scholar]
- 5.Kung HF, Redfield B, Treadwell BV, Eskin B, Spears C, Weissbach H. DNA-directed in vitro synthesis of beta-galactosidase. Studies with purified factors. J. Biol. Chem. 1977;252(19):6889–6894. doi: 10.1016/S0021-9258(17)39933-7. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 7.Awai T, Ichihashi N, Yomo T. Activities of 20 aminoacyl-tRNA synthetases expressed in a reconstituted translation system in Escherichia coli. Biochem. Biophys. Rep. 2015;3:140–143. doi: 10.1016/j.bbrep.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Haas W, Jackson K, Kuru E, Jewett MC, Fan ZH, et al. Cogenerating synthetic parts toward a self-replicating system. ACS Synth. Biol. 2017;6(7):1327–1336. doi: 10.1021/acssynbio.6b00342. [DOI] [PubMed] [Google Scholar]
- 9.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Ofengand, J., Del Campo, M. Modified nucleosides of Escherichia coli ribosomal RNA. EcoSal Plus.10.1128/ecosalplus.4.6.1 (2004). [DOI] [PubMed]
- 11.Murase Y, Nakanishi H, Tsuji G, Sunami T, Ichihashi N. In vitro evolution of unmodified 16S rRNA for simple ribosome reconstitution. ACS Synthetic Biology. 2018;7(2):576–583. doi: 10.1021/acssynbio.7b00333. [DOI] [PubMed] [Google Scholar]
- 12.Cui Z, Stein V, Tnimov Z, Mureev S, Alexandrov K. Semisynthetic tRNA complement mediates in vitro protein synthesis. J. Am. Chem. Soc. 2015;137:4404–4413. doi: 10.1021/ja5131963. [DOI] [PubMed] [Google Scholar]
- 13.Shepherd TR, Du L, Liljeruhm J, Samudyata J, Wang J, Sjödin MOD, et al. De novo design and synthesis of a 30-cistron translation-factor module. Nucleic Acids Res. 2017;45(18):10895–10905. doi: 10.1093/nar/gkx753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jørgensen F, Kurland CG. Processivity errors of gene expression in Escherichia coli. J. Mol. Biol. 1990;215(4):511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- 16.Sin C, Chiarugi D, Valleriani A. Quantitative assessment of ribosome drop-off in E. coli. Nucleic Acids Res. 2016;44(6):2528–2537. doi: 10.1093/nar/gkw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Na D, Lee D. RBSDesigner: Software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics. 2010;26(20):2633–2634. doi: 10.1093/bioinformatics/btq458. [DOI] [PubMed] [Google Scholar]
- 19.Seo SW, Yang J-S, Kim I, Yang J, Min BE, Kim S, et al. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013;15:67–74. doi: 10.1016/j.ymben.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Verma M, Choi J, Cottrell KA, Lavagnino Z, Thomas EN, Pavlovic-Djuranovic S, et al. A short translational ramp determines the efficiency of protein synthesis. Nat. Commun.10, 5774 (2019). [DOI] [PMC free article] [PubMed]
- 21.Dong H, Kurland CG. Ribosome mutants with altered accuracy translate with reduced processivity. J. Mol. Biol. 1995;248(3):551–561. doi: 10.1006/jmbi.1995.0242. [DOI] [PubMed] [Google Scholar]
- 22.Menninger JR. Peptidyl transfer RNA dissociates during protein synthesis from ribosomes of Escherichia coli. J. Biol. Chem. 1976;251(11):3392–3398. [PubMed] [Google Scholar]
- 23.Li J, Zhang C, Huang P, Kuru E, Forster-Benson ETC, Li T, et al. Dissecting limiting factors of the Protein synthesis Using Recombinant Elements (PURE) system. Translation. 2017;5(1):e1327006. doi: 10.1080/21690731.2017.1327006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillebrecht JR, Chong S. A comparative study of protein synthesis in in vitro systems: From the prokaryotic reconstituted to the eukaryotic extract-based. BMC Biotechnol. 2008;8:58. doi: 10.1186/1472-6750-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandiran V, Kramer G, Hardesty B. Expression of different coding sequences in cell-free bacterial and eukaryotic systems indicates translational pausing on Escherichia coli ribosomes. FEBS Lett. 2000;482(3):185–188. doi: 10.1016/S0014-5793(00)02017-2. [DOI] [PubMed] [Google Scholar]
- 26.Hurst GB, Asano KG, Doktycz CJ, Consoli EJ, Doktycz WL, Foster CM, et al. Proteomics-based tools for evaluation of cell-free protein synthesis. Anal. Chem. 2017;89(21):11443–11451. doi: 10.1021/acs.analchem.7b02555. [DOI] [PubMed] [Google Scholar]
- 27.Libicher K, Hornberger R, Heymann M, Mutschler H. In vitro self-replication and multicistronic expression of large synthetic genomes. Nat. Commun. 2020;11(1):904. doi: 10.1038/s41467-020-14694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu Y. ArfA recruits RF2 into stalled ribosomes. J. Mol. Biol. 2012;423(4):624–631. doi: 10.1016/j.jmb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Chadani Y, Ito K, Kutsukake K, Abo T. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 2012;86(1):37–50. doi: 10.1111/j.1365-2958.2012.08190.x. [DOI] [PubMed] [Google Scholar]
- 30.Chadani Y, Ono K, Ozawa S-I, Takahashi Y, Takai K, Nanamiya H, et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol. Microbiol. 2010;78(4):796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- 31.Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol. Microbiol. 2011;80(3):772–785. doi: 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- 32.Handa Y, Inaho N, Nameki N. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 2011;39(5):1739–1748. doi: 10.1093/nar/gkq1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Mandava CS, Cao W, Li X, Zhang D, Li N, et al. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 2015;22:906. doi: 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
- 34.Doerr A, de Reus E, van Nies P, van der Haar M, Wei K, Kattan J, et al. Modelling cell-free RNA and protein synthesis with minimal systems. Phys. Biol. 2019;16(2):025001. doi: 10.1088/1478-3975/aaf33d. [DOI] [PubMed] [Google Scholar]
- 35.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 36.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 37.Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, et al. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. U. S. A. 2013;110(38):15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou Y-M. Maintenance of protein synthesis reading frame by EF-P and m1G37-tRNA. Nat. Commun. 2015;6:7226. doi: 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mora L, Heurgué-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007;282(49):35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 40.Lilleorg S, Reier K, Remme J, Liiv A. The intersubunit bridge B1b of the bacterial ribosome facilitates initiation of protein synthesis and maintenance of translational fidelity. J. Mol. Biol. 2017;429(7):1067–1080. doi: 10.1016/j.jmb.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Chadani Y, Niwa T, Izumi T, Sugata N, Nagao A, Suzuki T, et al. Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing. Mol. Cell. 2017;68(3):528–39.e5. doi: 10.1016/j.molcel.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 42.Stögbauer T, Windhager L, Zimmer R, Rädler JO. Experiment and mathematical modeling of gene expression dynamics in a cell-free system. Integr. Biol. (Camb). 2012;4(5):494–501. doi: 10.1039/c2ib00102k. [DOI] [PubMed] [Google Scholar]
- 43.Van Nies P, Nourian Z, Kok M, van Wijk R, Moeskops J, Westerlaken I, et al. Unbiased tracking of the progression of mRNA and protein synthesis in bulk and inside lipid vesicles. ChemBioChem. 2013;14(15):1963–1966. doi: 10.1002/cbic.201300449. [DOI] [PubMed] [Google Scholar]
- 44.Borg A, Ehrenberg M. Determinants of the rate of mRNA translocation in bacterial protein synthesis. J. Mol. Biol. 2015;427(9):1835–1847. doi: 10.1016/j.jmb.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 45.Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc. Natl. Acad. Sci. 2012;109(1):131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazuta Y, Adachi J, Matsuura T, Ono N, Mori H, Yomo T. Comprehensive analysis of the effects of Escherichia coli ORFs on protein translation reaction. Mol. Cell Proteomics. 2008;7(8):1530–1540. doi: 10.1074/mcp.M800051-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuura T, Kazuta Y, Aita T, Adachi J, Yomo T. Quantifying epistatic interactions among the components constituting the protein translation system. Mol. Syst. Biol. 2009;5:297. doi: 10.1038/msb.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Gu L, Aach J, Church GM. Improved cell-free RNA and protein synthesis system. PLoS ONE. 2014;9(9):e106232. doi: 10.1371/journal.pone.0106232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazuta Y, Matsuura T, Ichihashi N, Yomo T. Synthesis of milligram quantities of proteins using a reconstituted in vitro protein synthesis system. J. Biosci. Bioeng. 2014;118(5):554–557. doi: 10.1016/j.jbiosc.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 50.Jewett MC, Forster AC. Update on designing and building minimal cells. Curr. Opin. Biotechnol. 2010;21(5):697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niwa T, Kanamori T, Ueda T, Taguchi H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc. Natl. Acad. Sci. U. S. A. 2012;109(23):8937–8942. doi: 10.1073/pnas.1201380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chizzolini F, Forlin M, Cecchi D, Mansy SS. Gene position more strongly influences cell-free protein expression from operons than T7 transcriptional promoter strength. ACS Synth. Biol. 2014;3(6):363–371. doi: 10.1021/sb4000977. [DOI] [PubMed] [Google Scholar]
- 53.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2013;16:38. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godino E, Noguera López J, Zarguit I, Doerr A, Jimenez M, Rivas G, Danelon C. Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. Commun. Biol. 2020;3:539. doi: 10.1038/s42003-020-01258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44(2):81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in the current study are available from the corresponding author upon reasonable request. This also includes the original .nd file (created in Mathematica version 11.3, Wolfram Research) used to generate the data displayed in the Supplementary Data file.